Abstract
H4N8 subtype avian influenza viruses were isolated from shorebirds in eastern Hokkaido. All the isolates shared >99.7% nucleotide homology, and all the viral genes except for PB1 were highly related to those of A/red-necked stint/Australia/1/04. Thus, the isolates were regarded as PB1 reassortants. The most similar PB1 gene was identified in A/mallard/New Zealand/1615-17/04 (H4N6) with nucleotide homology of 90.9%. BALB/c mice intranasally inoculated with the H4N8 isolates developed severe respiratory disease, which eventually led to death in some mice. Virus was isolated from the lungs, and viral antigen was detected in the lungs with pneumonia. Other H4 subtype viruses tested did not cause any symptoms in mice, although these viruses were also isolated from the lungs. The PB2 gene of the H4N8 isolates contains K482R, but not the E627K or D701N substitutions. The PB1-F2 gene of the isolates consists of a 101-amino acid unique sequence, but lacks the N66S mutation.
Keywords: avian influenza virus, reassortant, H4N8, wild bird, mouse, pathogenicity
Introduction
Influenza A virus contains 8 segments of RNA as a genome, which encodes 11 proteins including the 2 envelope proteins known as hemagglutinin (HA) and neuraminidase (NA). Sixteen HA subtypes and nine NA subtypes of influenza A viruses have been reported so far and all of the HA and NA subtypes have been found in avian influenza viruses (AIVs) isolated from wild aquatic birds. Thus, wild birds such as waterfowl and shorebirds are considered natural reservoirs for influenza A viruses (Wright et al., 2007). It is known that most AIVs have no clear impact on the health of wild birds and an individual bird can carry multiple subtypes of AIVs simultaneously (Kida et al., 1980; Webster et al., 1992). In a host cell infected with 2 or more AIV subtypes, reassortment of the viral segmented genes can generate novel influenza viruses (Hinshaw et al., 1980). Historically, H1, H2, and H3 subtypes have caused influenza pandemics in humans (Trifonov et al., 2009; Webster et al., 1997). Recent studies have suggested that the 1918 “Spanish flu” virus (H1N1) was likely to be entirely an avian-like virus that adapted to humans (Taubenberger et al., 2005) and studies of the viral strains responsible for the pandemics of 1957 (H2N2) and 1968 (H3N2) revealed that the viruses were generated by reassortment of 2 or 3 gene segments derived from AIV and others from human influenza virus (Fang et al., 1981; Gething et al., 1980; Kawaoka et al., 1989). Advance knowledge of the diversity of influenza A virus genetic features circulating in the nature is likely to be a critical component of better influenza pandemic preparedness.
A new human pandemic H1N1 virus appeared in April 2009, and in June of 2009, the World Health Organization (WHO) declared it the first influenza pandemic of the 21st century. It was found that the pandemic H1N1 virus contains a unique combination of gene segments originating from swine viruses of Eurasian and North American lineage. The latter virus is known to be a descendant of the triple-reassortant of swine, avian, and human influenza viruses (Trifonov et al., 2009). Molecular markers that were reported as predictive determinants for the viral adaptation to humans were not detected in the pandemic H1N1 viruses (Garten et al., 2009). This suggests that currently unrecognized molecular determinants might be responsible for virus transmission among humans. In addition, we have recognized that none of the AIVs responsible for transferring genes to the 1957, 1968, and 2009 pandemic viruses were highly pathogenic avian influenza (HPAI) viruses. We obviously must improve our understanding of the natural evolution of influenza viruses in order to prepare for the next influenza pandemics.
Surveillance studies of wild aquatic birds that carry AIVs which may play critical roles in viral evolution can provide valuable knowledge regarding natural virus evolution. We isolated viruses of the H4N8 subtype from shorebirds during the course of AIV surveillance in eastern Hokkaido, Japan. H4 subtype AIVs are frequently isolated from wild birds worldwide, and many viruses of the H4N8 subtype have been isolated in the USA, Canada, and Australia (Hanson et al., 2003; Hurt et al., 2006; Sharp et al., 1993). In contrast, this subtype is rarely isolated in Asian countries. In fact, despite substantial AIV surveillance efforts, only 1 H4N8 isolate has been obtained previously in Japan. The absence of H4N8 in Japan could be because surveillance has not been sufficient, but it is also possible that H4N8 viruses have only recently been introduced into Asian regions, and these viruses may be currently expanding their circulation area. Characterizing such viruses might provide important knowledge regarding the evolution of the influenza A virus. In this study, the H4N8 subtype virus isolated from shorebirds in Japan was characterized by virological and genetic methods. We also evaluated its pathogenesis in mice in comparison to several other H4 subtype viruses.
Results
Isolation of the H4N8 viruses from shorebirds
The original cloacal/fecal samples were analyzed by real-time reverse transcription-PCR (RRT-PCR) for the M gene of the influenza A virus. Six out of 497 samples gained threshold cycle (Ct) values between 40.43 and 43.82. Among these 6 samples, 5 resulted in successful AIV isolation in embryonated chicken eggs. Among the 5 isolates, 6KS0185 and 6KS0191 were found to originate from the fecal samples of slaty-backed gulls collected in July 2006 on Yururi Island. Another 3 isolates, 6KS0242, 6KS0261, and 6KS0279 were obtained from the cloacal swabs of red-necked stints. These samples were collected in September 2006 at Lake Komuke, which is located 125 miles north of Yururi Island. All 5 AIV isolates are an H4N8 subtype.
The virus titers of the H4N8 isolates were 4.25 TCID50/ml in MDCK cells in both the presence and absence of trypsin. The virus titers of A/red-necked stint/Australia/1/04 (H4N8) were also 4.25 TCID50/ml in the two conditions. In contrast, the virus titers of A/duck/Osaka/1/05 (H4N8), A/duck/Shiga/8/04 (H4N6), and A/duck/Czechoslovakia/56 (H4N6) in MDCK cells were 7.25, 4.58, and 5.58 in the presence of trypsin, but 5.25, 3.25, and 4.25 in the absence of trypsin, respectively.
Genetic and phylogenetic analysis of the H4N8 isolates
First, the full-length nucleotide sequences of the M gene were analyzed by the dideoxy method for the 5 isolates, and it was found that all of the isolates are 100% homologous in the M gene. The amino acid sequence at the cleavage site of the HA gene was -PEKASK/GLF-. This is characteristic of a virus with low pathogenicity. The second-generation sequencing was successfully conducted by the Viral Projects Team at the J. Craig Venter Institute (JCVI) for all of the gene segments of the 4 isolates except for 6KS0261. The sequencing results revealed 99.7–100.0% and 98.7–100.0% homology by segment among isolates by nucleotide and amino acid content, respectively.
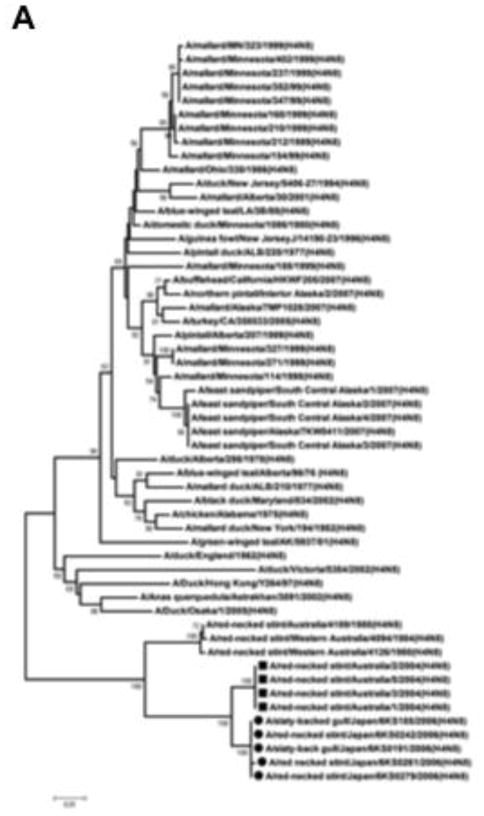
The BLAST homology search revealed that all of the gene segments except for PB1 of the H4N8 isolates were close to those of A/red-necked stint/Australia/1/04 (H4N8) with nucleotide sequence homology between 96.8–98.7% (Table 1). Results of a phylogenetic analysis showed that the 5 isolates indeed belonged to the Australian lineage cluster, in which all other viruses originate from the red-necked stint in Australia. In contrast, another large American lineage cluster includes H4N8 strains isolated from varieties of bird species other than the red-necked stint. These species are mainly dabbling ducks. The small cluster falling between the American and Australian clusters contains H4N8 strains originating from ducks in Canada, England and Asia; A/duck/Osaka/1/05 is included in this cluster (Fig. 1A).
Table 1.
Comparison of the viral gene sequences between the H4N8 viruses isolated in Japan and A/red-necked stint/Australia/1/04 (H4N8)a.
| Gene | Nucleotide identity (%)b with A/red-necked stint/Australia/1/04
|
|
|---|---|---|
| 6KS0185 | 6KS0279 | |
| PB2 | 98.3 (99.4) | 98.3 (99.4) |
| PB1 | 89.1 (97.2) | 89.1 (97.1) |
| PA | 96.8 (99.0) | 96.8 (99.0) |
| HA | 97.1 (97.7) | 97.0 (97.7) |
| NP | 98.7 (100.0) | 98.7 (100.0) |
| NA | 97.2 (97.5) | 97.0 (96.7) |
| Mc | 98.7 (100.0) | 98.7 (100.0) |
| NSd | 97.0 (94.3) | 97.0 (94.4) |
Full-length sequences obtained in this study (2nd generation sequencing) are compared with the sequences of A/red-necked stint/Australia/1/04 (GenBank accession numbers CY077632 to CY77640).
Identity in nucleotide and amino acid (in parenthesis).
Amino acid similarity was compared for the M1 protein.
Amino acid similarity was compared for the NS1 protein.
Fig. 1.
Phylogenetic analysis of the M and PB1 genes of the H4N8 viruses isolated from gulls and stints in Japan. Full-length nucleotide sequences of the viral genes were phylogenetically analyzed in comparison with the sequences available in GenBank. (A) Analysis of the M gene. All the strains in this figure are H4N8 subtype viruses. The new H4N8 isolates are related to the Australian lineage, in which all other H4N8 viruses have originated from red-necked stints in Australia. (B) Analysis of the PB1 gene. The PB1 genes of the new H4N8 isolates are genetically distant from that of A/red-necked stint/Australia/1/04. The H4N8 viruses isolated in this study are highlighted by circles, and A/red-necked-stint/Australia/1/04 (H4N8) and its related strains are highlighted with squares. The numbers in the tree represent bootstrap values (1,000 replicates).
In contrast, nucleotide homology in the PB1 gene is approximately 89% between the H4N8 isolates and A/red-necked stint/Australia/1/04 (Table 1). A BLAST search for the nucleotide sequence of the PB1 gene did not provide any strains with high sequence homology to the H4N8 isolates. In a phylogenetic analysis of the PB1 gene, the H4N8 isolates were dropped into a position that is far from A/red-necked stint/Australia/1/04 (H4N8). The strains close to the H4N8 isolates included A/mallard/New Zealand/1615-17/04 (H4N6), A/mallard/New Zealand/1365-355/05 (H7N7), and A/mallard/New Zealand/479-8/05 (H6N2) (Fig. 1B). However, the nucleotide sequence identity was approximately 90% between the H4N8 isolates of this study and the New Zealand strains (Table 2). A BLAST search of protein sequences revealed that the PB1 proteins of A/mallard/Ohio/64/89 (H6N8), A/duck/Ohio/470655/07 (H5N2), and A/mallard/Alaska/44430-031/08 (H4N6) were most similar to those of the new H4N8 isolates. The protein sequence identity of PB1 between the new H4N8 isolates and the American strains was 98.4%, but nucleotide homology was approximately 87% between the 2 groups (Table 2).
Table 2.
Comparison of the PB1gene and protein between the H4N8 isolate and virus strains available in GenBank.
| Virusa | Identity (%) with 6KS0185b |
|
|---|---|---|
| nucleotide | amino acid | |
| A/mallard/New Zealand/1615-17/04 (H4N6) | 90.9 | 97.6 |
| A/mallard/New Zealand/1365-355/05 (H7N7) | 90.4 | 97.5 |
| A/mallard/New Zealand/479-8/05 (H6N2) | 90.3 | 97.5 |
| A/red-necked stint/Australia/1/04 (H4N8) | 89.1 | 97.2 |
| A/mallard/Ohio/64/89 (H6N8) | 87.2 | 98.4 |
| A/duck/Ohio/470655/07 (H5N2) | 86.8 | 98.4 |
| A/mallard/Alaska/44430-031/08 (H4N6) | 86.7 | 98.4 |
Top 3 strains showing similarities to 6KS0185 either at the nucleotide or protein levels, and A/red-necked stint/Australia/1/04 are included. The highest similarity values are underlined.
Full-length sequences obtained by the 2nd generation sequencing in this study are compared to the sequences of other strains available in GenBank.
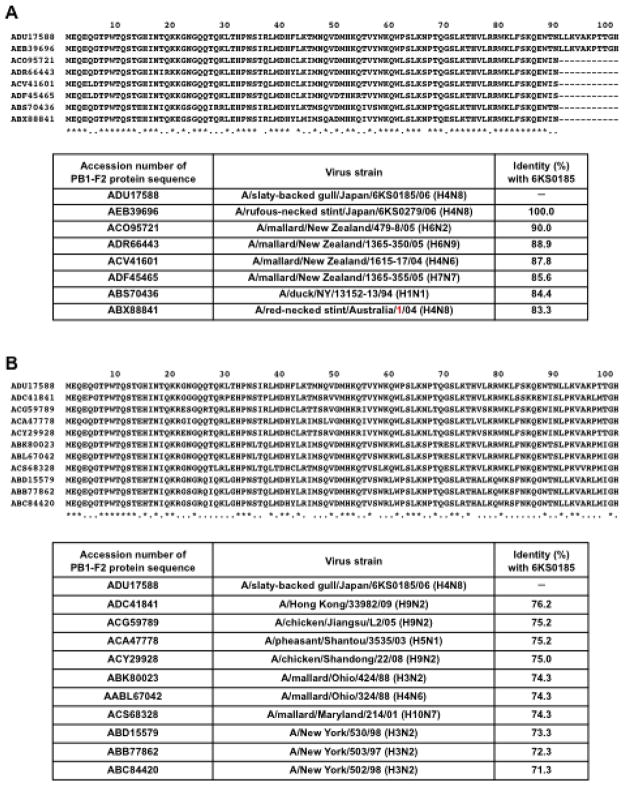
The identity of the PB1-F2 protein sequence was 83.3% between the new H4N8 isolates and A/red-necked-stint/Australia/1/04. The PB1-F2 protein has 101 amino acids in the new H4N8 isolates and 90 amino acids in A/red-necked stint/Australia/1/04. In a BLAST search of PB1-F2, A/mallard/New Zealand/479-8/05 (H6N2) was found be most similar to the new H4N8 isolates with 90.0% amino acid sequence homology followed by A/mallard/New Zealand/1365-350/05 (H6N9) (88.9%) and A/mallard/New Zealand/1615-17/04 (H4N6) (87.8%). This is despite the fact that PB1-F2 of the New Zealand strains consists of 90 amino acids (Fig. 2A). All of the PB1-F2 proteins with 101 amino acids found in GenBank showed even lower similarity to PB1-F2 of the new H4N8 isolates (Fig. 2B).
Fig. 2.
Comparison of the PB1-F2 protein sequence of the H4N8 isolates obtained in this study and the reference strains. (A) The PB1-F2 protein sequences of the H4N8 isolates (6KS0185 and 6KS0279) were aligned with sequences identified in a BLAST search. The strains of the top 6 in identity to 6KS0185 and 6KS0279 are shown. The PB1-F2 protein consists of 101 amino acids in the H4N8 isolates but 90 amino acids in other strains. (B) The PB1-F2 protein sequence of the H4N8 isolate 6KS0185 was aligned with other sequences of a length of 101 amino acids, which were identified in GenBank. Asterisks represent the homologous amino acids in all genes. Identities between the 6KS0185 protein and each protein are presented in the table below the sequence alignment.
Disease caused by intranasal inoculation of the H4N8 isolates in mice
Pathogenicity of the H4N8 isolates was examined in mice. All mice that were intranasally inoculated with 103 TCID50 of 6KS0185 (originating from gull) or 6KS0279 (originating from stint) showed general clinical signs of the disease such as ruffled fur, hunched back, and lethargy on 2 days post-infection (dpi). The mouse body weight decreased by 16.9–20.5% on 3 dpi, and typical symptoms of dyspnea such as abdominal breathing were observed in those mice. In the lungs of the mice euthanized on 3 dpi, the viral M gene was detected by RRT-PCR with Ct values ranging from 20.42 to 31.96.
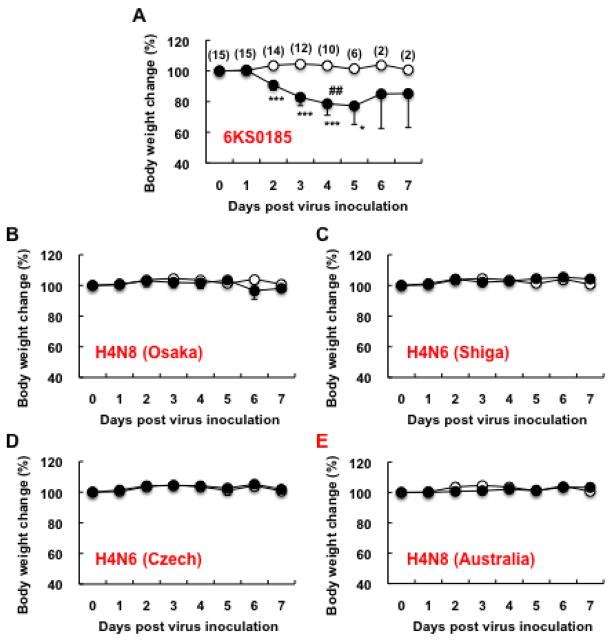
As shown in Fig. 3A, among the mice inoculated with different amounts of 6KS0185, a significant body weight reduction was observed in the mice inoculated with 103 TCID50 between 2 and 8 dpi. The mice that received 102 TCID50 of 6KS0185 had a slight but significant reduction in body weight between 4 and 6 dpi, but no changes were observed in mice that received 10 TCID50 of the virus. However, HI antibody against 6KS0185 was detected in all serum samples collected from the mice with a titer between 32 and 64 on 14 dpi (Fig. 3B).
Fig. 3.
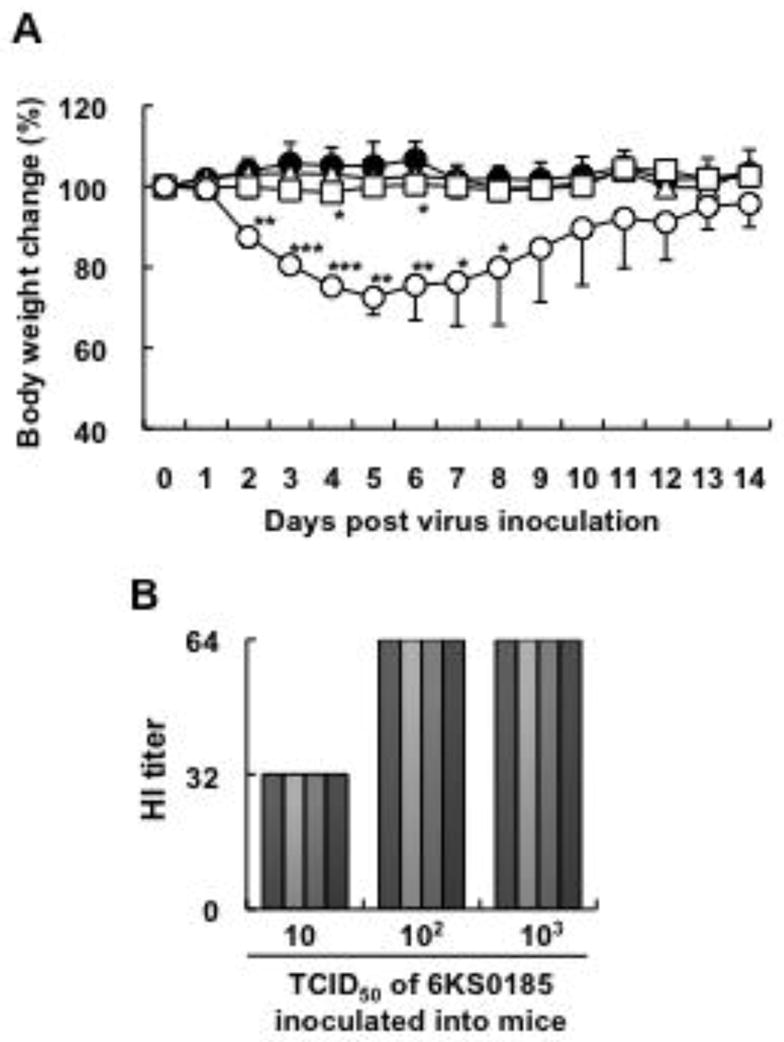
The mouse infection study of the H4N8 isolate 6KS0185 (n = 4). (A) Body weight changes in the mice that were intranasally inoculated with 103 (○), 102 (□), and 10 (△) TCID50 of 6KS0185. Control mice (●) received the same amount of PBS. The results are presented as mean ± S.D. Asterisks represent significant differences between the control and experimental groups. *p < 0.05, **p < 0.001, ***p < 0.0001 by Student’s t-test. (B) HI titers of the sera obtained from the mice inoculated with 6KS0185 on 14 dpi. Each column represents the HI titer of a mouse that received 6KS0185, and that of a control mouse receiving PBS.
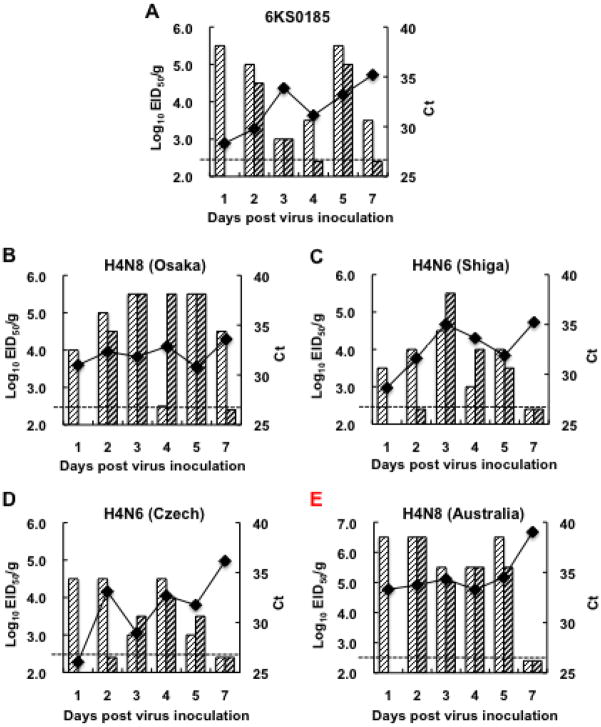
Tropism of the virus and the possibility of mouse-to-mouse transmission of the virus were investigated. Naïve mice housed in the same cage with the mice inoculated with 6KS0185 did not develop any clinical signs or weight loss indicative of disease (Fig. 4A). The viral M gene was not detected in all the tissue samples obtained from these mice between 1 and 7 dpi (data not shown). In contrast, all mice inoculated with the virus developed severe symptoms, and 2 of the 9 mice died on 3 dpi. The M gene was detected in both the lung and the nasal turbinate (NT) of all mice inoculated with 6KS0185 between 1 and 7 dpi. The average Ct values for the lung and NT samples ranged from 28.8 to 32.4, and 35.6 to 41.0, respectively (Figs. 4B and C). Virus originating from all of the lung samples was successfully isolated by egg inoculation. The virus titers in the lungs were between 4.5 and 6.5 log10 EID50/g on 1 dpi, and ranged from 3.0 to 5.5 after 2 dpi (Fig. 4B). In NT, the virus was isolated from only 2 samples, obtained on 1 and 2 dpi (Fig. 4C). Two brain samples obtained on 1 and 2 dpi and 1 spleen sample obtained on 1 dpi had Ct values above 41.0 in the RRT-PCR, and virus was not isolated from these samples (data not shown).
Fig. 4.
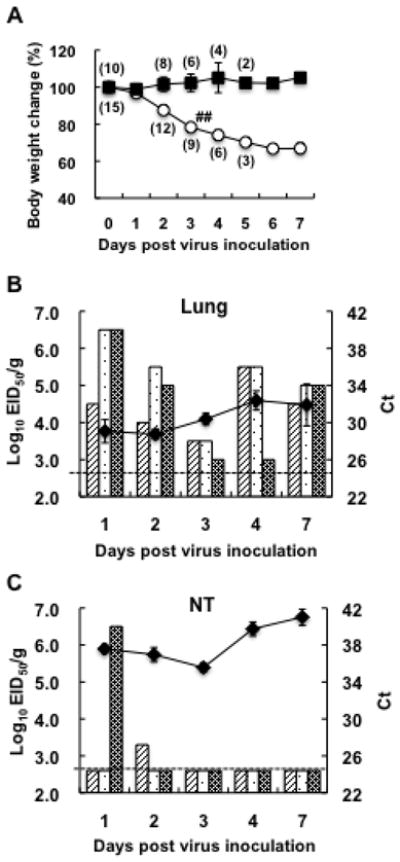
The tissue tropism in mice and mouse-to-mouse transmission of the H4N8 isolate. (A) Body weight changes of the mice inoculated with 103 TCID50 of 6KS0185 (○) and the contact mice (■). The results are presented as mean ± S.D. Numbers in parentheses indicate mouse numbers. One sharp mark represents 1 mouse death. Virus titers (EID50) and Ct values in the RRT-PCR for the viral M gene obtained for the lung and NT samples are presented in (B) and (C), respectively. Each column represents the virus titer of 1 mouse, and the broken line indicates a threshold level for virus detection. The line graph shows Ct values (mean ± S.D.) in the RRT-PCR. The graphs (B, C) do not include the results of the RRT-PCR for the control group, since the Ct values of those samples are “undetermined” in the assay. All the control samples were therefore regarded as negative (below the detection limit) in the RRT-PCR.
Comparison of pathogenicity in mice between the H4N8 isolates and other H4 subtype viruses
The pathogenicity of 6KS0185 in mice was compared with that of A/duck/Osaka/1/05 (H4N8), A/duck/Shiga/8/04 (H4N6), A/duck/Czechoslovakia/56 (H4N6) and A/red-necked stint/Australia/1/04 (H4N8). In the mice inoculated with 6KS0185, a drastic body weight loss was observed, and 2 of the 10 inoculated mice died on 4 dpi (Fig. 5A). In the mice inoculated with other viruses, a marked change in body weight was not observed (Figs. 5B, C, D and E). Nevertheless, the viral M gene was detected not only in the mice inoculated with 6KS0185 but also in mice inoculated with other H4 subtype viruses, and the virus was isolated from most of the samples collected between 1 and 7 dpi (Figs. 6A, B, C, D and E). In the lungs of the mice inoculated with 6KS0185, the virus titer was 4.5–5.5 log10 EID50/g on 1, 2, and 5 dpi, but lower on the other days (Fig. 6A). High titers (>5.5) of the virus were detected in the mice inoculated with A/duck/Osaka/1/05 between 3 and 5 dpi, in the mice inoculated with A/duck/Shiga/8/04 on 3 dpi, and in A/red-necked stint/Australia/1/04 on days 1, 2 and 5 dpi (Figs. 6B, C and E). In the lungs of the mice inoculated with A/duck/Czechoslovakia/56, the highest titer of 4.5 log10 EID50/g was detected on 1, 2, and 4 dpi (Fig. 6D). In all of the NT samples obtained from mice infected with 6KS0185, the viral M gene was detected, but the virus was only isolated from 1 sample obtained on 2 dpi. This is similar to the results shown in Fig. 4C. In the NT of the mice inoculated with A/duck/Shiga/8/04, A/duck/Czechoslovakia/56 and A/red-necked stint/Australia/1/04, the viral M gene was detected until 4 or 5 dpi; however, the viral M gene was not detected after 2 dpi in the NTs of the mice inoculated with A/duck/Osaka/1/05 (data not shown).
Fig. 5.
Body weight changes of the mice inoculated with the H4N8 isolate and the reference viruses. 103 TCID50 of 6KS0185 (A), A/duck/Osaka/1/05 (B), A/duck/Shiga/8/04 (C), A/duck/Czechoslovakia/56 (D) or A/red-necked-stint/Australia/1/04 (E) was intranasally inoculated into the mice. The results are presented as the mean ± S.D. Open circles show the results of the control mice, and filled circles show the results of the experimental mice. Numbers in parentheses indicate mouse numbers. One sharp mark represents 1 mouse death. *p < 0.05, ***p < 0.0001 by Student’s t-test.
Fig. 6.
Virus titers (EID50) and Ct values in RRT-PCR analysis of the viral M gene for the lungs obtained from the mice inoculated with the H4N8 isolate and the reference viruses. The mice were euthanized on the indicated days after inoculation (103 TCID50) with 6KS0185 (A), A/duck/Osaka/1/05 (B), A/duck/Shiga/8/04 (C), A/duck/Czechoslovakia/56 (D) or A/red-necked-stint/Australia/1/04 (E). Each column represents a virus titer of 1 mouse and the broken line indicates the threshold level for virus detection. The line graph presents the average of the Ct values in the RRT-PCR.
The lungs, tracheae, and other organs obtained on 4 and 5 dpi from the mice inoculated with different viruses were analyzed histopathologically and immunohistochemically for the detection of viral antigens. In the lungs of the mice infected with 6KS0185, severe pneumonia was observed, and the viral antigen was detected in the corresponding regions (Figs. 7B and C). The viral antigen was also detected in the tracheae although the histopathological change was moderate (data not shown). The viral antigen was not detected in any of the other organs investigated, including the spleen, liver, pancreas, intestine, kidney, heart, and brain of the 6KS0185-infected mice. In contrast, fewer histopathological changes were observed in the lungs of the mice inoculated with the other viruses, although the viral antigen was detected (Figs. 7D, E and F). The magnitude of the histopathological changes and the viral antigen detection in tracheae varied among the groups that were inoculated with different viruses. Tracheitis was most prominent in mice that received A/duck/Czechoslovakia/56, in which the viral antigen was concomitantly detected (data not shown).
Fig 7.
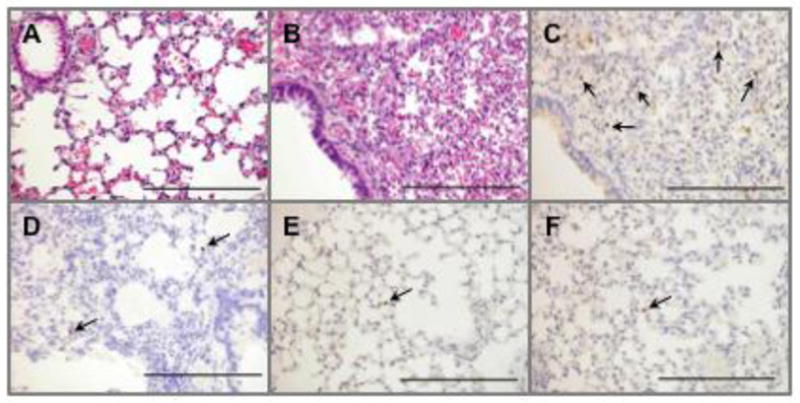
Histopathological and immunohistochemical analyses of the lungs obtained from the mice inoculated with the H4N8 isolate and the reference viruses. Hematoxylin and eosin staining of the lung of the control mouse (A) and the mouse infected with 6KS0185 (B). Detection of the viral antigen in the lungs of the mice inoculated with 6KS0185 (C), A/duck/Osaka/1/05 (D), A/duck/Shiga/8/04 (E), and A/duck/Czechoslovakia/56 (F). The tissue samples were collected from the mice 4 or 5 days after the intranasal inoculation of the viruses (103 TCID50). The results are of a representative mouse of each group (n = 4). Arrows indicate the viral antigen detected by polyclonal antibody against influenza A virus. The scale bar represents 200 μm.
Comparison of the viral protein sequences of the H4N8 isolates with the molecular determinants of viral pathogenicity
To understand the pathogenicity of the new H4N8 isolates in mice, the protein sequences of the new H4N8 isolate (6KS0185), A/red-necked-stint/Australia/1/04, A/duck/Osaka/1/05, and A/duck/Shiga/8/04 were compared with respect to viral molecular determinants of virulence in mammalian species that have been previously reported. The results are summarized in Table 3. The K482R substitution was identified in the PB2 protein of the new H4N8 isolate and A/rednecked-stint/Australia/1/04 (H4N8) but not in the other viruses. The new H4N8 isolates did not have E627K or D701N mutations in the PB2 gene or the N66S mutation in the PB1-F2 gene, nor did the virus recovered from the lungs of the mice infected with the new H4N8 isolates. However, as described above, PB1-F2 of the new H4N8 isolates was found to consist of a unique sequence of 101 amino acids (Fig. 2).
Table 3.
Comparison of the protein sequence between the H4N8 isolate and other H4 subtype viruses with the molecular determinants of virus pathogenicity in mammalian speciesa.
| Protein | Position | Virus
|
Pathogenicity
|
|||||
|---|---|---|---|---|---|---|---|---|
| 6KS0185 | H4N8/Australia | H4N8/Osaka | H4N6/Shiga | Low | High | Ref.b | ||
| PB2 | 271 | T | T | T | T | T | A | a) |
| 482 | R | R | K | K | K | R | b) | |
| 627 | E | E | E | E | E | K | c) | |
| 701 | D | D | D | D | D | N | d), e) | |
| 714 | S | S | S | S | S | R | d) | |
| PB1 | 13 | P | P | P | P | P | d) | |
| 538 | D | D | D | D | D | G | d) | |
| 678 | S | N | S | S | N | d) | ||
| PB1-F2 | 66 | N | N | N | N | N | S | f) |
| PA | 97 | T | T | T | C | T | I | g) |
| 550 | L | L | L | L | I | L | h) | |
| 615 | R | R | K | K | K | N | d) | |
| M1 | 139 | T | T | T | T | T | A | b) |
| NS1 | 92 | D | D | (ND)c | D | D | E | i) |
| C-terminus | GSEV | ESEV | (ND) | ESEV | RSEV | ESEV | j), k) | |
The protein sequences of 6KS0185, A/red-necked stint/Australia/1/04 (H4N8), A/duck/Osaka/1/05 (H4N8), and A/duck/Shiga/8/04 (H4N6) were compared with the molecular determinants of viral pathogenicity. The amino acids matched to the determinants for high pathogenicity are highlighted by gray shading.
References: a) Bussey et al., 2010; b) Brown and Bailly, 1999; c) Subbarao et al., 1993; d) Gabriel et al., 2005; e) Li et al., 2005; f) Conenello et al., 2007; g) Song et al., 2009; h) Rolling et al., 2009; i) Seo et al., 2002; j) Jackson et al., 2008; k) Obenauer et al., 2006.
Not determined.
Discussion
We isolated 5 strains of the H4N8 subtype virus from gulls and stints during an AIV surveillance project in the summer of 2006. This is only the second report on the isolation of an H4N8 subtype virus in Japan. A/duck/Osaka/1/05 was the only H4N8 subtype virus that had been previously isolated in this country. Genetic analyses revealed that the newly isolated H4N8 viruses were genetically similar to each other but not related to A/duck/Osaka/1/05 (Fig. 1A). This suggests that our H4N8 isolates and A/duck/Osaka/1/05 originated from different sources. A BLAST search revealed that all of the gene segments of the H4N8 isolates except for PB1 are highly related to A/red-necked stint/Australia/1/04 (H4N8) and its related strains (Table 1 and Fig. 1A). Among the 5 isolates, 2 originated from slaty-backed gulls on Yururi Island and the other 3 isolates were obtained from red-necked stints at Lake Komuke. Red-necked stints are migratory birds that fly from Siberia and the Russian Far East to Australia with stopovers in Japan and elsewhere. Slaty-backed gulls are either resident or migratory birds that migrate across eastern Asia and the Pacific Rim (Brazil, 2009). Therefore, it is possible that H4N8 viruses circulating among red-necked stints in Australia were transported to Japan by the stints, and were subsequently transmitted to slaty-backed gulls, possibly in Japan. In fact, phylogenetic analysis of the M gene revealed that the H4N8 viruses we isolated from gulls and stints in Japan are incorporated into the cluster with all other viruses originating from stints in Australia (Fig. 1A). To the best of our knowledge, this is the first report on the isolation of an H4N8 subtype virus from gull species. A series of A/red-necked stint/Australia/04 (H4N8) strains was isolated by Hurt et al. (2006) in 2004. This report indicated that the HA gene of the viruses is most closely related to A/budgerigar/Hokkaido/1/77 (H4N6), but the homology was only 87%. None of the other H4 subtype virus sequences available in the public databases had a particularly close genetic relationship with the Australian strains at the time their BLAST search was conducted. Here, we report that the new H4N8 viruses sampled in this study in Japan in 2006 are similar to the Australian strains isolated in 2004.
Unlike the other viral genes, the nucleotide identity of the PB1 gene was found to be only 89.1% between the H4N8 isolates and A/red-necked stint/Australia/1/04 (Table 1). This suggests that the H4N8 isolates are PB1 reassortants. Despite an extensive search in the GenBank database, we could not find any sequences that are closely related to the PB1 gene of the new isolates. The PB1 genes of the H6N8, H5N2, and H4N6 subtype viruses isolated from duck species in North America were found to have 98.4% homology at the protein level, but less than 90.0% homology at the nucleotide level. The PB1 genes of the New Zealand strains of H4N6, H7N7, and H6N2 subtypes showed nucleotide homology slightly higher than 90.0%, but the amino acid identity was less than 98.0% (Table 2). From these results, it is concluded that the PB1 genes of the H4N8 isolates are unique, and thus, we could not identify the geographical origin of the PB1 gene.
Another interesting feature was identified in PB1 from the new H4N8 isolates. None of the PB1-F2 protein sequences in GenBank showed homology higher than 90.0% to the new H4N8 isolates. The virus with the highest similarity for this gene was A/mallard/New Zealand/479-8/05 (H6N2) (90.0% homology), followed by the other New Zealand strains. Even more surprising, the open reading frame of PB1-F2 of the new H4N8 isolates consists of 101 amino acids, although PB1-F2 of all American and New Zealand strains described above consist of 90 amino acids (Fig. 2A). PB1-F2 was described in 2001 as the 11th protein of influenza A virus. This protein is translated from the +1 reading frame of the PB1 gene segment (Chen et al., 2001). The PB1-F2 protein is not required for virus replication, but is considered to contribute as a virulence factor of influenza A virus (Conenello et al., 2007; McAuley et al., 2010; Zamarin et al., 2006). The standard size of the PB1-F2 protein is known to be 90 amino acids but shorter and longer size variants of PB1-F2 have been reported (Krumbholz et al, 2011; McAuley et al., 2010). It was recently reported that the PB1-F2 variants of swine viruses have lengths between 8 and 90 amino acids (Krumbholz et al, 2011), and the H9N2 viruses that appear to be undergoing active evolution in China have variable lengths of the PB1-F2 protein (Huang et al., 2010). McAuley et al. (2010) reported on a possible relationship between the truncation or mutation in PB1-F2 and the attenuation of the virulent influenza viruses. However, there are no other studies that have reported the pathogenicity of a virus carrying a PB1 gene encoding a PB1-F2 that is longer, at 101 amino acids, except for the current study.
The H4N8 isolates originating from gulls and stints unexpectedly caused marked body weight reduction (>20%), and symptoms of severe respiratory disease, which eventually led to the death of some of the inoculated mice (Figs. 3A, 4A and 5A). In the lungs of the inoculated mice, the viral antigen was detected in the region of severe pneumonia (Figs. 7B and C). The viral gene was also detected by RRT-PCR in a small number of spleen and brain samples with high Ct values. This suggests the possibility of a systemic infection caused by the H4N8 isolates. However, the viral antigen was identified only in the lungs and tracheae. Therefore, the mouse deaths were attributed to respiratory disorders caused by infection with the H4N8 isolates. The quite rapid development of respiratory disease may raise a concern that another microorganism contained in the virus sample could cause the observed symptoms. However, since both the samples, originating from a gull (6KS0185) and a stint (6KS0279), caused the same disease, it was unlikely that something other than the H4N8 virus was the causative agent. A decrease of the virus titer on day 3 followed by a rebound increase of the titer was observed in the lungs of the mice infected with the H4N8 isolates (Figs. 4B and 6A). Such a phenomenon might be observed if the virus increased its virulence in the host. It has been reported that PB2 E627K mutation rapidly occurred in the mice inoculated with H5N1 avian influenza virus, which resulted in an increased virulence (Bogs et al., 2011; Mase et al., 2006). In the current study, we did not observe the PB2 mutation in mice infected with the H4N8 isolates. Further study is necessary to elucidate the mechanism of the virus titer rebound in mice: that would include studies searching candidates of genetic mutations related to the titer change and/or investigating host-related factors such as the immune response of the host towards the virus. It should be interesting to passage the virus in mice to determine whether virulence is increased by host specific adaptations. Mouse-to-mouse transmission was not observed in the H4N8 infection (Fig. 4). Since the mouse is not regarded as a suitable model for investigating viral transmission (O’Donnell and Subbarao, 2011), additional studies are required to evaluate transmission of the H4N8 isolates among mammalian species by using a more reliable animal model such as ferrets (Gustin et al., 2011) and guinea-pigs (Lowen et al., 2006). While the virus we identify in this study is not considered a pandemic threat, these additional studies could help clarify the boundaries existing between avian viruses normally considered low pathogenic and potential pandemic strains.
Driskell et al. (2010) also studied the pathogenicity of several subtypes of viruses isolated from wild birds including ducks and shorebirds such as turnstone, sanderling, and knot. These subtypes replicated well in MDCK cells without trypsin in a manner similar to the replication of the H4N8 isolates observed in the current study. Among the 28 strains Driskel et al. tested, most of the viruses (71%) replicated efficiently in mouse lungs to attain high titers without adaptation and induction of histopathological lesions in the lungs. However, the maximum reduction in body weight observed was less than 8%, and mouse deaths were not recorded. The results of previous studies and the current study suggest that the H4N8 viruses newly isolated from shorebirds are more virulent in mice than are the previously isolated viruses. Ability to replicate well in cell culture in the absence of trypsin has been regarded as a possible factor for the virus to acquire virulence in hosts, since proteolytic cleavage of the influenza virus hemagglutinin by protease is essential for viral infectivity and spread (Goto and Kawaoka, 1998; Tumpey et al., 2005). In the current study, both the new H4N8 isolate and A/red-necked stint/Australia/1/04 replicate in MDCK to similar titers in the presence and absence of trypsin. However, the new H4N8 isolate but not A/red-necked stint/Australia/1/04 caused disease in mice, indicating that trypsin-independent replication is not necessarily linked to the pathogenicity of this virus.
We also found that mice inoculated with A/duck/Osaka/1/05, A/duck/Shiga/8/04, and A/duck/Czechoslovakia/56 did not show any symptoms of disease, and there were no marked body weight reductions (Fig. 5). In the lungs of these mice, the viral gene was detected with Ct values similar to those of 6KS0185, and the virus was also isolated in similar titers (Fig. 6). These results indeed indicate that 6KS0185 as well as the other H4 subtype viruses tested in this study could efficiently replicate in mouse lungs without prior adaptation. These results also indicate that the efficiency of viral replication in the lung is not the sole determinant of the differences in virulence observed. There must be other factors involved in order for the virus to cause severe respiratory disease in the inoculated mice. Hurt et al. (2006) reported that strains of A/red-necked stint/Australia/2004 are not pathogenic to chickens. We also found that the H4N8 viruses we isolated are not pathogenic to chickens (data not shown). A lethal infection of A/chicken/Alabama/7395/75 (H4N8) in chickens has been reported (Slemons et al., 1991), and infection of pigs with H4 subtype viruses have also been reported (Karasin et al., 2000; Ninomiya et al., 2002). However, this is the first report on disease caused by H4N8 virus infection in a mammalian host. Moreover, the major differences between the new H4N8 isolates that are pathogenic in mice and the non-pathogenic A/red-necked-stint/Australia/1/04 are only in the PB1 gene.
Many studies have been performed to identify the molecular determinants responsible for viral pathogenicity in the host, and the accumulated data provide concrete evidence for many determinants, as described in recent review articles (Neumann et al., 2009; O’Donnell and Subbarao, 2011; Tscherne and Garcia-Sastre, 2011). The protein sequence of the viruses examined in this study was compared to the previously identified virulence determinants. The PB2 protein of the new H4N8 isolates contains the K482R substitution. However, A/red-necked-stint/Australia/1/04 also has the substitution, suggesting that the amino acid 482 in the PB2 protein is not likely to relate to the pathogenicity of the new H4N8 isolates. Other virulence determinants that have been reported, including PB2-627K, PB2-701N, and PB1-F2-66S were not identified in the proteins of the new H4N8 isolates (Table 3).
Amino acid positions for the receptor-binding sites of HA have been identified for H1, H2, and H3 subtypes, and certain positions have been identified as predictors of the receptor specificity, i.e., avian preference (SAα2,3) or human preference (SAα2,6) (Neumann et al., 2009; O’Donnell and Subbarao, 2011; Tscherne and Garcia-Sastre, 2011). However, no such information is currently available for the H4 subtype virus.
The PB1 gene of the H4N8 isolates was found to be unique, especially in PB1-F2, which has a length of 101 amino acids. Several PB1-F2 proteins of this length were identified in GenBank. The viruses containing PB1-F2 of this length can be divided into 2 groups. The first group includes viruses isolated in North America between 1988 and 2001. This group includes the H3N2, H4N6, and H10N7 subtype viruses from mallard, and many H3N2 subtype human viruses. The second group includes viruses isolated in China between 2003 and 2009. This group includes H9N2 viruses isolated from humans and chickens and the H5N1 virus from pheasants. All of the PB1-F2 proteins with lengths of 101 amino acids found in GenBank have amino acid homology lower than 80.0% to the new H4N8 viruses from stints and gulls in Japan (Fig. 2B). This result could be attributed to a shortage of amino acid sequence data for PB1-F2 currently available in GenBank. A more diverse global representation of PB1-F2 sequence of influenza A viruses in the public database may reveal more closely related viruses in the future.
Although the exact role of the PB1-F2 in viral infection has not been fully elucidated, expression of PB1-F2 has been reported to enhance viral pathogenicity in mouse models of influenza infection (Conenello et al., 2007; McAuley et al., 2007; Zamarin et al., 2006). Increased virulence caused by PB1-F2 seems to involve mechanisms such as: 1) mitochondrial targeting leading to apoptosis, 2) an increased inflammatory response, and 3) enhancement of transcription by an increase in the nuclear retention time (Chen et al., 2001; Conenello et al., 2007; Gibbs et al., 2003; Mazur et al., 2008; McAuley et al., 2010, 2007). Further characterization of PB1-F2 of the H4N8 isolates, which causes severe inflammatory disease in mice, may provide new information to improve our understanding of the role of PB1-F2 in diseases caused by the influenza A virus, and ultimately improve our understanding of the evolution of the PB1-F2 protein. Since molecular determinants of influenza virulence depend on the gene constellation and backbone, a study employing reverse genetics will be necessary to test the exact role of the PB1-F2 in the virus pathogenicity. The results obtained in these studies will help to define the virulence determinant in H4N8 viruses originating from gulls and stints.
Materials and methods
Sample collection
A total of 497 shorebird samples (369 cloacal and 128 fecal samples) were collected in 2006 between June and September in eastern Hokkaido, Japan. The locations included Lake Komuke (GPS coordinates 44° 15′ N, 143° 31′ E, n = 172), Kushiro Wildlife Center (43° 03′ N, 144° 17′ E, n = 162), and Yururi Island (43° 13′ N, 145° 36′ E, n = 128). Additional samples were obtained at three other locations. Cloacal swabs were collected by licensed bird banders during the banding procedure. The fecal samples were collected around the habitat area of the birds, and only fresh samples for which the species of origin was defined were included in this study. The bird species included black-faced bunting (n = 132), slaty-backed gull (n = 128), red-necked stint (n = 94), grey-tailed tattler (n = 27), greenshank (n = 15), Latham’s snipe (n = 15), Mongolian plover (n = 11), green wing teal duck (n = 9), and other non-duck species. Following the collection of samples in the field, each sample was stored in virus transport medium (VTM: M4RT, Remel Inc., Lenexa, KS) and kept in a cold chain followed by storage at −80°C after transportation to the laboratory. The VTM samples were subjected to both RRT-PCR for detection of the M gene of influenza A virus and virus isolation using embryonated chicken eggs as described below.
Cells and viruses
Madin-Darby canine kidney (MDCK) cells were cultured in Dulbecco’s Modified Eagle’s medium (DMEM: Nissui Pharmaceutical Co., Ltd., Tokyo, Japan) supplemented with 10% fetal bovine serum (FBS) and 2 mM L-glutamine. Cells were seeded onto 96-well tissue culture plates to evaluate viral titers. Upon virus inoculation, the cells were washed twice with the DMEM and the medium was replaced with virus growth medium (VGM) containing trypsin following protocol in the WHO Manual on Animal Influenza Diagnosis and Surveillance (WHO/CDS/CSR/NCS/2002.5 Rev. 1: WHO Manual). The sample to be tested was serially diluted (1:10) for the titration. Based on the cytopathic effect (CPE) observed 4 days post-inoculation (dpi), the 50% tissue culture infectious dose (TCID50) was calculated by the Behrens-Kärber method. The hemagglutination test using 0.5% chicken erythrocytes suspended in phosphate-buffered saline (PBS, pH 7.4) was performed on the cell culture supernatants to confirm that the observed CPE reflects the growth of the virus in the cells. In some experiments, VGM lacking trypsin was used to measure the virus titer.
A/duck/Osaka/1/05 (H4N8) and A/duck/Shiga/8/04 (H4N6) were kindly provided by the National Institute of Animal Health, Japan. A/duck/Czechoslovakia/56 (H4N6) was supplied by Dr. H. Kida at the OIE Reference Laboratory for HPAI at Hokkaido University, Japan. A/rednecked stint/Australia/1/04 (H4N8) was kindly provided by Dr. Aeron Hurt at the WHO Collaborating Centre for Reference and Research on Influenza and the Victorian Infectious Diseases Reference Laboratory. The viruses were propagated in the allantoic cavity of 10-day-old embryonated chicken eggs. Aliquots of the allantoic fluids containing the viruses were stored at −80°C until use.
Virus isolation
Virus isolation from cloacal/fecal samples was carried out in 10-day-old embryonated chicken eggs according to the WHO Manual with some modifications. Prior to egg inoculation, the thawed samples were mixed well and centrifuged at 1,000 × g for 10 min at 4°C. The supernatants were supplemented with antibiotics and antimycotics to achieve the following final concentrations: 1,000 U/ml penicillin, 1 mg/ml streptomycin, 100 μg/ml gentamicin, and 10μg/ml amphotericin B. These supernatants were kept at room temperature for 2 h. Then, 0.1 ml of the sample was inoculated into the allantoic cavity of each egg (2 eggs for each sample). After an incubation period of 4 days at 37°C, the eggs were chilled overnight at 4°C. Egg allantoic fluid from the initial inoculation (E1) was tested by a hemagglutination test. The E1 allantoic fluids with negative results in the test were used in a second egg inoculation followed by the hemagglutination test.
Allantoic fluid with hemagglutination activity were subjected to hemagglutination inhibition (HI) and neuraminidase inhibition (NI) tests performed according to the WHO Manual, to identify the influenza virus subtypes. The reference antisera against influenza A viruses and the reference viruses for the HI and NI tests were also provided by Dr. H. Kida.
RT-PCR and RRT-PCR
The viral RNA of the M gene of the influenza virus in the original and the allantoic fluid samples was detected by RRT-PCR as we previously reported (Bui et al., 2011). In brief, total RNA was extracted from the samples by a KingFisher purification system (Thermo Scientific, Waltham, MA) and a Magmax-96 AI/ND Viral RNA isolation kit (Thermo Scientific). First-strand cDNA was prepared using random hexamer primers (Invitrogen, Carlsbad, CA) and M-MLV reverse transcriptase (Invitrogen) under the following conditions: 25°C for 10 min, 37°C for 50 min, and 65°C for 10 min. Using Taqman Universal PCR Master mix (Applied Biosciences, Foster City, CA) RRT-PCR was carried out using an ABI PRISM Sequence Detection System 7900HT (Applied Biosciences) as follows: stage 1, 95°C for 10 min and stage 2, 45 cycles of 95°C for 15 sec and 60°C for 1 min. Samples with Ct below 40 were regarded as M gene positive, and samples with Ct over 40 were considered as suspect positives.
Nucleotide sequencing and phylogenetic analysis
Total RNA extracted from allantoic fluid (E1) containing AIV was transcribed into cDNA using the Uni12 primer (5′-agcraaagcagg-3′) and SuperScript III Reverse Transcriptase (Invitrogen) at 50°C for 60 min followed by 70°C for 10 min. Using the cDNAs as templates, a full length of the 8 viral gene segments was amplified as described by Hoffmann et al. (2001). The PCR products were separated by 1% agarose gel electrophoresis and purified using a QIAquick PCR Purification kit (Qiagen, Hilden, Germany). The purified PCR products were directly applied to the sequencing reaction using a BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems). Nucleotide sequencing was performed in an ABI PRISM 310 Genetic Analyzer (Applied Biosystems). The primer sets amplifying the PCR products were first used for the sequencing reaction, and then primer walking was conducted to read the full-length nucleotide sequence of the gene. Alternatively, the PCR products were subcloned into the pGEM-T Easy vector (Promega, Madison, WI, USA), and the plasmids obtained were used as templates for sequencing. Simultaneously, the cDNA samples were sent to JCVI as a partner submission to the National Institute of Allergy and Infectious Diseases-funded Influenza Genome Sequencing Project. International transportation of the cDNAs was performed according to the protocol permitted by the United States Department of Agriculture (USDA #108081). The sequence analysis for the samples was performed at the JCVI using 454 FLX/Roche and Solexa/Illumina systems. The output data from the JCVI were alto utilized in this study.
Sequence data were analyzed by GENETYX Ver. 9 (Genetyx Corp., Tokyo, Japan) and compared with other GenBank sequences that were identified in the BLAST homology searches (conducted in May 2011). The nucleotide sequences were aligned by Clustal W (Thompson et al., 1994), and the evolutionary distances were computed using the Tamura-Nei method. Phylogenetic trees were constructed using the neighbor-joining method (Saitou and Nei, 1987; Tamura et al., 2004) and bootstrap analysis (1,000 replicates), as well as the bootstrap interior branch test for phylogeny construction using Mega 4.0 software (Tamura et al., 2007).
Mouse infection study of the H4N8 isolates
To investigate the pathogenicity of the H4N8 viruses isolated in this study, a mouse infection study was performed using 8- to 10-week-old female BALB/c mice purchased from Clea Japan Inc. (Tokyo, Japan). All mouse studies were conducted in compliance with the institutional rules for the care and use of laboratory animals, and using protocols approved by the relevant committee at the University.
First, a preliminary study was performed using 2 groups of mice (3 mice/group). A group of mice received intranasal inoculations of 103 TCID50 of 6KS0185, (a H4N8 virus isolate obtained from a gull) and another group received inoculations of 6KS0279 (a H4N8 virus isolate obtained from a stint) in 50 μl of allantoic fluid (E1) under light ether anesthesia. Body weights and clinical signs of the mice were recorded daily afterwards. The mice were sacrificed on 3 dpi. The lungs excised from the mice were weighed and homogenized in DMEM containing penicillin (500 U/ml) and streptomycin (500 μg/ml) to prepare 10% homogenates. Lung homogenates were then subjected to RRT-PCR to amplify the M gene. Since the results obtained in the preliminary study were quite similar among the mice inoculated with 6KS0185 and 6KS0279, subsequent studies were performed only for 6KS0185. In the next experiment, 3 groups of mice (4 mice/group) received an intranasal inoculation of different concentrations of 6KS0185 (10, 102, or 103 TCID50), and 4 control mice received PBS. The body weights were monitored daily and blood samples were collected from the mice prior to sacrifice on 14 dpi. The sera obtained were subjected to the HI test measuring antibody against the virus inoculated.
Tissue tropism and mouse-to-mouse transmission of the H4N8 isolates
Tissue tropism and transmission of the H4N8 isolates were studied using a total of 25 mice. The mice were assigned to 5 cages with 5 mice in each cage. Three mice in each cage were intranasally inoculated with 103 TCID50 of 6KS0185, and the 2 mice that did not receive the virus were kept in the same cage. On 1, 2, 3, 4, and 7 dpi, all 5 mice were sacrificed in 1 of the 5 cages, and their lungs, NTs, spleens, and brains were collected. The tissue samples were subjected to a 10% homogenate preparation, except for the NT (5%) homogenates, followed by the RRT-PCR for the viral M gene and isolation of the virus. For virus isolation, homogenate samples were serially diluted 1:10 and inoculated into eggs. The 50% egg infectious dose (EID50) was calculated by the Behrens-Kärber method. Nucleotide sequence of the gene segments of the virus isolated in the lung samples was analyzed as described above.
In mice that developed symptoms of disease, the animals were maintained alive, unless found dead, until the intended euthanizing time points to follow the disease time course.
Comparison of the pathogenicity with other H4 subtype viruses
Six groups of 15 mice were used to compare the pathogenicity of 6KS0185 with that of other H4 subtype viruses including A/duck/Osaka/1/05 (H4N8), A/duck/Shiga/8/04 (H4N6), A/duck/Czechoslovakia/56 (H4N6), and A/red-necked stint/Australia/1/04 (H4N8). Five groups of mice received 103 TCID50 of the different viruses via the nasal route. Another group of mice served as controls and received the same amount of PBS. Body weights and clinical signs of disease were monitored daily for all of the mice. On 1 dpi, 1 mouse in each group was sacrificed, and between 2 and 7 dpi, 1 mouse in the control group and 2 mice in each experimental group were sacrificed each day. Lungs and NTs obtained from these mice were subjected to viral gene detection and virus isolation as described above. On 4 and 5 dpi, 2 additional mice were euthanized in each group for histopathological and immunohistochemical analysis of the lungs, tracheae, and other major organs. The organ samples were fixed in 10% neutralized buffered formalin solution. All samples were then embedded in paraffin, sectioned at 4 μm, and stained with hematoxylin and eosin. The immunoperoxidase technique was used to detect influenza virus antigens in formalin-fixed, paraffin-embedded samples. A Histofine Simple Stain MAX-PO(G) kit (Nichirei Inc., Tokyo, Japan) was used according to the manufacture’s instructions. Goat polyclonal antibody against influenza A virus (AB1074, Gt X Influenza A virus, Chemicon International, Temecula, CA, USA) was used as the primary antibody at 1:8,000. After staining, sections were counterstained with hematoxylin.
Nucleotide sequence accession numbers
The nucleotide sequences obtained in this study are available from GenBank under accession numbers CY089478 to CY089497 (dideoxy method), CY079283 to CY079298 and CY080223 to CY080238 (2nd generation method). The following published AIV sequences were retrieved from GenBank and included in the analyses of the current study: accession numbers CY028259 to CY028266, CY016625, CY039373, CY045365, CY061624, GQ923547, and HM193786.
Highlights.
We isolated H4N8 subtype avian influenza viruses from stints and gulls in Japan.
All the isolates were PB1 reassortants between Australian and New Zealand lineages.
The H4N8 isolates caused severe respiratory disease leading to death in mice.
The PB2 gene of the isolates contains K482R, but not the E627K or D701N substitution.
The PB1-F2 gene of the H4N8 isolates consists of a 101-amino acid unique sequence.
Acknowledgments
We would like to thank Florian Aldehoff, University of Alaska Fairbanks for his great help on sequence analysis and data management. We also would like to thank Eric Bortz, Mount Sinai School of Medicine for his help on the sequence analysis. We are also grateful to Sachiko Matsuda for technical assistance. This work was partially supported by grants from the Program of Founding Research Centers for Emerging and Reemerging Infectious Diseases, and by a Grant-in-Aid for Exploratory Research (19659115) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan. This work was also partially supported by the US National Institute of Allergy and Infectious Diseases (NIAID contracts HHSN266200700009C and HHSN266200700007C).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bogs J, Kalthoff D, Veits J, Pavlova S, Schwemmle M, Mänz B, Mettenleiter TC, Stech J. Reversion of PB2-627E to -627K during replication of an H5N1 Clade 2.2 virus in mammalian hosts depends on the origin of the nucleoprotein. J Virol. 2011;85:10691–10698. doi: 10.1128/JVI.00786-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazil M. Birds of East Asia. Princeton University Press; Princeton, New Jersey: 2009. [Google Scholar]
- Brown EG, Bailly JE. Genetic analysis of mouse-adapted influenza A virus identifies roles for the NA, PB1, and PB2 genes in virulence. Virus Res. 1999;61:63–76. doi: 10.1016/s0168-1702(99)00027-1. [DOI] [PubMed] [Google Scholar]
- Bui VN, Ogawa H, Karibe K, Matsuo K, Nguyen TH, Awad SSA, Minoungou GL, Xininigen, Saito K, Watanabe Y, Runstadler JA, Happ GM, Imai K. Surveillance of avian influenza virus in migratory water birds in eastern Hokkaido, Japan. J Vet Med Sci. 2011;73:209–215. doi: 10.1292/jvms.10-0356. [DOI] [PubMed] [Google Scholar]
- Bussey KA, Bousse TL, Desmet EA, Kim B, Takimoto T. PB2 residue 271 plays a key role in enhanced polymerase activity of influenza A viruses in mammalian host cells. J Virol. 2010;84:4395–4406. doi: 10.1128/JVI.02642-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Calvo PA, Malide D, Gibbs J, Schubert U, Bacik I, Basta S, O’Neill R, Schickli J, Palese P, Henklein P, Bennink JR, Yewdell JW. A novel influenza A virus mitochondrial protein that induces cell death. Nat Med. 2001;7:1306–1312. doi: 10.1038/nm1201-1306. [DOI] [PubMed] [Google Scholar]
- Conenello GM, Zamarin D, Perrone LA, Tumpey T, Palese P. A single mutation in the PB1-F2 of H5N1 (HK/97) and 1918 influenza A viruses contributes to increased virulence. PLoS Pathog. 2007;3:1414–1421. doi: 10.1371/journal.ppat.0030141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driskell EA, Jones CA, Stallknecht DE, Howerth EW, Tompkins SM. Avian influenza virus isolates from wild birds replicate and cause disease in a mouse model of infection. Virology. 2010;399:280–289. doi: 10.1016/j.virol.2010.01.005. [DOI] [PubMed] [Google Scholar]
- Fang R, Min Jou W, Huylebroeck D, Devos R, Fiers W. Complete structure of A/duck/Ukraine/63 influenza hemagglutinin gene: animal virus as progenitor of human H3 Hong Kong 1968 influenza hemagglutinin. Cell. 1981;25:315–323. doi: 10.1016/0092-8674(81)90049-0. [DOI] [PubMed] [Google Scholar]
- Gabriel G, Dauber B, Wolff T, Planz O, Klenk HD, Stech J. The viral polymerase mediates adaptation of an avian influenza virus to a mammalian host. Proc Natl Acad Sci U S A. 2005;102:18590–18595. doi: 10.1073/pnas.0507415102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garten RJ, Davis CT, Russell CA, Shu B, Lindstrom S, Balish A, Sessions WM, Xu X, Skepner E, Deyde V, Okomo-Adhiambo M, Gubareva L, Barnes J, Smith CB, Emery SL, Hillman MJ, Rivailler P, Smagala J, de Graaf M, Burke DF, Fouchier RA, Pappas C, Alpuche-Aranda CM, Lopez-Gatell H, Olivera H, Lopez I, Myers CA, Faix D, Blair PJ, Yu C, Keene KM, Dotson PDJ, Boxrud D, Sambol AR, Abid SH, St George K, Bannerman T, Moore AL, Stringer DJ, Blevins P, Demmler-Harrison GJ, Ginsberg M, Kriner P, Waterman S, Smole S, Guevara HF, Belongia EA, Clark PA, Beatrice ST, Donis R, Katz J, Finelli L, Bridges CB, Shaw M, Jernigan DB, Uyeki TM, Smith DJ, Klimov AI, Cox NJ. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science. 2009;325:197–201. doi: 10.1126/science.1176225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gething MJ, Bye J, Skehel J, Waterfield M. Cloning and DNA sequence of double-stranded copies of haemagglutinin genes from H2 and H3 strains elucidates antigenic shift and drift in human influenza virus. Nature. 1980;287:301–306. doi: 10.1038/287301a0. [DOI] [PubMed] [Google Scholar]
- Gibbs JS, Malide D, Hornung F, Bennink JR, Yewdell JW. The influenza A virus PB1-F2 protein targets the inner mitochondrial membrane via a predicted basic amphipathic helix that disrupts mitochondrial function. J Virol. 2003;77:7214–7224. doi: 10.1128/JVI.77.13.7214-7224.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto H, Kawaoka Y. A novel mechanism for the acquisition of virulence by a human influenza A virus. Proc Natl Acad Sci U S A. 1998;95:10224–10228. doi: 10.1073/pnas.95.17.10224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustin KM, Belser JA, Wadford DA, Pearce MB, Katz JM, Tumpey TM, Maines TR. Influenza virus aerosol exposure and analytical system for ferrets. Proc Natl Acad Sci U S A. 2011;108:8432–8437. doi: 10.1073/pnas.1100768108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson BA, Stallknecht DE, Swayne DE, Lewis LA, Senne DA. Avian influenza viruses in Minnesota ducks during 1998–2000. Avian Dis. 2003;47:867–871. doi: 10.1637/0005-2086-47.s3.867. [DOI] [PubMed] [Google Scholar]
- Hinshaw VS, Bean WJ, Webster RG, Sriram G. Genetic reassortment of influenza A viruses in the intestinal tract of ducks. Virology. 1980;102:412–419. doi: 10.1016/0042-6822(80)90108-7. [DOI] [PubMed] [Google Scholar]
- Hoffmann E, Stech J, Guan Y, Webster RG, Perez DR. Universal primer set for the full-length amplification of all influenza A viruses. Arch Virol. 2001;146:2275–2289. doi: 10.1007/s007050170002. [DOI] [PubMed] [Google Scholar]
- Huang Y, Hu B, Wen X, Cao S, Gavrilov BK, Du Q, Khan MI, Zhang X. Diversified reassortant H9N2 avian influenza viruses in chicken flocks in northern and eastern China. Virus Res. 2010;151:26–32. doi: 10.1016/j.virusres.2010.03.010. [DOI] [PubMed] [Google Scholar]
- Hurt AC, Hansbro PM, Selleck P, Olsen B, Minton C, Hampson AW, Barr IG. Isolation of avian influenza viruses from two different transhemispheric migratory shorebird species in Australia. Arch Virol. 2006;151:2301–2309. doi: 10.1007/s00705-006-0784-1. [DOI] [PubMed] [Google Scholar]
- Jackson D, Hossain MJ, Hickman D, Perez DR, Lamb RA. A new influenza virus virulence determinant: the NS1 protein four C-terminal residues modulate pathogenicity. Proc Natl Acad Sci U S A. 2008;105:4381–4386. doi: 10.1073/pnas.0800482105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karasin AI, Brown IH, Carman S, Olsen CW. Isolation and characterization of H4N6 avian influenza viruses from pigs with pneumonia in Canada. J Virol. 2000;74:9322–9327. doi: 10.1128/jvi.74.19.9322-9327.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaoka Y, Krauss S, Webster RG. Avian-to-human transmission of the PB1 gene of influenza A viruses in the 1957 and 1968 pandemics. J Virol. 1989;63:4603–4608. doi: 10.1128/jvi.63.11.4603-4608.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kida H, Yanagawa R, Matsuoka Y. Duck influenza lacking evidence of disease signs and immune response. Infect Immun. 1980;30:547–553. doi: 10.1128/iai.30.2.547-553.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumbholz A, Philipps A, Oehring H, Schwarzer K, Eitner A, Wutzler P, Zell R. Current knowledge on PB1-F2 of influenza A viruses. Med Microbiol Immunol. 2011;200:69–75. doi: 10.1007/s00430-010-0176-8. [DOI] [PubMed] [Google Scholar]
- Li Z, Chen H, Jiao P, Deng G, Tian G, Li Y, Hoffmann E, Webster RG, Matsuoka Y, Yu K. Molecular basis of replication of duck H5N1 influenza viruses in a mammalian mouse model. J Virol. 2005;79:12058–12064. doi: 10.1128/JVI.79.18.12058-12064.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowen AC, Mubareka S, Tumpey TM, Garcia-Sastre A, Palese P. The guinea pig as a transmission model for human influenza viruses. Proc Natl Acad Sci U S A. 2006;103:9988–9992. doi: 10.1073/pnas.0604157103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazur I, Anhlan D, Mitzner D, Wixler L, Schubert U, Ludwig S. The proapoptotic influenza A virus protein PB1-F2 regulates viral polymerase activity by interaction with the PB1 protein. Cell Microbiol. 2008;10:1140–1152. doi: 10.1111/j.1462-5822.2008.01116.x. [DOI] [PubMed] [Google Scholar]
- McAuley JL, Chipuk JE, Boyd KL, Van De Velde N, Green DR, McCullers JA. PB1-F2 proteins from H5N1 and 20th century pandemic influenza viruses cause immunopathology. PLoS Pathog. 2010;6:e1001014. doi: 10.1371/journal.ppat.1001014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mase M, Tanimura N, Imada T, Okamatsu M, Tsukamoto K, Yamaguchi S. Recent H5N1 avian influenza A virus increases rapidly in virulence to mice after a single passage in mice. J Gen Virol. 2006;87:3655–3659. doi: 10.1099/vir.0.81843-0. [DOI] [PubMed] [Google Scholar]
- McAuley JL, Hornung F, Boyd KL, Smith AM, McKeon R, Bennink J, Yewdell JW, McCullers JA. Expression of the 1918 influenza A virus PB1-F2 enhances the pathogenesis of viral and secondary bacterial pneumonia. Cell Host Microbe. 2007;2:240–249. doi: 10.1016/j.chom.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann G, Noda T, Kawaoka Y. Emergence and pandemic potential of swine-origin H1N1 influenza virus. Nature. 2009;459:931–939. doi: 10.1038/nature08157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninomiya A, Takada A, Okazaki K, Shortridge KF, Kida H. Seroepidemiological evidence of avian H4, H5, and H9 influenza A virus transmission to pigs in southeastern China. Vet Microbiol. 2002;88:107–114. doi: 10.1016/s0378-1135(02)00105-0. [DOI] [PubMed] [Google Scholar]
- Obenauer JC, Denson J, Mehta PK, Su X, Mukatira S, Finkelstein DB, Xu X, Wang J, Ma J, Fan Y, Rakestraw KM, Webster RG, Hoffmann E, Krauss S, Zheng J, Zhang Z, Naeve CW. Large-scale sequence analysis of avian influenza isolates. Science. 2006;311:1576–1580. doi: 10.1126/science.1121586. [DOI] [PubMed] [Google Scholar]
- O’Donnell CD, Subbarao K. The contribution of animal models to the understanding of the host range and virulence of influenza A viruses. Microbes Infect. 2011;13:502–515. doi: 10.1016/j.micinf.2011.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolling T, Koerner I, Zimmermann P, Holz K, Haller O, Staeheli P, Kochs G. Adaptive mutations resulting in enhanced polymerase activity contribute to high virulence of influenza A virus in mice. J Virol. 2009;83:6673–6680. doi: 10.1128/JVI.00212-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Seo SH, Hoffmann E, Webster RG. Lethal H5N1 influenza viruses escape host antiviral cytokine responses. Nat Med. 2002;8:950–954. doi: 10.1038/nm757. [DOI] [PubMed] [Google Scholar]
- Sharp GB, Kawaoka Y, Wright SM, Turner B, Hinshaw V, Webster RG. Wild ducks are the reservoir for only a limited number of influenza A subtypes. Epidemiol Infect. 1993;110:161–176. doi: 10.1017/s0950268800050780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slemons RD, Condobery PK, Swayne DE. Assessing pathogenicity potential of waterfowl-origin type A influenza viruses in chickens. Avian Dis. 1991;35:210–215. [PubMed] [Google Scholar]
- Song MS, Pascua PN, Lee JH, Baek YH, Lee OJ, Kim CJ, Kim H, Webby RJ, Webster RG, Choi YK. The polymerase acidic protein gene of influenza a virus contributes to pathogenicity in a mouse model. J Virol. 2009;83:12325–12335. doi: 10.1128/JVI.01373-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbarao EK, London W, Murphy BR. A single amino acid in the PB2 gene of influenza A virus is a determinant of host range. J Virol. 1993;67:1761–1764. doi: 10.1128/jvi.67.4.1761-1764.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tamura K, Nei M, Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci U S A. 2004;101:11030–11035. doi: 10.1073/pnas.0404206101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taubenberger JK, Reid AH, Lourens RM, Wang R, Jin G, Fanning TG. Characterization of the 1918 influenza virus polymerase genes. Nature. 2005;437:889–893. doi: 10.1038/nature04230. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trifonov V, Khiabanian H, Rabadan R. Geographic dependence, surveillance, and origins of the 2009 influenza A (H1N1) virus. N Engl J Med. 2009;361:115–119. doi: 10.1056/NEJMp0904572. [DOI] [PubMed] [Google Scholar]
- Tscherne DM, Garcia-Sastre A. Virulence determinants of pandemic influenza viruses. J Clin Invest. 2011;121:6–13. doi: 10.1172/JCI44947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumpey TM, Basler CF, Aguilar PV, Zeng H, Solorzano A, Swayne DE, Cox NJ, Katz JM, Taubenberger JK, Palese P, Garcia-Sastre A. Characterization of the reconstructed 1918 Spanish influenza pandemic virus. Science. 2005;310:77–80. doi: 10.1126/science.1119392. [DOI] [PubMed] [Google Scholar]
- Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol Rev. 1992;56:152–179. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster RG, Shortridge KF, Kawaoka Y. Influenza: interspecies transmission and emergence of new pandemics. FEMS Immunol Med Microbiol. 1997;18:275–279. doi: 10.1111/j.1574-695X.1997.tb01056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright PF, Neumann G, Kawaoka Y. Orthomyxoviruses. In: Knipe DM, Howley PM, editors. Fields Virology. 5. Lippincott Williams & Wilkins Co; Philadelphia: 2007. pp. 1691–1740. [Google Scholar]
- Zamarin D, Ortigoza MB, Palese P. Influenza A virus PB1-F2 protein contributes to viral pathogenesis in mice. J Virol. 2006;80:7976–7983. doi: 10.1128/JVI.00415-06. [DOI] [PMC free article] [PubMed] [Google Scholar]