Abstract
Cells sustain endogenous DNA damage at rates greater than 20,000 DNA lesions per cell per day. These damages occur largely as a result of the inherently unstable nature of DNA and the presence of reactive oxygen species within cells. The base excision repair system removes the majority of DNA lesions resulting from endogenous DNA damage. There are several enzymes that function during base excision repair. Importantly, there are over 100 germline single nucleotide polymorphisms in genes that function in base excision repair and that result in non-synonymous amino acid substitutions in the proteins they encode. Somatic variants of these enzymes are also found in human tumors. Variant repair enzymes catalyze aberrant base excision repair. Aberrant base excision repair combined with continuous endogenous DNA damage over time has the potential to lead to a mutator phenotype. Mutations that arise in key growth control genes, imbalances in chromosome number, chromosomal translocations, and loss of heterozygosity can result in the initiation of human cancer or its progression.
Keywords: Base excision repair, mutator phenotype, cancer
1. DNA is Damaged Continuously in Cells Through a Variety of Processes
Endogenous DNA damage occurs in cells at a rate greater than 20,000 DNA adducts per cell per day (for an extensive review see 1). The majority of these endogenous DNA adducts are non-bulky, and are repaired by the base excision repair (BER) pathway. Examples of endogenous DNA base damage include DNA hydrolysis, deamination, oxidation, and alkylation.
As Lindahl pointed out in 1993, 2 the N-glycosyl bond in DNA is very labile. Together with its presence in largely B-form DNA, which is fully hydrated, it is subject to hydrolysis, leading to the formation of abasic sites in DNA 2. Base residues of DNA also undergo hydrolytic deamination, an example of which is the deamination of cytosine to uracil 1, 2. Normal aerobic metabolism gives rise to reactive oxygen species (ROS), which damage DNA base residues (for an excellent review see 3). As the human immune system mounts an inflammatory response to pathogens, leukocytes (including macrophages) release ROS, which has been shown to result in base damage to DNA (for a review see 4). Lipid peroxidation also results in damage to DNA, generating a variety of damages including the etheno adducts. S-adenosylmethionine (SAM) functions in the methylation of DNA to regulate gene expression, but due to its reactive methyl group, it also participates in gratuitous methylation of DNA base residues 1, 5, 6. Abasic sites and oxidized or methylated base residues all can give rise to mutations if not properly repaired. Given the extremely high rate of endogenous DNA damage, even slightly deficient DNA repair would be expected to result in the accumulation of mutations. This has been clearly demonstrated in bacterial cells where various mutant combinations of BER enzymes lead to very high spontaneous mutation frequencies (see for example 7).
2. The BER Pathway Repairs Endogenous Base Damage
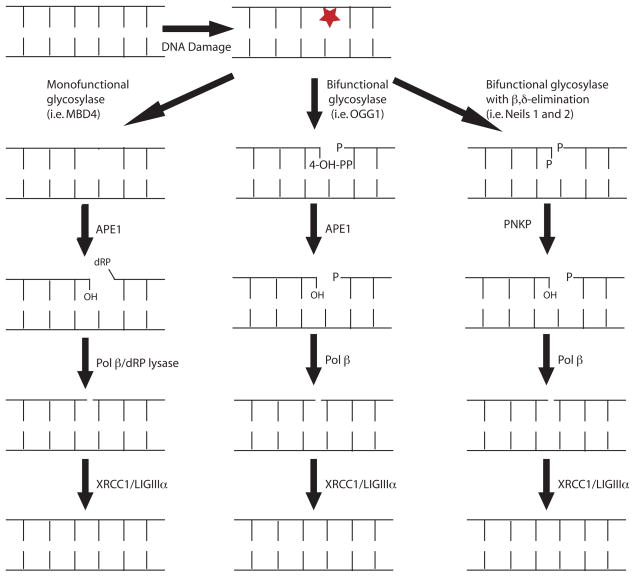
BER is an evolutionarily conserved DNA repair pathway that repairs non-bulky DNA base damage (for a review see 8). There are five basic steps that occur during BER, as shown in Figure 1. An overview of BER proteins is given in Table 2. First, the damaged base is recognized and excised by a DNA glycosylase, leaving an abasic site. Second, the abasic site is removed, either by the lyase activity of a DNA glycosylase or by the enzymatic activity of apurimidinic endonuclease I (APE1). Third, the 3′ and/or 5′ ends of the DNA break are modified to generate 3′OH and 5′P. Fourth, insertion of one or more nucleotides by a DNA polymerase fills the gap, although it has recently been shown that deletions can arise by direct ligation of the 3′OH and 5′P without gap filling 9. This suggests that initiation of the BER process can be mutagenic. The fifth general step of BER is ligation of the ends by a DNA ligase. Here, we will focus predominantly on short patch BER, in which Pol β usually inserts a single nucleotide during the gap-filling step. In long-patch BER greater than a single nucleotide is used to fill the gap. In this process, Pol β likely inserts the first base during gap filling and initiates strand displacement synthesis of the DNA 5′ to the gap. Replicative polymerases, along with PCNA fill the gap, and Flap endonuclease I (FEN1) processes the resulting 5′flap 10, 11.
Figure 1. The BER pathway.
There are five major steps in the BER pathway: 1) excision of the damaged base; 2) AP site incision; 3) modification of the ends of the DNA break; 4) gap filling; and 5) ligation of the nick. For a monofunctional glycosylase (i.e. MBD4), the glycosylase removes the damaged base leaving an abasic site. Ape1 incises the abasic site leaving a 3′ OH and 5′ dRP. Polβ removes the dRP group with its lyase activity and fills in the missing nucleotide. In the case of bifunctional glycosylases (i.e. OGG1), the damaged base is removed and the glycosylase incises the abasic site leaving a 3′ 4-OH pentenal phosphate (4-OH-PP) and 5′ phosphate. Ape1 modifies the end to 3′ OH and Polβ fills in the gap. In an Ape1-independent pathway, bifunctional glycosylases (i.e. Neils 1 and 2) remove the damaged base and incise the abasic site via with β,δ elimination, leaving 3′ and 5′ phosphates. PNKP modifies the 3′ end to an OH and Pol β fills in the gap. In the final step, the DNA is ligated by the XRCC1/LIGIIIα complex.
Table 2.
Summary of BER proteins.
| Gene Symbol | Gene | Function |
|---|---|---|
| APEX1 (APE1) | APEX nuclease (multifunctional DNA repair enzyme) 1 | binds to abasic site and cleaves backbone; remodels 3′ end in the case of bifunctional glycosylases |
| XRCC1 | X-ray complementing defective repair in Chinese hamster cells 1 | scaffold protein that helps recruit BER proteins |
| POLB | polymerase (DNA directed), beta | main polymerase in BER |
| POLL | polymerase (DNA directed), lambda | substitutes for Pol β in BER |
| PARP1 | poly (ADP-ribose) polymerase 1 | binds to breaks in DNA and recruits other BER proteins |
| LIG3 | ligase III, DNA, ATP-dependent | ligates the ends of the repaired DNA |
3. Enzymes and Proteins of the BER Pathway
3.1. DNA Glycosylases
Several DNA glycosylases are expressed in mammalian cells (for reviews see 8, 12). Here we will provide an overview of the DNA glycosylases and the adducts they act upon, as summarized in Table 1. In general, these enzymes recognize specific DNA adducts, flip the damaged base into their active sites, and cleave the N-glycosylic bond.
Table 1.
Summary of DNA Glycosylases
| Gene Symbol | Gene | Type of Glycosylase | Substrates |
|---|---|---|---|
| OGG1 | 8-oxoguanine DNA glycosylase | bifunctional | 8-oxoG:C/G/T; 8-oxoG-FapyG; |
| MUTYH | mutY homolog (E. coli) | monofunctional | 8-oxoG:A |
| TDG | thymine-DNA glycosylase | monofunctional | T:G; ethenoC:G |
| MBD4 | methyl-CpG binding domain protein 4 | monofunctional | T:G; 5-meCpG:TpG; O6-meG:T |
| UNG | uracil-DNA glycosylase | monofunctional | uracil |
| SMUG1 | single-strand-selective monofunctional uracil-DNA glycosylase 1 | monofunctional | U:G; 5-hydroxymethyluracil; 5-formyluracil |
| NTHL1 (NTH1) | nth endonuclease III-like 1 (E. coli) | bifunctional | oxidized pyrimidines, formamidopyrimidines, 5-formyluracil; incises AP sites |
| NEIL1 | nei endonuclease VIII-like 1 (E. coli) | bifunctional; APE-independent | oxidized pyrimidines, formamidopyrimidines, thymine glycol, hydantoin, guanidinohydantoin, spiroiminodihydantoin; removes lesions in single-stranded, bubble, and forked DNA structures |
| NEIL2 | nei endonuclease VIII-like 2 (E. coli) | bifunctional; APE-independent | oxidized pyrimidiines; removes lesions in single-stranded and bubble DNA structures |
| NEIL3 | nei endonuclease VIII-like 3 (E. coli) | bifunctional | hydantoin, guanidinohydantoin, spiroiminodihydantoin; removes lesions in single-stranded and bubble DNA structures |
| AAG (MPG) | N-methylpurine-DNA glycosylase | monofunctional | methylated bases (i.e. 7-meG, 3-meA); 1-N6-etheno-A, 1-N2-ethenoG, uracil |
One of the most common mutagenic adducts that results from oxidative DNA damage is 7,8-dihydro-8-oxoguanine (8-oxoG) 8. 8-oxoG can also be incorporated into DNA, but the 8-oxoGTP precursor is hydrolyzed by the MutT homologs (MTH) 1 or 2 13. Adenine pairs with 8-oxoG during DNA synthesis, ultimately resulting in transversions 14, 15. The DNA glycosylase 8-oxoguanine glycosylase (OGG1) excises both 8-oxoG and 2,6-diamino-4-hydroxy-5-formamidopyrimidine (FapyG) from 8-oxoG:C base pairs, whereas the MutYH DNA glycosylase excises adenine from the 8-oxoG:A mispair 16–18. OGG1 and MutYH are bifunctional DNA glycosylases that excise the damaged base and also use β-elimination to hydrolyze the DNA backbone.
Deamination of 5-methylcytosine (5-meC) to thymine is a common event that gives rise to the T:G mispair. In fact, the mutation rate of 5-meC to thymine is 10- to 50-fold higher than other transition mutations in humans 19–21. The Thymine DNA glycosylase (TDG) and Methyl-CpG-binding domain protein 4 (MBD4) DNA glycosylases excise thymine from T:G mispairs. TDG is also able to excise adducts such as ethenocytosine that arise as a byproduct of lipid peroxidation 22. The MBD4 enzyme, also known as MED1, has a domain that recognizes methylated and hemi-methylated CpG sites in DNA, as well as a glycosylase domain 19, 23. MBD4 prefers to bind to m5CpG:TpG mismatches, which arise as a result of deamination of 5-meC, and it catalyzes the excision of thymine from T:G or uracil from U:G mismatches 19. MBD4 also catalyzes the removal of T opposite O6-methylG, which can lead to futile cycling of excision of T and its incorporation opposite the G lesion by DNA polymerases, similar to what has been described for mismatch repair 24. Phosphorylated MBD4 promotes excision of methylated DNA bases 25. Interestingly, MBD4 interacts with MutL homolog 1 (MLH1) 26, and deletion of MBD4 leads to significant down-regulation of mismatch repair proteins 24, suggesting that the MBD4-initiated BER pathway and mismatch repair are associated in some way. Both of these glycosylases are monofunctional, excising only the damaged base.
In humans, there are two nuclear uracil DNA glycosylases, uracil DNA glycosylase 1 (UNG) and single-strand selective monofunctional uracil DNA glycosylase (SMUG1). UNG1 removes uracil during DNA replication 27, whereas SMUG1 likely excises uracil that arises from deamination of cytosine 28. SMUG1 also excises 5-hydroxymethyluracil and 5-formyluracil and other oxidized bases 28, 29. Both of these glycosylases are monofunctional.
Human NTH1, like all the EcoNth orthologs, possesses DNA-glycosylase/lyase activity on oxidized pyrimidines, formamidopyrimidines, 5-formyluracil and also incises AP sites 30–40. In E. coli, a backup activity to EcoNth for oxidized pyrimidines, EcoNei, was identified and characterized (see for example, 41). Its first eukaryotic homologs were found in humans and designated NEIL1, NEIL2 and NEIL 3 (NEI-like). NEIL1 and NEIL2 have been extensively characterized 42,43–45. An active form of mouse NEIL3 recently has been expressed and characterized 46. Like NTH1, NEIL1 recognizes oxidized pyrimidines, formadopyrimidines, and thymine residues oxidized in the methyl group 42–45,47, 48. Unlike hNTH1, NEIL1 recognizes both stereoisomers of thymine glycol 49–51. Thus far, the best substrates for hNEIL1 appear to be the hydantoin lesions, guanidinohydantoin (Gh), and spiroiminodihydantoin (Sp) 52 that are further oxidation products of 8-oxoG52. This also is true for NEIL3 46. NEIL1 is also capable of removing lesions from single-stranded DNA as well as from bubble and forked DNA structures 53, 54. NEIL2 prefers oxidized pyrimidines, but shows a greater preference than NEIL1 for lesions in single-stranded and bubble structures 53 as does mouse Neil3 53. Because the expression of NEIL1 is cell-cycle dependent 43, it acts on forked DNA structures 55 and it interacts with PCNA 53 and FEN-1 55; accordingly, it has been proposed that NEIL1 functions in replication associated repair. The expression of NEIL2 is not cell-cycle dependent 43; and because of its propensity for lesions in bubble structures, it has been proposed to act during transcription. The NTH1 and NEIL3 enzymes are bifunctional glycosylases that hydrolyze the DNA backbone by β-elimination. However, both NEIL1 and 2 are bifunctional enzymes that hydrolyze the DNA backbone using β, δ-elimination, resulting in APE1-independent downstream BER events.
Human 3-methyladenine DNA glycosylase (AAG) recognizes and excises a variety of methylated bases including 7-methylguanine, 3-methyladenine, and 1-N6-ethenoadenine from DNA 56, 57. This enzyme recently was shown to act on a variety of novel DNA substrates including 1-N2-ethenoguanine and uracil 58. AAG is a monofunctional DNA glycosylase.
3.2. Cleavage of the DNA Backbone
Cleavage of the DNA backbone is conducted by APE1 when a monofunctional glycosylase excises the damaged base, or with the lyase activity of a bifunctional DNA glycosylase.
Excision of a damaged base by a monofunctional DNA glycosylase leaves an abasic site. APE1 binds to this site and cleaves the phosphodiester backbone 5′ to the abasic site, resulting in a 3′OH and 5′deoxyribosephosphate (dRP) (for a review see 59). APE1 also has a 3′ diesterase or phosphatase activity that can remodel a 3′ end to generate a 3′-OH. The 8kDa amino terminus of Polβ has dRP lyase activity and functions to remove the 5-′ dRP group after cleavage of the phosphodiester backbone by APE1.
After a bifunctional DNA glycosylase excises the damaged base leaving an abasic site, hydrolysis of the phosphodiester backbone by β-elimination leaves a 3′ unsaturated aldehyde and a 5′ phosphate. The 3′ phosphodiesterase activity of APE1 acts to restore the 3′OH.
The bifunctional glycosylases NEIL1, NEIL2 and NEIL3 hydrolyze the DNA backbone via β,δ-elimination, which leaves a 5′ and a 3′ phosphate on the ends of the DNA. The 3′ phosphate is removed by polynucleotide kinase/phosphatase (PNKP) to generate a 3′ OH in an APE1-independent reaction 60.
3.3. The XRCC1 protein acts as a scaffold during BER
The X-ray Cross-Complementing Factor 1 (XRCC1) has no intrinsic enzymatic activity, but interacts with several proteins that function in single-strand break repair and in BER, including APE1, Polβ, and LIGIIIα (for a review see 61). Alteration of the XRCC1 gene results in strong sensitivity to alkylating agents 62, most likely because XRCC1 acts as a scaffold to recruit and facilitate interactions between proteins that repair single-strand breaks that result from cleavage of the phosphodiester backbone by APE1. During short-patch BER, the XRCC1-LIGIIIα complex is recruited to seal the nick generated by the action of APE1 and Polβ. The absence of XRCC1 in cells leads to a reduction in cellular levels of LIGIIIα, through an unknown mechanism.
3.4. DNA Polymerase Beta Fills the Gap
During short patch BER and after excision of the damaged base, cleavage of the DNA backbone, and end remodeling to generate 3′ OH and 5′P ends, Polβ fills in the gap, usually adding a single nucleotide, (for a review see 12). During long-patch BER, which appears to be a minor cellular repair pathway 63, Polβ initiates gap filling and completion of this process is performed by DNA polymerases δ and/or ε. In this case, the Fen1 flap endonuclease removes the 5′ dRP group after strand displacement synthesis has occurred 64. In both cases, the XRCC1/LIGIIIα complex catalyzes ligation of the resulting ends. In addition to Polβ, the closely related polymerase Polλ also has been shown to participate in BER 65. The substitution of Polλ for Polβ is dependent on the type of DNA damage; Polλ functions more readily in the repair of oxidative damage, but not alkylating damage 66. Similarly to Polβ, Polλ possesses both dRP lyase and polymerase activities. Polλ also interacts with the DNA glycosylases SMUG1 67, AAG, and OGG1 68 further implicating its role in BER.
4. Genetic Variation in BER Genes and Mutagenesis
Importantly, over 100 different germline polymorphisms exist within the normal population that are predicted to result in nonsynonymous amino substitutions within various BER proteins, including DNA glycoslyases, APE1, XRCC1, and Polβ exist within the normal population (www.genome.utah.edu/genesnps/). In addition, variants of several BER proteins also are found in tumors (for a review see 69). The majority of these variants are heterozygous. Thus, they may exert their effects via a dominant phenotype. Alternatively, as work from our lab indicates, a dominant phenotype is not necessary, as subtle variations in activity can lead to mutagenesis and cellular transformation 70, 71. These variants may not function as well as their wild type counterparts with the end result being aberrant BER. Because BER is a highly coordinated DNA repair pathway that involves several protein-protein interactions, we speculate that even subtle alterations in specific proteins that function in BER could result in mutagenesis and/or genomic instability. Here we define mutagenesis as small changes in the base sequence of DNA and genomic instability as large deletions, copy number variation, or chromosomal aberrations.
4.1. DNA Glycosylase Variants
Variant BER enzymes could lead to increased genomic instability and here, we outline our view of how genomic instability could occur as a result of the presence of an altered DNA glycosylase. Some glycosylase variants have been characterized and were shown to contain altered or low enzymatic activity (see for example 72), but a complete study of all known glycosylase variants has not been undertaken. Fewer damaged bases are likely to be removed in cells expressing variants with low substrate binding affinity or low enzymatic activity. Error-prone lesion bypass could result in incorrect base incorporation, leading to the accumulation of mutations. Should these mutations occur in key growth control genes, leading to an alteration in either the expression or function of a protein, cancer could result. Replication fork arrest by lesions per se -- or that result from a large number of lesions -- could be processed via homology dependent repair (HDR) or non-homologous end joining (NHEJ), which could lead to the accumulation of chromosomal aberrations. Although many chromosomal aberrations are incompatible with life, several of them are associated with human cancer. These include imbalances in chromosome number, translocations, and loss of heterozygosity. Slowing of fork elongation in repetitive sequences including fragile sites also has been shown to result in replication stress, likely resulting in the firing of additional origins of replication. This leads to higher than normal levels of single-stranded DNA, ultimately resulting in chromosomal translocations which are associated with cancer 73. The slowing of replication fork movement due to the presence of an unrepaired lesion could also lead to massive genomic instability. Low turnover by an enzyme with normal or increased substrate binding affinity would result in the removal of fewer damaged bases and perhaps an increase in protein-DNA complexes at the lesion site. Transcriptional mutagenesis has also been shown to result from mutagenic bypass of DNA lesions by RNA polymerases (see for example 74). Translation of mutated RNA transcripts has been shown to lead to phenotypic changes in cells.
DNA glycosylase variants could also possess altered substrate specificity, resulting in the excision of normal bases, or excision of damaged bases outside the normal sequence context. One example of such is the S326C OGG1 variant, ranging in allele frequency between 0.13 and 0.62 in the normal population. S326C has 2–6-fold decreased catalytic efficiency for the removal of 8-OxoG and abasic site cleavage, and is highly dependent upon the base opposite the lesion 75, 76. The decreased catalytic efficiency of this variant is most likely due to its decreased binding affinity for lesions in the DNA 75. Importantly, enzymatic turnover of the S326C variant is not stimulated by APE 1, as is the WT; in addition, S326C has a dimeric conformation, unlike WT, which is monomeric. The dimeric conformation is thought to interfere with the stimulation of S326C by APE1 75. A more subtle possibility is that the variants might exhibit altered sequence context specificity. It is known that the WT versions of these enzymes exhibit sequence context specificity (for examples see 52, 75). Variants could excise damaged bases with a higher rate in certain sequence contexts, for example, either nearest neighbors or dependence upon the base opposite the damage. Cells with variants exhibiting altered sequence context specificity might have increased mutagenesis within certain sequence contexts or non-B-DNA structures. Should this increased mutagenesis occur in key growth control genes, cancer could result.
Mutations in the MYH gene are associated with colon cancer that arises from genomic instability 77, 78. The MYH DNA glycosylase excises A opposite 8-oxoG, so a deficiency in the enzymatic activity of this protein leads to an accumulation of G to T transversions that appear to accumulate in genes, including APC and KRAS2. Importantly, loss of glycosylase activity has been shown in variants of MYH associated with colon cancer 79, 80.
DNA glycosylase-associated AP lyase activity could be altered in germline variants. A decrease in lyase activity would likely result in an increase in AP sites in the DNA. If not removed by APE1 due to structural or other constraints, AP site bypass by error prone polymerases could lead to mutation accumulation. Increased AP lyase activity could result in an accumulation of single-strand breaks (SSBs), which are converted to DSBs upon replication, or by clustering of the SSBs themselves. DSBs are repaired via HDR or NHEJ, the latter process of which may lead to chromosomal aberrations. Both mutagenesis and chromosomal aberrations can lead to cancer, as described above.
The variants might also have altered affinity for their protein partners, many of which are likely unknown; or they may interact aberrantly with other proteins with which they usually do not interact. This could result in sequestration of repair proteins that are needed for BER or other repair processes, ultimately leading to defective repair and mutagenesis. Aberrant protein interactions also could result in imprecise handoff to the next enzyme in the pathway (i.e. glycosylase to AP endonucleaset to polymerase) leading to unprotected accumulation of BER intermediates such as AP sites and breaks. This could also result in genomic instability. Most of the scenarios listed here predict that the cell will sustain increased DNA damage and genomic instability, which could lead to cancer should alterations in cellular growth control result.
4.2. APE1 Variants
APE1 germline variants that have amino acid alterations within the active site of the protein have been characterized. Some of these proteins exhibit reduced DNA cleavage activity 81. The activities of variants with alterations outside the active site are not known. Because APE1 interacts with a variety of other BER proteins, complete characterization is important. APE1 variants with low enzymatic activity would likely result in an increase in abasic sites in cells, which could lead to the accumulation of mutations via error-prone translesion bypass. Mutations in key growth control genes could lead to a malignant phenotype. Variants of APE1 with reduced specificity could cleave the phosphodiester backbone in the absence of abasic sites, resulting in gratuitous single-strand breaks. Compromised ability of an APE1 variant to remove a 3′-dRP would result in ends that are not ligated, which could lead to formation of a DSB or fork collapse during DNA replication, and ultimately, genomic instability (see for example 82). Should the genomic instability lead to translocations, loss of heterozygosity, or an imbalance in chromosome numbers, cancer could result.
APE1 interacts with Werner protein (WRN) and has been found to impede its helicase activity 83. APE1 physically associates with XRCC1, which results in an increased efficiency of its AP endonuclease and 3′phosphodiesterase activities 84 (and for a review see 85). APE1 physically associates with FEN1 and PCNA, suggesting that it may function in long patch BER 86. APE1 stimulates the glycosylase activity of OGG1 by facilitating product release in such a way as to perhaps inhibit the lyase activity of OGG1 87, 88. Demple and colleagues have shown that APE1 interacts with, and stimulates Polβ 89–91. Variant forms of APE1 might exhibit aberrant interactions with some of its protein partners; this could lead to genomic instability. Higher affinity for a protein partner could lead to its sequestration and inability to function in DNA repair. Lower affinity for a protein partner could lead to less modulation of its activity. For example, a low affinity APE1 protein that does not bind tightly to OGG1 would result in lower stimulation of its AP lyase activity. Ends unable to be ligated would accumulate and lead to genomic instability as described above. Finally, APE 1 undergoes posttranslational modification 85 and disruption of this could result in an enzyme that participates aberrantly in BER, leading to genomic instability.
4.3. XRCC1 Scaffold Variants
Chinese hamster ovary (CHO) cell lines that are XRCC1-deficient exhibit strong sensitivity to alkylating agents, increased sister chromatid exchanges and micronuclei, a decrease in the repair of single-strand breaks, and an increase in deletions in response to treatment with an alkylating agent (see for example 62, 92). Recent work also shows that the R280H human germline variant of XRCC1, previously suggested to be associated with deficient DNA repair 93, 94, exhibits decreased DNA binding and retention time at DNA breaks in vivo 95. These studies provide strong evidence that alterations of XRCC1 induce genomic instability.
It is important to point out that a potentially cytotoxic and mutagenic single-strand break is a central intermediate DNA substrate of each of the BER sub-pathways. XRCC1 is recruited by PolyADPribose polymerase 1 (PARP1) to the break, where it mediates interactions between key BER proteins, including Polβ and XRCC1-LIGIIIα. It follows that a decrease in retention at breaks by the R280H XRCC1 variant would not provide the most efficient scaffolding of these proteins at the site of breaks, leading to a decrease in their repair. Even a subtle decrease in break repair is likely to result in genomic instability, leading to cancer, as described above. For example, we have shown that an inability to fill in single nucleotide gaps by the E295K Polβ variant (see below) leads to massive genomic instability 82. The presence of unrepaired single-strand breaks can lead to replication fork collapse, the formation of double-strand breaks, and, if not cytotoxic, error-free or error-prone repair of these breaks. Error-prone repair by end joining pathways can lead to deletions, translocations, and loss of heterozygosity. The encounter of a single-strand break by the replication fork can also lead to slowing of elongation which has been shown to result in replication stress and additional firing of origins 96. Breaks present within fragile sites and repetitive DNA sequences that induce replication stress increase the levels of single-stranded DNA in cells, likely from aberrant lagging-strand replication. Processing of these regions of single-strandedness leads to chromosomal aberrations, including translocations and breaks (for a review see73), and can result in cancer.
4.4. PARP1 Is Important for Efficient Damage Processing
PolyADPribose polymerase 1 (PARP1) binds to DNA breaks, becomes activated by ADP ribosylation, and then recruits repair proteins to the site of DNA damage (for an excellent review see 97). Interestingly, cells deleted of PARP1 are moderately sensitive to alkylating agents, whereas treatment of PARP1+/+ cells with PARP inhibitors results in hypersensitivity to alkylating agents. This indicates that an enzymatically-inhibited PARP1 protein must be present in the cell to for it to be defective in the repair of breaks. There are several germline SNPS in the gene encoding PARP1, many of which result in nonsynonymous amino acid substitutions. Some of these variants may exhibit lower DNA binding, activation, or recruitment activity, similar to what occurs in the presence of PARP inhibitors. Hypersensitivity to alkylating agents is indicative of a defect in break repair, meaning that single-strand breaks remain in the DNA. As discussed above for XRCC1, this could lead to genomic instability via a double-strand break intermediate 98.
4.5. Polβ Variants
We have previously reported that mutations in Polβ are found in 30% of tumors examined 99, but are not found in matched normal tissues. Many of these are nonsynonymous point mutations. Thorough structural and biochemical analyses of Polβ have provided us with the ability to predict the outcomes of certain mutations based upon where they are found within the protein. Mutations that even slightly alter the positioning or folding of the protein may alter its catalytic activity, resulting in an aberrant phenotype. For example, the hydrophobic hinge located in the 31 kDa polymerase domain is required for the fidelity of polymerase due to its involvement in the conformational change that occurs during dNTP binding 100–103. Mutations in these hinge residues, such as I260, I174, or Y265, affect the enzyme’s ability to discriminate between correct and incorrect dNTPs and result in increased mutagenesis. When a mutation causes an increase in the mutation frequency, we hypothesize that this variant would make more errors during the repair of endogenous DNA damage compared to the WT enzyme. If these errors occur in genes that regulate important cellular processes such as cell cycle, proliferation, or apoptosis, this could lead to cancer progression.
Importantly, a mutator phenotype may also occur when a variant does not affect the overall mutation frequency, but instead increases certain types of mutations compared to WT. This idea is supported by our findings with the cancer-associated variant I260M. I260M has a unique mutational spectrum compared to the WT enzyme and was found to induce more transversions and frameshift mutations than the WT enzyme 71. Expression of I260M results in cellular transformation 70 that continued even after the expression was extinguished, suggesting that there is a mutational basis for this transformation, possibly due to the unique mutations induced by the variant.
Polβ has two catalytic activities: dRP lyase and polymerase activities. The dRP lyase activity lies within the N-terminal 8 kDa domain and also contains the DNA-binding domain which requires residues K41, K60, H34, R40, Y39, K68, K72, and R83 104. Sobol, et al., demonstrated that reconstitution of only the dRP lyase activity is able to rescue Polβ-deficient cells from DNA damage 105. We have also shown that a Leu to Pro mutation at residue 22 found in gastric cancer has no dRP lyase activity, is deficient in base excision repair, and is unable to complement Polβ null cells in response to DNA damaging agents 100. Although L22P is not directly involved in forming the DNA binding pocket, it has decreased DNA binding affinity. The mutation may alter the organization of the binding pocket, preventing Polβ from binding DNA efficiently and preventing polymerization from occurring. Hence, any mutations in the dRP lyase domain, despite whether or not they are in critical residues, can prevent the enzyme from participating in BER. These mutations could prevent the removal of the 5′-dRP group and the filling of the gap. They could also prevent Polβ from binding the DNA that would result in unrepaired lesions. These variants could result in an accumulation of BER intermediates leading to genomic instability. Unrepaired DNA can cause stalled and collapsed replication forks leading to the formation of double-strand breaks 106. These breaks could be repaired by HDR or the error- prone NHEJ, and chromosomal aberrations and fusions could accumulate, resulting in genomic instability.
The other role of Polβ in BER is to fill in the gap. Mutations within the 31 kDa polymerase domain can result in a protein with aberrant polymerase function. No base will be incorporated if the mutation renders the enzyme inactive. For example, the gastric cancer variant E295K binds DNA with the same affinity as WT Polβ, but lacks polymerase activity and does not participate in BER. The expression of E295K in Polβ-deficient cells does not rescue the cells from MMS-induced cytotoxicity 82. Since E295K is able to bind DNA with anaffinity similar to WT, it is likely that E295K is acting in a dominant negative fashion and it is able to compete with WT and prevent the repair of the damage. Our studies with E295K showed an increase in sister chromatid exchanges compared to wild type; this most likely is due to the lack of polymerase activity of E295K. If the variant is inactive and unable to bind DNA, it could still interact with BER proteins and sequester them from repairing other damage and indirectly cause an accumulation of damage. Even variants that have a lower rate of catalysis can be detrimental. If the variant binds the DNA and is slow to polymerize, this could cause the replication fork to slow or stall, resulting in double strand breaks that may lead to overall genomic instability.
It is important to note that a proper balance of Polβ expression must be maintained. As we discussed above, expression of non-functional or dysfunctional Polβ can cause deleterious effects. Likewise, over-expression of Polβ can be detrimental. Elevated levels of Polβ have been detected in tumors 107; this was suggested to be due to the larger need for Polβ as a result of the higher rate of DNA damage in the tumor. Alternatively, the up-regulation of Polβ could disrupt other cellular processes and cause more damage to the cell. One possibility is that Polβ could substitute for replicative polymerases. Polβ has a lower fidelity than replicative polymerases and has no known robust proofreading abilities 108 which ultimately could result in increased mutagenesis. Over-expression of Polβ interferes with normal replication and impairs replication fork progression without activating a checkpoint response 96. Genomic instability could occur by increasing chromosomal rearrangements and common fragile sites 96.
Polβ also interacts with proteins involved in BER (i.e. XRCC1, APE1, gycosylases, LigaseIIIα). If additional Polβ is available, it may not participate in BER, and instead sequester the other proteins leading to inefficient BER and thus increase BER intermediates that could result in genomic instability.
4.6. DNA LigaseIIIα and Lack of Variants
There are several germline polymorphisms within the DNA ligase III gene, according to the Environmental Genome Project (http://egp.gs.washington.edu/). However, only one of these is predicted to result in a non-synonymous substitution within the protein. This alteration is predicted to be well-tolerated, so it is unlikely to result in a phenotype associated with genomic instability. Because DNA ligases are critical to a number of DNA repair processes and as well as to replication, perhaps even the presence of subtle variations in a ligase would not be compatible with life.
5. Imbalances of BER Proteins Might Be Mutagenic
Spontaneous mutation rates have been known for some time to be significantly affected by imbalances in BER proteins, as shown in the classic study in yeast by Samson and colleagues 109. Over-expression of AAG1 and APE1 is associated with chronic inflammation and microsatellite instability in ulcerative colitis 110. These enzymes can be up-regulated at the transcriptional level, either as a result of induction by the presence of DNA damage that would be expected to be present during inflammation, or by posttranslational modification. Over-expression of the BER proteins could lead to the removal of normal, non-damaged bases, initiating gratuitous BER.
6. Mouse Models of Aberrant BER and Genomic Instability
Investigations into the links between BER genes, a mutator phenotype, and cancer in an organism have largely been initiated by knocking out, or deleting, genes that function in BER; and much of this field has been reviewed by Klungland 111. In general, deletion of DNA glycosylase genes results in accumulation of the lesions excised by the specific glyosylase studied; but genome instability and cancer rarely are observed. However, cancer and genome instability have been observed in mouse models in which at least two DNA glycosylases have been deleted (see for example 112, 113). Deletion of genes that encode proteins that act downstream of the DNA glycosylases usually results in embryonic or early onset lethality inmice. Some of these mice have aberrant neuronal phenotypes, suggesting the importance of BER in neuronal DNA maintenance. It is suggested that BER is critical for development and maintenance of certain types of cells and that deletion of BER genes is not compatible with life. Indeed, deletions of BER genes are rarely, if ever, observed to occur in the germline of humans. Thus, it is suggested that knock-in mouse models, harboring specific BER polymorphisms, might act as better models for the study of genomic instability, cancer, and possibly other human diseases that might arise from aberrant BER.
7. Genetics and Environment Contribute to Genomic Instability and Cancer
Although children suffer from cancer, the highest percentage of people diagnosed with cancer are adults, many of whom are older adults. This, along with the existence of many low frequency germline SNPs in BER genes, indicates that a genetic predisposition, combined with long-term environmental exposure, lead to a mutator phenotype and cancer. In the case of BER, the exposures likely result from endogenous damage due to the inherently unstable nature of DNA, metabolism of oxygen, and aberrant DNA methylation. Subtle deficiencies in BER of these endogenous lesions can lead to genomic instability and cancer after several years of exposure combined with subtly aberrant BER. The mutations arising from aberrant BER need to accumulate in genes and regulatory regions that are critical for growth control, genome stability, and damage signaling in order for mutagenesis, genomic instability, and tumor initiation to occur. For example, many of the polymorphic variants of Polβ we have studied possess subtle mutator phenotypes that lead to cellular transformation after many passages in tissue culture (for example see 70).
8. Conclusions
Endogenous DNA damage occurs at very high rates in human cells. These lesions normally are repaired and the DNA sequence is restored. However, in the presence of environmental stress and BER polymorphic variants, BER is aberrant and mutations accumulate in the genome which have the potential to lead to genomic instability and cancer.
Acknowledgments
Support for the preparation of this article was provided by CA129186 from the National Cancer Institute (J.B.S.), and T32 CA009259 from the National Cancer Institute (A.A.N.) an PO1 CA090689 from the National Cancer Institute (S.S.W. and J.B.S.)
Abbreviations
- BER
base excision repair
- ROS
reactive oxygen species
- SAM
S-adenosylmethionine
- APE1
apurimidinic endonuclease I
- MTH
MutT homolog
- OGG1
DNA glycosylase 8-oxoguanine glycosylase
- FapyG
2,6-diamino-4-hydroxy-5-formamidopyrimidine
- 5-meC
5-methylcytosine
- MBD4
methyl-CpG-binding domain protein 4
- TDG
thymine DNA glycosylase
- MLH1
MutL homolog 1
- UNG
uracil DNA glycosylase 1
- SMUG1
single-strand selective monofunctional uracil DNA glycosylase
- AAG
3-methyladenine DNA glycosylase
- DRP
5′deoxyribosephosphate
- XRCC1
X-Ray Cross-Complementing Factor 1
- LIGIIIα
DNA ligaseIIIα
- Polβ
DNA polymerase beta
- Polλ
DNA polymerase lambda
- HDR
homology dependent repair
- NHEJ
non-homologous end joining
- SSBs
single-strand breaks
- DSBs
double-strand breaks
- PARP1
polyADPribose polymerase 1
References
- 1.De Bont R, van Larebeke N. Endogenous DNA damage in humans: a review of quantitative data. Mutagenesis. 2004;19:169–185. doi: 10.1093/mutage/geh025. [DOI] [PubMed] [Google Scholar]
- 2.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 3.Sedelnikova OA, Redon CE, Dickey JS, Nakamura AJ, Georgakilas AG, Bonner WM. Role of oxidatively induced DNA lesions in human pathogenesis. Mutat Res. 2010;704:152–159. doi: 10.1016/j.mrrev.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hussain SP, Hofseth LJ, Harris CC. Radical causes of cancer. Nat Rev Cancer. 2003;3:276–285. doi: 10.1038/nrc1046. [DOI] [PubMed] [Google Scholar]
- 5.Rydberg B, Lindahl T. Nonenzymatic methylation of DNA by the intracellular methyl group donor S-adenosyl-L-methionine is a potentially mutagenic reaction. Embo J. 1982;1:211–216. doi: 10.1002/j.1460-2075.1982.tb01149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stern LL, Mason JB, Selhub J, Choi SW. Genomic DNA hypomethylation, a characteristic of most cancers, is present in peripheral leukocytes of individuals who are homozygous for the C677T polymorphism in the methylenetetrahydrofolate reductase gene. Cancer Epidemiol Biomarkers Prev. 2000;9:849–853. [PubMed] [Google Scholar]
- 7.Blaisdell JO, Hatahet Z, Wallace SS. A novel role for Escherichia coli endonuclease VIII in prevention of spontaneous G-->T transversions. J Bacteriol. 1999;181:6396–6402. doi: 10.1128/jb.181.20.6396-6402.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barnes DE, Lindahl T. Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annu Rev Genet. 2004;38:445–476. doi: 10.1146/annurev.genet.38.072902.092448. [DOI] [PubMed] [Google Scholar]
- 9.Lyons DM, O’Brien PJ. Human base excision repair creates a bias toward -1 frameshift mutations. J Biol Chem. 2010;285:25203–25212. doi: 10.1074/jbc.M110.118596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Asagoshi K, Liu Y, Masaoka A, et al. DNA polymerase beta-dependent long patch base excision repair in living cells. DNA Repair (Amst) 2010;9:109–119. doi: 10.1016/j.dnarep.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Asagoshi K, Tano K, Chastain PD, 2nd, et al. FEN1 functions in long patch base excision repair under conditions of oxidative stress in vertebrate cells. Mol Cancer Res. 2010;8:204–215. doi: 10.1158/1541-7786.MCR-09-0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Svilar D, Goellner EM, Almeida KH, Sobol RW. Base Excision Repair and lesion-dependent sub-pathways for repair of oxidative DNA damage. Antioxid Redox Signal. 2010 doi: 10.1089/ars.2010.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maki H, Sekiguchi M. MutT protein specifically hydrolyses a potent mutagenic substrate for DNA synthesis. Nature. 1992;355:273–275. doi: 10.1038/355273a0. [DOI] [PubMed] [Google Scholar]
- 14.Moriya M, Grollman AP. Mutations in the mutY gene of Escherichia coli enhance the frequency of targeted G:C-->T:a transversions induced by a single 8-oxoguanine residue in single-stranded DNA. Mol Gen Genet. 1993;239:72–76. doi: 10.1007/BF00281603. [DOI] [PubMed] [Google Scholar]
- 15.Wood ML, Dizdaroglu M, Gajewski E, Essigmann JM. Mechanistic studies of ionizing radiation and oxidative mutagenesis: genetic effects of a single 8-hydroxyguanine (7-hydro-8-oxoguanine) residue inserted at a unique site in a viral genome. Biochemistry. 1990;29:7024–7032. doi: 10.1021/bi00482a011. [DOI] [PubMed] [Google Scholar]
- 16.Girard PM, Guibourt N, Boiteux S. The Ogg1 protein of Saccharomyces cerevisiae: a 7,8-dihydro-8-oxoguanine DNA glycosylase/AP lyase whose lysine 241 is a critical residue for catalytic activity. Nucleic Acids Res. 1997;25:3204–3211. doi: 10.1093/nar/25.16.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bjoras M, Luna L, Johnsen B, et al. Opposite base-dependent reactions of a human base excision repair enzyme on DNA containing 7,8-dihydro-8-oxoguanine and abasic sites. Embo J. 1997;16:6314–6322. doi: 10.1093/emboj/16.20.6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohtsubo T, Nishioka K, Imaiso Y, et al. Identification of human MutY homolog (hMYH) as a repair enzyme for 2-hydroxyadenine in DNA and detection of multiple forms of hMYH located in nuclei and mitochondria. Nucleic Acids Res. 2000;28:1355–1364. doi: 10.1093/nar/28.6.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hendrich B, Hardeland U, Ng HH, Jiricny J, Bird A. The thymine glycosylase MBD4 can bind to the product of deamination at methylated CpG sites. Nature. 1999;401:301–304. doi: 10.1038/45843. [DOI] [PubMed] [Google Scholar]
- 20.Duncan BK, Miller JH. Mutagenic deamination of cytosine residues in DNA. Nature. 1980;287:560–561. doi: 10.1038/287560a0. [DOI] [PubMed] [Google Scholar]
- 21.Sved J, Bird A. The expected equilibrium of the CpG dinucleotide in vertebrate genomes under a mutation model. Proc Natl Acad Sci U S A. 1990;87:4692–4696. doi: 10.1073/pnas.87.12.4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abu M, Waters TR. The main role of human thymine-DNA glycosylase is removal of thymine produced by deamination of 5-methylcytosine and not removal of ethenocytosine. J Biol Chem. 2003;278:8739–8744. doi: 10.1074/jbc.M211084200. [DOI] [PubMed] [Google Scholar]
- 23.Bellacosa A. Role of MED1 (MBD4) Gene in DNA repair and human cancer. J Cell Physiol. 2001;187:137–144. doi: 10.1002/jcp.1064. [DOI] [PubMed] [Google Scholar]
- 24.Cortellino S, Turner D, Masciullo V, et al. The base excision repair enzyme MED1 mediates DNA damage response to antitumor drugs and is associated with mismatch repair system integrity. Proc Natl Acad Sci U S A. 2003;100:15071–15076. doi: 10.1073/pnas.2334585100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim MS, Kondo T, Takada I, et al. DNA demethylation in hormone-induced transcriptional derepression. Nature. 2009;461:1007–1012. doi: 10.1038/nature08456. [DOI] [PubMed] [Google Scholar]
- 26.Bellacosa A, Cicchillitti L, Schepis F, et al. MED1, a novel human methyl-CpG-binding endonuclease, interacts with DNA mismatch repair protein MLH1. Proc Natl Acad Sci U S A. 1999;96:3969–3974. doi: 10.1073/pnas.96.7.3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nilsen H, Stamp G, Andersen S, et al. Gene-targeted mice lacking the Ung uracil-DNA glycosylase develop B-cell lymphomas. Oncogene. 2003;22:5381–5386. doi: 10.1038/sj.onc.1206860. [DOI] [PubMed] [Google Scholar]
- 28.Masaoka A, Matsubara M, Hasegawa R, et al. Mammalian 5-formyluracil-DNA glycosylase. 2. Role of SMUG1 uracil-DNA glycosylase in repair of 5-formyluracil and other oxidized and deaminated base lesions. Biochemistry. 2003;42:5003–5012. doi: 10.1021/bi0273213. [DOI] [PubMed] [Google Scholar]
- 29.Matsubara M, Masaoka A, Tanaka T, Terato H, Ohyama Y, Ide H. Identification and characterization of mammalian 5-formyluracil-DNA glycosylase. Nucleic Acids Res Suppl. 2003:233–234. doi: 10.1093/nass/3.1.233. [DOI] [PubMed] [Google Scholar]
- 30.Hilbert TP, Boorstein RJ, Kung HC, et al. Purification of a mammalian homologue of Escherichia coli endonuclease III: identification of a bovine pyrimidine hydrate-thymine glycol DNAse/AP lyase by irreversible cross linking to a thymine glycol-containing oligoxynucleotide. Biochemistry. 1996;35:2505–2511. doi: 10.1021/bi952516e. [DOI] [PubMed] [Google Scholar]
- 31.Hilbert TP, Chaung W, Boorstein RJ, Cunningham RP, Teebor GW. Cloning and expression of the cDNA encoding the human homologue of the DNA repair enzyme, Escherichia coli endonuclease III. J Biol Chem. 1997;272:6733–6740. doi: 10.1074/jbc.272.10.6733. [DOI] [PubMed] [Google Scholar]
- 32.Aspinwall R, Rothwell DG, Roldan-Arjona T, et al. Cloning and characterization of a functional human homolog of Escherichia coli endonuclease III. Proc Natl Acad Sci U S A. 1997;94:109–114. doi: 10.1073/pnas.94.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sarker AH, Ikeda S, Nakano H, et al. Cloning and characterization of a mouse homologue (mNthl1) of Escherichia coli endonuclease III. J Mol Biol. 1998;282:761–774. doi: 10.1006/jmbi.1998.2042. [DOI] [PubMed] [Google Scholar]
- 34.Ikeda S, Biswas T, Roy R, et al. Purification and characterization of human NTH1, a homolog of Escherichia coli endonuclease III. Direct identification of Lys-212 as the active nucleophilic residue. J Biol Chem. 1998;273:21585–21593. doi: 10.1074/jbc.273.34.21585. [DOI] [PubMed] [Google Scholar]
- 35.Asagoshi K, Odawara H, Nakano H, et al. Comparison of substrate specificities of Escherichia coli endonuclease III and its mouse homologue (mNTH1) using defined oligonucleotide substrates. Biochemistry. 2000;39:11389–11398. doi: 10.1021/bi000422l. [DOI] [PubMed] [Google Scholar]
- 36.Asagoshi K, Yamada T, Okada Y, et al. Recognition of formamidopyrimidine by Escherichia coli and mammalian thymine glycol glycosylases. Distinctive paired base effects and biological and mechanistic implications. J Biol Chem. 2000;275:24781–24786. doi: 10.1074/jbc.M000576200. [DOI] [PubMed] [Google Scholar]
- 37.Eide L, Luna L, Gustad EC, et al. Human endonuclease III acts preferentially on DNA damage opposite guanine residues in DNA. Biochemistry. 2001;40:6653–6659. doi: 10.1021/bi0028901. [DOI] [PubMed] [Google Scholar]
- 38.Marenstein DR, Ocampo MT, Chan MK, et al. Stimulation of human endonuclease III by Y box-binding protein 1 (DNA-binding protein B). Interaction between a base excision repair enzyme and a transcription factor. J Biol Chem. 2001;276:21242–21249. doi: 10.1074/jbc.M101594200. [DOI] [PubMed] [Google Scholar]
- 39.Marenstein DR, Chan MK, Altamirano A, et al. Substrate specificity of human endonuclease III (hNTH1). Effect of human APE1 on hNTH1 activity. J Biol Chem. 2003;278:9005–9012. doi: 10.1074/jbc.M212168200. [DOI] [PubMed] [Google Scholar]
- 40.Miyabe I, Zhang QM, Kino K, et al. Identification of 5-formyluracil DNA glycosylase activity of human hNTH1 protein. Nucleic Acids Res. 2002;30:3443–3448. doi: 10.1093/nar/gkf460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Melamede RJ, Hatahet Z, Kow YW, Ide H, Wallace SS. Isolation and characterization of endonuclease VIII from Escherichia coli. Biochemistry. 1994;33:1255–1264. doi: 10.1021/bi00171a028. [DOI] [PubMed] [Google Scholar]
- 42.Bandaru V, Sunkara S, Wallace SS, Bond JP. A novel human DNA glycosylase that removes oxidative DNA damage and is homologous to Escherichia coli endonuclease VIII. DNA Repair (Amst) 2002;1:517–529. doi: 10.1016/s1568-7864(02)00036-8. [DOI] [PubMed] [Google Scholar]
- 43.Hazra TK, Izumi T, Boldogh I, et al. Identification and characterization of a human DNA glycosylase for repair of modified bases in oxidatively damaged DNA. Proc Natl Acad Sci U S A. 2002;99:3523–3528. doi: 10.1073/pnas.062053799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hazra TK, Kow YW, Hatahet Z, et al. Identification and characterization of a novel human DNA glycosylase for repair of cytosine-derived lesions. J Biol Chem. 2002;277:30417–30420. doi: 10.1074/jbc.C200355200. [DOI] [PubMed] [Google Scholar]
- 45.Wallace SS, Bandaru V, Kathe SD, Bond JP. The enigma of endonuclease VIII. DNA Repair (Amst) 2003;2:441–453. doi: 10.1016/s1568-7864(02)00182-9. [DOI] [PubMed] [Google Scholar]
- 46.Liu M, Bandaru V, Bond JP, et al. The mouse ortholog of NEIL3 is a functional DNA glycosylase in vitro and in vivo. Proc Natl Acad Sci U S A. 2010;107:4925–4930. doi: 10.1073/pnas.0908307107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hu J, de Souza-Pinto NC, Haraguchi K, et al. Repair of formamidopyrimidines in DNA involves different glycosylases: role of the OGG1, NTH1, and NEIL1 enzymes. J Biol Chem. 2005;280:40544–40551. doi: 10.1074/jbc.M508772200. [DOI] [PubMed] [Google Scholar]
- 48.Zhang QM, Yonekura S, Takao M, Yasui A, Sugiyama H, Yonei S. DNA glycosylase activities for thymine residues oxidized in the methyl group are functions of the hNEIL1 and hNTH1 enzymes in human cells. DNA Repair (Amst) 2005;4:71–79. doi: 10.1016/j.dnarep.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 49.Ocampo-Hafalla MT, Altamirano A, Basu AK, et al. Repair of thymine glycol by hNth1 and hNeil1 is modulated by base pairing and cis-trans epimerization. DNA Repair (Amst) 2006;5:444–454. doi: 10.1016/j.dnarep.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 50.McTigue MM, Rieger RA, Rosenquist TA, Iden CR, De Los Santos CR. Stereoselective excision of thymine glycol lesions by mammalian cell extracts. DNA Repair (Amst) 2004;3:313–322. doi: 10.1016/j.dnarep.2003.11.009. [DOI] [PubMed] [Google Scholar]
- 51.Miller H, Fernandes AS, Zaika E, et al. Stereoselective excision of thymine glycol from oxidatively damaged DNA. Nucleic Acids Res. 2004;32:338–345. doi: 10.1093/nar/gkh190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Krishnamurthy N, Zhao X, Burrows CJ, David SS. Superior removal of hydantoin lesions relative to other oxidized bases by the human DNA glycosylase hNEIL1. Biochemistry. 2008;47:7137–7146. doi: 10.1021/bi800160s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dou H, Theriot CA, Das A, et al. Interaction of the human DNA glycosylase NEIL1 with proliferating cell nuclear antigen. The potential for replication-associated repair of oxidized bases in mammalian genomes. J Biol Chem. 2008;283:3130–3140. doi: 10.1074/jbc.M709186200. [DOI] [PubMed] [Google Scholar]
- 54.Dou H, Mitra S, Hazra TK. Repair of oxidized bases in DNA bubble structures by human DNA glycosylases NEIL1 and NEIL2. J Biol Chem. 2003;278:49679–49684. doi: 10.1074/jbc.M308658200. [DOI] [PubMed] [Google Scholar]
- 55.Hegde ML, Theriot CA, Das A, et al. Physical and functional interaction between human oxidized base-specific DNA glycosylase NEIL1 and flap endonuclease 1. J Biol Chem. 2008;283:27028–27037. doi: 10.1074/jbc.M802712200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.O’Connor TR. Purification and characterization of human 3-methyladenine-DNA glycosylase. Nucleic Acids Res. 1993;21:5561–5569. doi: 10.1093/nar/21.24.5561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Singer B, Antoccia A, Basu AK, et al. Both purified human 1, N6-ethenoadenine-binding protein and purified human 3-methyladenine-DNA glycosylase act on 1, N6-ethenoadenine and 3-methyladenine. Proc Natl Acad Sci U S A. 1992;89:9386–9390. doi: 10.1073/pnas.89.20.9386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee CY, Delaney JC, Kartalou M, et al. Recognition and processing of a new repertoire of DNA substrates by human 3-methyladenine DNA glycosylase (AAG) Biochemistry. 2009;48:1850–1861. doi: 10.1021/bi8018898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Demple B, Harrison L. Repair of oxidative damage to DNA: enzymology and biology. Annu Rev Biochem. 1994;63:915–948. doi: 10.1146/annurev.bi.63.070194.004411. [DOI] [PubMed] [Google Scholar]
- 60.Wiederhold L, Leppard JB, Kedar P, et al. AP endonuclease-independent DNA base excision repair in human cells. Mol Cell. 2004;15:209–220. doi: 10.1016/j.molcel.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 61.Caldecott KW. XRCC1 and DNA strand break repair. DNA Repair (Amst) 2003;2:955–969. doi: 10.1016/s1568-7864(03)00118-6. [DOI] [PubMed] [Google Scholar]
- 62.Thompson LH, Brookman KW, Dillehay LE, et al. A CHO-cell strain having hypersensitivity to mutagens, a defect in DNA strand-break repair, and an extraordinary baseline frequency of sister-chromatid exchange. Mutat Res. 1982;95:427–440. doi: 10.1016/0027-5107(82)90276-7. [DOI] [PubMed] [Google Scholar]
- 63.Sobol RW, Horton JK, Kuhn R, et al. Requirement of mammalian DNA polymerase-beta in base-excision repair. Nature. 1996;379:183–186. doi: 10.1038/379183a0. [DOI] [PubMed] [Google Scholar]
- 64.Dianov GL, Sleeth KM, Dianova II, Allinson SL. Repair of abasic sites in DNA. Mutat Res. 2003;531:157–163. doi: 10.1016/j.mrfmmm.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 65.Braithwaite EK, Prasad R, Shock DD, Hou EW, Beard WA, Wilson SH. DNA polymerase lambda mediates a back-up base excision repair activity in extracts of mouse embryonic fibroblasts. J Biol Chem. 2005;280:18469–18475. doi: 10.1074/jbc.M411864200. [DOI] [PubMed] [Google Scholar]
- 66.Tano K, Nakamura J, Asagoshi K, et al. Interplay between DNA polymerases beta and lambda in repair of oxidation DNA damage in chicken DT40 cells. DNA Repair (Amst) 2007;6:869–875. doi: 10.1016/j.dnarep.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Braithwaite EK, Kedar PS, Lan L, et al. DNA polymerase lambda protects mouse fibroblasts against oxidative DNA damage and is recruited to sites of DNA damage/repair. J Biol Chem. 2005;280:31641–31647. doi: 10.1074/jbc.C500256200. [DOI] [PubMed] [Google Scholar]
- 68.Braithwaite EK, Kedar PS, Stumpo DJ, et al. DNA Polymerases beta and lambda Mediate Overlapping and Independent Roles in Base Excision Repair in Mouse Embryonic Fibroblasts. PLoS One. 2010:5. doi: 10.1371/journal.pone.0012229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sweasy JB, Lang T, DiMaio D. Is base excision repair a tumor suppressor mechanism? Cell Cycle. 2006;5:250–259. doi: 10.4161/cc.5.3.2414. [DOI] [PubMed] [Google Scholar]
- 70.Sweasy JB, Lang T, Starcevic D, et al. Expression of DNA polymerase {beta} cancer-associated variants in mouse cells results in cellular transformation. Proc Natl Acad Sci U S A. 2005;102:14350–14355. doi: 10.1073/pnas.0505166102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dalal S, Hile S, Eckert KA, Sun KW, Starcevic D, Sweasy JB. Prostate-cancer-associated I260M variant of DNA polymerase beta is a sequence-specific mutator. Biochemistry. 2005;44:15664–15673. doi: 10.1021/bi051179z. [DOI] [PubMed] [Google Scholar]
- 72.Roy LM, Jaruga P, Wood TG, McCullough AK, Dizdaroglu M, Lloyd RS. Human polymorphic variants of the NEIL1 DNA glycosylase. J Biol Chem. 2007;282:15790–15798. doi: 10.1074/jbc.M610626200. [DOI] [PubMed] [Google Scholar]
- 73.Durkin SG, Glover TW. Chromosome fragile sites. Annu Rev Genet. 2007;41:169–192. doi: 10.1146/annurev.genet.41.042007.165900. [DOI] [PubMed] [Google Scholar]
- 74.Saxowsky TT, Meadows KL, Klungland A, Doetsch PW. 8-Oxoguanine-mediated transcriptional mutagenesis causes Ras activation in mammalian cells. Proc Natl Acad Sci U S A. 2008;105:18877–18882. doi: 10.1073/pnas.0806464105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hill JW, Evans MK. Dimerization and opposite base-dependent catalytic impairment of polymorphic S326C OGG1 glycosylase. Nucleic Acids Res. 2006;34:1620–1632. doi: 10.1093/nar/gkl060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dherin C, Radicella JP, Dizdaroglu M, Boiteux S. Excision of oxidatively damaged DNA bases by the human alpha-hOgg1 protein and the polymorphic alpha-hOgg1(Ser326Cys) protein which is frequently found in human populations. Nucleic Acids Res. 1999;27:4001–4007. doi: 10.1093/nar/27.20.4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jones S, Emmerson P, Maynard J, et al. Biallelic germline mutations in MYH predispose to multiple colorectal adenoma and somatic G:C-->T:A mutations. Hum Mol Genet. 2002;11:2961–2967. doi: 10.1093/hmg/11.23.2961. [DOI] [PubMed] [Google Scholar]
- 78.Jones S, Lambert S, Williams GT, Best JM, Sampson JR, Cheadle JP. Increased frequency of the k-ras G12C mutation in MYH polyposis colorectal adenomas. Br J Cancer. 2004;90:1591–1593. doi: 10.1038/sj.bjc.6601747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pope MA, Chmiel NH, David SS. Insight into the functional consequences of hMYH variants associated with colorectal cancer: distinct differences in the adenine glycosylase activity and the response to AP endonucleases of Y150C and G365D murine MYH. DNA Repair (Amst) 2005;4:315–325. doi: 10.1016/j.dnarep.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 80.Chmiel NH, Livingston AL, David SS. Insight into the functional consequences of inherited variants of the hMYH adenine glycosylase associated with colorectal cancer: complementation assays with hMYH variants and pre-steady-state kinetics of the corresponding mutated E. coli enzymes. J Mol Biol. 2003;327:431–443. doi: 10.1016/s0022-2836(03)00124-4. [DOI] [PubMed] [Google Scholar]
- 81.Hadi MZ, Coleman MA, Fidelis K, Mohrenweiser HW, Wilson DM., 3rd Functional characterization of Ape1 variants identified in the human population. Nucleic Acids Res. 2000;28:3871–3879. doi: 10.1093/nar/28.20.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lang T, Dalal S, Chikova A, DiMaio D, Sweasy JB. The E295K DNA polymerase beta gastric cancer-associated variant interferes with base excision repair and induces cellular transformation. Mol Cell Biol. 2007;27:5587–5596. doi: 10.1128/MCB.01883-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ahn B, Harrigan JA, Indig FE, Wilson DM, 3rd, Bohr VA. Regulation of WRN helicase activity in human base excision repair. J Biol Chem. 2004;279:53465–53474. doi: 10.1074/jbc.M409624200. [DOI] [PubMed] [Google Scholar]
- 84.Vidal AE, Boiteux S, Hickson ID, Radicella JP. XRCC1 coordinates the initial and late stages of DNA abasic site repair through protein-protein interactions. Embo J. 2001;20:6530–6539. doi: 10.1093/emboj/20.22.6530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fan J, Wilson DM., 3rd Protein-protein interactions and posttranslational modifications in mammalian base excision repair. Free Radic Biol Med. 2005;38:1121–1138. doi: 10.1016/j.freeradbiomed.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 86.Dianova, Bohr VA, Dianov GL. Interaction of human AP endonuclease 1 with flap endonuclease 1 and proliferating cell nuclear antigen involved in long-patch base excision repair. Biochemistry. 2001;40:12639–12644. doi: 10.1021/bi011117i. [DOI] [PubMed] [Google Scholar]
- 87.Hill JW, Hazra TK, Izumi T, Mitra S. Stimulation of human 8-oxoguanine-DNA glycosylase by AP-endonuclease: potential coordination of the initial steps in base excision repair. Nucleic Acids Res. 2001;29:430–438. doi: 10.1093/nar/29.2.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pascucci B, Maga G, Hubscher U, et al. Reconstitution of the base excision repair pathway for 7,8-dihydro-8-oxoguanine with purified human proteins. Nucleic Acids Res. 2002;30:2124–2130. doi: 10.1093/nar/30.10.2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wong D, Demple B. Modulation of the 5′-deoxyribose-5-phosphate lyase and DNA synthesis activities of mammalian DNA polymerase beta by apurinic/apyrimidinic endonuclease 1. J Biol Chem. 2004;279:25268–25275. doi: 10.1074/jbc.M400804200. [DOI] [PubMed] [Google Scholar]
- 90.Bennett RA, Wilson DM, 3rd, Wong D, Demple B. Interaction of human apurinic endonuclease and DNA polymerase beta in the base excision repair pathway. Proc Natl Acad Sci U S A. 1997;94:7166–7169. doi: 10.1073/pnas.94.14.7166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wong D, DeMott MS, Demple B. Modulation of the 3′-->5′-exonuclease activity of human apurinic endonuclease (Ape1) by its 5′-incised Abasic DNA product. J Biol Chem. 2003;278:36242–36249. doi: 10.1074/jbc.M306065200. [DOI] [PubMed] [Google Scholar]
- 92.Russo D, Fronza G, Ottaggio L, et al. XRCC1 deficiency influences the cytotoxicity and the genomic instability induced by Me-lex, a specific inducer of N3-methyladenine. DNA Repair (Amst) 2010;9:728–736. doi: 10.1016/j.dnarep.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Takanami T, Nakamura J, Kubota Y, Horiuchi S. The Arg280His polymorphism in X-ray repair cross-complementing gene 1 impairs DNA repair ability. Mutat Res. 2005;582:135–145. doi: 10.1016/j.mrgentox.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 94.Pachkowski BF, Winkel S, Kubota Y, Swenberg JA, Millikan RC, Nakamura J. XRCC1 genotype and breast cancer: functional studies and epidemiologic data show interactions between XRCC1 codon 280 His and smoking. Cancer Res. 2006;66:2860–2868. doi: 10.1158/0008-5472.CAN-05-3388. [DOI] [PubMed] [Google Scholar]
- 95.Berquist BR, Singh DK, Fan J, et al. Functional capacity of XRCC1 protein variants identified in DNA repair-deficient Chinese hamster ovary cell lines and the human population. Nucleic Acids Res. 2010;38:5023–5035. doi: 10.1093/nar/gkq193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pillaire MJ, Betous R, Conti C, et al. Upregulation of error-prone DNA polymerases beta and kappa slows down fork progression without activating the replication checkpoint. Cell Cycle. 2007;6:471–477. doi: 10.4161/cc.6.4.3857. [DOI] [PubMed] [Google Scholar]
- 97.Horton JK, Wilson SH. Hypersensitivity phenotypes associated with genetic and synthetic inhibitor-induced base excision repair deficiency. DNA Repair (Amst) 2007;6:530–543. doi: 10.1016/j.dnarep.2006.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Heacock ML, Stefanick DF, Horton JK, Wilson SH. Alkylation DNA damage in combination with PARP inhibition results in formation of S-phase-dependent double-strand breaks. DNA Repair (Amst) 2010;9:929–936. doi: 10.1016/j.dnarep.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Starcevic D, Dalal S, Sweasy JB. Is there a link between DNA polymerase beta and cancer? Cell Cycle. 2004;3:998–1001. [PubMed] [Google Scholar]
- 100.Dalal S, Chikova A, Jaeger J, Sweasy JB. The Leu22Pro tumor-associated variant of DNA polymerase beta is dRP lyase deficient. Nucleic Acids Res. 2008;36:411–422. doi: 10.1093/nar/gkm1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Starcevic D, Dalal S, Jaeger J, Sweasy JB. The hydrophobic hinge region of rat DNA polymerase beta is critical for substrate binding pocket geometry. J Biol Chem. 2005;280:28388–28393. doi: 10.1074/jbc.M502178200. [DOI] [PubMed] [Google Scholar]
- 102.Starcevic D, Dalal S, Sweasy J. Hinge residue Ile260 of DNA polymerase beta is important for enzyme activity and fidelity. Biochemistry. 2005;44:3775–3784. doi: 10.1021/bi047956x. [DOI] [PubMed] [Google Scholar]
- 103.Yamtich J, Starcevic D, Lauper J, et al. Hinge residue I174 is critical for proper dNTP selection by DNA polymerase beta. Biochemistry. 2010;49:2326–2334. doi: 10.1021/bi901735a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Prasad R, Batra VK, Yang XP, et al. Structural insight into the DNA polymerase beta deoxyribose phosphate lyase mechanism. DNA Repair (Amst) 2005;4:1347–1357. doi: 10.1016/j.dnarep.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 105.Sobol RW, Prasad R, Evenski A, et al. The lyase activity of the DNA repair protein beta-polymerase protects from DNA-damage-induced cytotoxicity. Nature. 2000;405:807–810. doi: 10.1038/35015598. [DOI] [PubMed] [Google Scholar]
- 106.Saleh-Gohari N, Bryant HE, Schultz N, Parker KM, Cassel TN, Helleday T. Spontaneous homologous recombination is induced by collapsed replication forks that are caused by endogenous DNA single-strand breaks. Mol Cell Biol. 2005;25:7158–7169. doi: 10.1128/MCB.25.16.7158-7169.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Srivastava DK, Husain I, Arteaga CL, Wilson SH. DNA polymerase beta expression differences in selected human tumors and cell lines. Carcinogenesis. 1999;20:1049–1054. doi: 10.1093/carcin/20.6.1049. [DOI] [PubMed] [Google Scholar]
- 108.Sweasy JB. Fidelity mechanisms of DNA polymerase beta. Prog Nucleic Acid Res Mol Biol. 2003;73:137–169. doi: 10.1016/s0079-6603(03)01005-5. [DOI] [PubMed] [Google Scholar]
- 109.Xiao W, Samson L. In vivo evidence for endogenous DNA alkylation damage as a source of spontaneous mutation in eukaryotic cells. Proc Natl Acad Sci U S A. 1993;90:2117–2121. doi: 10.1073/pnas.90.6.2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hofseth LJ, Khan MA, Ambrose M, et al. The adaptive imbalance in base excision-repair enzymes generates microsatellite instability in chronic inflammation. J Clin Invest. 2003;112:1887–1894. doi: 10.1172/JCI19757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Larsen E, Meza TJ, Kleppa L, Klungland A. Organ and cell specificity of base excision repair mutants in mice. Mutat Res. 2007;614:56–68. doi: 10.1016/j.mrfmmm.2006.01.023. [DOI] [PubMed] [Google Scholar]
- 112.Xie Y, Yang H, Cunanan C, et al. Deficiencies in mouse Myh and Ogg1 result in tumor predisposition and G to T mutations in codon 12 of the K-ras oncogene in lung tumors. Cancer Res. 2004;64:3096–3102. doi: 10.1158/0008-5472.can-03-3834. [DOI] [PubMed] [Google Scholar]
- 113.Chan MK, Ocampo-Hafalla MT, Vartanian V, et al. Targeted deletion of the genes encoding NTH1 and NEIL1 DNA N-glycosylases reveals the existence of novel carcinogenic oxidative damage to DNA. DNA Repair (Amst) 2009;8:786–794. doi: 10.1016/j.dnarep.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]