Capsule Summary
This study identifies transcriptional phenotypes of sputum samples from normal volunteers and atopic asthmatics exposed to ozone. Network analyses suggest that asthmatics elevate immune signaling following oxidative stress, while nonasthmatics attempt to mitigate the ozone-induced response.
Keywords: Asthma, Ozone, Induced Sputum, Profiling, Oxidative Stress
Respiratory complications caused by ozone (O3) represent a significant public health burden1–4. Ozone and a range of other pollutants are thought to exert their effects via oxidative stress responses in the lower airway. Despite studies suggesting that pulmonary oxidative insults by air pollutants lead to deleterious health outcomes, treatment options for environmentally-induced oxidative stress are limited, partially due to the lack of understanding of exactly how these pollutants affect susceptible populations.
Our group recently reported that atopic asthmatics (AA) had enhanced sputum inflammatory responses compared to normal volunteers (NV) after a 2 hour, 0.4 ppm O3 exposure, characterized by increases in macrophage TLR4 expression and concentrations of IL-1β, IL-6, and IL-8 in sputum samples5. This report expands on those results by examining gene expression profiles of sputum inflammatory cells from NV and AA volunteers as a means of identifying additional mechanisms that may explain the differences in inflammatory response between these two cohorts, and to confirm the airway inflammatory phenotype in a larger cohort of AA and NV.
Subject recruitment, sample collection and analysis techniques are identical to those we have recently reported5. Demographic data from a total of 51 subjects (34 NV, 17 AA) are presented in Table I. Statistical tests include paired t-tests to assess a specific cohort response to O3 for cell differentials; non-parametric paired Wilcoxon signed rank test to assess O3 responses for induced sputum cytokines; and non-parametric t tests (Mann-Whitney Test) to examine baseline differences in cytokines or cell differentials between AA and NV.
Table I.
Characteristics of Participants
| Clinical Characteristic | Normal Volunteer | Atopic Asthmatic |
|---|---|---|
| n | 34 | 17 |
| Age (y), mean (SD) | 24.2 (3.9) | 24.4 (5.5) |
| Gender | 20 Female / 14 Male | 10 Female / 7 Male |
| Race | 27 White; 4 African American; 3 Asian | 13 White; 3 African American 1 Asian |
| Pre O3 | Post O3 | Pre O3 | Post O3 | |
|---|---|---|---|---|
| Induced Sputum Cellularity | ||||
| % Neutrophils, Mean (SD) | 40.9 (19.3) | 64.3 (15.7)* | 35.8 (19.4) | 58.4 (23.4)* |
| % Eosinophils, Mean (SD) | 0.12 (0.2) | 0.5 (0.2) | 1.95 (2)** | 2.9 (4.7) |
| % Macrophages, Mean (SD) | 55.6 (18.7) | 32.1 (14.8) | 59.6 (19.7) | 34.2 (20.7) |
| Induced Sputum Cytokines (pg/ml sputum) | ||||
| IL-1β, Median (25%–75%) | 208 (162–691) | 221 (77–475) | 648+ (244–1247) | 1121++ (295–2018) |
| IL-8, Median (25%–75%) | 4694 (1892–11373) | 3622 (837–14071) | 12496^ (8347–25669) | 23278++ (7558–46020) |
| IL-6, Median (25%–75%) | 110 (46–172) | 123 (39–233) | 32 (18–138) | 139# (31–666) |
| IL-10, Median (25%–75%) | 36 (0–307) | 7 (0–240) | 0** (0–6) | 0 (0–53) |
p<0.001 comparing pre O3 to post O3 values
p<0.01 comparing baseline values of AA to NV
p=0.03 comparing baseline values of AA to NV
p=0.03 comparing pre O3 to post O3 values
p=0.02 comparing baseline values of AA to NV
p=0.01 comparing pre O3 to post O3 values
Data for induced sputum cell differentials and cytokines from 9 NV and 6 AA were added to our previously published cohort5. The composition of sputum cellularity was similar between the two cohorts, with the exception of increased sputum eosinophils in the AA cohort at baseline compared to NV (Table I). AA continued to show evidence of increased levels of IL-1β, IL-6, and IL-8 after O3 exposure (Table I), with reduced levels of IL-10 and increased levels of IL-1β and IL-8 at baseline. The larger sample size supports the notion that despite similarities in sputum cellularity, AA display increased levels of airway pro-inflammatory cytokines after O3 exposure compared to NV.
Of the 51 volunteers described above, adequate mRNA was recovered from 18 NV and 13 AA for microarray analysis using a custom-designed microarray representing more than 2000 immune response gene targets6. Raw expression data were first normalized using the down-weighting loess fit7 to remove systematic ratio-intensity dependence. The normalized red/green expression ratio was log2 transformed and uploaded to Significant Analysis of Microarrays software package8 in R (version 2.10) to identify differentially expressed genes. To control for multiple comparisons, we used false discovery rate (FDR), estimated by 5000 permutations for each target.
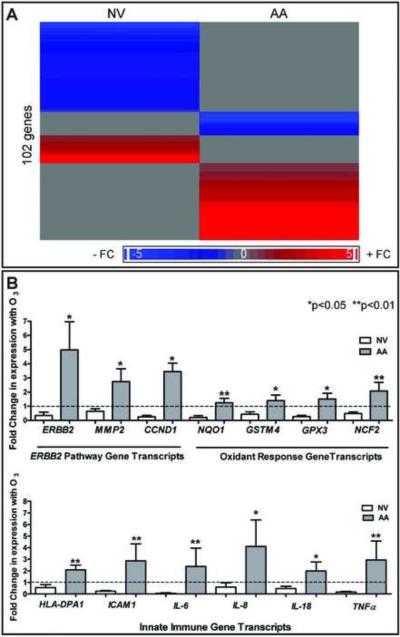
At FDR = 0.05, we found a total of 102 genes that showed differential expression after O3 exposure compared to baseline conditions (Figure 1A). Fifty-five genes had significant expression changes with O3 exposure in NV; 47 genes had significant changes in gene expression with O3 exposure in AA. Only one gene overlapped between the two cohorts, aurora kinase B.
Figure 1. O3-induced transcript profiles in NV and AA.
A) Heat map of O3-modulated genes in NV and AA with low (blue) and high (red) expression. B) Changes with O3 in PCR-amplified ERBB2 pathway, oxidant response and innate immune transcripts relative to ACTB (n=6 NV, 8 AA). Mean changes in gene expression with O3 (± SEM). Nonparametric t testing (Mann Whitney testing) was used to compare differences in NV and AA.
In order to identify potential biological pathways affected by O3 exposure, the differentially expressed gene profiles for each cohort were overlaid onto protein-protein maps enabled through the Ingenuity Pathways' Knowledge Base6. Statistical significance of each network was calculated using a Fischer's exact test. This test generated a p-value signifying the probability that each network was associated with the mRNA targets by chance alone.
For each of the two gene lists, significant networks were identified. The set of 55 genes that showed O3-induced changes in NV could be integrated into four networks ranging in significance from 10−16 to 10−44. The most significant NV network is enriched for proteins involved in the ERBB2 (also known as HER-2) pathway, with most of the signaling predicted to be downregulated in this pathway (Table E1). The 47 differentially expressed genes associated with the O3 challenge in AA were integrated into four networks ranging in significance from 10−9 to 10−45. The most significant AA network is enriched for genes involved in immune response signaling such as NFκB (Table E2).
Of the genes that showed the most statistically significant changes in gene expression, confirmatory quantitative PCR was performed on induced sputum mRNA from 6 NV and 8 AA using previously published methods9. A comparative CT approach was used to discern changes in relative expression due to O3 with β-Actin as the housekeeping gene and pre-O3 as untreated control (Figure 1B).
The expression levels of HER-2 pathway genes such as ERBB2, cyclin D1 (CCND1), and matrix metalloprotease II (MMP2) were significantly increased in AA compared to NV (Figure 1B). Furthermore, AA had significantly increased expression of genes involved in cellular detoxification after oxidative stress (GPX3, GSTM4, NQO1) and those involved in neutrophil oxidative burst (NCF2) compared to NV (Figure 1B). This is consistent with the idea that asthmatics have a differential oxidative stress response following O3 exposure compared to NV. The expression levels of several innate immune genes, such as the MHC-II molecule HLA-DPA1, the integrin ICAM-1, and the pro-inflammatory cytokines IL-6, IL-8, IL-18, and TNFα were significantly increased in AA compared to NV upon exposure to O3 (Figure 1B).
In summary, gene expression profiles for sputum cells are distinctly different in NV and AA despite similar neutrophil and macrophage proportions after acute O3 exposure. Compared to NV, AA showed increased immune signaling, elevated pro-inflammatory cytokines, and up-regulated expression of the HER-2 gene network, indicative of enhanced epithelial cell proliferation. These data suggest that unlike NV, AA cannot limit epithelial cell proliferative responses following O3 -induced oxidative stress. HER-2 blockade has been found to reduce lung fibrosis and remodeling in a model of bleomycin-induced lung injury10. Limitations of this study include small sample size and mixed sputum cellularity that prevented cell specific determination of oxidative stress responses in these populations. Future studies will focus on dissecting the contributions of distinct cell types on determining responses to O3 in a larger cohort of individuals, as well as including baseline measures of inflammation. This information is important for the judicious development of therapeutics that will prevent or reduce the harmful effects of oxidant lung injury.
Acknowledgments
Funding Sources: MLH is supported by NIH KL2RR025746; WJB, NEA, JPYT, and DBP are supported by NIAID U19AI077437; RCF and JR are supported by P30ES010126. This work was also funded by CR 83346301 from the US Environmental Protection Agency.
Abbreviations
- O3
Ozone
- AA
atopic asthmatic
- NV
normal volunteer
- ppm
part per million
- IL-1β
interleukin -1 beta
- IL-8
interleukin-8
- IL-6
interleukin-6
- TLR4
toll-like receptor 4
- TLR2
toll-like receptor 2
- TNFα
tumor necrosis factor alpha
- ICAM1
Inter-Cellular Adhesion Molecule 1
- HLA-DP1
major histocompatibility complex, class II, DP
- GSTM4
glutathione S-transferase mu 4
- GPX3
glutathione peroxidase 3
- NCF2
neutrophil cytosolic factor 2
- ERBB2
v-erb-b2 erythroblastic leukemia viral oncogene homolog 2, neuro/glioblastoma derived oncogene homolog (avian)
- CCND1
cyclin D-1
- MMP2
matrix metalloprotease II
- NQO1
NAD(P)H dehydrogenase, quinone 1
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures: N. E. Alexis has contracts with Centocor Pharmacueticals, GlaxoSmithKline and MedImmune. D. B. Peden has been a consultant to GlaxoSmithKline and Aquinox Pharmaceuticals and a contract with MedImmune. J. PY. Ting had a one-year contract with GlaxoSmithKline (Philadelphia). The rest of the authors have declared that they have no conflicts of interest.
References
- 1.Bell ML, McDermott A, Zeger SL, Samet JM, Dominici F. Ozone and short-term mortality in 95 US urban communities, 1987-2000. JAMA. 2004;292:2372–8. doi: 10.1001/jama.292.19.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chuang GC, Yang Z, Westbrook DG, Pompilius M, Ballinger CA, White CR, et al. Pulmonary ozone exposure induces vascular dysfunction, mitochondrial damage, and atherogenesis. Am J Physiol Lung Cell Mol Physiol. 2009;297:L209–16. doi: 10.1152/ajplung.00102.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jerrett M, Burnett RT, Pope CA, 3rd, Ito K, Thurston G, Krewski D, et al. Long-term ozone exposure and mortality. N Engl J Med. 2009;360:1085–95. doi: 10.1056/NEJMoa0803894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stafoggia M, Forastiere F, Faustini A, Biggeri A, Bisanti L, Cadum E, et al. Susceptibility factors to ozone-related mortality: a population-based case-crossover analysis. Am J Respir Crit Care Med. 2010;182:376–84. doi: 10.1164/rccm.200908-1269OC. [DOI] [PubMed] [Google Scholar]
- 5.Hernandez ML, Lay JC, Harris B, Esther CR, Jr., Brickey WJ, Bromberg PA, et al. Atopic asthmatic subjects but not atopic subjects without asthma have enhanced inflammatory response to ozone. J Allergy Clin Immunol. 2010;126:537–44. e1. doi: 10.1016/j.jaci.2010.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alexis NE, Brickey WJ, Lay JC, Wang Y, Roubey RA, Ting JP, et al. Development of an inhaled endotoxin challenge protocol for characterizing evoked cell surface phenotype and genomic responses of airway cells in allergic individuals. Ann Allergy Asthma Immunol. 2008;100:206–15. doi: 10.1016/S1081-1206(10)60444-9. [DOI] [PubMed] [Google Scholar]
- 7.Dudoit S, Yang YH, Callow MJ, Speed TP. Statistical methods for identifying differentially expressed genes in replicated cDNA microarray experiments. Statistica Sinica. 2002;12:111–39. [Google Scholar]
- 8.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–21. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brickey WJ, Alexis NE, Hernandez ML, Reed W, Ting JP, Peden DB. Sputum inflammatory cells from patients with allergic rhinitis and asthma have decreased inflammasome gene expression. J Allergy Clin Immunol. 2011;128:900–3. doi: 10.1016/j.jaci.2011.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Faress JA, Nethery DE, Kern EF, Eisenberg R, Jacono FJ, Allen CL, et al. Bleomycin-induced pulmonary fibrosis is attenuated by a monoclonal antibody targeting HER2. J Appl Physiol. 2007;103:2077–83. doi: 10.1152/japplphysiol.00239.2007. [DOI] [PubMed] [Google Scholar]