Abstract
Background
Several organisms display dormancy and developmental arrest at embryonic stages. Long-term survival in the dormant form is usually associated with desiccation, orthodox plant seeds and Artemia cysts being well documented examples. Several aquatic invertebrates display dormancy during embryonic development and survive for tens or even hundreds of years in a hydrated form, raising the question of whether survival in the non-desiccated form of embryonic development depends on pathways similar to those occurring in desiccation tolerant forms.
Methodology/Principal Findings
To address this question, Illumina short read sequencing was used to generate transcription profiles from the resting and amictic eggs of an aquatic invertebrate, the rotifer, Brachionus plicatilis. These two types of egg have very different life histories, with the dormant or diapausing resting eggs, the result of the sexual cycle and amictic eggs, the non-dormant products of the asexual cycle. Significant transcriptional differences were found between the two types of egg, with amictic eggs rich in genes involved in the morphological development into a juvenile rotifer. In contrast, representatives of classical “stress” proteins: a small heat shock protein, ferritin and Late Embryogenesis Abundant (LEA) proteins were identified in resting eggs. More importantly however, was the identification of transcripts for messenger ribonucleoprotein particles which stabilise RNA. These inhibit translation and provide a valuable source of useful RNAs which can be rapidly activated on the exit from dormancy. Apoptotic genes were also present. Although apoptosis is inconsistent with maintenance of prolonged dormancy, an altered apoptotic pathway has been proposed for Artemia, and this may be the case with the rotifer.
Conclusions
These data represent the first transcriptional profiling of molecular processes associated with dormancy in a non-desiccated form and indicate important similarities in the molecular pathways activated in resting eggs compared with desiccated dormant forms, specifically plant seeds and Artemia.
Introduction
Living organisms have evolved remarkable mechanisms for survival in unpredictable environments. Under extreme conditions that do not allow the maintenance of homeostasis, they may enter a state of reduced metabolism or developmental arrest, waiting for improved conditions that will allow the resumption of life activities, development and reproduction. Many organisms, from prokaryotes to mammals, have evolved the capacity to enter dormancy and exit from it. Surprisingly, common pathways have been identified, in spite of the diversity and complexity in the survival strategies in organisms displaying dormancy [1]–[3]. The term “dormant” or “dormancy” refers to a temporary suspension of visible metabolic activity or arrested development and encompasses the phenomena of diapause, quiescence or cryptobiosis [4]–[6]. Most often long-term dormancy is associated with desiccation [7].
Several organisms display dormancy and developmental arrest at embryonic developmental stages, including mammals [8]. Plants, rotifers, daphnids, copepods and Artemia are well known examples where dormancy is developmentally programmed in the form of seeds, resting eggs, ephippia or cysts [9]. In orthodox plant seeds, tolerance to desiccation is a programmed phase of embryological development which is regulated by abscisic acid and other hormones and initiated by maternal factors rather than environmental signals [10], [11]. During maturation, seeds undergo a series of declining water concentrations and acquire mechanisms for coping with osmotic adjustments (by accumulating carbohydrates), reactive oxidative species (by producing anti-oxidants) and structural changes by the production of small heat shock and Late Embryonenesis Abundant (LEA) proteins [1], [12]. Survival in a desiccated form is closely associated with the formation of an intracellular “glassy” matrix, with high intracellular viscosity restricting molecular mobility and life activities [1], [6]. Invertebrate desiccated cysts of Artemia, display some similar adaptations to those of plant seeds [13], [14]. Conversely, aquatic organisms, such as rotifers, cladocerans or copepods producing resting eggs or ephippia can survive for several decades or hundreds of years in a non-desiccated form [15]–[19], raising the question whether survival in the non-desiccated form of embryonic dormancy depends on pathways similar to those occurring in desiccation tolerant forms.
At least 15 monogonont rotifer species, including the rotifer Brachionus plicatilis, are known to enter dormancy or diapause through the formation of resting eggs by sexual reproduction [20]. The parthenogenetic (amictic) phase dominates the monogonont life cycle in the absence of males, but following certain environmental cues, sexual reproduction (the mictic phase) takes place via meiosis leading to the formation of resting eggs [9]. The dormant (or diapausing) resting egg is an encased embryo, differing morphologically from the non-dormant asexually (amictic) produced egg [20]–[22] (Figure 1) and in this form, can retain viability for decades [18]. Following specific cues, the resting eggs hatch after an obligatory dormant period and populate the water body through asexual reproduction [9]. The water content of resting eggs is around 70% [23] and in the natural environment they occur in the sediments of the littoral zone or saline ponds [16], [18]. However, rotifer resting eggs can also retain their viability after desiccation and lyophilisation, when their water content is reduced to ∼7% [23].
Figure 1. Comparison between amictic and resting eggs.
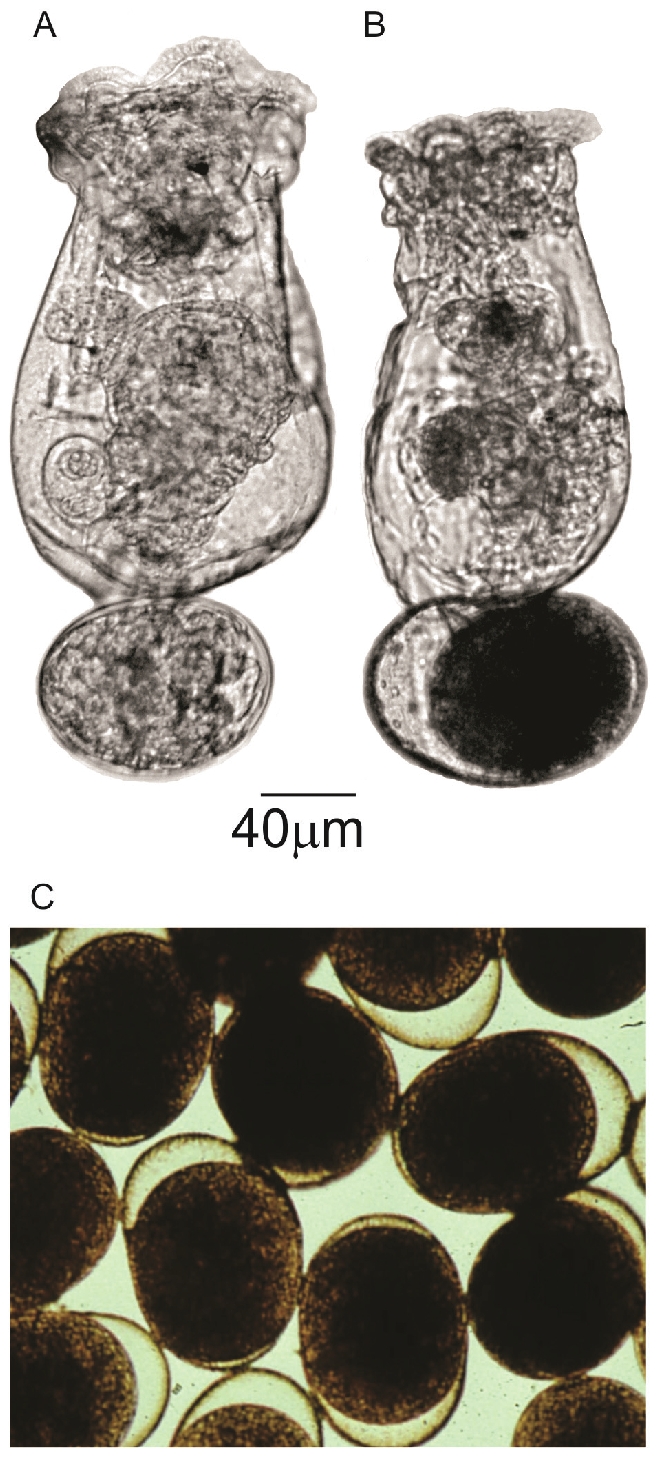
Photographs of: A: An amictic female carrying an amictic egg with a developing embryo, B: A mictic female carrying a resting egg with an encased dormant embryo, and C: Resting eggs.
Genomic resources for developmentally arrested invertebrate embryos displaying dormancy are scarce with the exception of Artemia [24], [25] and the current study is the first for non-desiccated dormant embryos. In this study, gene expression profiles were generated for both B. plicatilis resting and amictic eggs using Illumina short read sequencing mapped against an existing EST transcriptome backbone [26]. This was a single time point replicated assay. Resting eggs first appeared to be carried by mictic females, 3–5 days after hatching and the resting eggs were collected 14–25 days after the hatching of the parental resting eggs. Amictic eggs were collected at the same time. The expression profiles were analysed to identify unique functions characteristic of each state. The genes identified in the resting egg transcription profiles were also compared to current data in other species, such as Artemia and plant seeds, to further understand potentially common processes involved in the maintenance of dormant forms.
Results and Discussion
Illumina short read sequencing was used to generate transcription profiles of B. plicatilis resting and amictic eggs (details in Table S1). These two types of egg have very different life histories, with the resting eggs the result of the sexual cycle and are the dormant or diapausing form, whilst the amictic eggs are the parthenogenetic diploid products of the asexual cycle (Figure 1; also Figure 1 in [26]). The resting eggs can survive for decades in the dormant form [18], the original reference culture for this project dates back to 1981 (see Materials and Methods). Viability of the resting eggs used for RNA extractions analysed in this article were tested after 8 weeks of collection, having been stored in the dark at 4°C. Between 47–49% of the resting eggs hatched (Table S2). The proportion of viable eggs was probably higher, as previous results showed that resting eggs that do not hatch in the first attempt, hatch later under slightly different conditions [27], [28].
An analysis of the most commonly expressed transcripts in both forms identified striking similarities, but also some major differences (Table 1; Figure 2). There were commonalities represented by transcripts putatively involved in protein turnover, energy production in the form of genes involved in the mitochondrial respiratory chain and ATP synthesis and cytoskeletal elements. Hence there was a certain level of metabolic activity in both types of egg. Transcripts of the mitochondrial transport protein (voltage dependent anion channel or porin) were present in numerous copies in both types of egg, however, exact assignment of function is difficult as this is a multifunctional protein, crucial in many aspects of cell life and death. This protein forms a pore in the mitochondria and is the effective gatekeeper for entry and exit of metabolites, including ATP between this organelle and the cytosol. It also has important roles in protection against oxidative stress and the regulation of apoptosis [29]. Hence, this molecule could potentially be performing different functions in each of the egg forms.
Table 1. The ten most commonly expressed transcripts in the two egg forms.
| Signature clone | Sequence match | Accession number | Expect | Function |
| Resting eggs | ||||
| sb103P0019I07_F.ab1 | Ubiquitin carboxyl-terminal hydrolase | B4MJN2 | 0.11 | Protein turnover |
| sbs02P0001I19_F.ab1 | Voltage dependent anion-selective channel | E3TCS5 | 2.8E-12 | Transport |
| rotifera-CL1Contig18 | ATP synthase lipid binding protein | C1BPP0 | 1.1E-14 | ATP synthesis |
| rotifera-CL5Contig1 | HSP27 | E5DVQ8 | 1.1E-35 | Chaperone/stress protein |
| rotifera-CL90Contig1 | Actin-related protein 2/3 complex | E2COG1 | 1.8E-05 | Cytoskeleton |
| rotifera-CL485Contig2 | CAMK/TSSK protein kinase | E1FMN8 | 1.7E-21 | Cell differentiation |
| rotifera-CL1Contig28 | Late Embryogenesis Abundant protein | D5IOZ6 | 9.4E-98 | Desiccation tolerance |
| rotifera-CL2Contig3 | Cytochrome C oxidase, subunit III | B1GYK1 | 8.9E-98 | Electron transport |
| rotifera-CL57Contig1 | Ferritin-like protein | C6JUM7 | 3.6E-25 | Antioxidant |
| sbs01P0015E02R_.ab1 | Peptidyl-prolyl cis-trans isomerise-like | Q9H2H8 | 7.6E-12 | Accelerates protein folding |
| Amictic eggs | ||||
| sb103P0019I07_F.ab1 | Ubiquitin carboxyl-terminal hydrolase | B4MJN2 | 0.11 | Protein turnover |
| rotifera-CL2Contig5 | Cytochrome C oxidase, subunit III | B1GYK1 | 4.9E-96 | Electron transport |
| rotifera-CL485Contig2 | cAMP-dependent protein catalytic sub-unit | E0VNF8 | 3.7E-21 | Protein phosphorylation |
| rotifera-CL105Contig1 | Tubulin ß chain | P11833 | 2.9E-216 | Cytoskeleton |
| rotifera-CL2998Contig1 | Ankyrin domain protein | C0QTZ9 | 2.0E-15 | Transcriptional activator |
| rotifera-CL1Contig3 | Selenophosphate synthetase | Q3HR35 | 9.0E-116 | Cell cycle progression |
| rotifera-CL3Contig3 | ATPsynthase F0 subunit 6 | E2D7B0 | 1.6E-36 | ATP synthesis |
| rotifera-CL52Contig1 | Translation elongation factor 1α | C6L868 | 1.1E-231 | Protein biosynthesis |
| rotifera-CL2956Contig1 | NADH dehydrogenase subunit 3 | B1GYK2 | 9.6E-12 | Electron transport |
| sbs02P0001I19_F.ab1 | Voltage dependent anion-selective channel | E3TCS5 | 2.8E-12 | Transport |
Representative clones have been annotated with putative functions. Different members of gene families (e.g. cytochrome c oxidase sub-units) have only been uniquely represented to provide a wider overview of the most common functions associated with each egg form. All expect values are less than 1.0e-12 except for that of ubiquitin carboxyl-terminal hydrolase (0.11). This was a very short, but accurate match, which is reflected in the high expect value. All assignments were manually verified.
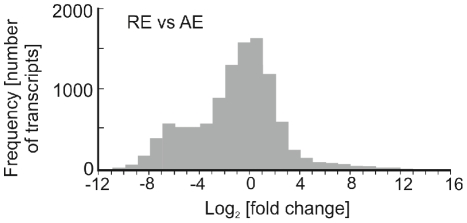
Figure 2. Two-way analysis comparing the number of transcripts (reads) and their respective fold change, between resting eggs (RE) and amictic eggs (AE).
Each column shows the number of reads at the respective fold change. The reads in columns corresponding with range of −1 and +1 fold change, did not differ significantly between resting eggs and amictic eggs.
An overview of the differences between the two types of embryo, can be obtained by examining those transcripts differentially expressed in each library via GO enrichment. These data revealed that resting egg transcripts showed cellular components and biological processes associated with protein turnover and degradation and molecular functions associated with, again, protein degradation, but also oxidoreductase activity (Table 2). The enriched functions linked to amictic eggs showed a very different composition indicating active cytoskeletal remodelling and cellular communication (Table 3). Molecular functions that predominated were associated with calcium ions, actin and cytoskeletal protein binding and kinase activity, whilst cell communication and signal transduction were the most significant biological processes.
Table 2. GO enrichment results of resting egg transcripts, showing most significant results for cellular component (denoted by C preceding the GO ID), biological process (B) and molecular function (M).
| GO ID | Description | ratio | p-value | q-value |
| C:0000502 | Proteasome complex | 5.71 | 4.20E-21 | 8.08E-18 |
| M:0004298 | Threonine endopeptidase activity | 5.92 | 2.32E-19 | 1.48E-16 |
| C:0005839 | Proteasome core complex | 5.92 | 2.32E-19 | 1.48E-16 |
| B:030163 | Protein catabolic process | 3.14 | 1.63E-16 | 7.85E-14 |
| B:0006511 | ubiquitin-dependent protein catabolic process | 3.35 | 3.33E-15 | 8.01E-13 |
| B:0043632 | modification-dependent macromolecule catabolic process | 3.35 | 3.33E-15 | 8.01E-13 |
| B:0019941 | modification-dependent protein catabolic process | 3.35 | 3.33E-15 | 8.01E-13 |
| B:0051603 | Proteolysis involved in cellular protein catabolic process | 3.35 | 3.33E-15 | 8.01E-13 |
| C:005829 | cytosol | 2.88 | 5.67E-15 | 1.21E-12 |
| B:0044257 | Cellular protein catabolic process | 3.26 | 1.19E-14 | 2.29E-12 |
| B:0043285 | Biopolymer catabolic process | 2.57 | 4.25E-13 | 7.43E-11 |
| C:0044445 | Cytosolic part | 3.36 | 9.81E-11 | 1.57E-08 |
| B:0009057 | Macromolecule catabolic process | 2.18 | 1.58E-10 | 2.33E-08 |
| B:0044265 | Cellular macromolecule catabolic process | 2.18 | 4.93E-09 | 6.76E-07 |
| B:0009056 | Catabolic process | 1.58 | 7.97E-06 | 1.02E-03 |
| C:0030529 | Ribonucleoprotein complex | 1.59 | 1.44E-05 | 1.73E-03 |
| B:0044248 | Cellular catabolic process | 1.60 | 1.68E-05 | 1.90E-03 |
| M:0016491 | oxidoreductase_activity | 1.40 | 2.13E-05 | 2.28E-03 |
| C:0032991 | Macromolecular complex | 1.29 | 2.44E-05 | 2.46E-03 |
| B:0043284 | Biopolymer biosynthetic process | 2.78 | 4.15E-05 | 3.98E-03 |
Table 3. GO enrichment results of amictic egg transcripts, showing most significant results for cellular component (denoted by C preceding the GO ID), biological process (B) and molecular function (M).
| GO ID | Description | ratio | p-value | q-value |
| C:0005856 | cytoskeleton | 1.87 | 1.07E-17 | 2.69E-14 |
| C:0016020 | membrane | 1.32 | 2.02E-16 | 2.54E-13 |
| M:0008092 | Cytoskeletal protein binding | 2.34 | 8.39E-16 | 7.02E-13 |
| M:0005509 | Calcium ion binding | 1.82 | 1.45E-14 | 9.12E-12 |
| B:0007154 | Cell communication | 1.46 | 1.15E-12 | 5.78E-10 |
| C:0044425 | Membrane part | 1.32 | 3.44E-12 | 1.44E-09 |
| C:0042995 | Cell projection | 2.11 | 5.16E-12 | 1.85E-09 |
| C:0044430 | Cytoskeletal part | 1.85 | 6.48E-12 | 2.04E-09 |
| M:0003779 | Actin binding | 2.39 | 9.91E-12 | 2.77E-09 |
| B:0007165 | Signal transduction | 1.43 | 3.18E-10 | 7.99E-08 |
| C:0031224 | Intrinsic to membrane | 1.33 | 4.39E-10 | 1.00E-07 |
| C:0005886 | Plasma membrane | 1.60 | 1.38E-09 | 2.88E-07 |
| C:0016021 | Integral to membrane | 1.32 | 1.58E-09 | 3.05E-07 |
| B:0051674 | Localization of cell | 2.31 | 1.04E-08 | 1.64E-06 |
| B:0006928 | Cell motility | 2.31 | 1.04E-08 | 1.64E-06 |
| C:0019861 | flagellum | 2.31 | 1.04E-08 | 1.64E-06 |
| B:0032501 | Multicellular organismal process | 1.47 | 2.27E-08 | 3.35E-06 |
| C:0015630 | Microtubule cytoskeleton | 1.76 | 3.25E-08 | 4.54E-06 |
| M:0016301 | Kinase activity | 1.43 | 6.02E-08 | 7.95E-06 |
| B:0007010 | Cytoskeleton organization and biogenesis | 1.71 | 2.78E-07 | 3.49E-05 |
In-depth analysis aided by manual verification revealed greater detail on the cellular activities of resting and amictic eggs. Full details of these transcripts with associated BLAST sequence similarity data and putatively assigned functions are given in Tables S3 and S4, with an extraction of the function data summarised in Table 4. These are discussed in more detail below.
Table 4. List of putative functions assigned to transcripts differentially up-regulated in resting and amictic eggs (extracted from data in Tables S3 and S4).
| Putative function | Resting eggs | Amictic eggs |
| Antioxidant | 29 | |
| Chaperone systems | 12 | |
| Transcription factor | 8 | |
| Oxidoreductase | 7 | |
| Nucleotide biosynthesis | 7 | |
| Late Embryogenesis Abundant protein | 4 | |
| Apoptosis | 4 | |
| Translation | 4 | |
| Chromosome structure, replication and transcriptional regulation | 3 | |
| Trehalose biochemical pathway | 3 | |
| Cell adhesion/neuronal function | 2 | |
| Protein damage repair | 2 | |
| Amino acid metabolism | 2 | |
| DNA replication | 2 | |
| Mitochondrial distribution | 2 | |
| RNA metabolism | 2 | |
| Cell proliferation | 2 | |
| Protein modification | 1 | |
| Accelerate protein folding | 1 | |
| Molecular facilitator | 1 | |
| Metabolism | 12 | 2 |
| Transporter | 8 | 1 |
| Protein turnover | 8 | 1 |
| Signalling molecule | 8 | 8 |
| Immune function | 6 | 3 |
| Lipid metabolism | 4 | 7 |
| Energy production | 3 | 2 |
| Transcription | 2 | 1 |
| Cell cycle progression/development | 2 | 5 |
| Cell differentiation | 1 | 3 |
| Neurosecretion | 1 | |
| Cell adhesion | 2 | |
| Cilia component | 3 | |
| Development | 5 | |
| Proteolytic enzyme | 8 | |
| Cytoskeleton/muscle | 26 |
Transcripts up-regulated in resting eggs
Putatively identified transcripts in this list (Table 2) share many features in common with previous studies on dormancy and diapause of desiccated forms such as Artemia cysts and plant seeds [12], [13], in particular the preponderance of antioxidants, oxidoreductases, chaperones and late embryogenesis abundant proteins (LEAs).
Antioxidants
These are represented by database matches to ferritin, peroxiredoxin, catalase, glutathione-s-transferase and the superoxide dismutases. Although these genes have been shown to have other functions, such as increased lifespan [30] and iron sequestration [31], a major role of these enzymes is their action against reactive oxygen species (ROS). DJ-1, in particular has numerous functions including transcriptional regulation, protease activity and mitochondrial regulation, but given the biological context, allied to the catalogue of other up-regulated genes, the most probable function is protection against oxidative stress [32]. ROS are toxic in all life stages as they directly damage DNA, lipids and proteins, but are especially problematic for dormant forms. In plant seeds desiccation causes loss of control mechanisms that maintain low ROS concentrations and therefore antioxidant activity has great importance [33]. However, recently it has been shown that ROS play an important role in cell signalling particularly via post-translational modification of cysteine residues. Biological processes affected include chemotaxis, cell proliferation and neurogenesis [34] and it may well be that a certain level of ROS activity is beneficial to the resting eggs.
Oxidative stress leads to the accumulation of toxic compounds such as aldehydes that serve as intermediates in metabolism but may also be toxic to cells and therefore their amount must be tightly regulated. The higher expression of aldo-keto reductases may facilitate increased oxidative and heat stress tolerance [35], and protection of mitochondria against oxidative stress is suggested by higher levels of retinol dehydrogenase 13 (Table S3) [36].
Chaperones
Many proteins of the cellular chaperone systems are expressed ubiquitously in the normal cell state to aid in the folding of native polypeptides and their translocation to different cellular compartments [37], [38]. However, during the stress response, they may be up-regulated to further assist mis-folded proteins to attain or regain their native states and also target degraded proteins and regulate their removal from the cell, thus preventing the formation of cytotoxic aggregates [39]. The fact that elements of this system are up-regulated in resting eggs, supports the supposition that this state imposes a “stress” on rotifer cellular homeostasis. This is exemplified by the up-regulation of the HSP70 pathway (HSP70 with DnaJ co-chaperones and the HSC70/HSP90 organising protein), which has long been associated with the cellular stress response [37]. In Artemia cysts, the small heat shock protein p26 is present in high abundance and thought to play a major role in combating stressful conditions. This is a multifunctional molecule which possesses molecular chaperone activity, inhibits cell division by disruption of the mitotic apparatus, influences transcription levels and inhibits apoptosis (reviewed in [13]). All functions of which are essential for survival in the dormant state. The highest sequence similarity results for the small rotifer heat shock proteins identified here were to Artemia HSP21, but sequence homologies between small HSPs are generally low. To date, no orthologue of artemin, the ferritin superfamily member found exclusively associated with diapause and desiccation in Artemia [40], has been identified in any other species including the rotifer. This is a multifunctional protein that also acts as a chaperone depending on cellular requirements [13]. Small heat shock proteins may play a particularly important role in dormant organisms, as they act as ATP-independent chaperones, thus preventing protein aggregation in stressful conditions without the expenditure of important ATP reserves [41].
The specific expression of genes encoding small heat shock proteins and HSP70 have also been reported in association with dormancy in plant seeds [2], [42] and diapause in insects [13].
Oxidoreductases
This category comprises a range of different enzyme families including the cytochrome P450s, alcohol dehydrogenases and selenium binding proteins. These proteins play critical roles in the intermediate metabolism of many cellular substrates such as lipids, amino acids and carbohydrates, but are also major players in detoxification processes and degradation of xenobiotics, in the latter case, this is particularly true of the cytochrome P450 family. They are involved in the regulation of hormones, act as signalling molecules and sense the redox status in metabolic or transcriptional processes. Therefore they regulate vital cellular processes [43], [44]. Which role predominates in resting eggs cannot be determined purely from transcriptional profiles and certainly for such complex multifunctional families, the answer will almost certainly be a combination of functions. The CYP450 family is associated with synthesis of abcisic acid (P450 707) promoting dormancy in plant seeds [11] and dauer formation in C. elegans [45]. It is also up-regulated during diapause in the insect, Allonemobius socius [46]. Cytochrome p450s accumulate in Artemia cysts [47] indicating once again that potentially similar mechanisms may be operating in both the rotifer and Artemia dormant states.
Late embryogenesis abundant proteins (LEAs) and trehalose
These proteins were originally identified in plant seeds during the late stages of embryonic development [48] and are associated with desiccation tolerance throughout the life cycle of all major plant taxa [12]. They have subsequently been described in a number of naturally anhydrobiotic organisms and differentiated into seven groups, the most numerous of which are the Group 3 LEAs, which are the main ones found in non-plant species (for review see [49]). They are a family of highly hydrophilic proteins which are thought to act as a “molecular shield” playing a role in anti-aggregation and protein stabilisation [49]. Two Group 3 LEAs have previously been characterized in B. plicatilis and Q-PCR analyses showed their expression was significantly up-regulated in resting eggs and females with resting eggs [26], [50]. The presence of these LEAs in the transcription profile of resting eggs clearly substantiates this previous result and confirms their importance in both the maintenance of this physiologically “inert” state and desiccation tolerance, if this occurs. Although in general rotifer resting eggs are not desiccated, given particularly adverse conditions they can survive in a desiccated state. This could potentially explain the presence of the LEA proteins and also DNA repair enzymes as a preparative measure.
The LEAs are the only desiccation tolerance proteins identified in B. plicatilis to date. Trehalose is the classic anhydroprotectant (first identified in Artemia cysts [51], [52]), which stabilises biomacromolecules and the membrane bi-layer structure under stressful conditions [53]. However the transcription profiles only indicate the presence of trehalase in resting eggs. This is the enzyme which breaks down trehalose. To date, only very small amounts of trehalose have been measured in B. plicatilis resting eggs [54] with no up-regulation of trehalose production [26]. This is further verified by our data. The presence of trehalase could indicate that the small amount of trehalose present in resting eggs is actually being broken down to glucose. This could then be used as a primary energy source, prior to lipid utilisation from the lipid bodies described below. Non-reducing sugars such as trehalose, sucrose and oligosaccharides, in relatively high concentrations, are characteristic of orthodox plant seeds and provide an increased tolerance to abiotic stress [2].
Additional putative functions
Whilst resting eggs may be regarded in physiological terms as relatively inert or dormant, the expression profile data indicate otherwise. There are putative gene identifications in many categories, including those associated with transcription factors, cell division and cell proliferation, production of new RNAs and proteins via transcription, translation and RNA metabolism, protein turnover and general cellular metabolism. These cannot all be discussed in detail, but certain transcripts warrant highlighting as potential candidates for further investigation, either in their own right or because of the indication of biochemical pathway activation in the resting egg.
Apoptosis and cellular differentiation
Stressful conditions often lead to programmed cell death. There are three transcripts present; BAP-31, an integral ER membrane protein, misato homolog 1 and peptidase C14, a caspase catalytic subunit. All are key players in apoptosis, however processes concerned with cell death are not conducive to long-term survival and recovery during dormancy. In Artemia, whilst a similar complement of genes is present in the apoptotic pathway, these are not subject to the same control mechanisms as mammalian systems [55]. Potentially this is the case here, especially as it has been found that caspases in other invertebrates (e.g. C. elegans and Drosophila) indicate that they may be involved in non-lethal roles such as cellular differentiation processes [56]; cell proliferation, adhesion and differentiation [57]. Indeed there are other transcripts present which are involved in cellular differentiation and proliferation processes in invertebrates. These include headcase which is involved in adult morphogenesis in Drosophila [58], ependymin, a cell adhesion molecule up-regulated during regeneration in the echinoderm Holothuria glaberrima [59]; inorganic pyrophosphatase associated with larval development in the round worm Ascaris lumbricoides [60] and tetraspanin, which has been termed a molecular facilitator [61] and is expressed in diverse developmental stages in the tobacco hornmoth Manduca sexta [62]. There are also transcripts present which have been implicated in neuronal functions, such as the serine/threonine protein phosphatase which shows highest sequence similarity to PPEF-1 in Homo sapiens and sulfotransferase [63]. The presence of these transcripts is particularly interesting as they imply continued morphological development in the egg.
Lipid metabolism
Also of significant interest for future research are genes potentially involved in lipid metabolism. Methyltransferase-like 7 transcripts are involved in lipid droplet formation [64]. Dormancy has been associated with a function for lipids and lipid metabolism and lipid droplets were reported to occur in rotifer resting eggs [22]. Lipid metabolic pathways are up-regulated in the dormant dauer stage of C. elegans [65] and lipids serve as the main energy source during hibernation [66]. Unexpectedly Pauwels et al [67] did not find differences in the triglycerol content between subcutaneous and dormant eggs in Daphnia magna, however, newborn Brachionus calyciflorus females hatching from resting eggs had many more lipid droplets in their tissues than similar sized newborn females from parthenogenetic eggs [68]. Cathepsin L- protease was found essential for yolk processing and degradation during embryogenesis in C. elegans [69] and possibly may also have a function in eggs, as several transcripts for cathepsin-L-like cysteine proteinase were found to be up-regulated in resting eggs but also in amictic eggs.
To add to this novel identification is the presence of other genes which potentially protect lipid stores. The Krüppel-like transcription factor; Copeb-prov protein is up-regulated during lipid peroxidation in rats [70] and certain glutathione-s-transferases in Drosophila are known to be involved in combating oxidative stress and metabolism of endogenously formed lipid peroxidation products [71]. Transcripts for a selenium binding protein were up-regulated in resting eggs. Selenoproteins function as antioxidants and can decrease lipid peroxidation in Drosophila [72] and a selenoprotein was found to be up-regulated during diapause in female Culex pipiens [73].
Defence and protection
Hydrated resting eggs have specific external protective layers that are not found in amicitic eggs (Figure 1B) [22]. They are exposed during their long dormant period to bacteria and fungi that may affect their survival and reduce hatching [74]. A few transcripts associated with the immune function were up-regulated in resting eggs. These include Toll-like receptor 3 that could serve against viral infection [75], peptidase C14, a caspase mediating programmed death [76], F-box and WD-40, associated with the cellular proteolytic machinery and dauer development [46] and lanthionine synthetase C-like protein, representing a family associated peptide modifying enzyme components in eukaryotic cells and immune response to protozoan infection in an oyster [77]. F-box/WD-40 was also expressed in Artemia diapause destined embryos [47]. The higher expression of genes related to defence and protection was demonstrated for dormant plant seeds [42].
Finally, of particular interest in this list of differentially expressed genes was the presence of transcripts involved in the production of messenger ribonucleoprotein particles. These were reported in both Artemia and plant seeds and are associated with stored mRNA pools. These stabilise RNA and inhibit translation, providing a viable source of useful RNAs which can be immediately activated on the exit from dormancy [78], [79].
Transcripts up-regulated in amictic eggs
Although there were some shared functional categories between the transcripts differentially expressed in resting and amictic eggs, such as metabolism, signalling, immune function etc, the relative balance of these in each type of egg differs and there was a very distinct partitioning of key genes and functions (Table 4, Table S4).
Cytoskeleton
By far the vast majority of identifications were transcripts potentially involved in the cytoskeleton and development and functioning of muscle. These included smoothelin, a muscle cell specific cytoskeletal protein, which is developmentally regulated in vertebrates [80]. The phenotype of smooth muscle cells varies depending on origin, location or function and can change during maturation. Smooth muscle cells express a combination of proteins characteristic of the degree of maturation and smoothelin is one of these markers. Transcripts were also present for calponin, a component of smooth muscle and numerous myosins. The latter are an extensive gene family, the functions of which are largely unknown but have been implicated in large repertoire of cellular functions including cell migration, cell adhesion, organelle transport, receptor mediated and fluid phase endocytosis, mRNA transport and transcription, along with their interactions with the cytoskeleton (e.g. [81]. Increased activity of the cytoskeleton has direct links to and indeed, a requirement for, cell signalling. In the list of differentially expressed genes in amictic eggs (Table S4), calmodulin, in particular, mediates the control of a large number of enzymes and other proteins by calcium. It directly interacts with another transcript (enkurin) present in this dataset. Likewise, the CAPSL protein transcript contains an EF hand domain, which is characteristic of signalling molecules including calmodulin.
Morphological development
The preponderance of cytoskeletal genes in the transcription profile of amictic eggs accords with the morphological development of the egg into a juvenile rotifer. This is substantiated by the detection of transcripts specifically involved cell proliferation and adhesion such as ependymin and countin, which regulates cell adhesion and controls cell numbers [82]. There were also transcripts active in development, the prime example being notch. This gene family is highly conserved throughout the animal kingdom and is involved in diverse developmental and physiological pathway lineage decisions, boundary segregations, neurogenesis and cell fates [83]. There were signals of increasing neuronal development with the identification of innexins. These are involved in neurogenesis and are involved in neuronal development in the leech [84]. Of particular note was the presence of the prohormone vasotocin. Relatively little is known about the functioning of neurosecretory cell types in invertebrates. Database mining shows that annelids and molluscs have preserved a large fraction of vertebrate-type neuropeptides, including vasotocin. However, this hormone is not ubiquitous to invertebrates, being absent in both Drosophila or C. elegans. This prohormone is expressed in neurosecretory cells and involved in the neuronal development of annelid worms [85]. The identification of this transcript in the rotifer is clearly of interest in terms of organism development, but will also be of more general interest to evolutionary biologists. Complementary to the work on Artemia, was the detection of a transcript with high sequence similarity to SPARC (secreted protein, acidic, rich in cysteine): also termed osteonectin and BM-40. This is a calcium binding glycoprotein expressed in extracellular matrices of various cell types undergoing morphogenesis, development, remodelling and wound healing. This gene was identified in Artemia and whilst faintly expressed in embryos, was considerably up-regulated in prenauplii and nauplii [86]. Whilst a developmental role is proposed for this protein in Artemia, phylogenetic analysis showed distinct separation of this sequence into vertebrates and invertebrates, suggesting that there are, as yet, novel unidentified functions in invertebrates.
Lipid metabolism
Finally, was the identification of a group of transcripts potentially involved in lipid metabolism. While such genes were identified in resting eggs (which have already been described), the amictic eggs contained a different sub-set. Calveolin is one of a limited group of proteins associated with lipid bodies [87]. Endogenous calveolin moves to lipid bodies in response to the accumulation of lipids and hence its presence substantiated findings of lipid body accumulation in rotifers [68]. However, the cellular location of this protein is not static; in regenerating liver calveolins move from the plasma membrane to newly formed lipid bodies suggesting a role for calveolin in lipid transport to/from lipid bodies and general intra cellular homeostasis of lipids [87]. Additionally, in the differentially expressed genes was the presence of an ecdysteroid-regulated 16 kDa protein. This molecule contains a lipid recognition domain and has been shown to be involved in protein uptake in larval fat body in Drosophila [88]. Also present was a delta 6 fatty acid desaturase. This is a component of the lipid metabolic pathway that catalyzes biosynthesis of highly unsaturated fatty acids from precursor essential polyunsaturated fatty acids such as linoleic acid. These support previously published results on synthesis of unsaturated acids in rotifers [89]. Highly unsaturated fatty acids play pivotal roles in many biological functions. Combined with the results of lipid metabolism genes in resting eggs, these identifications indicated the potential dynamic nature of lipid metabolism in rotifer life history stages, providing candidate genes for future analyses of the requirements for entry and exit into dormancy of this species.
Comparative analyses with other species
Considering the results, the most surprising, was the identification of numerous transcripts in the resting eggs that encode genes for essential biochemical pathways (Table 4). This is counter-intuitive, given the knowledge that B. plicatilis resting eggs can remain dormant for tens of years and still remain viable [16], [18], [19]. However if this biochemical turnover were in a continual state then energy stores would rapidly become depleted and the eggs inviable, a situation clearly at odds with long term survival in the dormant state. While there are no reports on metabolic rates of rotifer dormant eggs, the decrease in embryo size with time in sediments supports the notion that dormant or diapausing eggs are metabolically active albeit at a very low level [19]. Also observations showing survival in resting eggs collected from pond sediments, indicates a dependency of long-term survival on environmental conditions [19]. However there are a small number of examples of hydrated resting eggs from marine and freshwater systems which exhibit long term survival in this dormant form [90]. In particular is the example of Artemia, where intensive investigations of the metabolic activity of hydrated eggs under anoxic conditions failed to reveal any measureable metabolism over four years [91]. The mechanism by which these species can survive and remain viable under such conditions for long periods of time is, as yet, unknown.
It is also curious that there was an increased complexity of functions associated with resting eggs compared to amictic eggs. One reason to explain why there is transcriptional activity associated with the resting eggs is that when they are formed, they are not “mature” enough to survive a dormant period. This is substantiated by findings that during this maturation period the eggs cannot be hatched and will not survive the dormant period, similar to plant seeds [27], [42]. It is suggested that during this obligatory diapause stage, metabolic changes can occur, but to date there is no description of their nature. These transcription profiles are potentially the first snapshot of this period.
Artemia
In response to adverse conditions, embryos of the Artemia enter an anhydrobiotic dormant state in the form of a cyst. This is associated with developmental arrest and cessation of virtually all metabolism and DNA, RNA and protein synthesis [92]. The organism may remain in this ametabolic state for decades, but still remain viable if the appropriate environmental signals are applied [93]. On rehydration, the cysts almost instantaneously resume development and hatch as naupilus larvae. Dormancy in this species has been studied at various levels for almost 30 years and is one of the best studied systems in this regard. In particular work has concentrated in two inter-linked areas: metabolic arrest and lack of activation of programmed cell death (apoptosis).
Transcription and translation studies showed that dormant embryos contain significant amounts of mRNA. Comparisons of active and dormant organisms indicated that both states contained similar quantities of translatable mRNA, hence there was no net degradation of mRNA pools during dormancy. The conclusion being that control was at the translation level with an arrest of protein synthesis [94], [95]. The exact nature of how this is effected in the embryo (in terms of cell signalling) has yet to be accurately defined, but there are almost certainly several factors involved.
Early measurements on nucleotide metabolism showed an 80% reduction in ATP levels in first hour of embryonic anoxia [96]. This dramatic reduction in cellular energy levels could potentially explain the rapid in vivo arrest, without the detail of the mechanisms. Dormant embryos contain large quantities of messenger ribonucleoprotein particles [78] and it is thought that their association with the stored mRNA pool performs two roles: mRNA stabilisation and inhibition of translation [79]. This is potentially allied to the finding of an extension of mRNA half-lives and transcription initiation reduced to approximately 90% of normoxic levels in short-term treated anoxic embryos [97]. p26, a small heat shock protein is also mobilised from the cytoplasm to the nucleus during anoxia and reversed under normoxia [98]. This molecule possesses molecular chaperone activity, but has also been implicated in transcriptional arrest [99]. No doubt, it is a combination of factors that lead to the successful induction of dormancy, but with regard to the maintenance of this state, programmed cell death must also be inhibited.
The latter is an automatic reaction of mammalian cells to an environmental challenge. This produces an uncoupling of respiration and release of cytochrome c in the mitochondria, with a sudden increase in permeability of the inner mitochondrial membrane to solutes with molecular masses of up to 1500 Da. This is known as the mammalian mitochondrial permeability transition. Cytochrome c in mitochondria is essential for oxidative phosphorylation, but its release into cytoplasm initiates assembly of the apoptosome, the molecular machinery activating the caspase 9 pathway and programmed cell death. Mitochondrial permeability and caspase activation in Artemia are not subject to the same control mechanisms as mammalian systems [55]. Artemia caspase activation is not dependant on cytochrome c, but is regulated (at least partly) by nucleotide concentrations. Therefore similarly to metabolic arrest, lack of caspase activation is consistent with energetic trade-offs occurring as a result of rapid and dramatic energy limitation [55]. Putative transcripts for BAP-31 and a caspase were identified in the resting eggs and these are obvious candidates for future studies to further our understanding of cell death processes and their control in different species.
Plant seeds
The relationship of the resting eggs to plant seeds, although sharing similarities with Artemia, is proving slightly more complex to dissect. Like Artemia, dry mature seeds contain a large number of stored mRNAs and again translation is thought to be the major level of control [100]. These transcriptionally competent mRNAs are not thought to be actively transcribed in vivo due to severely reduced moisture levels [101]. Whilst over half the stored mRNA species are remnants from embryogenesis and seed maturation, the others provide important RNA templates for protein synthesis during the early stages of germination. Indeed the most highly expressed 2–3% of mRNAs in stored seeds encode functions associated with metabolism, protein synthesis and degradation [101]. Hence when conditions are suitable for germination, the cellular machinery can rapidly produce the required proteins with minimal investment in transcription. However, several recent studies have detected low levels of both transcription and translation in plant seeds (reviewed in [102]). Seeds can exist in many states of partial imbibition, therefore “dry”, in at least some plant species, may mean low-hydrated with regional compartmentalisation of hydration states. This could mean that seeds maintain a number of more hydrated areas with limited capacity for complex cellular interactions. How relevant this is to the final germination process is still under evaluation, but offers tangible opportunities for laser micro-dissection studies [102].
The work described here and many of the reference studies involve analyses of mRNA transcription profiles. These are undertaken with the a priori hypothesis that such profiles and changes in levels of transcript expression are indicative of the general functioning and responses of the organism under study. How changes in gene transcript abundances translate into production of proteins and whole animal functions is an area of considerable interest and clearly impact on the relevance of our conclusions. A recent investigation in a prokaryote (Mycoplasma pneumonia) indicated that regulation of gene expression was largely decoupled from protein dynamics, but this did depend on which genes were analysed, with strong correlations recorded for some, e.g. heat shock proteins [103]. The situation in eukaryotes may differ. Earlier work on whole animals showed a general correlation of mRNA synthesis with protein production, but the timing and magnitude of protein production was not consistently predicted by mRNA concentration [104]. A more extensive tissue-specific study noted that differences in gene expression reflected well-established tissue-specific metabolic requirements, suggesting that measures of gene expression accurately reflected changes in proteins and their phenotypic effects [105]. More recent data have shown that expression levels between orthologous proteins and mRNAs, were generally positively correlated [106], in fact, much better than previously thought with around 40% of the variance in protein abundance explained by mRNA levels [107]. This new understanding has been possible due to methodological improvements and the ability to survey increasingly large numbers of transcripts/proteins in the same experiment [107]. Almost certainly, our abilities to undertake such fine-scale studies will increase in the future, enabling us to disentangle the complex relationship between mRNAs, proteins and whole animal functioning.
Summary
Amictic eggs showed a consolidated transcriptional profile associated with cell proliferation, cytoskeletal remodelling and development, which matches the physiological changes that the egg undergoes as it matures into a juvenile rotifer. However resting eggs revealed a more a complex transcriptional profile associated with many different cellular processes. Knowledge of rotifer biology and comparisons with other species indicate several possible reasons for this observation. The first of which is that the resting eggs are still maturing. Secondly there is abundant evidence from both Artemia and plant seeds that there is a preparative storing of useful transcripts with a cessation of protein synthesis, so that on emergence from dormancy the organism only has to activate translation for the egg to develop, not both processes of transcription and translation (which is far more energetically costly, when nutrients are limiting). It is particularly interesting to note that the transcription factors identified were all present in the resting egg state and are often associated with cell proliferation and differentiation in other species (e.g. MYND and SET domains, and Krüppel-like factors [108], [109].
Resting eggs represent forms entering dormancy during embryonic development but are not necessarily desiccated. The main characteristic of this form of dormancy is the suspension of development. In forms undergoing desiccation such as Artemia cysts or orthodox plant seeds, metabolic activity will be dramatically reduced due to the intracellular glassy state and high viscosity. It is not clear how metabolism is suspended in hydrated forms, such as rotifer, cladoceran and crustacean resting eggs but its duration may depend on energy stores such as lipids, protection against invasive microorganisms and possibly an altered programmed cell death pathway. There are clearly critical adaptations to survival similar to other species undergoing dormancy or diapause, producing abundant amounts of chaperones, antioxidants, and LEA proteins but more studies are needed to unveil the regulation of these processes in dormant embryos of invertebrates. Indeed, although this rotifer study does represent a substantial increase in expression data related to the dormancy process, it is a single time point assay and to understand what may be going on more detailed sampling may be required in the future. The transcriptional basis of dormancy seems to differ substantially between species, reflecting the multiple evolutionary origins and life strategies. Yet, dormancy always involves reduced metabolism, up-regulation of stress resistance and upholding of cellular or protein structure. Investigations into long term survival in hydrated embryos could lead to developing methods for long term preservation of hydrated cells, in analogy with achievements in the dry preservation of human cells, which stemmed from investigations into long-term survival and desiccation tolerance of Artemia cysts [110].
Materials and Methods
Rotifer samples
Brachionus plicatilis rotifers were hatched from resting eggs produced in the laboratory from rotifers collected at a seaside pond in Atlit, (40 km south of Haifa, Israel) in 1981. Some of the resting eggs were hatched in 2003 and resting eggs produced from them were stored in the laboratory at 4°C and hatched in 2005. Resting eggs that were hatched in 2005 served to produce several batches of resting eggs that were stored at 4°C. Four different cultures were started, originating from four different batches of resting eggs. These eggs were hatched in an illuminated culture room (at 25°C) and the salinity of the seawater culture medium was 10 ppt. In general, rotifers were cultured as previously described in [26], [111]. Resting eggs first appeared to be carried by mictic females, 3–5 days after hatching and the resting eggs were collected 14–25 days after hatching of the parental resting eggs. In order to assess the viability of the resting eggs that were collected for RNA extraction, resting eggs from two replicate cultures, were hatched after 0, 2, 4, 6 and 8 weeks of collection. Between 47–49% of the resting eggs hatched after 8 weeks of storage in the dark at 4°C (Table S2). The proportion of viable eggs is probably higher, as previous results showed that resting eggs that do not hatch in the first attempt hatch later, under slightly different conditions [27].
Collection of resting eggs
Resting eggs were collected from the bottom layer of the culture flasks. Aliquots of 10 ml collected from bottom layer were placed in a large petri-dish and resting eggs were picked manually and removed from the amictic eggs and rotifers found in this layer. RNA was extracted immediately. Four biological replicate experiments were performed, with each replicate originating from resting eggs produced by a different culture. About 5,000 resting eggs were used for RNA extraction from each biological replicate sample.
Collection of amictic eggs
Samples of amictic females (FA) were manually picked from each culture, when the resting-egg production rate was at its peak. An amictic female was recognized by the relatively large opaque eggs it carried and only females with eggs were manually collected. Rotifers for each replicate were suspended in fresh sterile seawater (10 ppt) and sieved with 60 µm plankton nets, washed with sterile diluted sea-water medium (10 ppt) and re-suspended in 10–15 ml of sterile seawater (10 ppt) in a 15 ml round bottom disposable vial. The amictic eggs (AE) were collected by intensive up and down movement with a five ml pipette tip (‘re-pipetting”), of five ml samples containing amictic females. In this way, the amictic eggs were shed from the females and could be collected from the bottom of the vial. They were manually cleaned from newborn females that continuously hatched from the eggs. Hatching of the amictic eggs was stopped by adding ethanol (10% final concentration), whereby the eggs settled at the bottom of a vial, the supernatant was removed, and the sample was frozen in liquid nitrogen. The eggs were stored at −70°C until RNA extraction. Four biological replicate experiments were performed with approximately 5,000 eggs used for RNA extraction from each biological replicate sample.
Library construction and sequencing
RNA was extracted from resting and amictic eggs using TRIzol reagent (Invitrogen) according to manufacturer's instructions. Total RNA was extracted from each one of the four biological replicates of amictic eggs and four biological replicates of resting eggs. A pooled RNA sample was formed from equal amounts of RNA from each replicate of resting eggs and similarly, a pooled RNA sample was formed from equal amounts of RNA from each replicate of amictic eggs. After measurement and quality control 1.5 µg of total RNA (from the pooled RNA sample of resting egg sample or pooled RNA sample of amictic egg RNA) was used as template for cDNA synthesis, by the Mint cDNA synthesis kit (Evrogen, Russia) following the manufacturer instructions. The amplified cDNA was purified (QIAquick PCR Purification Kit, Qiagen). The cDNA was measured and quality checked by gel electrophoreses. An amount of at least 8 µg of cDNA per replicate with a size distribution between 0.5–3 kbp was obtained. Library generation and sequencing were performed using the Illumina sequencing platform (Genome Analyzer, Illumina) according to the manufacturer's instructions for shotgun sequencing. Each of the two sequenced libraries represented approximately 15,000 individuals for each of the amictic eggs and resting eggs. A full description of the sequencing protocols is described in [111]. We previously reported on the construction and Illumina sequencing of 13 libraries from different life stages of Brachionus plicatilis and the current study focuses on two of these libraries [111].
Analyses of data from Illumina sequencing
Sequences were extracted using Gerald from the Illumina Pipeline v1.0 with all possible basecalls and with standard QC turned on (chastity filter > = 0.6). The sequences (reads) from the libraries were aligned against rotifer ∼18,000 EST sequences [26] using SOAP [112] allowing 2 mismatches within 32 bp. Prior to that, reads containing more than two Ns were filtered and discarded. For each library, the sum of all aligned reads of their specific contig was calculated (equal to transcripts in this case), not keeping track of the uniqueness. Because the number of total reads varied between libraries (due to technical differences stemming from PCR amplification, library preparation etc.), a simple scaling method was used to facilitate comparisons between them (described in [111]). The analysis for differential expression was carried out using R [113] with standard chi-square test, Fisher exact test and a self implemented version of Audic and Claverie bayesian approach [114]. A transcript was considered to be changed differentially if it matched the following cases:
state1/state2< = 0.5 OR state1/state2> = 2.0
#{reads.state1}> = 50 OR #{reads.state2}> = 50
one of the three statistical test p-values< = 0.01
The backbone contigs (and therefore short read mappings) were annotated via database searches using Blastx [115] against the NCBI database with matches annotated for all scores with an expect score in excess of 1e-10. All transcript matches presented in the results were manually verified. Sequences with a database match were then further annotated using GO [116]. Further sequence manipulation was carried out using the EMBOSS suite of programmes [117]. In order to classify the genes by functional categories of GO terms, enrichment analysis was performed using the hypergeometric distribution function. Multiple test corrections of p-values were performed by q value calculations using the R-package “qvalue”. Significant-thresholds for selections were p-value< = 0.05, q-value< = 0.1. Verification of the two transcription profiles were performed across the biological replicates using Q-PCR, evaluated and described for 13 libraries [111] and is shown here for the libraries formed from amictic eggs and resting eggs (Figure S1). Files containing the reads have been submitted to EBI (ERA051893).
Supporting Information
Q-PCR validation of Illumina sequencing results for resting eggs (RE, top panel) and amictic eggs (AE, lower panel). The relative abundance of twelve genes (list of primers is shown in [111]), was normalized using the equation: ratio = (Etarget)CP_target/(Eef1a)CP_reference where E = 10-1/slop [118]. The median was calculated for all transcript ratios and the transcript ratios were displayed as the log2[{transcript ratio of a sample}/{median of transcript ratios of all samples}]. It was very difficult to find one gene that could serve as a reference for all samples as ATP synthase (atps) changed between amictic eggs and resting eggs and elongation factor 1 alfa (ef1a) changed between females and males. Therefore, the relative abundance of transcripts was normalized to ef1a in the comparison made with the eggs samples but atps was used for comparisons between female and male samples. The expression levels obtained with real-time PCR were compared with the expression levels obtained from the number of Illumina reads for each corresponding gene. Regression analysis revealed high correlation between expression values obtained by real-time RT-PCR and number of reads for each transcript for the resting eggs (Person r = 0.980) and amictic eggs (Pearson r = 0.957). Negative values in real-time PCR indicate that the expression level of a specific transcript, was lower than that of the median. For Illumina reads, a negative value indicates that the number of reads assigned to a specific gene transcript were lower than the median value of the reads in a specific library.
(TIFF)
Basic information obtained by Illumina sequencing technology of libraries constructed from amictic (AE) and resting (RE) eggs. The total number of reads was circa 16 and 14 million for the amictic and resting egg libraries, respectively. Analysis of the data was performed as described in [111] and included scaling of the data, aligning of the reads with the sequences of EST data base obtained previously [26] and filtering of ESTs with less than 50 reads per EST. Only 69% and 73% of the final ESTs showed significant matches against proteins using Blastx sequence similarity searching.
(RTF)
Hatching dynamics of resting eggs collected from two batches that served for RNA extraction.
(RTF)
Differentially expressed transcripts in resting eggs with associated BLAST sequence similarity data and putatively assigned functions.
(XLS)
Differentially expressed transcripts in amictic eggs with associated BLAST sequence similarity data and putatively assigned functions.
(XLS)
Acknowledgments
We thank Mrs. Irena Pekarski and Ms. Aliza Hadani (Israel Oceanographic and Limnological Research, Haifa, Israel) for technical assistance in raising rotifers and collection of samples and Hana Bernard (Israel Oceanographic and Limnological Research, Haifa, Israel) for graphical assistance.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This study was financially supported by the European Community (NEST # 012674; Sleeping Beauty). The analyses were completed by the team from the British Antarctic Survey within the Ecosystems Adaptations and Physiology Work Package. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Berjak P. Unifying perspectives of some mechanisms basic to desiccation tolerance across life forms. Seed Sci Res. 2006;16:1–15. [Google Scholar]
- 2.Leprince O, Buitink J. Desiccation tolerance: From genomics to the field. Plant Sci. 2010;179:554–564. [Google Scholar]
- 3.Lubzens E, Cerdà J, Clark MS. Topics in Current Genetics: Dormancy and Resistance in Harsh Environments. Pub. Springer, Heidelberg, Dordrecht, London, New York; 2010. 283 Volume. 21. [Google Scholar]
- 4.Keilin D. The problem of anabiosis or latent life: history and current concept. Proc R Soc Lond B. 1959;150:149–191. doi: 10.1098/rspb.1959.0013. [DOI] [PubMed] [Google Scholar]
- 5.Clegg JS. Cryptobiosis - a peculiar state of biological organization. Comp Biochem Physiol B. 2001;128:613–624. doi: 10.1016/s1096-4959(01)00300-1. [DOI] [PubMed] [Google Scholar]
- 6.Ricci C. Dormancy patterns in rotifers. Hydrobiologia. 2001;446:1–11. [Google Scholar]
- 7.Alpert P. Constraints of tolerance: why are desiccation-tolerant organisms so small or rare? J Exp Biol. 2006;209:1575–1584. doi: 10.1242/jeb.02179. [DOI] [PubMed] [Google Scholar]
- 8.Lopes FL, Desmarais JA, Murphy BD. Embryonic diapause and its regulation. Reprod. 2004;128:669–678. doi: 10.1530/rep.1.00444. [DOI] [PubMed] [Google Scholar]
- 9.Denekamp NY, Suga T, Hagiwara A, Reinhardt R, Lubzens E. A role for molecular studies in unveiling the pathways for formation of rotifer resting eggs and their survival during dormancy. In: Lubzens E, Cerdà J, Clark MS, editors. Dormancy and Resistance in Harsh Environments. Pub Springer-Verlag, Berlin Heidelberg; 2010. pp. 109–132. [Google Scholar]
- 10.Bewley JD, Black M. 1994. 445 Seeds: Physiology of Development and Germination (2nd ed. New York: Plenum Press)
- 11.Finkelstein R, Reeves W, Ariizumi T, Steber C. Molecular aspects of seed dormancy. Annu Rev Plant Biol. 2008;59:387–415. doi: 10.1146/annurev.arplant.59.032607.092740. [DOI] [PubMed] [Google Scholar]
- 12.Berjak P, Pammenter NW. Recent progress towards the understanding of desiccation tolerance. In: Adkins SW, Ashmore SE, Navie SC, editors. Seeds: Biology, Development and Ecology. Wallingford: Cabi Publishing-CAB Int; 2007. pp. 17–27. [Google Scholar]
- 13.MacRae TH. Gene expression, metabolic regulation and stress tolerance during diapause. Cell Mol Life Sci. 2010;67:2405–2424. doi: 10.1007/s00018-010-0311-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hengherr S, Schill RO, Clegg JS. Mechanisms associated with cellular desiccation tolerance in the animal extremophile Artemia. Physiol Biochem Zool. 2011;84:249–257. doi: 10.1086/659314. [DOI] [PubMed] [Google Scholar]
- 15.Carvalho GR, Wolf HG. Resting eggs of lake Daphnia.1. Distribution, abundance and hatching of eggs collected from various depths in lake sediments. Freshw Biol. 1989;22:459–470. [Google Scholar]
- 16.Marcus NH, Lutz R, Burnett W, Cable P. Age, viability, and vertical distribution of zooplankton resting eggs from an anoxic basin - Evidence of an egg bank. Limnol Oceanogr. 1994;39:154–158. [Google Scholar]
- 17.Hairston NG, Vanbrunt RA, Kearns CM, Engstrom DR. Age and survivorship of diapausing eggs in a sediment egg bank. Ecol. 1995;76:1706–1711. [Google Scholar]
- 18.Kotani T, Ozaki M, Matsuoka K, Snell TW, Hagiwara A. Reproductive isolation among geographically and temporally isolated marine Brachionus strains. Hydrobiologia. 2001;446:283–290. [Google Scholar]
- 19.Garcia-Roger EM, Carmona MJ, Serra M. Patterns in rotifer diapausing egg banks: Density and viability. J Exp Mar Biol Ecol. 2006;336:198–210. [Google Scholar]
- 20.Gilbert JJ. Timing of diapauses in monogonont rotifers: mechanisms and strategies. In: Alekseev VR, De Stasio B, Gilbert JJ, editors. Diapause in aquatic invertebrates: Theory and human use. Monographiae Biologicae, volume 84. Pub. Springer, Dordrecht, The Netherlands; 2007. pp. 11–27. [Google Scholar]
- 21.Wurdak E, Gilbert JJ, Jagels R. Resting egg ultrastructure and formation of the shell in Asplanchna sieboldi and Brachionus calyciflorus. Arch Hydrobiol Beih. 1977;8:298–302. [Google Scholar]
- 22.Wurdak ES, Gilbert JJ, Jagels R. Fine structure of resting eggs of rotifers Brachionus calyciflorus and Asplanchna sieboldi. Trans American Microscop Soc. 1978;97:49–72. [PubMed] [Google Scholar]
- 23.Hagiwara A. Use of resting eggs for mass preservation of marine rotifers. Tech Rep Jap Sea Ranching Progr. 1996;24:109–120. (in Japanese) [Google Scholar]
- 24.Chen WH, Ge XM, Wang WW, Yu J, Hu SN. A gene catalogue for post-diapause development of an anhydrobiotic arthropod Artemia franciscana. BMC Genomics. 2009;10:9. doi: 10.1186/1471-2164-10-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qui Z, MacRae TH. A molecular overview of diapauses in embryos of the crustacean, Artemia franciscana. In: Lubzens E, Cerdà J, Clark MS, editors. Dormancy and Resistance in Harsh Environments. Pub. Springer-Verlag, Berlin Heidelberg; 2010. pp. 165–187. [Google Scholar]
- 26.Denekamp NY, Thorne MAS, Clark MS, Kube M, Reinhardt R, et al. Discovering genes associated with dormancy in the monogonont rotifer Brachionus plicatilis. BMC Genomics. 2009;10:108. doi: 10.1186/1471-2164-10-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Minkoff G, Lubzens E, Kahan D. Environmental factors affecting hatching of rotifer (Brachionus plicatilis) resting eggs. Hydrobiologia. 1983;104:61–69. [Google Scholar]
- 28.Hagiwara A, Hoshi N, Kawahara F, Tominaga K, Hirayama K. Resting eggs of the marine rotifer Brachionus plicatilis Müller: Development, and effect of irradiation on hatching. Hydrobiologia. 1995;313/314:223–229. [Google Scholar]
- 29.Shoshan-Barmatz V, De Pinto V, Zweckstetter M, Raviv Z, Keinan N, et al. VDAC, a multi-functional mitochondrial protein regulating cell life and death. Mol Asp Med. 2010;31:227–285. doi: 10.1016/j.mam.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 30.Kaneko G, Yoshinaga T, Yanagawa Y, Kinoshita S, Tsukamoto K, et al. Molecular characterization of Mn-superoxide dismutase and gene expression studies in dietary restricted Brachionus plicatilis rotifers. Hydrobiologia. 2005;546:117–123. [Google Scholar]
- 31.Torti FM, Torti SV. Regulation of ferritin genes and protein. Blood. 2002;99:3505–3516. doi: 10.1182/blood.v99.10.3505. [DOI] [PubMed] [Google Scholar]
- 32.Li HM, Taira T, Maita C, Ariga H, Iguchi-Ariga SMM. Protection against nonylphenol-induced cell death by DJ-1 in cultured Japanese medaka (Oryzias latipes) cells. Toxicol. 2006;228:229–238. doi: 10.1016/j.tox.2006.08.040. [DOI] [PubMed] [Google Scholar]
- 33.Kranner I, Birtic S. A modulating role for antioxidants in desiccation tolerance. Integr Comp Biol. 2005;45:734–740. doi: 10.1093/icb/45.5.734. [DOI] [PubMed] [Google Scholar]
- 34.Dickinson BC, Chang CJ. Chemistry and biology of reactive oxygen species in signalling or stress responses. Nature Chem Biol. 2011;7:504–511. doi: 10.1038/nchembio.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Turoczy Z, Kis P, Torok K, Cserhati M, Lendvai A, et al. Overproduction of a rice aldo-keto reductase increases oxidative and heat stress tolerance by malondialdehyde and methylglyoxal detoxification. Plant Mol Biol. 2011;75:399–412. doi: 10.1007/s11103-011-9735-7. [DOI] [PubMed] [Google Scholar]
- 36.Belyaeva OV, Korkina OV, Stetsenko AV, Kedishvili NY. Human retinol dehydrogenase 13 (RDH13) is a mitochondrial short-chain dehydrogenase/reductase with a retinaldehyde reductase activity. FEBS J. 2008;275:138–147. doi: 10.1111/j.1742-4658.2007.06184.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Feder ME, Hofmann GE. Heat-shock proteins, molecular chaperones, and the stress response: Evolutionary and ecological physiology. Ann Rev Physiol. 1999;61:243–282. doi: 10.1146/annurev.physiol.61.1.243. [DOI] [PubMed] [Google Scholar]
- 38.Hartl FU, Hayer-Hartl M. Protein folding - Molecular chaperones in the cytosol: from nascent chain to folded protein. Science. 2002;295:1852–1858. doi: 10.1126/science.1068408. [DOI] [PubMed] [Google Scholar]
- 39.Parsell DA, Lindquist S. The function of heat shock proteins in stress tolerance - Degradation and reactivation of damaged proteins. Ann Rev Genet. 1993;27:437–496. doi: 10.1146/annurev.ge.27.120193.002253. [DOI] [PubMed] [Google Scholar]
- 40.Chen T, Villeneuve TS, Garant KA, Amons R, MacRae TH. Functional characterization of artemin, a ferritin homolog synthesized in Artemia embryos during encystment and diapause. FEBS J. 2007;274:1093–1101. doi: 10.1111/j.1742-4658.2007.05659.x. [DOI] [PubMed] [Google Scholar]
- 41.Kumsta C, Jakob U. Redox-regulated Chaperones. Biochem. 2009;48:4666–4676. doi: 10.1021/bi9003556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cadman CSC, Toorop PE, Hilhorst HWM, Finch-Savage WE. Gene expression profiles of Arabidopsis Cvi seeds during dormancy cycling indicate a common underlying dormancy control mechanism. Plant J. 2006;46:805–822. doi: 10.1111/j.1365-313X.2006.02738.x. [DOI] [PubMed] [Google Scholar]
- 43.Kavanagh K, Jörnvall H, Persson B, Oppermann U. The SDR superfamily: functional and structural diversity within a family of metabolic and regulatory enzymes. Cell Mol Life Sci. 2008;65:3895–3906. doi: 10.1007/s00018-008-8588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jörnvall H. MDR and SDR gene and protein superfamilies. Cell Mol Life Sci. 2008;65:3875–3878. doi: 10.1007/s00018-008-8586-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fielenbach N, Antebi A. C. elegans dauer formation and the molecular basis of plasticity. Genes Dev. 2008;22:2149–2165. doi: 10.1101/gad.1701508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reynolds JA, Hand SC. Embryonic diapause highlighted by differential expression of mRNAs for ecdysteroidogenesis, transcription and lipid sparing in the cricket Allonemobius socius. J Exp Biol. 2009;212:2074–2083. doi: 10.1242/jeb.027367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qiu Z, Tsoi SCM, MacRae TH. Gene expression in diapause-destined embryos of the crustacean, Artemia franciscana. Mech Dev. 2007;124:856–867. doi: 10.1016/j.mod.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 48.Galau GA, Hughes DW, Dure L. Abscisic acid induction of cloned cotton Late Embryogenesis-Abundant (LEA) messenger RNAs. Plant Mol Biol. 1986;7:155–170. doi: 10.1007/BF00021327. [DOI] [PubMed] [Google Scholar]
- 49.Tunnacliffe A, Hincha DK, Leprince O, Macherel D. LEA proteins: Versatility of form and function. In: Lubzens E, Cerdà J, Clark MS, editors. Dormancy and Resistance in Harsh Environments. Pub. Springer-Verlag, Berlin Heidelberg; 2010. pp. 91–108. [Google Scholar]
- 50.Denekamp NY, Reinhardt R, Kube M, Lubzens E. Late Embryogenesis Abundant (LEA) proteins in nondesiccated, encysted, and diapausing embryos of rotifers. Biol Repro. 2010;82:714–724. doi: 10.1095/biolreprod.109.081091. [DOI] [PubMed] [Google Scholar]
- 51.Dutrieu J. Observations biochimique et physiologiques sur le développement d'Artemia salina, Leach. Archives de zoologie Experimentale et Génerale. 1960;99:1–134. [Google Scholar]
- 52.Clegg JS. Artemia cysts as a model for the study of water in biological systems. Method Enzymol. 1986;127:230–239. doi: 10.1016/0076-6879(86)27019-6. [DOI] [PubMed] [Google Scholar]
- 53.Crowe JH, Crowe LM. Preservation of mammalian cells - learning nature's tricks. Nat Biotechnol. 2000;18:145–146. doi: 10.1038/72580. [DOI] [PubMed] [Google Scholar]
- 54.Caprioli M, Katholm AK, Melone G, Ramlov H, Ricci C, et al. Trehalose in desiccated rotifers: a comparison between a bdelloid and a monogonont species. Comp Biochem Physiol A. 2004;139:527–532. doi: 10.1016/j.cbpb.2004.10.019. [DOI] [PubMed] [Google Scholar]
- 55.Hand SC, Menze MA. Mitochondria in energy-limited states: mechanisms that blunt the signalling of cell death. J Exp Biol. 2008;211:1829–1840. doi: 10.1242/jeb.000299. [DOI] [PubMed] [Google Scholar]
- 56.Abraham MC, Shaham S. Death without caspases, caspases without death. Trends Cell Biol. 2004;14:184–193. doi: 10.1016/j.tcb.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 57.Busso D, Dominguez C, Perez-Acle T, Moreno RD. Life-giving caspases: revealing new roles during mouse embryo preimplantation development. Int J Dev Biol. 2010;54:857–865. doi: 10.1387/ijdb.092921db. [DOI] [PubMed] [Google Scholar]
- 58.Weaver TA, White RAH. Headcase, an imaginal specific gene required for adult morphogenesis in Drosophila melanogaster. Development. 1995;121:4149–4160. doi: 10.1242/dev.121.12.4149. [DOI] [PubMed] [Google Scholar]
- 59.Suarez-Castillo EC, Medina-Ortiz WE, Roig-Lopez JL, Garcia-Arraras JE. Ependymin, a gene involved in regeneration and neuroplasticity in vertebrates, is overexpressed during regeneration in the echinoderm Holothuria glaberrima. Gene. 2004;334:133–143. doi: 10.1016/j.gene.2004.03.023. [DOI] [PubMed] [Google Scholar]
- 60.Islam MK, Miyoshi T, Kasuga-Aoki H, Isobe T, Arakawa T, et al. Inorganic pyrophosphatase in the roundworm Ascaris and its role in the development and molting process of the larval stage parasites. Eur J Biochem. 2003;270:2814–2826. doi: 10.1046/j.1432-1033.2003.03658.x. [DOI] [PubMed] [Google Scholar]
- 61.Maecker HT, Todd SC, Levy S. The tetraspanin superfamily: Molecular facilitators. FASEB J. 1997;11:428–442. [PubMed] [Google Scholar]
- 62.Todres E, Nardi JB, Robertson HM. The tetraspanin superfamily in insects. Insect Mol Biol. 2000;9:581–590. doi: 10.1046/j.1365-2583.2000.00222.x. [DOI] [PubMed] [Google Scholar]
- 63.Falany CN, Xie XW, Wang J, Ferrer J, Falany JL. Molecular cloning and expression of novel sulphotransferase-like cDNAs from human and rat brain. Biochem J. 2000;346:857–864. [PMC free article] [PubMed] [Google Scholar]
- 64.Zehmer JK, Bartz R, Bisel B, Liu PS, Seemann J, et al. Targeting sequences of UBXD8 and AAM-B reveal that the ER has a direct role in the emergence and regression of lipid droplets. J Cell Sci. 2009;122:3694–3702. doi: 10.1242/jcs.054700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang J, Kim SK. Global analysis of dauer gene expression in Caenorhabditis elegans. Development. 2003;130:1621–1634. doi: 10.1242/dev.00363. [DOI] [PubMed] [Google Scholar]
- 66.Dark J. Annual lipid cycles in hibernators: Integration of physiology and behavior. Annu Rev Nutr. 2005;25:469–497. doi: 10.1146/annurev.nutr.25.050304.092514. [DOI] [PubMed] [Google Scholar]
- 67.Pauwels K, Stoks R, Verbiest A, De Meester L. Biochemical adaptation for dormancy in subitaneous and dormant eggs of Daphnia magna. Hydrobiologia. 2007;594:91–96. [Google Scholar]
- 68.Gilbert JJ. Females from resting eggs and parthenogenetic eggs in the rotifer Brachionus calyciflorus: lipid droplets, starvation resistance and reproduction. Freshwater Biol. 2004;49:1505–1515. [Google Scholar]
- 69.Britton C, Murray L. Cathepsin L protease (CPL-1) is essential for yolk processing during embryogenesis in Caenorhabditis elegans. J Cell Sci. 2004;117:5133–5143. doi: 10.1242/jcs.01387. [DOI] [PubMed] [Google Scholar]
- 70.Stärkel P, Sempoux C, Leclercq I, Herin M, Deby C, et al. Oxidative stress, KLF6 and transforming growth factor-beta up-regulation differentiate non-alcoholic steatohepatitis progressing to fibrosis from uncomplicated steatosis in rats. J Hepatol. 2003;39:538–546. doi: 10.1016/s0168-8278(03)00360-x. [DOI] [PubMed] [Google Scholar]
- 71.Sawicki R, Singh SP, Mondal AK, Benes H, Zimniak P. Cloning, expression and biochemical characterization of one Epsilon-class (GST-3) and ten Delta-class (GST-1) glutathione S-transferases from Drosophila melanogaster, and identification of additional nine members of the Epsilon class. Biochem J. 2003;370:661–669. doi: 10.1042/BJ20021287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Morozova N, Forry EP, Shahid E, Zavacki AM, Harney JW, et al. Antioxidant function of a novel selenoprotein in Drosophila melanogaster. Genes Cells. 2003;8:963–971. doi: 10.1046/j.1365-2443.2003.00687.x. [DOI] [PubMed] [Google Scholar]
- 73.Robich RM, Rinehart JP, Kitchen LJ, Denlinger DL. Diapause-specific gene expression in the northern house mosquito, Culex pipiens L., identified by suppressive subtractive hybridization. J Insect Physiol. 2007;53:235–245. doi: 10.1016/j.jinsphys.2006.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Balompapueng MD, Hagiwara A, Nozaki Y, Hirayama K. Preservation of resting eggs of the euryhaline rotifer Brachionus plicatilis O. F. Muller by canning. Hydrobiologia. 1997;358:163–166. [Google Scholar]
- 75.Zheng L, Zhang L, Lin H, McIntosch MT, Malacrida AR. Toll-like receptors in invertebrate innate immunity. Invertebrate Survival J. 2005;2:105–113. [Google Scholar]
- 76.Vermelho AB, Branquinha MH, D'Avila-Levy CM, dos Santos ALS, de Souza Dias EP, et al. Biological roles of peptidases in trypanosomatids. Open Parasitol J. 2010;4:5–23. [Google Scholar]
- 77.Tanguy A, Guo XM, Ford SE. Discovery of genes expressed in response to Perkinsus marinus challenge in Eastern (Crassostrea virginica) and Pacific (C. gigas) oysters. Gene. 2004;338:121–131. doi: 10.1016/j.gene.2004.05.019. [DOI] [PubMed] [Google Scholar]
- 78.Slegers H, Deherdt E, Kondo M. Non polysomal poly(A)-containing messenger ribonucleoproteins of cryptobiotic gastrulae of Artemia salina. Eur J Biochem. 1981;117:111–120. doi: 10.1111/j.1432-1033.1981.tb06309.x. [DOI] [PubMed] [Google Scholar]
- 79.Hofmann GE, Hand SC. Comparison of messenger RNA pools in active and dormant Artemia franciscana embryos - Evidence for translational control. J Exp Biol. 1992;164:103–116. doi: 10.1242/jeb.164.1.103. [DOI] [PubMed] [Google Scholar]
- 80.Deruiter MC, Rensen SSM, Coolen G, Hierck BP, Bergwerff M, et al. Smoothelin expression during chicken embryogenesis: Detection of an embryonic isoform. Dev Dyn. 2001;221:460–463. doi: 10.1002/dvdy.1156. [DOI] [PubMed] [Google Scholar]
- 81.Cameron RS, Liu CD, Mixon AS, Pihkala JPS, Rahn RJ, et al. Myosin16b: the COOH-tail region directs localization to the nucleus and overexpression delays S-phase progression. Cell Motil Cytoskeleton. 2007;64:19–48. doi: 10.1002/cm.20162. [DOI] [PubMed] [Google Scholar]
- 82.Tang L, Ammann R, Gao T, Gomer RH. A cell number-counting factor regulates group size in Dictyostelium by differentially modulating cAMP-induced cAMP and cGMP pulse sizes. J Biol Chem. 2001;276:27663–27669. doi: 10.1074/jbc.M102205200. [DOI] [PubMed] [Google Scholar]
- 83.Bray SJ. Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol. 2006;7:678–689. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- 84.Dykes IM, Macagno ER. Molecular characterization and embryonic expression of innexins in the leech Hirudo medicinalis. Dev Genes Evol. 2006;216:185–197. doi: 10.1007/s00427-005-0048-1. [DOI] [PubMed] [Google Scholar]
- 85.Tessmar-Raible K, Raible F, Christodoulou F, Guy K, Rembold M, et al. Conserved sensory-neurosecretory cell types in annelid and fish forebrain: Insights into hypothalamus evolution. Cell. 2007;129:1389–1400. doi: 10.1016/j.cell.2007.04.041. [DOI] [PubMed] [Google Scholar]
- 86.Tanaka S, Nambu F, Nambu Z. Isolation of a cDNA encoding a putative SPARC from the brine shrimp, Artemia franciscana. Gene. 2001;268:53–58. doi: 10.1016/s0378-1119(01)00419-x. [DOI] [PubMed] [Google Scholar]
- 87.Pol A, Martin S, Fernandez MA, Ferguson C, Carozzi A, et al. Dynamic and regulated association of caveolin with lipid bodies: Modulation of lipid body motility and function by a dominant negative mutant. Mol Biol Cell. 2004;15:99–110. doi: 10.1091/mbc.E03-06-0368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Burmester T, Antoniewski C, Lepesant JA. Ecdysone-regulation of synthesis and processing of Fat Body Protein 1, the larval serum protein receptor of Drosophila melanogaster. Eur J Biochem. 1999;262:49–55. doi: 10.1046/j.1432-1327.1999.00315.x. [DOI] [PubMed] [Google Scholar]
- 89.Lubzens E, Marko A, Tietz A. De novo synthesis of fatty acids in the rotifer, Brachionus plicatilis. Aquaculture. 1985;47:27–37. [Google Scholar]
- 90.Marcus NH, Boero F. Minireview: The importance of benthic-pelagic coupling and the forgotten role of life cycles in coastal systems. Limnol Oceanogr. 1998;43:763–768. [Google Scholar]
- 91.Clegg JS. Embryos of Artemia franciscana survive four years of continuous anoxia: The case for complete metabolic rate depression. J Exp Biol. 1997;200:467–475. doi: 10.1242/jeb.200.3.467. [DOI] [PubMed] [Google Scholar]
- 92.Clegg JS, Golub AL. Protein synthesis in Artemia salina embryos: II. Resumption of RNA and protein synthesis upon cessation of dormancy in the encysted gastrula. Dev Biol. 1969;19:178–200. doi: 10.1016/0012-1606(69)90054-2. [DOI] [PubMed] [Google Scholar]
- 93.Warner AH, Clegg JS. Diguanosine nucleotide metabolism and the survival of Artemia embryos during years of continuous anoxia. Eur J Biochem. 2001;268:1568–1576. [PubMed] [Google Scholar]
- 94.Hofmann GE, Hand SC. Global arrest of translation during invertebrate quiescence. Proc Natl Acad Sci U S A. 1994;91:8492–8496. doi: 10.1073/pnas.91.18.8492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hardewig I, Anchordoguy TJ, Crawford DL, Hand SC. Profiles of nuclear and mitochondrial encoded mRNAs in developing and quiescent embryos of Artemia franciscana. Mol Cell Biochem. 1996;158:139–147. doi: 10.1007/BF00225840. [DOI] [PubMed] [Google Scholar]
- 96.Stocco DM, Warner AH, Beers PC. Effect of anoxia on nucleotide metabolism in encysted embryos of brine shrimp. Dev Biol. 1972;27:479–493. doi: 10.1016/0012-1606(72)90187-x. [DOI] [PubMed] [Google Scholar]
- 97.Van Breukelen F, Maier R, Hand SC. Depression of nuclear transcription and extension of mRNA half-life under anoxia in Artemia franciscana embryos. J Exp Biol. 2000;203:1123–1130. doi: 10.1242/jeb.203.7.1123. [DOI] [PubMed] [Google Scholar]
- 98.Clegg JS, Jackson SA, Warner AH. Extensive intracellular translocations of a major protein accompany anoxia in embryos of Artemia franciscana. Exp Cell Res. 1994;212:77–83. doi: 10.1006/excr.1994.1120. [DOI] [PubMed] [Google Scholar]
- 99.Clegg JS, Jackson SA, Liang P, Macrae TH. Nuclear cytoplasmic translocations of protein P26 during aerobic anoxic transitions in embryos of Artemia franciscana. Exp Cell Res. 1995;219:1–7. doi: 10.1006/excr.1995.1197. [DOI] [PubMed] [Google Scholar]
- 100.Rajjou L, Gallardo K, Debeaujon I, Vandekerckhove J, Job C, et al. The effect of alpha-amanitin on the Arabidopsis seed proteome highlights the distinct roles of stored and neosynthesized mRNAs during germination. Plant Physiol. 2004;134:1598–1613. doi: 10.1104/pp.103.036293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Nakabayashi K, Okamoto M, Koshiba T, Kamiya Y, Nambara E. Genome-wide profiling of stored mRNA in Arabidopsis thaliana seed germination: epigenetic and genetic regulation of transcription in seed. Plant J. 2005;41:697–709. doi: 10.1111/j.1365-313X.2005.02337.x. [DOI] [PubMed] [Google Scholar]
- 102.Holdsworth MJ, Finch-Savage WE, Grappin P, Job D. Post-genomics dissection of seed dormancy and germination. Trends Plant Sci. 2008;13:7–13. doi: 10.1016/j.tplants.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 103.Maier T, Schmidt A, Güell M, Kühner S, Gavin A-C, et al. Quantification of mRNA and protein and integration with protein turnover in a bacterium. Mol Syst Biol. 2011;7:511. doi: 10.1038/msb.2011.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Buckley BA, Gracey AY, Somero GN. The cellular response to heat stress in the goby Gillichthys mirabilis: a cDNA microarray and protein-level analysis. J Exp Biol. 2006;209:2660–2677. doi: 10.1242/jeb.02292. [DOI] [PubMed] [Google Scholar]
- 105.Whitehead A, Crawford DL. Variation in tissue-specific gene expression among natural populations. Genome Biol. 2005;6:R13. doi: 10.1186/gb-2005-6-2-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rees BB, Andacht T, Skripnikova E, Crawford DL. Population proteomics: Quantitative variation within and among populations in cardiac protein expression. Mol Biol Evol. 2011;28:1271–1279. doi: 10.1093/molbev/msq314. [DOI] [PubMed] [Google Scholar]
- 107.Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, et al. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 108.Gottlieb PD, Pierce SA, Sims RJ, Yamagishi H, Weihe EK, et al. Bop encodes a muscle-restricted protein containing MYND and SET domains and is essential for cardiac differentiation and morphogenesis. Nature Genet. 2002;31:25–32. doi: 10.1038/ng866. [DOI] [PubMed] [Google Scholar]
- 109.Slavin DA, Koritschoner NP, Prieto CC, Lopez-Diaz FJ, Chatton B, et al. A new role for the Kruppel-like transcription factor KLF6 as an inhibitor of c-Jun proto-oncoprotein function. Oncogene. 2004;23:8196–8205. doi: 10.1038/sj.onc.1208020. [DOI] [PubMed] [Google Scholar]
- 110.Hand SC, Menze MA, Borcar A, Patil Y, Covi JA, et al. Metabolic restructuring during energy-limited states: Insights from Artemia franciscana embryos and other animals. J Insect Physiol. 2011;57:584–594. doi: 10.1016/j.jinsphys.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Denekamp NY, Reinhardt R, Albrecht MW, Drungowski M, Kube M, et al. The expression pattern of dormancy-associated genes in multiple life-history stages in the rotifer Brachionus plicatilis. Hydrobiologia. 2011;662:51–63. [Google Scholar]
- 112.Li RQ, Li YR, Kristiansen K, Wang J. SOAP: short oligonucleotide alignment program. Bioinformatics. 2008;24:713–714. doi: 10.1093/bioinformatics/btn025. [DOI] [PubMed] [Google Scholar]
- 113.Ihaka R, Gentleman R. R: A language for data analysis. J Comp Graphic Stat. 1996;5:299–314. [Google Scholar]
- 114.Audic S, Claverie JM. The significance of digital gene expression profiles. Genome Res. 1997;7:986–995. doi: 10.1101/gr.7.10.986. [DOI] [PubMed] [Google Scholar]
- 115.Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, et al. Gene Ontology: tool for the unification of biology. Nature Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Rice P, Longden I, Bleasby A. EMBOSS: The European molecular biology open software suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 118.Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in realtime PCR. Nucl Acid Res. 2002;30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Q-PCR validation of Illumina sequencing results for resting eggs (RE, top panel) and amictic eggs (AE, lower panel). The relative abundance of twelve genes (list of primers is shown in [111]), was normalized using the equation: ratio = (Etarget)CP_target/(Eef1a)CP_reference where E = 10-1/slop [118]. The median was calculated for all transcript ratios and the transcript ratios were displayed as the log2[{transcript ratio of a sample}/{median of transcript ratios of all samples}]. It was very difficult to find one gene that could serve as a reference for all samples as ATP synthase (atps) changed between amictic eggs and resting eggs and elongation factor 1 alfa (ef1a) changed between females and males. Therefore, the relative abundance of transcripts was normalized to ef1a in the comparison made with the eggs samples but atps was used for comparisons between female and male samples. The expression levels obtained with real-time PCR were compared with the expression levels obtained from the number of Illumina reads for each corresponding gene. Regression analysis revealed high correlation between expression values obtained by real-time RT-PCR and number of reads for each transcript for the resting eggs (Person r = 0.980) and amictic eggs (Pearson r = 0.957). Negative values in real-time PCR indicate that the expression level of a specific transcript, was lower than that of the median. For Illumina reads, a negative value indicates that the number of reads assigned to a specific gene transcript were lower than the median value of the reads in a specific library.
(TIFF)
Basic information obtained by Illumina sequencing technology of libraries constructed from amictic (AE) and resting (RE) eggs. The total number of reads was circa 16 and 14 million for the amictic and resting egg libraries, respectively. Analysis of the data was performed as described in [111] and included scaling of the data, aligning of the reads with the sequences of EST data base obtained previously [26] and filtering of ESTs with less than 50 reads per EST. Only 69% and 73% of the final ESTs showed significant matches against proteins using Blastx sequence similarity searching.
(RTF)
Hatching dynamics of resting eggs collected from two batches that served for RNA extraction.
(RTF)
Differentially expressed transcripts in resting eggs with associated BLAST sequence similarity data and putatively assigned functions.
(XLS)
Differentially expressed transcripts in amictic eggs with associated BLAST sequence similarity data and putatively assigned functions.
(XLS)