Abstract
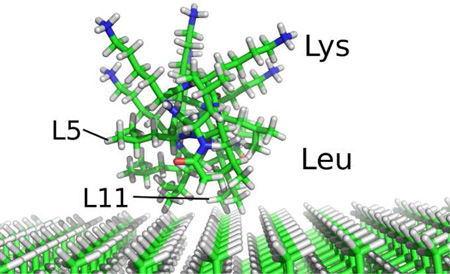
The artificial amphiphilic peptide LKα14 adopts a helical structure at interfaces, with opposite orientation of its leucine (L, hydrophobic) and lysine (K, hydrophilic) side chains. When adsorbed onto surfaces, different residue side chains necessarily have different proximities to the surface, depending on both their position in the helix and the composition of the surface itself. Deuterating the individual leucine residues (isopropyl-d7) permits the use of solid-state deuterium NMR as a site-specific probe of side chain dynamics. In conjunction with SFG as a probe of the peptide binding face, we demonstrate that the mobility of specific leucine side chains at the interface is quantifiable in terms of their surface proximity.
Development of biocompatible surfaces is a major focus of the materials and tissue engineering communities1, particularly for using peptide or protein coatings in recreating a natural extracellular matrix to direct wound repair or tissue development and homeostasis.2 Many of the current materials used in biomedical materials and tissue-engineering scaffolds are hydrophobic.3 However, proteins often lose functionality when adsorbed onto hydrophobic surfaces, so a major concern in these applications is retention of native structure and/or dynamics. Deuterium solid state nuclear magnetic resonance (NMR) is a versatile, nonperturbing probe for those parameters4.
Degrado and Lear first demonstrated synthetic peptides that adopt a chosen structure and orientation at interfaces using alternating periods of leucine (L) and lysine (K) residues5. At hydrophobic surfaces, rather than unfolding, these use a recognition method to stabilize a given secondary structure where the planar environment restricts the conformational degrees of freedom. Previously, we used NMR to explore a structural determination approach using one such 14-residue “LK” peptide,
LKα14: Ac-LKKLLKLLKKLLKL-OH
wherein dipolar recoupling and double-quantum NMR techniques were used to measure phi and psi Ramachandran angles in adjacent carbonyl-carbonyl pairs, thus quantifying the local secondary structure of the peptide while adsorbed onto a hydrophobic polystyrene (PS) surface6.
The interactions of multiple amphiphilic peptides with PS and silica surfaces has been studied also by Somorjai and coworkers7 using sum frequency generation (SFG) vibrational spectroscopy, demonstrating LKα14 adsorption onto PS with the leucine side chains alongside and lysine side chains opposite that surface.
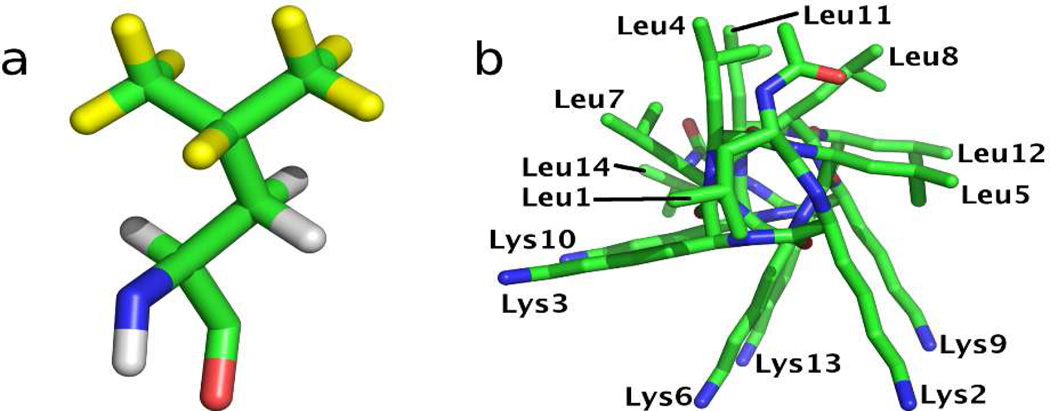
Here, we demonstrate how solid state deuterium NMR can be used to define site-specifically and quantifiably, the perturbations of amino acid side chain dynamics induced by peptide adsorption onto a hydrophobic surface. In this initial study we use a series of selectively deuterated LKα14 peptides, where L-leucine, isopropyl-2H7 (d7Leu) (see Figure 1a) was incorporated at individual sites in the peptide’s primary sequence.
Figure 1.
(a) Structure of d7Leu, with deuterated sites highlighted in yellow. (b) End-on view of LKα14 helical structure with residues labeled.
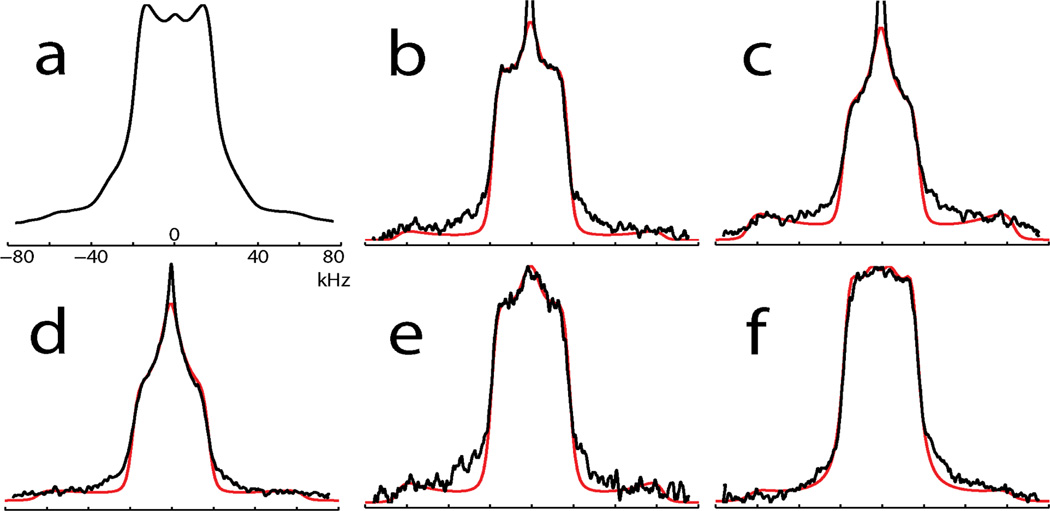
Figure 2a shows the deuterium NMR spectrum of unadsorbed, lyophilized LKα14 with d7Leu incorporated at position L8. Figures 2b-e are spectra of PS-physisorbed and subsequently lyophilized LKα14 with d7Leu incorporated selectively at L5, L8, L11, and L14. Figure 2f is the spectrum of LKα14 with d7Leu incorporated at L8 but adsorbed onto 14 nm diameter colloidal gold particles coated with a self-assembled monolayer (SAM) of carboxyl-terminated alkane thiolates, COOH(CH2)16SH.
Figure 2.
2H NMR spectra (black lines) and simulations (red lines, b-f only) of (a) free LKα14, d7Leu8; polystyrene-adsorbed LKα14 samples labeled at (b) L5, (c) L8, (d) L11, (e) L14; (f) L8 bound to carboxyl-functionalized gold nanoparticles.
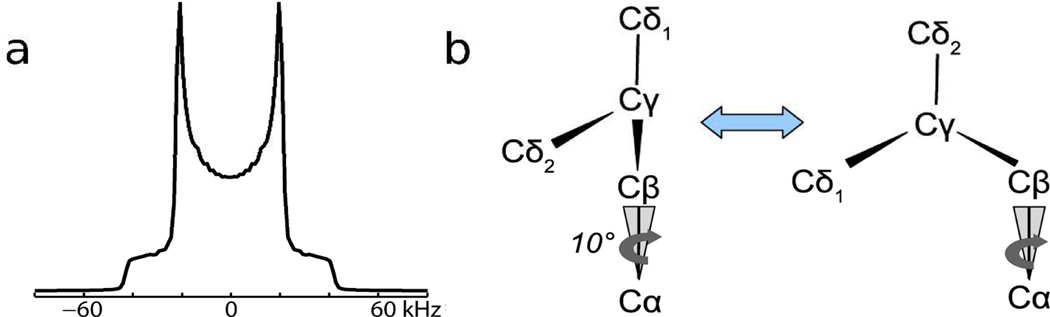
A striking aspect of the spectra in Figure 2 is that none resemble the deuterium NMR spectrum of polycrystalline d7Leu, represented by a simulation in Figure 3a, which consists primarily of an axially symmetric, Pake-type powder pattern with an effective QCC of about 50 kHz, corresponding to the six deuterons of the rapidly rotating δ1 and δ2 CD3 groups. The γ-methine deuteron generates a weaker pattern with QCC≈170 kHz having low experimental sensitivity, not shown in Figure 3a.
Figure 3.
(a) Simulation of polycrystalline d7Leu. (b) Two-site jump model with 10° half-angle cone motion used for simulations.
Figure 2c represents PS-adsorbed LKα14 deuterated at L8, so any difference between Figure 2c and the spectrum of unbound LKα14 in Figure 2a is assumed due to the influence of the PS surface on the leucyl side chain dynamics. Similarly, the spectral pattern variation observed in Figures 2b-e is presumably due to variation in dynamics as a function of side chain proximity to the PS surface. Finally, as Figure 2f represents the spectrum of LKα14 deuterated at L8 and bound to the surface of a carboxyl-terminated SAM on colloidal gold, the spectral pattern variation between Figures 2c and 2f is likely due to the differential impact on leucyl side chain of adsorption onto a non-polar PS surface versus adsorption onto the polar surface of the SAM.
The dynamics of leucyl side chains has been studied and quantified by Torchia and coworkers8 in a 2H NMR study of helical collagen fibrils with L-leucine, 2H10 uniformly incorporated. The temperature variation of the deuterium lineshape was modeled as variation in the rate of a two site exchange between two conformations of the leucine side chain observed in crystallographic studies of leucine monomer and leucyl peptides9. To simulate the spectral patterns in Figure 2, we similarly assume here an exchange between the two conformers shown in Figure 3b. The site exchange corresponds to an angular change of about 110° for the Cβ-Cγ bond axis. The rapid rotational motions of the CD3 groups are treated simply with an effective QCC. The exchange rate and a priori conformer populations were varied in the simulations. In addition, to account for motion of the peptide chain itself, the Cα-Cβ bond was treated as moving on the surface of a cone.
Simulation results are shown with spectral data superimposed in Figure 2, and in the table below. Although Figures 2b-e show features of a dynamically-induced η=1 spectral powder pattern, a residual HDO signal limits our ability to simulate exactly the central feature of these spectra.
Table 1.
Simulation parameters for spectra seen in Figure 2.
| L5 | L8(PS) | L11 | L14 | L8(SAM) | |
|---|---|---|---|---|---|
| P1/P2 | 4/6 | 4/6 | 4/6 | 3.5/6.5 | 3/7 |
| kex(s−1) | 3×105 | 6×105 | 1×106 | 3×105 | 3×105 |
| kcone(s−1) | 2×103 | 2×103 | 2×103 | 3×103 | 6×103 |
| QCCeff(kHz) | 49 | 49 | 49 | 51 | 43 |
Simulation results shown in the table indicate that the rate of exchange between side chain conformers, kex, is the model parameter that varies most significantly with the series Figure 2b-f. The rate kex is greatest for L8 and L11, whose side chains are located closest to the PS surface, while kex for L5 and L14 are significantly smaller. There is much less variation in all other model parameters for the different leucine sites of LKα14 on PS. Interestingly, the side chain exchange rate kex of L8 on a carboxyl-terminated SAM surface is identical to L5 and L14, and differs from L8 on PS by a factor of two.
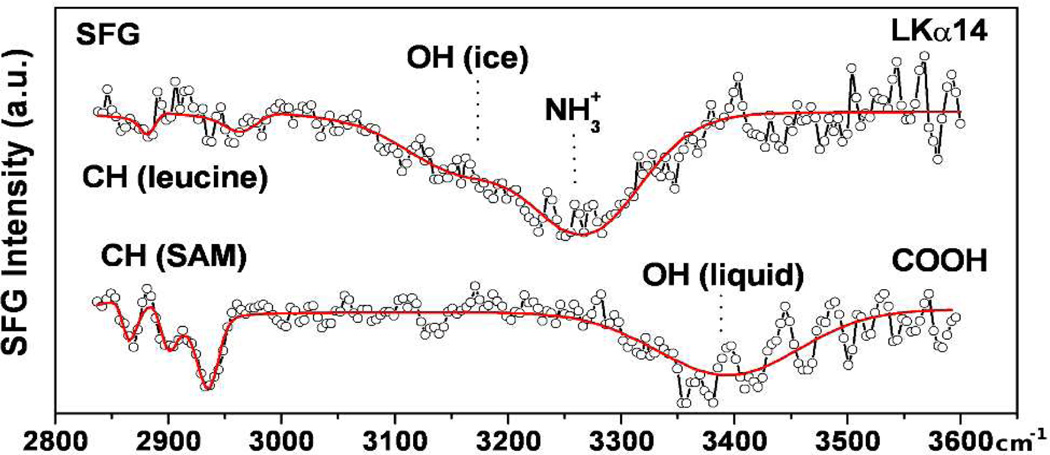
Verification of the peptide orientation on carboxylated surfaces was performed with SFG. Figure 4 presents spectra of a monolayer of carboxyl-terminated alkanethiolates (identical to those used in Figure 2e) on gold-coated CaF2 windows before and after adsorption of LK peptides.
Figure 4.
SFG spectra of COOH-functionalized gold surfaces alone (lower trace) and after adsorption of LKα14 (upper trace).
The spectrum of the bare SAM surface consists only of the methylene stretching modes at 2866 cm−1 and 2933 cm−1, with the carboxyl-adjacent methylene unit at 2901 cm−1, and loosely bound water near 3400 cm−1.10 Upon peptide adsorption, weak CH3 resonances related to leucine side chains appear (2961 cm−1 and 2880 cm−1). These modes are 180° phase shifted with respect to the gold background (appearing as a dip in the spectrum) showing the leucines are pointing away from the surface.11 A band near 3200 cm−1 is related to tetrahedrally coordinated water molecules.10 We assign the feature near 3265 cm−1 to ordered lysine NH3+ groups binding the SAM substrate.
This study indicates that dynamics of the isopropyl moiety of leucine side chains are extremely sensitive to surface proximity and surface functionalization. The factor of 2–3 change in kex observed between the L8/L11 versus L5/L14 sites in LKα14 on PS surfaces, is observed in collagen leucine lineshapes only as a result of a 40°C change in temperature8. The L8(SAM) data and SFG orientation data mutually demonstrate that a change in surface functionalization in turn changes the peptide orientation and consequently the surface proximities of amino acid side chains. The collagen study found the existence of a mobile interface between individual helical protein components of the fibers12, characterized in part by quantifying the dynamics of leucine side chains as a function of temperature. This study uses multiple, complementary spectroscopies to observe a similarly mobile interface between a peptide structure and bulk surfaces, with mobility quantifiable in terms of NMR exchange dynamics of leucine side chains with varying surface proximities, and suggests that the combination of NMR and SFG on such systems can provide significantly more information than either alone.
Supplementary Material
Acknowledgements
The authors acknowledge support from NIH RO1 GM074511, RO1 DE12554 and P41 EB002027. T.W. thanks the DFG for a research fellowship. We thank Sirnegada Techane for supplying the functionalized gold nanoparticles.
Footnotes
Supporting Information. Details of sample preparation, NMR and SFG experiments, and fitting results for Figure 4. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.(a) Healy KE. Curr. Opin. Solid State & Mat. Sci. 1999;4:381–387. [Google Scholar]; (b) Angelova N, Hunkeler D. Trends Biotech. 1999;17:409–421. doi: 10.1016/s0167-7799(99)01356-6. [DOI] [PubMed] [Google Scholar]; (c) Langer R. Acc. Chem. Res. 2000;33:94–101. doi: 10.1021/ar9800993. [DOI] [PubMed] [Google Scholar]; (d) Ratner BD, Bryant SJ. Annu. Rev. Biomed. Eng. 2004;6:41–75. doi: 10.1146/annurev.bioeng.6.040803.140027. [DOI] [PubMed] [Google Scholar]; (e) Vasita R, Shanmugam K, Katti DS. Curr. Top. Med. Chem. 2008;8:341–353. doi: 10.2174/156802608783790893. [DOI] [PubMed] [Google Scholar]
- 2.(a) Borkenhagen M, Clemence JF, Sigrist H, Aebischer PJ. Biomed. Mat. Res. 1998;40:392–400. doi: 10.1002/(sici)1097-4636(19980603)40:3<392::aid-jbm8>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]; (b) Rezania A, Healy KE. Biotech. Progress. 1999;15:19–32. doi: 10.1021/bp980083b. [DOI] [PubMed] [Google Scholar]; (c) Gilbert M, Shaw WJ, Long JR, Nelson K, Drobny GP, Giachelli CM, Stayton PS. J. Biol. Chem. 2000;275:16213–16218. doi: 10.1074/jbc.M001773200. [DOI] [PubMed] [Google Scholar]; (d) Venugopal J, Low S, Choon AT, Ramakrishna S. J. Biomed. Mater. Res. 2008;84B:34–48. doi: 10.1002/jbm.b.30841. [DOI] [PubMed] [Google Scholar]
- 3.(a) Drumheller PD, Elbert DL, Hubbell JA. Biotech. and Bioeng. 1994;43:772–780. doi: 10.1002/bit.260430812. [DOI] [PubMed] [Google Scholar]; (b) Neff JA, Caldwell KD, Tresco PA. J. Biomed. Mat. Res. 1998;40:511–519. doi: 10.1002/(sici)1097-4636(19980615)40:4<511::aid-jbm1>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]; (c) Tong YW, Shoichet MS. J. Biomat. Sci. – Polymer Ed. 1998;9:713–729. doi: 10.1163/156856298x00109. [DOI] [PubMed] [Google Scholar]; (d) Ostuni E, Grzybowski BA, Mrksich M, Roberts CS, Whitesides GM. Langmuir. 2003;19:1861–1872. [Google Scholar]
- 4.(a) Palmer AG, III, Williams J, McDermott A. J. Phys. Chem. 1996;100:13293–13310. [Google Scholar]; (b) Gardner KH, Kay LE. Annu. Rev. Biophys. Biomol. Struc. 1998;27:357–406. doi: 10.1146/annurev.biophys.27.1.357. [DOI] [PubMed] [Google Scholar]; (c) Bechinger B, Kinder R, Helmle M, Vogt TCB, Harzer U, Schinzel S. Biopolymers. 1999;51:174–190. doi: 10.1002/(SICI)1097-0282(1999)51:3<174::AID-BIP2>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 5.Degrado WF, Lear JD. J. Am. Chem. Soc. 1985;107:7684–7689. [Google Scholar]
- 6.Long JR, Oyler N, Drobny GP, Stayton PS. J. Am. Chem. Soc. 2002;124:6297–6303. doi: 10.1021/ja011624n. [DOI] [PubMed] [Google Scholar]
- 7.(a) Mermut O, Phillips DC, York RL, McCrea KR, Ward RS, Somorjai GA. J. Amer. Chem. Soc. 2006;128:3598–3607. doi: 10.1021/ja056031h. [DOI] [PubMed] [Google Scholar]; (b) Phillips DC, York RL, Mermut O, McCrea KR, Ward RS, Somorjai GA. J. Phys. Chem. C. 2007;111:255–261. [Google Scholar]
- 8.Batchelder LS, Sullivan CE, Jelinski LW, Torchia DA. Proc. Natl. Acad. Sci. USA. 1982;79:386–389. doi: 10.1073/pnas.79.2.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.(a) Benedetti C. In: Goodman M, Meienhofer J, editors. Proceedings of the Fifth American Peptides Symposium; New York: Wiley; 1977. pp. 257–274. [Google Scholar]; (b) Janin J, Wodak S, Levitt M, Maigret B. J. Mol. Biol. 1978;125:357–386. doi: 10.1016/0022-2836(78)90408-4. [DOI] [PubMed] [Google Scholar]
- 10.(a) Chi SC, Coi SC. Langmuir. 2005;21:11765–11772. doi: 10.1021/la050847e. [DOI] [PubMed] [Google Scholar]; (b) Du Q, Freysz E, Shen YR. Phys. Rev. Lett. 1994;72:238–241. doi: 10.1103/PhysRevLett.72.238. [DOI] [PubMed] [Google Scholar]
- 11.(a) Watry MR, Richmond GL. J. Phys. Chem. B. 2002;106:12517–12523. [Google Scholar]; (b) Ward RN, Davies PB, Bain CD. J. Phys. Chem. 1993;97:7141–7143. [Google Scholar]
- 12.(a) Jelinski LW, Torchia DA. J. Mol. Biol. 1980;138:255–272. doi: 10.1016/0022-2836(80)90286-7. [DOI] [PubMed] [Google Scholar]; (b) Jelinski LW, Sullivan CE, Torchia DA. Nature. 1980;284:531–534. doi: 10.1038/284531a0. [DOI] [PubMed] [Google Scholar]; (c) Jelinski LW, Sullivan CE, Batchelder LS, Torchia DA. Biophys. J. 1980;10:515–529. doi: 10.1016/S0006-3495(80)84987-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.