Abstract
Billions of cells die via apoptosis every day and are swiftly and efficiently removed. When a phagocyte engulfs an apoptotic cell, it essentially doubles its cellular contents, raising the question of how a phagocyte may manage the excess metabolic load. This review discusses phagocyte cellular metabolism, the digestion of the ingested apoptotic cell and the impact of these processes on engulfment.
The prompt and specific recognition of dying cells and their subsequent removal is an integral part of a number of biological processes, including developmental morphogenesis in embryonic life, and tissue turnover/regeneration in adults (Nagata et al., 2010). The turnover rate in humans has been estimated to be approximately a million cells per second each day. The dying cells are recognized and engulfed by both professional phagocytes (macrophages and immature dendritic cells) as well as non-professional phagocytes (such as fibroblasts). Defects in removal of apoptotic cells has often been linked to development of inflammation and autoimmune diseases (Nagata et al., 2010).
The first step in the engulfment process is the migration of the phagocyte in response to soluble find-me signals released by the apoptotic cells (e.g. lipid lysophosphatidylcholine (LPC); fraktalkine/CX3CL1, sphingosine 1-phosphate (S1P) (Gregory and Pound, 2011). Phagocyte engulfment receptors then recognize specific ‘eat-me signals’ exposed on the surface of apoptotic cells (Nagata et al., 2010). The most extensively characterized eat-me signal is the lipid phosphatidylserine (PtdSer), which can be recognized either directly, (via receptors such as Brain Angiogenesis Inhibitor 1, T-cell immunoglobulin domain and mucin domain 4, and Stabilin-2) or indirectly via bridging molecules (such as MFG-E8, Gas6 or protein S) (Nagata et al., 2010). Phagocytic receptor engagement activates signaling pathways that result in the activation of Rac1, a Rho family GTPase, leading to the physical internalization of the dying cell by the phagocyte. The fourth step, and the focus of this review, is the process of digesting the contents of the ingested corpse. When one cell engulfs another, it is akin to a neighbor moving all of his or her belongings into our house. The phagocyte essentially doubles its content of lipids, carbohydrates, protein, nucleotides and other cellular materials from the apoptotic cell. Hence, an interesting, but largely unanswered question is, how does the phagocyte handle the excess metabolic burden acquired from the ingested corpse? What are the metabolic changes that occur within the phagocyte such that it can continue its ‘normal’ functions while processing the extra metabolites? Also, it appears that there are fewer phagocytes in a tissue than there are dying cells, in which case a given phagocyte likely eats multiple targets sequentially. In this scenario, how does the ingested cargo regulate the phagocytic machinery and continued clearance by phagocytes? Some of the answers to these questions are beginning to emerge, as detailed below.
Regulation of phagocyte lipid metabolism
Some of the major players involved in cellular lipid homeostasis are peroxisome-proliferator activated receptors (PPARs) and liver x-receptor (LXR), members of the nuclear receptor superfamily. PPARs (isoforms α, β/δ, and γ) and LXR (isoforms α, β) are ligand-activated transcriptional activators that have varied tissue expression. PPARs and LXR heterodimerize with retinoid X receptors (RXRs) following binding to ligands such as fatty acids and oxysterols, and recruit co-activators to induce transcription of a variety of genes involved in lipid and cholesterol metabolism.
Studies over the past few years suggest specific roles for PPARs and LXR in the clearance of apoptotic cells. Macrophages exposed to apoptotic cells experience a rapid increase in PPARγ activation, as measured by binding to PPAR response elements. Functionally, macrophages derived from PPARγ−/− mice have been shown to engulf fewer apoptotic cells compared to wild-type cells (Roszer et al., 2011). Antagonists of PPARγ also impair engulfment, while PPARγ agonists promote phagocytosis of apoptotic targets by immature dendritic cells (Majai et al., 2010). Interestingly, PPARγ does not appear to be required for opsonized bacterial phagocytosis, suggesting specificity for the signaling induced within phagocytes by apoptotic cell engagement. In vivo and ex vivo studies have shown similar impaired phagocytic clearance in LXRαβ−/−, PPARδ−/−, and RXRα−/− mice (Gonzalez et al., 2009; Mukundan et al., 2009; Roszer et al., 2011). How does stimulation of PPARs or LXR regulate engulfment? Existing evidence suggests that LXR and PPAR activation during apoptotic cell clearance leads to the upregulation of phagocytic receptors (e.g. Mer) and opsonins (Gas6, MFG-E8, C1qb) by the phagocyte (Gonzalez et al., 2009; Mukundan et al., 2009) (Figure).
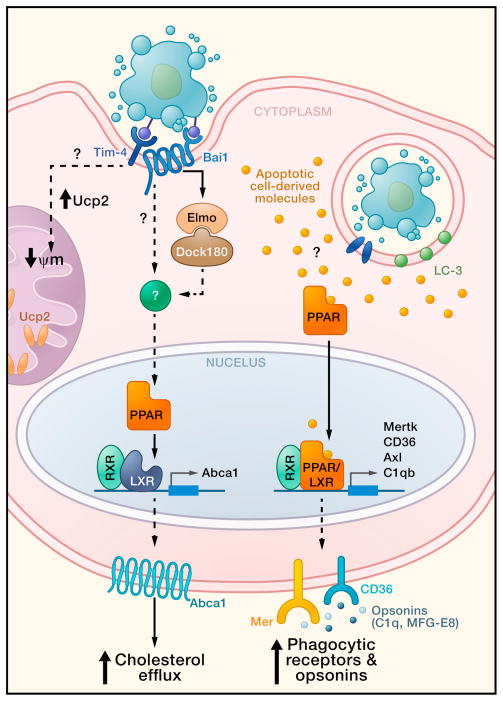
Figure 1. Metabolic factors and signaling pathways within the phagocyte involved in apoptotic cell clearance.
Engulfment of apoptotic cells activates several pathways that may affect phagocyte metabolism and promote continued corpse uptake. First, apoptotic cell engulfment leads to phagocyte PPAR and LXR activation, with their ligands possibly derived from ingested cargo. PPARs and LXR subsequently increase transcription of phagocytic receptors (e.g. Mer and CD36) and opsonins, (e.g. C1qb and MFG-E8) (Gonzalez et al., 2009, Mukundan et al., 2009). In a second pathway, tickling of engulfment receptors by phosphatidylserine exposed on the apoptotic cell leads to increased Abca1 expression and cholesterol efflux, via PPAR and LXR activity. Lastly, phagocytes increase Ucp2 expression in the mitochondria following engulfment of apoptotic cells, leading to decreased mitochondrial membrane potential; the decrease in mitochondrial membrane potential in turn helps to promote continued apoptotic cell uptake. Phagocytic receptor Tim-4 may participate in crosstalk with the mitochondrial uncoupling proteins. Phagosome containing apoptotic cell may also be decorated with the LC-3, a marker of macroautophagy that has been shown to facilitate phagosome maturation and degradation.
PPAR and LXR family members also play important roles in mediating lipid homeostasis. One mechanism by which phagocytes may regulate their lipid content is through enhancement of their basal cholesterol efflux during engulfment. The enhanced cholesterol efflux induced by apoptotic cells was dependent on the exposed phosphatidylserine on apoptotic cells binding to engulfment receptors on phagocytes, and involved the increased expression of the lipid transporter ABCA1, which appears to require LXRαβ activity (Kiss et al., 2006). Improper cholesterol efflux has been shown to exacerbate atherosclerosis and deficiency of certain engulfment receptors such as Mer has been linked to a predisposition to atherosclerosis (Tabas, 2010). The dual role of LXRαβ in cholesterol homeostasis and apoptotic cell clearance insinuates a possible linkage between metabolic signaling and continued clearance by the phagocyte, as well as connections to certain pathogenic sequelae.
Overall, the existing data suggest that the phagocytic process leads to changes within phagocytes, conferring properties ranging from enhanced engulfment to cholesterol homeostasis that are mediated at least in part by the PPAR and LXR families via the sensing of lipids (Gonzalez et al., 2009; Mukundan et al., 2009). It has long been known that the PPARs and LXR also dampen inflammation. Intriguingly, one of the hallmarks of apoptotic cell engulfment (compared to phagocytosis of pathogens) is the ‘immunologically silent’ removal of corpses with active production of anti-inflammatory cytokines. In this context, it is interesting that mice lacking PPARs and LXR developed signs of autoimmunity, including autoantibodies to nuclear antigens and glomerulonephritis (Gonzalez et al., 2009; Mukundan et al., 2009; Roszer et al., 2011), phenotypes often seen in mice with impaired apoptotic cell removal (Nagata et al., 2010). Inflammation is also considered a key component of atherosclerosis progression, and apoptotic cells as well as a necrotic core are part of late stage atherosclerotic plaques (Tabas, 2010). Furthermore, loss of PPARs and LXR expression, and the engulfment receptor Mer, worsen atherosclerosis in mouse models. What is unclear at this point is how the PtdSer at the cell surface ‘talks’ to the PPAR and LXR proteins, and how this translates to alterations on phagocyte lipid metabolism. Further studies on the signaling pathways that connect phagocytosis of apoptotic cells to the regulation of phagocyte lipid metabolism may yield therapeutic targets for atherosclerosis and diseases associated with improper or uncontrolled inflammation.
Links between glucose metabolism and phagocytosis
Surprisingly, few studies have directly examined the role of glucose metabolism in phagocytosis. Single nucleotide polymorphisms in the gene coding for Elmo1, a key Rac-regulator required for corpse removal in the testes (Elliott et al., 2010), have been associated with increased risk for diabetic nephropathy (Pezzolesi et al., 2009). While intriguing, whether defects in apoptotic cell clearance contribute to the development and pathogenesis of type 2 diabetes remains to be determined. Nevertheless, when considering that glycolysis is a major catabolic pathway that supplies cellular energy, the possibility arises that the degradation products of the ingested corpses could modify phagocyte glucose levels, and affect cellular glucose transport and synthesis. These events, in turn, may have a wider impact on glucose homeostasis on an organismal level.
Glycolytic metabolism has recently been of particular interest in the cancer biology field. Tumor cells often switch from oxidative phosphorylation to glycolysis while growing under hypoxic conditions, a transition known as the Warburg Effect (Cairns et al., 2011). This alteration in energy metabolism appears to confer a growth and invasiveness advantage to cancer cells, likely via enhanced resistance to apoptosis and increased proliferation rates (Cairns et al., 2011). Since many solid tumors have tumor-associated macrophages (TAMs) (Gregory and Pound, 2011), TAMs can be exposed to a partial hypoxic environment when they clear apoptotic cancer cells. It will be informative to determine whether phagocytes also switch energy utilization pathways following phagocytosis of apoptotic cells, and whether the switch bestows similar growth benefits upon the phagocyte. One pathway that may be of interest is the pentose phosphate pathway, in which the products NADPH and pentoses are used for amino acid and nucleic acid synthesis, which may be utilized for cellular growth purposes (i.e. anabolic). There could also be an extensive series of changes within the tumor microenvironment depending on how well dying cells are cleared within a tumor, as clearance of apoptotic cells results in production of anti-inflammatory cytokines that may dampen anti-tumor responses. Hence, corpse removal within a tumor may be a double-edged sword as the ingested apoptotic cells may serve as energy sources for TAMs, allowing for continued corpse uptake, yet simultaneously suppressing the immune system. It is noteworthy that culturing phagocytes in a different nutrient environment, such as low glucose, tended to promote phagocytosis of apoptotic cells, while a high glucose environment reduced corpse uptake (Park et al., 2011). The ingestion of apoptotic cells by phagocytes in particular microenvironments, with altered nutrient availability, could have important implications for tumor progression and other pathologies.
Phagocyte mitochondria, engulfment capacity, and the autophagy machinery
Lipids and carbohydrates represent the two principal cellular energy sources. Their catabolism and subsequent derivatives feed into the tricarboxylic acid (TCA) cycle and/or the electron transport chain (ETC) of the mitochondria. The ETC consists of four protein complexes embedded in the mitochondrial inner membrane. A series of redox reactions occurs in the ETC in which electrons are passed along to sequential acceptors. The transport of H+ across the inner membrane creates an electrochemical gradient (termed mitochondrial membrane potential or MMP), which drives protons through the ATP synthase, generating ATP for cellular use. Additional organic compounds produced by the TCA cycle and the ETC are also used in other cellular processes such as amino acid synthesis and nitrogen transportation.
A recent study showed that the MMP in phagocytes increases initially after ingestion of apoptotic cells, but decreases over time(Park et al., 2011); however, ATP levels within the phagocyte were largely unchanged, suggesting that proton transport and ATP production were disengaged (Park et al, 2011). One family of mitochondrial proteins that that can ‘uncouple’ the electrochemical gradient from ATP generation are the uncoupling proteins (Ucp), located in the inner mitochondrial membrane. Park et al show that Ucp2 levels increase in phagocytes after incubation with apoptotic cells, and that loss of Ucp2, resulted in an increase in mitochondrial membrane potential, and negatively affected phagocytic ability. Again, similar to the PPAR and LXR requirement, Ucp2 function was specific to apoptotic cell removal, and had little effect on phagocytosis of synthetic targets, zymosan particles, or E. coli.
Autophagy (also referred to as macroautophagy), the process by which the cells may generate energy in nutrient-starved conditions through selective and regulated digestion of internal components, has received much attention recently (Mizushima and Komatsu, 2011). Given that the mitochondria is a foci of energy production, the increase in mitochondrial membrane potential within the engulfing phagocyte may help generate ATP needed to facilitate plasma membrane remodeling and to support varied phosphorylation events as part of the engulfment process. Interestingly, components of the autophagic machinery, in particular, light chain 3 (LC3) have been found on phagosomes containing apoptotic cells (Florey et al., 2011; Martinez et al., 2011) (Figure). This event, termed LC-3 associated phagocytosis (LAP), was initially observed on toll-like receptor (TLR) activated phagosomes of phagocytes that have engulfed particles containing TLR ligands (Sanjuan et al., 2007; Lee et al., 2010). LAP differs from classical autophagy in that the hallmark autophagic double-membrane structure is absent; instead, the phagosome containing the apoptotic cell is a single-membrane and decorated with LC-3. The LC-3 containing phagosomes mature and degrade the internalized cargo much more rapidly than non-LC-3 containing phagosomes (Martinez et al., 2011; Sanjuan et al., 2007). Some autophagic proteins, such as Atg7, Atg5, Beclin-1, and Vps34, but not the Ulk1-Atg13-FIP200 pre-initiation complex, are required for LAP (Martinez et al., 2011; Florey et al., 2011). Interestingly, Vps34 is also essential for proper phagosome maturation in the context of corpse removal. Whether or not the consequences of LAP- or non-LAP-associated apoptotic cell clearance resembles that of autophagy where the degraded products are used to fuel cellular functions remains to be determined. Nevertheless, given the energy demands of the engulfment process as well as the documented involvement of the mitochondria, the engulfment of apoptotic cells is likely a metabolic stress on the phagocyte, requiring the phagocyte to utilize either itself or the ingested apoptotic cell as energy sources for continued clearance. Indeed, phagocytes engulfing apoptotic cells, but not synthetic beads, have an increased rate of fatty acid oxidation (Park et al., 2011), suggesting that this may be the case, although whether the products are used for generating additional energy is not clear. It will be interesting to determine whether LAP is always a component of apoptotic cell phagocytosis and if LAP plays a role in mediating the digestion and conversion of the engulfed cargo into usable energy.
The withdrawal of certain growth factors, such as insulin- growth factor like 1 (IGF-1) or nerve growth factor, induces autophagy. Interestingly, macrophages deficient in the growth factor progranulin, as well as C. elegans mutants lacking the progranulin homologue display better engulfment capacity (Kao et al., 2011), suggesting a connection between growth factor withdrawal and phagocytosis. More broadly, general nutrient sensing may be relevant in modulating engulfment of dying cells. Recently, thioglycollate-elicited peritoneal macrophages, following ingestion of apoptotic cells, were shown to increase AMP-activated protein kinase (AMPK) activity, a kinase involved in sensing cellular ATP levels (Bae et al., 2011). Pharmacological activation of AMPK was shown to augment phagocytic ability, which was in this case proposed to be related to an AMPK-mediated increase in microtubule and actin dynamics. Typically, AMPK phosphorylation leads to changes in cellular lipid and glucose metabolism, and in mitochondrial biogenesis. Whereas the cellular metabolic consequences of AMPK activation were not explored by Bae et al (2011), the connection between this key energy sensor to enhanced phagocytosis is intriguing. Given the relationship between growth factors and autophagy, it will be of interest to determine whether and how these growth factors affect nutrient-sensing responses and potentially impact apoptotic cell clearance in a tissue or in an altered nutrient state (e.g. hypoxic tumor environment). AMPK activation was not seen in bone-marrow derived macrophages exposed to apoptotic cells (Park et al., 2011), raising the possibility that there may be differential post-engulfment effects based on the activation status of certain phagocytes. This idea may even be extended to the question of whether professional phagocytes and non-professional phagocytes respond differently in digestion of ingested corpses.
Future directions
The field of apoptotic cell clearance has only recently turned its attention to the intricacies of how a phagocyte may digest and dispose off the excess material acquired from the ingested dying cells. Key unanswered questions center on the mechanism(s) underlying how engulfed apoptotic cells and/or the engulfment process activate various metabolic pathways. In the case of lipid homeostasis involving PPARs and LXR, it is possible that lipid derivatives from ingested apoptotic cells serve as the trigger for receptor activation and subsequent downstream effects (Figure). Degraded apoptotic cell lipids can be shuttled out of phagosomes into the cytoplasm, where they can bind to certain cytoplasmic PPARs and LXR, inducing PPAR and LXR activation and translocation to the nucleus for transcriptional activation and/or repression activities. Alternatively, phagosomes may dock to peroxisomes for further fatty acid processing and activation of PPARs.. Use of time lapse imaging with appropriate resolution, and dyes that mark the various compartments and components may help shed light on how metabolites from the apoptotic cells are distributed for processing following phagocytosis.
Phagocytosis of apoptotic cells appears to trigger the expression of phagocytic receptors, i.e. resembling a positive feedback loop (Gonzalez et al., 2009; Mukundan et al., 2009). This in turn would lead to further metabolic loading within the phagocyte, should it choose to engulf more dying targets. Therefore, changes in the phagocyte must encompass those associated with the ramping up and/or modification of metabolic steps, as well as those aimed at promoting the continued clearance of apoptotic cells. For example, PGC1α, a mitochondrial biogenesis regulator has been shown to function downstream of PPARs, but how the number of mitochondria within a phagocyte or the processing of metabolites by these mitochondria might be regulated by PPARs during engulfment is unclear. Ucp2 has been implicated in modulating metabolism by promoting fatty acid and glutamine metabolism and decreasing glycolysis-derived pyruvate catabolism (Pecqueur et al., 2008). Whether these other processes may also be partially responsible for the effects of Ucp2 on the ability of phagocytes to continue to ingest apoptotic cells will need further investigation.
A systematic approach to target all the metabolic changes within a phagocyte might reveal ‘stress points’ within the system during engulfment, such as managing target numbers (e.g. one versus multiple apoptotic cells), or target size (e.g. a macrophage eating another same-sized dying macrophage in an atherosclerotic plaque versus a much smaller dying thymocyte during T cell development in the thymus). Another area worthy of more detailed examination is how the phagocytes manage contents derived from apoptotic versus necrotic cells. A first level indication of this was seen when phagocytes incubated with necrotic cells failed to upregulate cholesterol efflux while they did so with apoptotic cells uptake (Kiss et al., 2006). Since atherosclerotic plaques have necrotic cores, the uptake of necrotic cells may negatively affect continued clearance by phagocytes and in turn potentially contribute to disease. Lastly, understanding how phagocytes process the excess metabolic load following engulfment might uncover new information on how surplus metabolites could be controlled at a single cell level, and in turn, reveal potential new metabolic steps for therapeutic intervention in conditions such as diabetes and obesity.
Acknowledgments
We thank J.M. Kinchen for helpful comments and discussions. This work was supported by the grants from the National Institutes of General Medical Sciences and the American Asthma Foundation (to K.S.R.) and pre-doctoral support via the T32 Training Grants from the NIAID and NIGMS (to C.Z.H.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- Bae HB, Zmijewski JW, Deshane JS, Tadie JM, Chaplin DD, Takashima S, Abraham E. FASEB J. 2011 doi: 10.1096/fj.11-190587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns RA, Harris IS, Mak TW. Nat Rev Cancer. 2011;11:85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- Elliott MR, Zheng S, Park D, Woodson RI, Reardon MA, Juncadella IJ, Kinchen JM, Zhang J, Lysiak JJ, Ravichandran KS. Nature. 2010;467:333–337. doi: 10.1038/nature09356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florey O, Kim SE, Sandoval CP, Haynes CM, Overholtzer M. Nat Cell Biol. 2011;13:1335–1343. doi: 10.1038/ncb2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez N, Bensinger SJ, Hong C, Beceiro S, Bradley MN, Zelcer N, Deniz J, Ramirez C, Diaz M, Gallardo G, de Galarreta CR, Salazar J, Lopez F, Edwards P, Parks J, Andujar M, Tontonoz P, Castrillo A. Immunity. 2009;31:245–258. doi: 10.1016/j.immuni.2009.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory CD, Pound JD. J Pathol. 2011;223:177–194. doi: 10.1002/path.2792. [DOI] [PubMed] [Google Scholar]
- Kao AW, Eisenhut RJ, Martens LH, Nakamura A, Huang A, Bagley JA, Zhou P, de LA, Neukomm LJ, Cabello J, Farese RV, Jr, Kenyon C. Proc Natl Acad Sci U S A. 2011;108:4441–4446. doi: 10.1073/pnas.1100650108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss RS, Elliott MR, Ma Z, Marcel YL, Ravichandran KS. Curr Biol. 2006;16:2252–2258. doi: 10.1016/j.cub.2006.09.043. [DOI] [PubMed] [Google Scholar]
- Lee HK, Mattei LM, Steinberg BE, Alberts P, Lee YH, Chervonsky A, Mizushima N, Grinstein S, Iwasaki A. Immunity. 2010;32:227–239. doi: 10.1016/j.immuni.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majai G, Gogolak P, Ambrus C, Vereb G, Hodrea J, Fesus L, Rajnavolgyi E. J Leukoc Biol. 2010;88:981–991. doi: 10.1189/jlb.0310144. [DOI] [PubMed] [Google Scholar]
- Martinez J, Almendinger J, Oberst A, Ness R, Dillon CP, Fitzgerald P, Hengartner MO, Green DR. Proc Natl Acad Sci U S A. 2011;108:17396–17401. doi: 10.1073/pnas.1113421108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Komatsu M. Cell. 2011;147:728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- Mukundan L, Odegaard JI, Morel CR, Heredia JE, Mwangi JW, Ricardo-Gonzalez RR, Goh YP, Eagle AR, Dunn SE, Awakuni JU, Nguyen KD, Steinman L, Michie SA, Chawla A. Nat Med. 2009;15:1266–1272. doi: 10.1038/nm.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata S, Hanayama R, Kawane K. Cell. 2010;140:619–630. doi: 10.1016/j.cell.2010.02.014. [DOI] [PubMed] [Google Scholar]
- Park D, Han CZ, Elliott MR, Kinchen JM, Trampont PC, Das S, Collins S, Lysiak JJ, Hoehn KL, Ravichandran KS. Nature. 2011;477:220–224. doi: 10.1038/nature10340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecqueur C, Bui T, Gelly C, Hauchard J, Barbot C, Bouillaud F, Ricquier D, Miroux B, Thompson CB. FASEB J. 2008;22:9–18. doi: 10.1096/fj.07-8945com. [DOI] [PubMed] [Google Scholar]
- Pezzolesi MG, Katavetin P, Kure M, Poznik GD, Skupien J, Mychaleckyj JC, Rich SS, Warram JH, Krolewski AS. Diabetes. 2009;58:2698–2702. doi: 10.2337/db09-0641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roszer T, Menendez-Gutierrez MP, Lefterova MI, Alameda D, Nunez V, Lazar MA, Fischer T, Ricote M. J Immunol. 2011;186:621–631. doi: 10.4049/jimmunol.1002230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjuan MA, Dillon CP, Tait SW, Moshiach S, Dorsey F, Connell S, Komatsu M, Tanaka K, Cleveland JL, Withoff S, Green DR. Nature. 2007;450:1253–1257. doi: 10.1038/nature06421. [DOI] [PubMed] [Google Scholar]
- Tabas I. Nat Rev Immunol. 2010;10:36–46. doi: 10.1038/nri2675. [DOI] [PMC free article] [PubMed] [Google Scholar]