Abstract
How long organisms may live is not entirely written in their genes. Recent findings reveal that epigenetic factors that regulate histone methylation, a type of chromatin modification, can affect lifespan. The reversible nature of chromatin modifications suggests that therapeutic targeting of chromatin regulators could be used to extend lifespan and healthspan. This review describes the epigenetic regulation of lifespan in diverse model organisms, focusing on the role and mode of action of chromatin regulators that affect two epigenetic marks – H3K4me3 and H3K27me3 – in longevity.
Regulation of longevity
Longevity is regulated by both genetic and environmental factors. Genetic mutations in a variety of pathways, including the insulin/IGF-1 pathway, can extend lifespan in various organisms (Table 1) [1, 2]. Environmental stimuli can also affect lifespan, either negatively or positively. Stress stimuli (e.g. DNA damage) are known to accelerate aging. Dietary restriction (DR) – restriction in food intake without malnutrition – extends lifespan and delays age-related diseases in a wide range of species [3]. In Caenorhabditis elegans, lifespan extension by different DR regimens is mediated by signaling pathways (e.g. TOR, AMPK) and downstream transcription factors (e.g. FOXA, FOXO, NRF2), which culminate in changes in gene expression profiles (Table 1) [3–8]. Signaling pathways and transcription factors regulate longevity in a conserved manner (Table 1) and, interestingly, recent evidence in mammalian cells indicates that several of these conserved longevity pathways can modulate chromatin states in mammalian cells [9–12]. These observations raise the exciting possibility that environmental stimuli that affect lifespan may do so by modulating chromatin states. Changes in chromatin state are likely to be more persistent than transient alterations in signaling pathways and transcription factor activity, which could have important implications for how environmental stimuli, dietary regimens or specific drugs, may affect lifespan even when applied transiently.
Table 1.
Major pathways that regulate longevity
| Pathway | Characteristics | Change that extend lifespan? |
Conservation |
|---|---|---|---|
| Insulin/IGF-1 | Endocrine signaling | Inhibition | Worms, flies, mice, humans |
| TOR | Nutrient/aminoacid sensing | Inhibition | Yeast, worms, flies, mice |
| AMPK | Nutrient/energy sensing | Overexpression | Worms, mice (indirect study) |
| SIR2 | NAD+ dependent histone deacetylase | Overexpression | Yeast, worms (in some cases), flies (in some cases) |
| Mitochondrial electron-transport chain | Respiration | Inhibition | Worms, flies, mice |
| Germline stem cells | Reproduction | Inhibition | Worms, flies |
Examples of conserved pathways that are known to regulate lifespan in different model organisms (see [1] for a comprehensive review).
Chromatin state and longevity
Chromatin state is governed by a series of modifications that include DNA methylation and histone modification [13–15]. These modifications are often termed ‘epigenetic’ [13], as they do not affect the genetic sequence per se. Post-translational modifications of the core histones, also known as ‘histone marks’, include acetylation, methylation, phosphorylation and ubiquitylation. Specific histone marks are associated with different chromatin states [13]. For example, trimethylation at lysine 4 of histone 3 (H3K4me3) is generally associated with gene expression [15]. Trimethylation at lysine 27 of histone 3 (H3K27me3) is generally associated with transcriptional repression [15]. Histone marks may represent a key intermediary step between signaling pathways that extend lifespan and the expression of specific genes involved in longevity. Histone modifications are reversibly catalyzed by specific enzymes, such as acetyltransferases, deacetylases, methyltransferases and demethylases [16, 17]. Thus, the enzymes that reversibly regulate histone marks may affect the expression of specific genes involved in longevity, and as such, represent interesting therapeutic targets to help prevent or treat age-dependent diseases, including neurodegenerative diseases and cancer.
Until recently, studies on chromatin regulators and lifespan had mostly focused on histone acetylases and deacetylases, in particular the SIR2 deacetylase family (Table 1) [18–20]. In the past couple of years, a series of studies has revealed that histone methyltransferases and demethylases also affect lifespan. This review synthesizes these recent findings and describes the role and mode of action of regulators of histone methylation in longevity in diverse model systems. While the main focus will be on H3K4me3 regulators, we will also examine the regulation and importance of other methylation marks in longevity.
H3K4me3 modifiers affect longevity in C. elegans
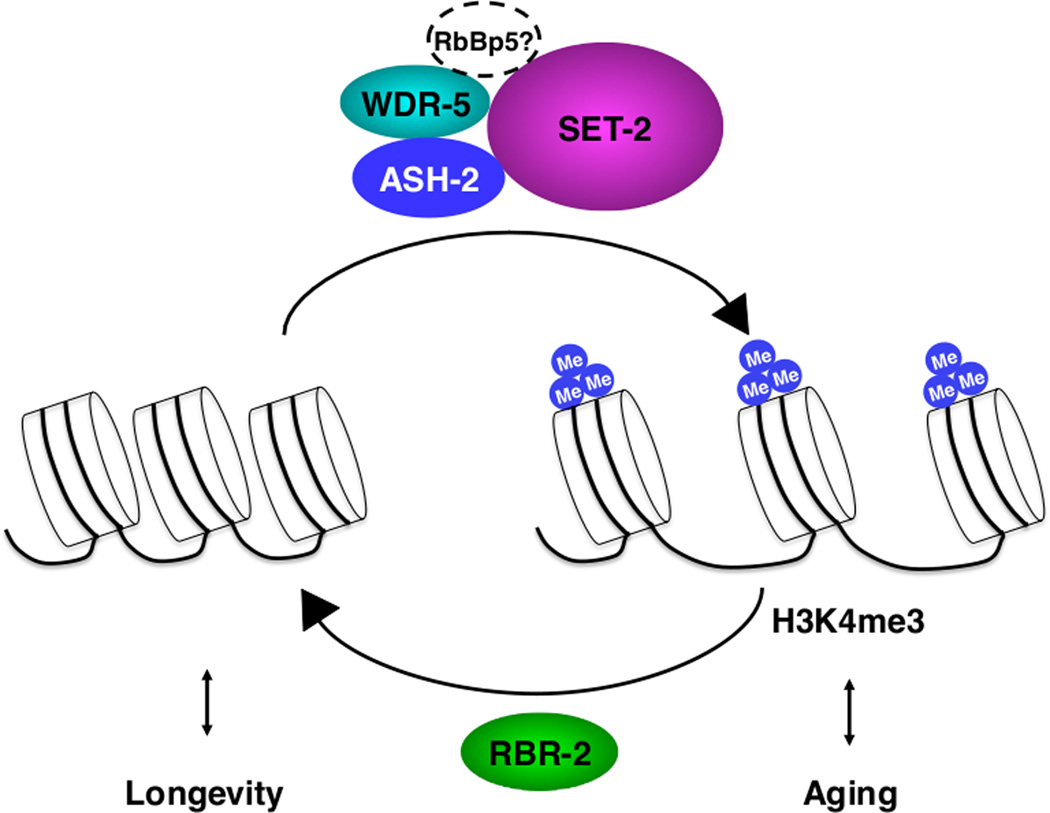
Histone methylation is catalyzed by protein complexes composed of a methyltransferase and other proteins that are important for the recruitment of the complex to specific genomic loci. Three major complexes are responsible for generating H3K4me3 in mammals: the COMPASS complex, the Trithorax complex and the Trithorax-related complex [21–27]. These H3K4me3 regulatory complexes are known to be important for development and stem cell function [24, 28, 29], but until recently their role in longevity had not been studied. Recent work indicates that the complex composed of ASH-2, WDR-5 and the H3K4me3 methyltransferase SET-2 affects lifespan in C. elegans. Worm SET-2 is phylogenetically related to methytransferases in the COMPASS complex (Figure 1). Knocking-down ASH-2, WDR-5 or SET-2 in fertile worms significantly extends worm lifespan (Table 2) [30]. Knock-down or mutation of ASH-2, WDR-5 or SET-2 reduces global H3K4me3 levels at the larval L3 stage [30], consistent with other studies [31–34], suggesting that ASH-2, WDR-5 and SET-2 affect lifespan by modulating H3K4me3 levels. ASH-2, WDR-5 and SET-2 function in the same genetic pathway to impact lifespan [30], although whether they physically interact remains to be established. Deficiency in RBR-2 – an H3K4me3 demethylase with homology to human JARID1A/KDM5A [35] – increases H3K4me3 levels [30, 35], and reduces lifespan in either wild-type worms or worms that are deficient for ASH-2, SET-2 or WDR-5 (Table 2) [30]. Importantly, overexpression of RBR-2 is sufficient to extend lifespan in C. elegans (Table 2) [30]. Thus, RBR-2 counteracts the ASH-2 complex in longevity and H3K4me3 levels (Figure 2). Overall, these studies suggest that limiting the H3K4me3 level is beneficial for longevity in C. elegans, perhaps because it reduces the transcription of genes that would normally lead to aging. The ASH-2/WDR-5/SET-2 complex might also affect longevity by acting via yet-to-be identified non-histone targets.
Figure 1. Phylogenetic analysis of C. elegans SET-2 and its homologs in yeast, flies and humans.
Worm SET-2 is phylogenetically closer to SET1 in flies and SET1A/SET1B in humans than it is to Trithorax (MLL1/2) or Trithorax-related families (MLL3/4). The phylogenetic tree was constructed with ClustalW2 using full-length protein sequences.
Table 2.
Histone methylation regulators in aging and longevity
| C. elegans | Drosophila | Mammals | |
|---|---|---|---|
| H3K4me3 | |||
| Change with age? | NT | decrease ♀ (in the brain) | change in landscape (in the brain) |
| KD or mutation of methyltransferase | increase lifespan ⚥ (COMPASS complex) | no effect on lifespan ♂ (Trx complex) | NT |
| OE of methyltransferase | NT | NT | NT |
| KD or mutation of demethylase | decrease lifespan ⚥ | decrease lifespan ♂ | NT |
| OE of demethylase | increase lifespan ⚥ | NT | NT |
| H3K4me1/me2 | |||
| Change with age? | NT | NT | NT |
| KD or mutation of methyltransferase | NT | NT | NT |
| OE of methyltransferase | NT | NT | NT |
| KD or mutation of demethylase | increase lifespan ⚥ | NT | NT |
| OE of demethylase | NT | NT | NT |
| H3K27me3 | |||
| Change with age? | decrease ⚥ | NT | NT |
| KD or mutation of methyltransferase | NT | increase lifespan ♂ | NT |
| OE of methyltransferase | NT | NT | NT |
| KD or mutation of demethylase | increase lifespan ⚥ | NT | NT |
| OE of demethylase | NT | NT | NT |
Specific histone methylation marks are altered during aging, and regulators of some of these marks affect lifespan. NT: not tested; KD: knock-down; OE: overexpression.
Figure 2. Regulation of longevity by H3K4me3 modifiers in C. elegans.
The ASH-2 H3K4me3 complex, consisting of ASH-2, SET-2, WDR-5 and possibly RbBp5, may promote aging by trimethylating H3K4. Note that the knock-down of the worm homologue of RbBP5, F21H12.1, has been shown to extend lifespan [84], although its impact on H3K4me3 is unknown. Conversely, the RBR-2 demethylase may promote longevity by demethylating H3K4me3.
The H3K4 demethylase LSD1 has also been found to affect worm lifespan (Table 2) [36]. Trimethylation of H3K4 first requires mono (me1)- and di (me2)-methylation [15]. LSD1 demethylates H3K4me1/me2 and triggers transcriptional repression of downstream target genes [37]. Knock-down of lsd-1 or suppression of lsd-1 mRNA by LiCl treatment extends lifespan in worms by 25% and 46%, respectively [36, 38]. It might appear surprising that deficiencies in LSD-1, an H3K4me1/me2 demethylase, extend longevity [36], while deficiencies in RBR-2, an H3K4me3 demethylase, shorten lifespan [30]. RBR-2 and LSD-1 regulate lifespan in an opposite manner perhaps because they have non-overlapping age-related gene targets, act in distinct tissues or function at different times of life. Exploring the combined effects of the ASH-2 complex, RBR-2 and LSD-1 on worm lifespan will provide insight into the respective contribution of H3K4me1, H3K4me2 and H3K4me3 to lifespan regulation. Overall, these results indicate that regulators of H3K4 methylation affect lifespan in C. elegans, which raises the question of where they act in the organism to control longevity.
H3K4me3 modifiers in the germline
The ASH-2 and RBR-2 H3K4me3 regulatory complexes appear to promote somatic longevity by acting mostly in the germline [30]. Both the ASH-2 protein and the H3K4me3 mark are highly enriched in the germline and newly fertilized eggs [30, 33, 34, 39]. ASH-2 deficiency no longer extends lifespan in worm mutants with an underdeveloped germline or in mutants that cannot produce fertilized eggs [30]. Thus, the presence of an intact germline is necessary for the ASH-2 complex to regulate lifespan. Furthermore, RNAi knock-down of ASH-2 or SET-2 still extends lifespan in a worm mutant strain that is mostly insensitive to RNAi in the soma [30]. Overexpression of RBR-2 specifically in the germline is sufficient to prolong lifespan [30]. These findings suggest that the ASH-2 complex and RBR-2 regulate lifespan by acting in the germline.
Do H3K4me3 regulatory complex deficiencies extend lifespan because they decrease fertility? Theories of aging have indeed postulated that resources are allocated to either reproduction or maintenance of the soma. Impairing reproduction may thus lead to better maintenance of the soma and lifespan extension [40, 41]. This does not appear to be the case for lifespan extension by deficiencies in H3K4me3 regulators, however. In the conditions used for lifespan assays, ASH-2 knock-down does not affect C. elegans fertility, as measured by germline morphology, germline cell number, number of eggs laid and brood size [30]. Furthermore, at the temperature used in lifespan assays (20°C), wdr-5 mutants develop normally and do not display a statistically significant increase in sterility [30], although these mutants do exhibit a slight increase in producing dead eggs [34]. Similarly, neither set-2 mutants nor rbr-2 mutants show significant fertility defects at 20°C [30, 34]. Thus, longevity due to deficiencies in ASH-2 complex members is unlikely to be due to a trade-off of resource allocation to reproduction versus maintenance of the soma. Nevertheless, it has to be noted that under more drastic conditions, these H3K4me3 regulators do show fertility defects. At 25°C, wdr-5 mutants exhibit brood size reduction, endomitotic oocytes and abnormal DNA morphology in their germ cells [34], and ash-2 genetic mutants have reduced fertility at 20°C and are sterile at 25°C [33]. Thus, there might still be an unidentified connection between the regulation of lifespan and germline stability by members of the ASH-2 complex.
Timing of H3K4me3 regulation
Knowing that the ASH-2 complex plays an important role in longevity regulation, when does it serve to control H3K4me3 levels? Determining the time of action of H3K4me3 regulatory complexes is an important step in deciphering the mechanisms underlying lifespan extension. Recent studies show that ASH-2, WDR-5 and SET-2 are necessary for H3K4 trimethylation in both the developing and adult germline in C. elegans [30, 33, 34]. For example, wdr-5 and set-2 mutant worms exhibit marked decreases in H3K4me2 and H3K4me3 in the developing and adult germline [30, 33, 34]. The decrease in H3K4me2/me3 marks in wdr-5 and set-2 mutant worms is particularly pronounced in the germline stem cell (GSC) region in adult worms [33, 34]. While genetic evidence suggests a role for the H3K4me3 regulatory complex in fertilized eggs rather than in GSC [30], the regulation of H3K4me3 in the GSC could indirectly participate in lifespan regulation by affecting germline stability.
Surprisingly, the importance of ASH-2 for H3K4me3 regulation differs in development versus adulthood. During development, ASH-2 knock-down significantly reduces H3K4me3 levels, and this reduction is particularly noticeable in the germline [30]. In contrast, in adult worms, ASH-2 knock-down does not significantly alter H3K4me3, in either the soma or the germline [33, 34] (Eric L. Greer and A.B., unpublished data). Thus, ASH-2 may interact with SET-2 during development, but form a complex with other methyltransferases in adults and in the soma. Overall, these observations suggest that ASH-2, WDR-5 and SET-2 regulate H3K4me3 in a largely overlapping manner, although each of these proteins may take part in other H3K4me3 regulatory complexes in a particular cell type or at a specific stage of life. As H3K4me3 is generally associated with gene expression [42, 43], the time- and tissue-specificity of H3K4me3 regulation may have crucial consequences on the expression of longevity-related genes.
Target genes of the H3K4me3-regulating complex
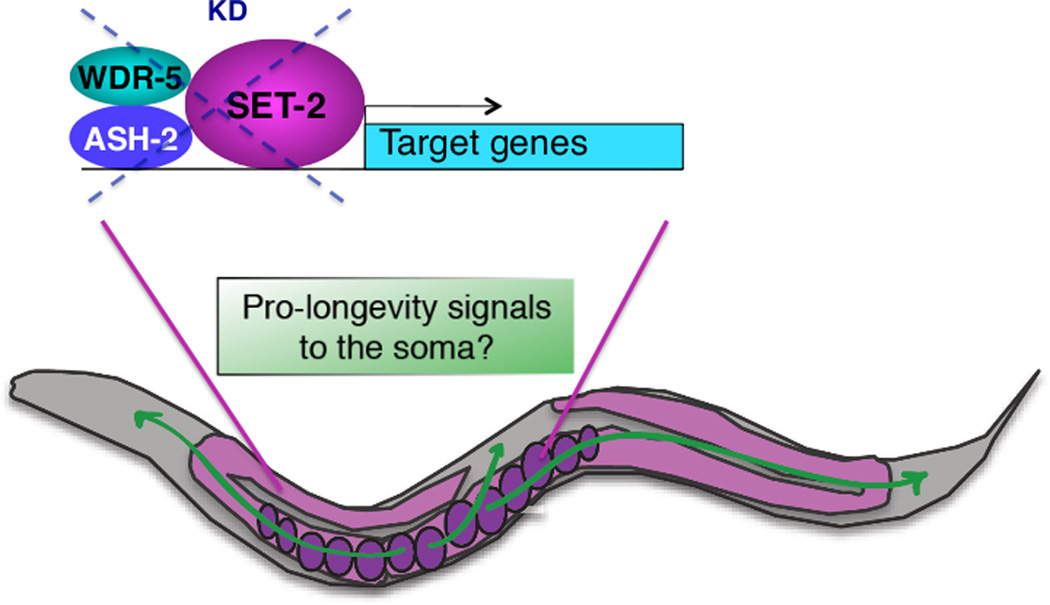
Do deficiencies in members of the H3K4me3 regulatory complex in worms affect all transcriptionally active genes or only a specific subset of genes? Whole-genome microarray analysis identified hundreds of genes regulated by ASH-2 at two stages of life (220 genes at the L3 larval stage and 847 genes at day 8 of life) [30], indicating that ASH-2 does not globally affect transcription, but rather regulates a relatively restricted number of genes. The vast majority of ASH-2-regulated genes are dependent on the presence of an intact germline [30]. Interestingly, a significant number of ASH-2 regulated genes at both ages are involved in lifespan determination [30]. ASH-2 target genes are also enriched for genes exhibiting differential expression with age [30, 44]. Together, these results suggest that ASH-2 deficiency and the subsequent decline in H3K4me3 may silence ‘pro-aging’ genes in the germline, which would serve as a signal to promote the longevity of the soma (Figure 3). Genetic evidence, using worm mutants that cannot produce fertilized eggs but are otherwise phenotypically wild-type, suggests that ASH-2 requires the presence of fertilized eggs to regulate the lifespan of the soma [30]. Thus, ASH-2 target genes could regulate the production of hormonal or metabolic signals by fertilized eggs, before the eggshell forms. These signals could diffuse into the soma, thereby preventing aging of somatic cells.
Figure 3. Interplay between the germline and the soma in the regulation of longevity by ASH-2 complex deficiency.
In adult worms, longevity by deficiencies in the ASH-2 complex requires an intact germline with continuous egg production. It is possible that unknown ‘pro-longevity’ signals are emitted from the fertilized eggs when ASH-2 complex members are deficient, thereby delaying aging of the soma. KD: knock-down.
It is possible that deficiencies in H3K4me3 regulators still cause a decrease in global transcription that is not captured by microarray studies because such experiments cannot measure absolute differences in gene expression [45]. Thus, more work will be needed to gain quantitative insight into the genes regulated by ASH-2 and other members of the complex. A general decrease in global transcription upon ASH-2 deficiency could lead to a reduction in global protein synthesis. Because reduced protein synthesis has been found to extend lifespan in worms and other species ([46–49], for a review see [50]), a decrease in global transcription/translation could also explain, at least in part, the beneficial effects of ASH-2 deficiency on lifespan. Thus, the mechanisms by which the H3K4me3 regulatory complex affects lifespan are not entirely clear. Comparison across species could give key insight into the role and mode of action of H3K4me3 complexes in longevity.
H3K4 methylation regulators in Drosophila aging
The role of the H3K4me3 demethylase RBR-2 in promoting longevity appears to be conserved in Drosophila (Table 2) [51]. Overexpression of Lid, which is the only fly orthologue of the worm RBR-2 (Figure 4), reduces H3K4me3 levels, whereas deficiencies in Lid result in elevated global H3K4me3 [52, 53]. Consistent with the worm studies [30], deficiencies in Lid shortened male fly lifespan by 18% [51]. Lid does not affect the lifespan of female flies, suggesting that this H3K4me3 demethylase may impact longevity in a sex-specific manner.
Figure 4. Phylogenetic analysis of C. elegans RBR-2 and its homologs in yeast, flies and humans.
The phylogenetic tree was constructed with ClustalW2 using full-length protein sequences.
In contrast, Trx, one of the three fly H3K4me3 methyltransferases, does not affect the lifespan of male flies (Table 2) [54]. Whether Trx affects female lifespan and global H3K4me3 levels was not examined in this study. Fly Trx does not have an ortholog in worms [55]. Thus, different H3K4me3 methyltransferase complexes (COMPASS, Trithorax and Trithorax-related) may have opposing roles in longevity, perhaps because they act in different tissues. Alternatively, different H3K4me3 methyltransferases may regulate lifespan in a sex-specific manner. Finally, different species may have variable requirements for chromatin regulators during aging, perhaps because different tissues are rate-limiting for longevity in each species or because of different metabolic requirements.
H3K4me3 in mammals
While the importance of H3K4me3 regulators in mammalian longevity has not yet been addressed, intriguing age-dependent changes in H3K4me3 landscape have been observed in the human brain (Table 2) [56]. Genome-wide H3K4me3 landscape in FACS-isolated neurons from the prefrontal cortex of individuals ranging between 0.5 to 69 years of age revealed that neurons from infants (<1 year) exhibit a larger number of loci with H3K4me3 peaks (~600 loci) than neurons from old adults (>60 years) (~100 loci) [56]. The subset of infant-specific, H3K4me3-marked genes is enriched in neurogenesis, neuronal growth and differentiation genes, likely reflecting the cellular plasticity of the developing brain. Notably, a subset of loci also displays selective H3K4me3 in old neurons but not in infant neurons, although this smaller set of genes do not show significant enrichment for particular categories of genes [56]. Collectively, these results suggest that the H3K4me3 epigenome landscape is altered as a function of age in neurons. However, in the absence of intermediary ages (young and middle-age adults), it is difficult to distinguish if H3K4me3 changes result from aging per se, or whether they are a consequence of the end of development.
It is also exciting to note that several H3K4me3 regulators have been implicated in the ability of stem cells to self-renew and differentiate, processes that are critical for proper tissue regeneration and maintenance throughout lifespan [28]. For example, mammalian WDR5, a critical component of the ASH2L complex, promotes mouse embryonic stem cell (ESC) pluripotency by activating the expression of self-renewal genes [57]. However, it is important to note that WDR5 could regulate ESC pluripotency via proteins other than MLLtypes of H3K4me3 methyltransferases, as WDR5 has been found to interact with a number of other proteins [58–60]. MLL1 – an H3K4me3 methyltransferase with phylogenetic similarities to Drosophila Trx (Figure 1) – is essential for the maintenance of fetal and adult hematopoietic stem cells (HSC) [61, 62], and for the production of neurons from post-natal neural stem cells (NSC) [63]. Furthermore, the H3K4me1/me2 demethylase LSD1 [37] promotes adult NSC proliferation via interaction with the NSC specific transcriptional regulator TLX and by repressing pro-differentiation genes [64]. RBP2 (also known as JARID1A or KDM5A), the H3K4me3 demethylase orthologous to RBR-2 (Figure 4), mediates transcriptional repression during mouse ESC differentiation by associating with polycomb complex PRC2 [65]. Thus, regulators of H3K4me3 appear to play important roles in regulating stem cell properties. Both positive and negative regulators of H3K4me3 appear to be associated with defects in stem cell properties, suggesting that appropriate amounts of this mark are likely essential for stem cells. The ability of H3K4me3 regulators to properly coordinate stem cell renewal and differentiation of stem cells in mammals may be essential to the regenerative potential and longevity of tissues.
In mammalian cells, H3K4me3 has been shown to engage in cross-talk with other marks, in particular the repressive mark H3K27me3 [66], which raises the important question of whether other methylation marks also affect longevity.
The repressive mark H3K27me3 in C. elegans and Drosophila longevity and in mammalian stem cell function
In worms, attenuation of the H3K27me3 demethylase UTX-1 by RNAi or heterozygous mutation extends lifespan (Table 2) [38, 67]. RNAi knockdown of UTX-1 in worms leads to elevated global H3K27me3 levels [38, 67, 68]. Contrary to the ASH-2/WDR-5/SET-2 complex, UTX-1 regulates lifespan independently of the germline, suggesting that this demethylase principally acts in the soma to regulate lifespan [38, 67]. Furthermore, UTX-1 regulates lifespan in a manner that depends on the insulin-FoxO pathway [38, 67]. Interestingly, H3K27me3 levels strikingly drop with age [38]. Consistent with this observation, utx-1 mRNA levels increase with age in worms [67]. Together, these studies suggest that preserving high levels of H3K27me3 by inhibiting UTX-1 may be critical for maintaining youthfulness. These data are also consistent with the loss of epigenetic control of repressed chromatin observed during aging in a number of species [69, 70].
In flies, heterozygous mutations in the complex that methylates H3K27, notably the PRC2 components E(z) and ESC, reduce global H3K27me3 levels and extend the longevity of males (females were not tested) (Table 2) [54]. Mutations in these PRC2 components lead to partial derepression of PRC2 target genes (i.e. Abd-B and Odc1), which may mediate the effect of PRC2 on longevity [54]. Heterozygous mutation of the H3K4me3 methyltransferase Trx reverts the long lifespan of PRC2 mutants, and indirectly leads to a modest restoration of global H3K27me3 levels and PRC2-dependent target gene expression [54], suggesting that the H3K4me3 Trx complex counteracts the H3K27me3 PRC2 complex to regulate lifespan. These results also indicate that, contrary to the model put forward in worms, excessive chromatin repression may actually shorten lifespan in flies. Why does deficiency in the H3K27 demethylase (in hermaphrodite worms) and in the H3K27 methyltransferases (in male flies) both result in lifespan extension? This discrepancy could be attributed to species-specific differences, perhaps due to the amount of proliferative cells versus differentiated cells present in the organism. Alternatively, H3K27me3 levels may not affect lifespan in the same manner, depending on sex, tissue or time of action. Target genes of H3K27me3 complexes may differ depending on the specific regulators that deposit it. It is interesting to note that in mammalian cells, UTX is part of the Trithorax-related complexes, but not a part of the COMPASS or Trithorax complexes [71, 72]. Thus, each complex may have a specific role in longevity, even though they target similar marks, perhaps because the genomic loci these marks affect are different. Finally, it cannot be excluded that PRC complexes and UTX demethylases have other substrates than H3K27, and that these substrates may differ in worms and flies.
In mammals, H3K27me3 regulators also play important roles in stem cell aging. BMI1, a member of the mammalian H3K27me3 polycomb complex 1 (PRC1), is necessary for the self-renewal of adult stem cells, such as HSCs [73–76] and NSCs [77–79], in large part through repression of the p16INK4a/p19ARF locus [80–82]. Perturbations in H3K27me3 and H3K4me3 regulators both result in defects in stem cell proliferation, suggesting that a delicate balance between activation and repression of distinct sets of target genes is required during the stem cell aging process. This balance might be particularly important in the context of ‘bivalent domains’, which are regions of the chromatin containing both H3K4me3 and H3K27me3. Bivalent domains have been suggested to mark regulatory regions of genes that are ‘poised’ for differentiation in ESC and potentially other stem cells [66]. Dpy-30, a core component of the mammalian SET1/MLL H3K4me3-regulating complexes, has been shown to be critical for mouse ESC fate specification by modulating H3K4 methylation in these bivalent domains [83]. It will be interesting to determine whether and how bivalent domains are affected during aging in pools of adult stem cells.
Other methylation marks in worm, fly and mammalian longevity
Interestingly, additional methyltransferases and demethylases have emerged from C. elegans lifespan screens, including the potential methyltransferases SET-9 [30, 84], SET-15 [30, 84], SET-6 [30], SET-4 [30], BLMP-1 [30] and the demethylase T26A5.5 [38]. While the exact marks regulated by these enzymes are not yet known in worms, it will be interesting to test the epistatic interactions between H3K4me3 regulators and these other chromatin modifiers.
Several methylation marks have recently been shown to change with age in Drosophila. ChIP-chip of several histone marks in 10-day (young) and 40-day (old) female fly heads revealed an age-dependent loss in activating marks (H3K4me3 and H3K36me3), and an increase in repressive marks (H3K9me3) [85], consistent with the notion that in flies, repressive marks may promote aging. The nuclear distribution of the H3K9me3 mark is also altered in older flies, presumably because this repressive mark aberrantly diffuses into the euchromatin in old individuals. The age-dependent increase in global H3K9me3 does not appear to be accompanied by a change in gene expression at these loci, as measured by microarrays [85]. Thus, the age-dependent increase in H3K9me3 may not be sufficient to alter gene expression. Alternatively, regions of H3K9me3 that might have led to transcriptional silencing may not have been covered by the ChIP-chip genomic arrays (e.g. repetitive regions). Finally, microarrays may underestimate changes in gene expression, particularly when they are global.
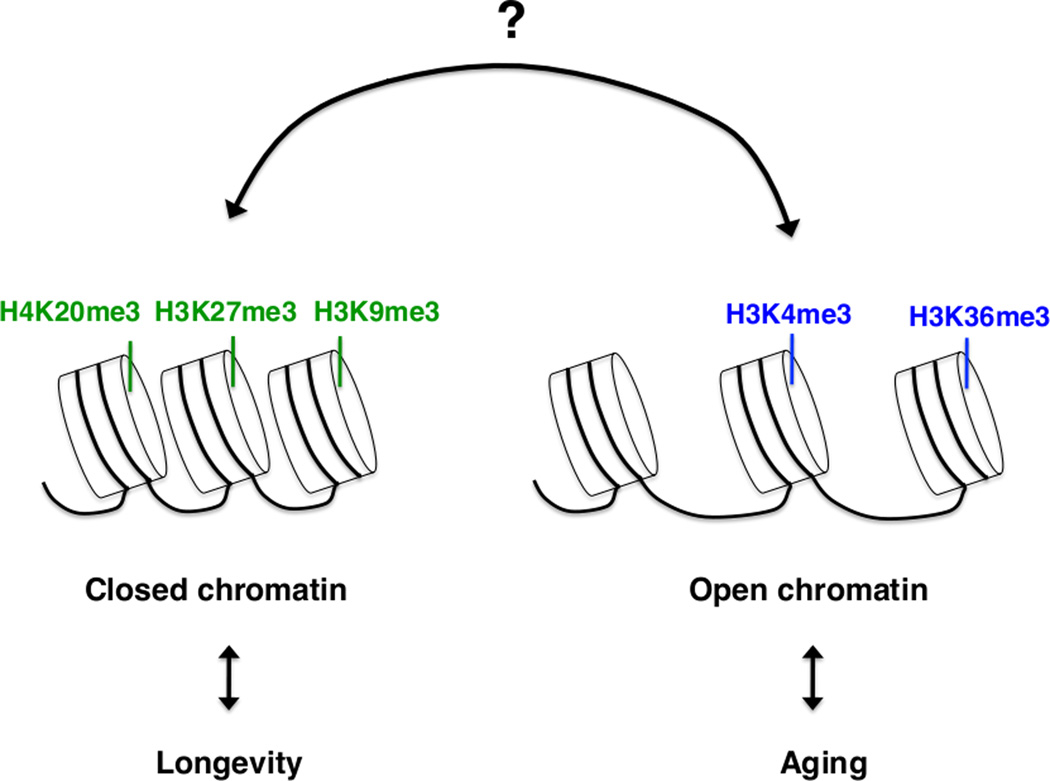
In mammals, H4K20me3 – a mark associated with constitutive heterochromatin [86] – increases with age in rat livers [87], as well as in cells derived from human patients with Hutchinson-Gilford progeria, a premature aging syndrome [88]. An elevation of repressive marks with age suggests that excessive transcriptional repression may contribute to the aging process. This conclusion, however, is in contrast with the finding that cells isolated from patients with Hutchinson-Gilford progeria exhibit reduction or complete loss of heterochromatin mark H3K9me3 [89]. Thus, more work is needed to determine how repressive and activating marks change with age, and to explore if these changes are a cause or a consequence of aging. An increasing number of histone marks have been found to change during aging, suggesting that different histone marks may act together, in a network, to affect longevity (Figure 5).
Figure 5. Potential interactions between histone methylation marks in longevity.
Several different histone methylation marks are regulated during aging in specific tissues (e.g. liver, brain) or the entire organism from worms to mammals. Histone regulators affect aging, at least in worms and flies. Complex interactions between different marks associated with active or repressed chromatin may participate in lifespan regulation.
Concluding remarks
The emerging evidence for the role of histone methylation regulators in longevity in invertebrates raises a series of questions. First, it will be interesting to test whether environmental interventions such as DR or stress stimuli could mediate longevity by altering histone methylation. It will be particularly informative to test whether regulators of histone methylation, similar to DR, could delay certain aspects of the aging process and slow the occurrence of age-related diseases.
Understanding the mechanisms underlying lifespan extension by regulators of histone methylation will also be crucial. Do these regulators act by specifically altering genes involved in aging or longevity? Or do they induce more global changes, for example in protein translation or in metabolism, which could in turn impact longevity? To address these questions, it will be important to identify the target genes regulated by histone methylation regulatory complexes and examine whether some of these genes are particularly pivotal in mediating the longevity phenotype of histone methylation regulators. It will be intriguing to identify the nature of the signal between fertilized eggs and soma for longevity due to H3K4me3 regulator deficiencies in worms.
Regulators of histone marks are strikingly conserved across species. Thus, dissecting the role of the different complexes in longevity in higher metazoans will be particularly important. It will be interesting to test whether interplay between fertilized embryos and mothers for longevity also occurs in other species. The fact that methyltransferases that modify the same H3K4me3 histone mark (e.g. SET-2 in worms and Trx in flies) have different effects on lifespan in these species suggests that the answer is likely to be complex. A factor that may contribute to the differences between organisms is the ratio of proliferative cells, like stem cells and germ cells, to post-mitotic differentiated cells. Another factor that could contribute to differences between species is that changes in chromatin state may affect global metabolism, and that the requirement for metabolic states could vary in different species.
An outstanding question is whether longevity induced by deficiencies in the H3K4me3 complex in the parental generation can be inherited in the subsequent generations, even if the H3K4me3 complex is no longer mutated in descendants. Strikingly, lifespan extension due to deficiencies in members of the H3K4me3 regulatory complex only in parents can be inherited epigenetically for up to three generations [90], providing the first example of transgenerational epigenetic inheritance of longevity. Mutation of one of the H3K4me1/me2 LSD1 demethylases, spr-5, in C. elegans, leads to progressive epigenetic germline mortality over many generations [91], raising the possibility that changes in this mark may also impact lifespan in a transgenerational manner. Taken together, these data imply that changes in chromatin in one generation could impact the lifespan of subsequent generations.
A greater understanding of the role of chromatin regulators in longevity and the molecular mechanisms by which they act will likely provide new avenues to extend healthspan and delay the onset of age-related diseases.
Acknowledgments
We thank Bérénice A. Benayoun, Eric L. Greer, Jana P. Lim, Travis J. Maures, and Elizabeth A. Pollina for critical reading of the manuscript and helpful suggestions. This work was supported by NIH R01-AG31198 grant to A.B. S.H. was supported by an NSF graduate fellowship and a Stanford graduate fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no conflict of interest.
References
- 1.Kenyon CJ. The genetics of ageing. Nature. 2010;464:504–512. doi: 10.1038/nature08980. [DOI] [PubMed] [Google Scholar]
- 2.Kapahi P, et al. With TOR, less is more: a key role for the conserved nutrient-sensing TOR pathway in aging. Cell Metab. 2010;11:453–465. doi: 10.1016/j.cmet.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mair W, Dillin A. Aging and survival: the genetics of life span extension by dietary restriction. Ann. Rev. of Biochem. 2008;77:727–754. doi: 10.1146/annurev.biochem.77.061206.171059. [DOI] [PubMed] [Google Scholar]
- 4.Greer E, Brunet A. The Handbook of the Biology of Aging. 7th edn. Academic Press; 2011. The Genetic Network of Life-Span Extension by Dietary Restriction. [Google Scholar]
- 5.Panowski SH, et al. PHA-4/Foxa mediates diet-restriction-induced longevity of C. elegans. Nature. 2007;447:550–555. doi: 10.1038/nature05837. [DOI] [PubMed] [Google Scholar]
- 6.Bishop NA, Guarente L. Two neurons mediate diet-restriction-induced longevity in C. elegans. Nature. 2007;447:545–549. doi: 10.1038/nature05904. [DOI] [PubMed] [Google Scholar]
- 7.Greer EL, et al. An AMPK-FOXO pathway mediates longevity induced by a novel method of dietary restriction in C. elegans. Curr. Biol. 2007;17:1646–1656. doi: 10.1016/j.cub.2007.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greer EL, Brunet A. Different dietary restriction regimens extend lifespan by both independent and overlapping genetic pathways in C. elegans. Aging Cell. 2009;8:113–127. doi: 10.1111/j.1474-9726.2009.00459.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hatta M, Cirillo LA. Chromatin opening and stable perturbation of core histone: DNA contacts by FoxO1. J. Biol. Chem. 2007;282:35583–35593. doi: 10.1074/jbc.M704735200. [DOI] [PubMed] [Google Scholar]
- 10.Lupien M, et al. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell. 2008;132:958–970. doi: 10.1016/j.cell.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cirillo LA, et al. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol. Cell. 2002;9:279–289. doi: 10.1016/s1097-2765(02)00459-8. [DOI] [PubMed] [Google Scholar]
- 12.Bungard D, et al. Signaling kinase AMPK activates stress-promoted transcription via histone H2B phosphorylation. Science. 2010;329:1201–1205. doi: 10.1126/science.1191241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 14.Li B, et al. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 15.Shilatifard A. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Ann. Rev. Biochem. 2006;75:243–269. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- 16.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 17.Mosammaparast N, Shi Y. Reversal of histone methylation: biochemical and molecular mechanisms of histone demethylases. Ann. Rev. Biochem. 2010;79:155–179. doi: 10.1146/annurev.biochem.78.070907.103946. [DOI] [PubMed] [Google Scholar]
- 18.Kaeberlein M, et al. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes & Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dang W, et al. Histone H4 lysine 16 acetylation regulates cellular lifespan. Nature. 2009;459:802–807. doi: 10.1038/nature08085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kawahara TL, et al. SIRT6 links histone H3 lysine 9 deacetylation to NF-kappaB-dependent gene expression and organismal life span. Cell. 2009;136:62–74. doi: 10.1016/j.cell.2008.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller T, et al. COMPASS: a complex of proteins associated with a trithorax-related SET domain protein. Proc. Natl. Acad. Sci. U. S. A. 2001;98:12902–12907. doi: 10.1073/pnas.231473398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Papoulas O, et al. The Drosophila trithorax group proteins, BRM, ASH1 and ASH2 are subunits of distinct protein complexes. Development. 1998;125:3955–3966. doi: 10.1242/dev.125.20.3955. [DOI] [PubMed] [Google Scholar]
- 23.Couture JF, et al. Molecular recognition of histone H3 by the WD40 protein WDR5. Nat. Struct. Mol. Biol. 2006;13:698–703. doi: 10.1038/nsmb1116. [DOI] [PubMed] [Google Scholar]
- 24.Eissenberg JC, Shilatifard A. Histone H3 lysine 4 (H3K4) methylation in development and differentiation. Dev. Biol. 2010;339:240–249. doi: 10.1016/j.ydbio.2009.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wysocka J, et al. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell. 2005;121:859–872. doi: 10.1016/j.cell.2005.03.036. [DOI] [PubMed] [Google Scholar]
- 26.Dou Y, et al. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat. Struct. Mol. Biol. 2006;13:713–719. doi: 10.1038/nsmb1128. [DOI] [PubMed] [Google Scholar]
- 27.Mohan M, et al. The COMPASS family of H3K4 methylases in Drosophila. Mol. Cell. Biol. 2011;31:4310–4318. doi: 10.1128/MCB.06092-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bibikova M, et al. Unraveling epigenetic regulation in embryonic stem cells. Cell Stem Cell. 2008;2:123–134. doi: 10.1016/j.stem.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 29.Pollina EA, Brunet A. Epigenetic regulation of aging stem cells. Oncogene. 2011;30:3105–3126. doi: 10.1038/onc.2011.45. [DOI] [PubMed] [Google Scholar]
- 30.Greer EL, et al. Members of the H3K4 trimethylation complex regulate lifespan in a germline-dependent manner in C. elegans. Nature. 2010;466:383–387. doi: 10.1038/nature09195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simonet T, et al. Antagonistic functions of SET-2/SET1 and HPL/HP1 proteins in C. elegans development. Dev. Biol. 2007;312:367–383. doi: 10.1016/j.ydbio.2007.09.035. [DOI] [PubMed] [Google Scholar]
- 32.Xu L, Strome S. Depletion of a novel SET-domain protein enhances the sterility of mes-3 and mes-4 mutants of Caenorhabditis elegans. Genetics. 2001;159:1019–1029. doi: 10.1093/genetics/159.3.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiao Y, et al. Caenorhabditis elegans chromatin-associated proteins SET-2 and ASH-2 are differentially required for histone H3 Lys 4 methylation in embryos and adult germ cells. Proc. Natl. Acad. Sci. U. S. A. 2011;108:8305–8310. doi: 10.1073/pnas.1019290108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li T, Kelly WG. A role for Set1/MLL-related components in epigenetic regulation of the Caenorhabditis elegans germ line. PLoS Genet. 2011;7 doi: 10.1371/journal.pgen.1001349. e1001349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Christensen J, et al. RBP2 belongs to a family of demethylases, specific for tri-and dimethylated lysine 4 on histone 3. Cell. 2007;128:1063–1076. doi: 10.1016/j.cell.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 36.McColl G, et al. Pharmacogenetic analysis of lithium-induced delayed aging in Caenorhabditis elegans. J. Biol. Chem. 2008;283:350–357. doi: 10.1074/jbc.M705028200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shi Y, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 38.Maures TJ, et al. H3K27 demethylase UTX-1 regulates C. elegans lifespan in a germline-independent, insulin-dependent, manner. Aging cell. 2011 doi: 10.1111/j.1474-9726.2011.00738.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang S, et al. Lineage specific trimethylation of H3 on lysine 4 during C. elegans early embryogenesis. Dev. Biol. 2011;355:227–238. doi: 10.1016/j.ydbio.2011.04.010. [DOI] [PubMed] [Google Scholar]
- 40.Kirkwood TB, Austad SN. Why do we age? Nature. 2000;408:233–238. doi: 10.1038/35041682. [DOI] [PubMed] [Google Scholar]
- 41.Kirkwood TB, Rose MR. Evolution of senescence: late survival sacrificed for reproduction. Phil. Trans. R. Soc. Lond. B. 1991;332:15–24. doi: 10.1098/rstb.1991.0028. [DOI] [PubMed] [Google Scholar]
- 42.Santos-Rosa H, et al. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- 43.Schneider R, et al. Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nat. Cell Biol. 2004;6:73–77. doi: 10.1038/ncb1076. [DOI] [PubMed] [Google Scholar]
- 44.Budovskaya YV, et al. An elt-3/elt-5/elt-6 GATA transcription circuit guides aging in C. elegans. Cell. 2008;134:291–303. doi: 10.1016/j.cell.2008.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wold B, Myers RM. Sequence census methods for functional genomics. Nat. Methods. 2008;5:19–21. doi: 10.1038/nmeth1157. [DOI] [PubMed] [Google Scholar]
- 46.Hsieh CC, Papaconstantinou J. Akt/PKB and p38 MAPK signaling, translational initiation and longevity in Snell dwarf mouse livers. Mech. Ageing Dev. 2004;125:785–798. doi: 10.1016/j.mad.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 47.Kaeberlein M, et al. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science. 2005;310:1193–1196. doi: 10.1126/science.1115535. [DOI] [PubMed] [Google Scholar]
- 48.Kapahi P, et al. Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway. Curr. Biol. 2004;14:885–890. doi: 10.1016/j.cub.2004.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hansen M, et al. Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans. Aging Cell. 2007;6:95–110. doi: 10.1111/j.1474-9726.2006.00267.x. [DOI] [PubMed] [Google Scholar]
- 50.Tavernarakis N. Ageing and the regulation of protein synthesis: a balancing act? Trends Cell Biol. 2008;18:228–235. doi: 10.1016/j.tcb.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 51.Li L, et al. Essential functions of the histone demethylase Lid. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001221. e1001221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eissenberg JC, et al. The trithorax-group gene in Drosophila little imaginal discs encodes a trimethylated histone H3 Lys4 demethylase. Nat. Struct. Mol. Biol. 2007;14:344–346. doi: 10.1038/nsmb1217. [DOI] [PubMed] [Google Scholar]
- 53.Secombe J, et al. The Trithorax group protein Lid is a trimethyl histone H3K4 demethylase required for dMyc-induced cell growth. Gene & Dev. 2007;21:537–551. doi: 10.1101/gad.1523007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Siebold AP, et al. Polycomb Repressive Complex 2 and Trithorax modulate Drosophila longevity and stress resistance. Proc. Natl. Acad. of Sci. U. S. A. 2010;107:169–174. doi: 10.1073/pnas.0907739107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Smith E, et al. The super elongation complex (SEC) and MLL in development and disease. Genes & Dev. 2011;25:661–672. doi: 10.1101/gad.2015411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cheung I, et al. Developmental regulation and individual differences of neuronal H3K4me3 epigenomes in the prefrontal cortex. Proc. Natl. Acad. Sci. U. S. A. 2010;107:8824–8829. doi: 10.1073/pnas.1001702107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ang YS, et al. Wdr5 mediates self-renewal and reprogramming via the embryonic stem cell core transcriptional network. Cell. 2011;145:183–197. doi: 10.1016/j.cell.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Suganuma T, et al. ATAC is a double histone acetyltransferase complex that stimulates nucleosome sliding. Nat. Struct. & Mol. Biol. 2008;15:364–372. doi: 10.1038/nsmb.1397. [DOI] [PubMed] [Google Scholar]
- 59.Thompson BA, et al. CHD8 is an ATP-dependent chromatin remodeling factor that regulates beta-catenin target genes. Mol. Cell. Biol. 2008;28:3894–3904. doi: 10.1128/MCB.00322-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mendjan S, et al. Nuclear pore components are involved in the transcriptional regulation of dosage compensation in Drosophila. Mol. Cell. 2006;21:811–823. doi: 10.1016/j.molcel.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 61.Jude CD, et al. Unique and independent roles for MLL in adult hematopoietic stem cells and progenitors. Cell Stem Cell. 2007;1:324–337. doi: 10.1016/j.stem.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McMahon KA, et al. Mll has a critical role in fetal and adult hematopoietic stem cell self-renewal. Cell Stem Cell. 2007;1:338–345. doi: 10.1016/j.stem.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 63.Lim DA, et al. Chromatin remodelling factor Mll1 is essential for neurogenesis from postnatal neural stem cells. Nature. 2009;458:529–533. doi: 10.1038/nature07726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sun G, et al. Histone demethylase LSD1 regulates neural stem cell proliferation. Mol. Cell. Biol. 2010;30:1997–2005. doi: 10.1128/MCB.01116-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pasini D, et al. Coordinated regulation of transcriptional repression by the RBP2 H3K4 demethylase and Polycomb-Repressive Complex 2. Genes & Dev. 2008;22:1345–1355. doi: 10.1101/gad.470008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 67.Jin C, et al. Histone Demethylase UTX-1 Regulates C. elegans Life Span by Targeting the Insulin/IGF-1 Signaling Pathway. Cell Metab. 2011;14:161–172. doi: 10.1016/j.cmet.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 68.Fisher K, et al. Methylation and demethylation activities of a C. elegans MLL-like complex attenuate RAS signalling. Dev. Biol. 2010;341:142–153. doi: 10.1016/j.ydbio.2010.02.023. [DOI] [PubMed] [Google Scholar]
- 69.Lund J, et al. Transcriptional profile of aging in C. elegans. Curr. Biol. 2002;12:1566–1573. doi: 10.1016/s0960-9822(02)01146-6. [DOI] [PubMed] [Google Scholar]
- 70.Bennett-Baker PE, et al. Age-associated activation of epigenetically repressed genes in the mouse. Genetics. 2003;165:2055–2062. doi: 10.1093/genetics/165.4.2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Issaeva I, et al. Knockdown of ALR (MLL2) reveals ALR target genes and leads to alterations in cell adhesion and growth. Mol. Cell. Biol. 2007;27:1889–1903. doi: 10.1128/MCB.01506-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cho YW, et al. PTIP associates with MLL3- and MLL4-containing histone H3 lysine 4 methyltransferase complex. J. Biol. Chem. 2007;282:20395–20406. doi: 10.1074/jbc.M701574200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Park IK, et al. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature. 2003;423:302–305. doi: 10.1038/nature01587. [DOI] [PubMed] [Google Scholar]
- 74.Lessard J, Sauvageau G. Bmi-1 determines the proliferative capacity of normal and leukaemic stem cells. Nature. 2003;423:255–260. doi: 10.1038/nature01572. [DOI] [PubMed] [Google Scholar]
- 75.Oguro H, et al. Differential impact of Ink4a and Arf on hematopoietic stem cells and their bone marrow microenvironment in Bmi1-deficient mice. J. Exp. Med. 2006;203:2247–2253. doi: 10.1084/jem.20052477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Iwama A, et al. Enhanced self-renewal of hematopoietic stem cells mediated by the polycomb gene product Bmi-1. Immunity. 2004;21:843–851. doi: 10.1016/j.immuni.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 77.Molofsky AV, et al. Bmi-1 dependence distinguishes neural stem cell self-renewal from progenitor proliferation. Nature. 2003;425:962–967. doi: 10.1038/nature02060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fasano CA, et al. shRNA knockdown of Bmi-1 reveals a critical role for p21-Rb pathway in NSC self-renewal during development. Cell Stem Cell. 2007;1:87–99. doi: 10.1016/j.stem.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 79.Zencak D, et al. Bmi1 loss produces an increase in astroglial cells and a decrease in neural stem cell population and proliferation. J. Neurosci. 2005;25:5774–5783. doi: 10.1523/JNEUROSCI.3452-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jacobs JJ, et al. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature. 1999;397:164–168. doi: 10.1038/16476. [DOI] [PubMed] [Google Scholar]
- 81.Molofsky AV, et al. Bmi-1 promotes neural stem cell self-renewal and neural development but not mouse growth and survival by repressing the p16Ink4a and p19Arf senescence pathways. Genes & Dev. 2005;19:1432–1437. doi: 10.1101/gad.1299505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Molofsky AV, et al. Increasing p16INK4a expression decreases forebrain progenitors and neurogenesis during ageing. Nature. 2006;443:448–452. doi: 10.1038/nature05091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jiang H, et al. Role for Dpy-30 in ES cell-fate specification by regulation of H3K4 methylation within bivalent domains. Cell. 2011;144:513–525. doi: 10.1016/j.cell.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hamilton B, et al. A systematic RNAi screen for longevity genes in C. elegans. Genes & Dev. 2005;19:1544–1555. doi: 10.1101/gad.1308205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wood JG, et al. Chromatin remodeling in the aging genome of Drosophila. Aging cell. 2010;9:971–978. doi: 10.1111/j.1474-9726.2010.00624.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schotta G, et al. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes & Dev. 2004;18:1251–1262. doi: 10.1101/gad.300704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sarg B, et al. Postsynthetic trimethylation of histone H4 at lysine 20 in mammalian tissues is associated with aging. J. Biol. Chem. 2002;277:39195–39201. doi: 10.1074/jbc.M205166200. [DOI] [PubMed] [Google Scholar]
- 88.Shumaker DK, et al. Mutant nuclear lamin A leads to progressive alterations of epigenetic control in premature aging. Proc. Natl. Acad. Sci. U. S. A. 2006;103:8703–8708. doi: 10.1073/pnas.0602569103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Scaffidi P, Misteli T. Reversal of the cellular phenotype in the premature aging disease Hutchinson-Gilford progeria syndrome. Nat. Med. 2005;11:440–445. doi: 10.1038/nm1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Greer EL, et al. Transgenerational epigenetic inheritance of longevity in Caenorhabditis elegans. Nature. 2011 doi: 10.1038/nature10572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Katz DJ, et al. A C. elegans LSD1 demethylase contributes to germline immortality by reprogramming epigenetic memory. Cell. 2009;137:308–320. doi: 10.1016/j.cell.2009.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]