Abstract
The transcriptional regulator DosR from M. tuberculosis plays a crucial role in the virulence to dormancy transition of the pathogen. DosR can be activated by DosT and DosS, two histidine kinases with heme-containing sensor GAF domains, capable of diatomic ligand binding, To investigate the initial processes occurring upon ligand dissociation, we performed ultrafast time-resolved absorption spectroscopy of the isolated sensor domains ligated with O2, NO and CO. The results reveal a relatively closed heme pocket for both proteins. For DosT the yield of O2 escape from the heme pocket on the picoseconds timescale upon photodissociation was found to be very low (1.5%), similar to other heme-based oxygen sensor proteins, implying that this sensor acts as an effective O2 trap. Remarkably, this yield is an order of magnitude higher in DosS (18%). For CO, by contrast, the fraction of CO rebinding within the heme pocket is higher in DosS. Experiments with mutant DosT sensor domains and molecular dynamics simulations indicate an important role in ligand discrimination of the distal tyrosine, present in both proteins, which forms a hydrogen bond with heme-bound O2. We conclude that despite their similarity, DosT and DosS display ligand-specific different primary dynamics during the initial phases of intra-protein signaling. The distal tyrosine, present in both proteins, plays an important role in these processes.
Mycobacterium tuberculosis is a widespread pathogen that is thought to have latently infected as much as one third of the human population worldwide, and to cause over 9 million new cases of tuberculosis each year (1). It has the capacity to switch from an actively replicating form to a dormant non-active form. The latter form can be maintained for extended periods of time within the host and is highly resistant to drugs, a combination of properties that largely contributes to the difficulty in eradicating tuberculosis.
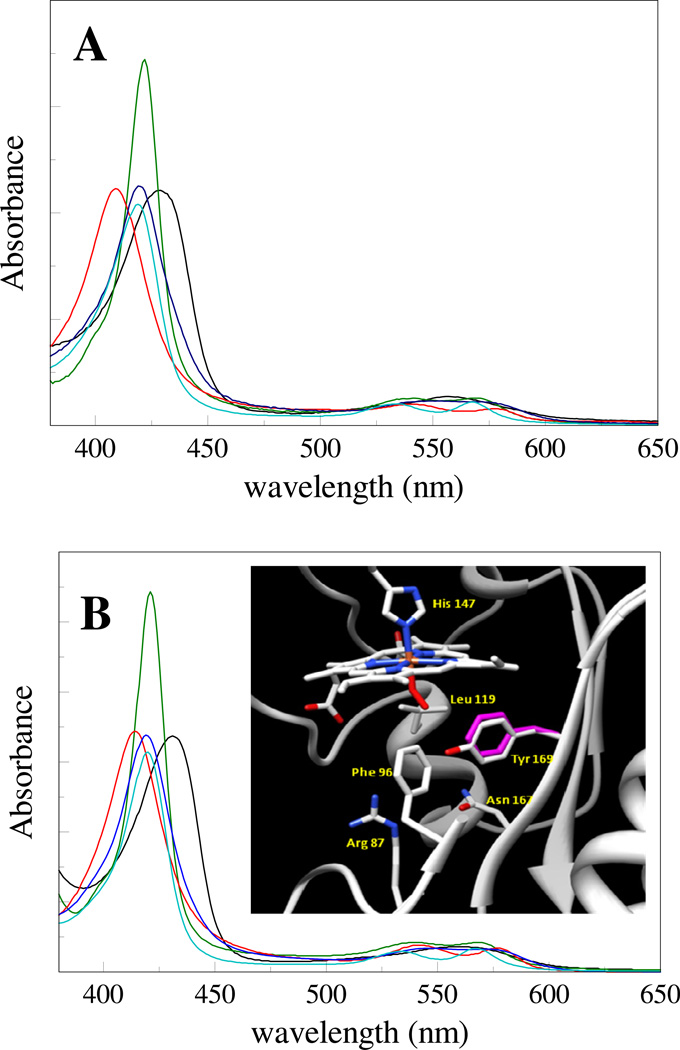
The transition between the two states is activated by stimuli including hypoxia (2) and exposure to NO (3). A crucial role in this transition is played by the transcriptional regulator DosR that induces the expression of a number of genes required for entry to dormancy (4). This response regulator is part of a two-component system that also comprises the signaling histidine kinases DosT and DosS (5), each of which can transfer phosphate to DosR. DosT and DosS show high similarity and both contain, apart from the C-terminal enzymatic domains, two GAF domains, one of which (the N-terminal GAF A domain) is a heme-based sensor domain (6, 7). DosT and DosS appear not to be redundant, but active in the initial and final phases of the transition to dormancy, respectively (8, 9). The sensor domains of DosT and DosS carry hemes that, in the ferrous state, are able to bind the external ligands O2, NO, and CO and are 5-coordinate in the absence of these ligands (6, 7, 10–12). Both kinases are inactive in the ferrous O2-bound state and active in other ferrous forms (7, 12). The autoxidation rates of the proteins are very low (7, 11). The proteins have been proposed to act as oxygen (or hypoxia) sensor (7, 11) or nitric oxide sensor (DosS) (13); alternatively for DosS a role as redox sensor has also been proposed (12, 14). The ligand binding and ligand discrimination properties of both proteins are somewhat different (7). Crystal structures of the GAF A domains of DosT and DosS have been reported, for the former including the oxycomplex (14, 15) (Inset Fig. 1B). A specific role for a distal tyrosine present in both proteins (Tyr 169 in DosT) that is in hydrogen bonding interaction with heme-bound molecular oxygen has been evoked for the ligand sensing, discrimination and transfer processes, although at least in DosS the oxygen binding properties are not very sensitive to its replacement (16). In the close environment of this tyrosine residue a few amino acids are not conserved; they may play a role in the different reactivities of the two sensor proteins.
Fig. 1.
Ground-state absorption spectra of different ligation states of GAF domains of DosS (A) and DosT (B): ferrous unliganded (black), ferrous CO-bound (green), ferrous O2-bound (red), ferrous NO-bound (blue) and ferric NO-bound (cyan). Inset: Structure of the distal heme pocket of the oxycomplex of the DosT GAF A domain (PDB 2VZW). The position of Phe169 in the Y169F mutant protein, as modelled by our molecular dynamics simulations, is also represented, in violet. Figure produced with UCSF Chimera (32).
The molecular mechanism of intra-protein signal transmission from the heme pocket in the GAF A sensor domain to the kinase domain is unknown. The initial phases following binding or dissociation of the ligand to/from the heme are presumably triggered by structural changes of the heme or its close environment. Heme-ligand dissociation processes can be synchronized using short photodissociation light pulses, and therefore ultrafast spectroscopy is a powerful technique to study these early events (17). Femtosecond transient visible absorption spectroscopy has been used to study the reactivity of dissociated ligands within the sensor domains of various heme-based gas sensor proteins including the NO receptor guanylate cyclase (18), the CO sensor CooA (19) and the oxygen sensors FixL and EcDos (20–25). These studies have revealed a high reactivity of these sensors, in particular for the physiologically sensed ligand, as witnessed by efficient heme-ligand geminate recombination or low dissociation yields (17). Combined with site-directed mutagenesis approaches, insight in the structural origin of these features has been obtained, especially for the oxygen sensor FixL (20, 21). In the present work, we compare ligand dissociation and geminate rebinding properties on the picosecond and early nanosecond timescale in the isolated DosS and DosT sensor domains from M. tuberculosis in order to get insight into the ligand dynamics and intra-protein signalling. The results reveal a relatively closed heme pocket for both proteins and remarkable differences in oxygen dissociation properties. The origin of these features and possible functional implications are discussed based on our studies on genetically modified DosT heme domains and molecular dynamics simulations.
Materials and Methods
The DosS GAF A domain from Mb. tuberculosis was expressed and purified as described (10) and suspended in 50 mM sodium phosphate buffer, pH 7.6 and 200 mM NaCl. The 444 bp DNA fragment corresponding to the sequence of the DosT GAF A domain from M. tuberculosis, comprising amino acids 61 to 208, was optimized for expression in Escherichia coli and cloned into the vector pQE 80 (QIAGEN) using BamHI and PstI restriction sites. The construct was confirmed by DNA sequencing, and the corresponding polypeptide was expressed after induction with 1mM IPTG in E. coli strain BL 21 in 1 liter of LB medium containing 1mg/liter ampicillin. The resulting 6xHis-tagged polypeptide was purified from cell-free extracts by affinity gravity-flow chromatography on Ni-NTA agarose (Marcherey-Nagel), followed by size exclusion chromatography on Econo-Pac 10 DG columns (Bio-Rad). The polypeptides corresponding to the WT and mutant DosT GAF A domains were suspended in 50 mM Tris, pH 7.6 and up to 250 mM NaCl. The desired amino acid substitutions were introduced following the QuikChange site-directed mutagenesis protocol (Stratagene) and confirmed by DNA sequencing prior to further analyses.
The experiments were carried out at a protein concentration of ~40 µM in 1-mm pathlength gastight optical cells. All samples were degassed in the measuring cells. For the CO-bound forms, the samples were reduced with sodium dithionite and subsequently equilibrated with 1 atm CO. For the ferric NO-bound form, the samples were equilibrated with 0.1 atm NO. The ferrous NO-bound form was generated for DosS, by adding sodium nitrite and sodium dithionite (both 2 mM), and for DosT by reducing with sodium dithionite and equilibration with 0.01 atm NO for 1h. For DosS, the oxycomplex was generated by reduction with a slight excess of dithionite and equilibration with 1 atm O2. The WT DosT protein after isolation and purification was in the oxycomplex form (see Results section). The oxycomplex of the mutant DosT proteins was generated by reduction with ascorbate (5 mM), using ruthenium hexamine (10 µM) as a mediator in some cases, and exposure to air or 1 atm. O2. For the R87A DosT mutant only ~80% oxycomplex formation could be achieved this way and 20% remained in the deoxy form. This incomplete feature was taken into account when calculating the quantum yield of oxygen escape.
Multicolor femtosecond absorption experiments were performed as described (26) with a 30 fs pump pulse centered at 565 nm and a <30 fs white light continuum probe pulse, at a repetition rate of 30 Hz. Full spectra of the test and reference beams were recorded using a combination of a polychromator and a CCD camera. The sample was continuously moved perpendicular to the beams to ensure sample renewal between shots.
Molecular dynamics simulations on the oxycomplex of the WT and Y169F sensor domain of DosT were performed using CHARMM (27) (version 33b2) with the parameter set of version 22. The models were based on the oxycomplex subunit of the crystal structure of the GAF A domain of WT DosT (PDB entry 2VZW) (15). The model includes 149 residues (60–208), one heme, one dioxygen ligand, 11 sodium ions and 3337 water molecules surrounding the protein. Nonbonded interactions were gradually set to zero between 10 and 14 Å. After energy minimization of the model, the Langevin equations of motion of the atoms were resolved using the LEAP integrator at 300 K, with 1 fs time steps over 2.5 ns. The simulation protocol was similar to that in Ref. (20). Rms deviations from the X-ray structure remained less than 1.6 Å during the simulations. The Y169F model was generated by deleting the Tyr169 Oη atom and switching Tyr169 to Phe169 in the coordinates input files. After energy minimization, the position of the Phe was found to be very close to that of Tyr169 in the WT structure (Fig. 1B, inset). The same protocol was used to simulate the two models.
O2 dissociation was simulated by deletion of the bond between the heme iron and the oxygen ligand from the bond list and simultaneous switching of heme parameters from six-coordinate to five-coordinate. Independent starting conditions of the model before ligand dissociation were obtained by prolongation of the free dynamic phase of the O2-bound form by subsequent periods of 250 ps. We performed nine dioxygen dissociations for each model. Structural coordinates were saved every 1 ps, and every 10 fs during 2.5 ps after each dissociation event.
Results
Ground-state absorption spectra
The ground-state spectra of the different ligation states studied in the time-resolved experiments below, as well as the spectrum of the ferrous unliganded state, are shown in Fig. 1 for the GAF A domains of M. tuberculosis DosS (Fig. 1A) and DosT (Fig. 1B) wild-type proteins. These spectra are similar to those reported previously for isolated GAF domains and full-length proteins (7, 10, 13, 15, 16). The spectrum of the DosS oxycomplex (obtained after reduction with a small excess of dithionite) was also very similar to the FMN-reduced spectrum reported by Cho et al. (14) that was assigned to a modified form of the ferrous deoxy-complex. The spectra of the different ligations states are similar for both complexes, but small spectral differences are present.
We found that the oxycomplex of the DosT GAF domain was directly formed upon purification. The spectrum of this form, with distinct peaks in the α band region at 577 and 542 nm and a Soret peak at 414 nm, is very similar to that of other 5-coordinate O2-binding heme proteins, including DosS, FixL and myoglobin (11, 23). It differs substantially however, from the spectrum of the isolated full-length protein reported by Kumar et al. (12) that, although assigned to the oxycomplex, appears as a mixture of the ferric form and the ferrous oxycomplex.
CO complex
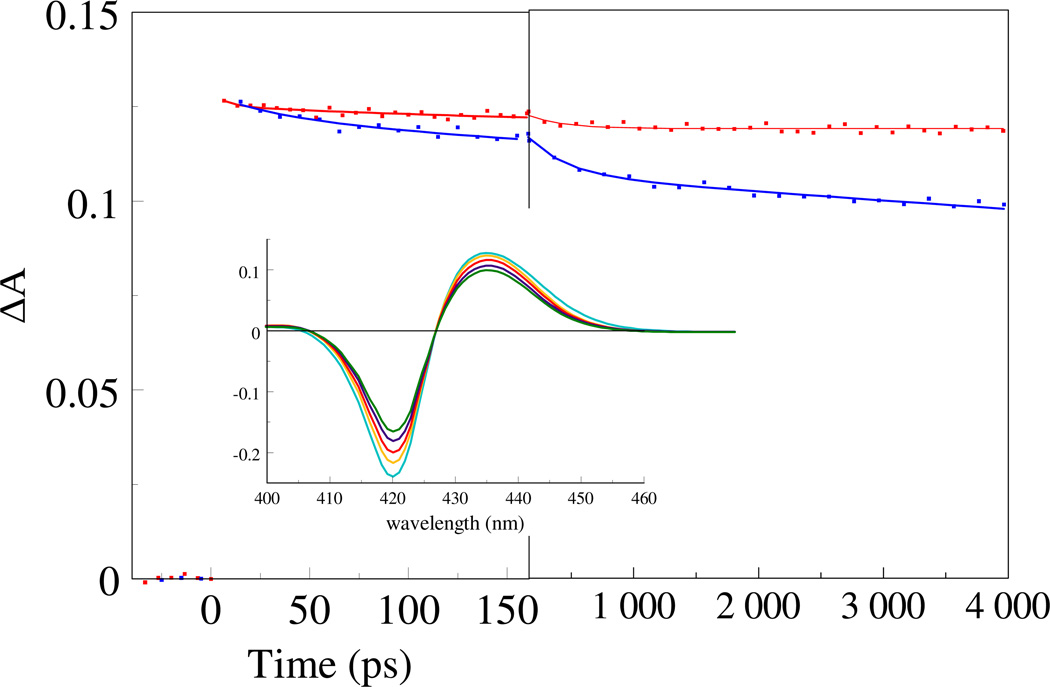
In the oxygen transport and storage proteins hemoglobin and myoglobin, dissociated CO does not rebind to a significant extent on the timescale prior to ~100 ns, but in sensor proteins like FixL, EcDos (24, 25), and CooA (19) modest to strong rebinding has been observed. Dissociation of CO from the GAF domain of DosS leads to geminate rebinding of only ~7% of CO, in a single phase with a time constant of 280 ps (Fig. 2, Table 1). In DosT, the main rebinding phase occurs with a similar time constant, 310 ps, but the rebinding during the time window of our experiment is three times more extensive (~23% has rebound at 4 ns) and additional rebinding phases with time constants of 33 ps and >4 ns (the limited time window does not allow determination of the precise extent and rate of this phase) are observed. These results suggest that the heme pocket is more tightly closed in DosT and/or that the dissociated CO is maintained in a more favorable position for rebinding.
Fig. 2.
Kinetics of CO rebinding to the GAF domains of DosS (red) and DosT (blue), measured at 435 nm, the maximum of the transient absorption spectra. Inset Transient spectra of DosT-CO at delay times of 5 ps (cyan), 45 ps (orange), 200 ps (red), 1 ns (blue) and 4 ns (green).
Table 1.
Fits of CO rebinding kinetics. The time constants τi were extracted from global fits of the data. The amplitudes Ai refer to the transient absorption maxima at ~435 nm and are normalized. A∞ refers to the non-rebinding fraction (τ>>4 ns).
| τ1 (ps) | τ2 (ps) | τ3 (ps) | A1 | A2 | A3 | A∞ | |
|---|---|---|---|---|---|---|---|
| DosS WT | 280 | 0.07 | 0.93 | ||||
| DosT WT | 33 | 310 | >4000 a | 0.04 | 0.13 | 0.83 a | a |
| Y169F | 100 | >4000 a | 0.38 | 0.62a | a | ||
| R87H | 88 | 1000 | 0.04 | 0.08 | 0.88 | ||
| R87A | 12 | 150 | 2200 | 0.10 | 0.19 | 0.10 | 0.61 |
| N167T | 330 | 3700 | 0.09 | 0.11 | 0.80 | ||
| N167W | 270 | 2500 | 0.05 | 0.20 | 0.75 | ||
| R87H/N167T | 26 | 400 | 2800 | 0.04 | 0.02 | 0.15 | 0.79 |
Due to the limited time window a precise time constant for this phase and the extent of any symptotic phase (A∞) could not be determined.
For both proteins, spectral evolution within a few picoseconds also occurs (not shown), which can be ascribed to heme photophysics (28). After decay of these phases, the spectra (Fig. 2, Inset) become similar to the steady state deoxy-minus-CO spectra.
NO complexes
Dissociation of the heme-NO bond in the ferrous (Fe2+) NO-complex leads to almost complete rebinding in a multiexponential manner in both proteins (Fig. 3A, Table 2), with a fastest and dominant phase of ~5 ps. Such features are observed in many heme proteins and are thought to reflect recombination from distinct conformational states (17). The kinetics in both proteins is similar, with overall rebinding in DosT somewhat faster.
Fig. 3.
Kinetics of NO rebinding to the ferrous (A) and ferric (B) GAF domains of DosS (red) and DosT (blue), measured at the bleaching maxima at 418 nm (A) and 420 nm (B). The Insets show transient spectra for ferrous DosT-NO (A) at delay times of 0.4 ps (blue), 3 ps (black), 9 ps (green), 50 ps (red) and 300 ps (cyan) and ferric DosS-NO (B) at delay times of 0.8 ps (blue), 3 ps (black), 20 ps (green), 50 ps (red) and 300 ps (cyan).
Table 2.
Fits of NO rebinding kinetics. The time constants τi were extracted from global fits of the data. The amplitudes Ai refer to the bleaching minima and are normalized. A∞ refers to the non-rebinding fraction (τ>4 ns).
| τ1 (ps) | τ2 (ps) | τ3 (ps) | A1 | A2 | A3 | A∞ | |
|---|---|---|---|---|---|---|---|
| Ferrous | |||||||
| DosS WT | 4.6 | 16 | 380 | 0.65 | 0.27 | 0.07 | 0.01 |
| DosT WT | 5.7 | 41 | 0.79 | 0.19 | 0.02 | ||
| Ferric | |||||||
| DosS WT | 8 | 47 | 0.23 | 0.70 | 0.07 | ||
| DosT WT | 17 | 64 | 0.64 | 0.35 | 0.01 | ||
NO can also be dissociated from the ferric (Fe3+) heme-NO complexes. In contrast to the ferrous complexes, in the difference spectrum this gives rise to a shift feature of the Soret band to lower wavelengths with a broad maximum below 400 nm (Fig. 3B, Inset), as the Soret absorption of 5-coordinate ferric heme consists of a broad band with a maximum around 395 nm (29), compared to the ~420 nm absorption maximum of the ferric heme-NO (6-coordinate) complex. We note that the transient spectra are quite different from the steady state ferric-minus-ferric NO spectra (not shown), as for both, DosT and DosS, the stable ferric protein is in the aquamet form with a Soret absorption around 406 nm (10, 15). The small induced absorption at the red side of the bleaching, observed at short delay times, is ascribed to excited state decay and heme cooling.
NO rebinding to the ferric heme occurs on the tens of picoseconds time scale, slower than to ferrous heme (a similar observation was made for horseradish peroxidase (30)) and in a non-exponential way (Fig. 3B). The kinetics are again similar for DosT and DosS, with the latter being somewhat slower and with a higher probability for NO to escape from the heme pocket (further rebinding on the timescale 300 ps-4 ns does not occur, not shown). Altogether, for the CO and NO complexes the rebinding kinetics is qualitatively similar for both sensor domains, but somewhat slower for DosS.
O2 complex
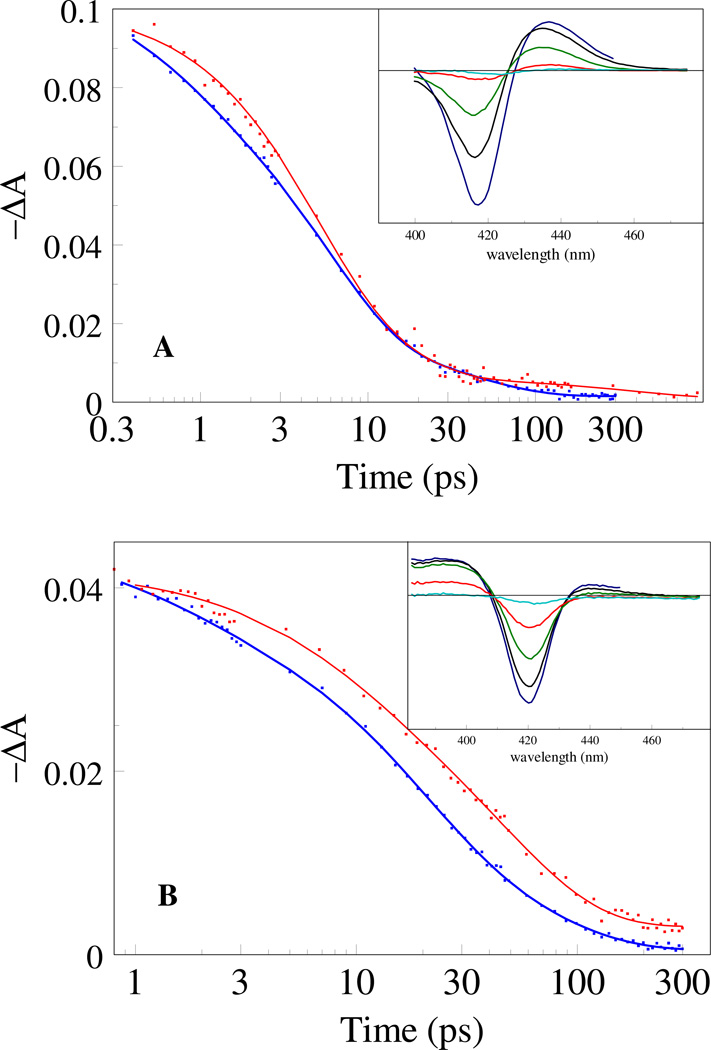
Excitation of the oxycomplex of DosT initially leads to a highly asymmetric, red-shifted spectrum, with a weak induced absorption lobe that mostly disappears with a time constant of ~ 5 ps, and the remaining spectrum being more symmetric (Fig. 4B,C). The initial difference spectrum is strongly perturbed with respect to the steady-state deoxy-minus-O2 spectrum, in a way very similar as in the bacterial oxygen sensor FixL (23). Based on our transient Raman analysis of the latter complex (21), we assign the analogous state in DosT predominantly to a thermally excited 6-coordinate O2-bound state rather than an actual O2-dissociated state, and the corresponding decay to thermal cooling. In DosS, the initial spectrum is also rather asymmetric. However, after decay of the 5-ps phase, the amplitude of the remaining spectrum, which is close in shape to the steady-state deoxy-minus-O2 spectrum, is strikingly higher compared to DosT, indicating that the dissociation yield on the timescale > 5 ps is much higher in DosS.
Fig. 4.
Transient spectra after excitation of the O2 complex of GAF domains of DosS (A) and DosT (B) at delay times of 0.5 ps (straight line) and 50 ps (dashed line). C: Normalized kinetics at 435 nm for DosS (straight line) and DosT (dashed line).
When we determined the quantum yield (QY) of dissociated O2 per absorbed photon at t = 50 ps using MbCO as a reference (21, 31), we indeed found a value for DosS of ~0.18 that is almost an order of magnitude higher than that for DosT (0.03, see Table 3). For comparison, the corresponding value for Mb is 0.26 (31). We conclude that the protein environment of the heme in DosT strongly prevents O2 from dissociation.
Table 3.
Quantum yield (QY) of O2 dissociation and O2 rebinding kinetics. The QY is determined at t=50 ps. For the WT proteins and the higher QY DosT mutants kinetics on extended to 4 ns were obtained. The time constants were extracted from global fits of the data. The amplitudes A (rebinding fraction) and A∞ (non-rebinding fraction, τ>4 ns) refer to the bleaching minima and are normalized to the signal remaining after the initial ~5 ps phase, which is thought not to reflect O2 rebinding (see text). nd, not determined.
| QY | τ (ps) | A | A∞ | ||
|---|---|---|---|---|---|
| DosS WT | 0.18 | - | 0 | 1 | |
| DosT WT | 0.03 | 110 | 0.5 | 0.5 | |
| Y169F | 0.19 | 140 | 0.4 | 0.6 | |
| R87H | 0.02 | nd | |||
| R87A | 0.04 | nd | |||
| N167T | 0.02 | nd | |||
| N167W | 0.09 | - | 0 | 1 | |
| R87H/N167T | 0.03 | nd | |||
On the timescale 50 ps-4 ns no O2 recombination was observed for DosS, indicating that the ~18% dissociated O2 can escape the protein. By contrast, for DosT about half of the ~3% dissociated O2 rebinds in ~100 ps (see below), further reducing the escape yield of O2 from the protein. This difference in O2 escape yield suggests differences in signalling reactivity of the proteins upon thermal heme-O2 dissociation (see Discussion).
Mutant DosT GAF A domains
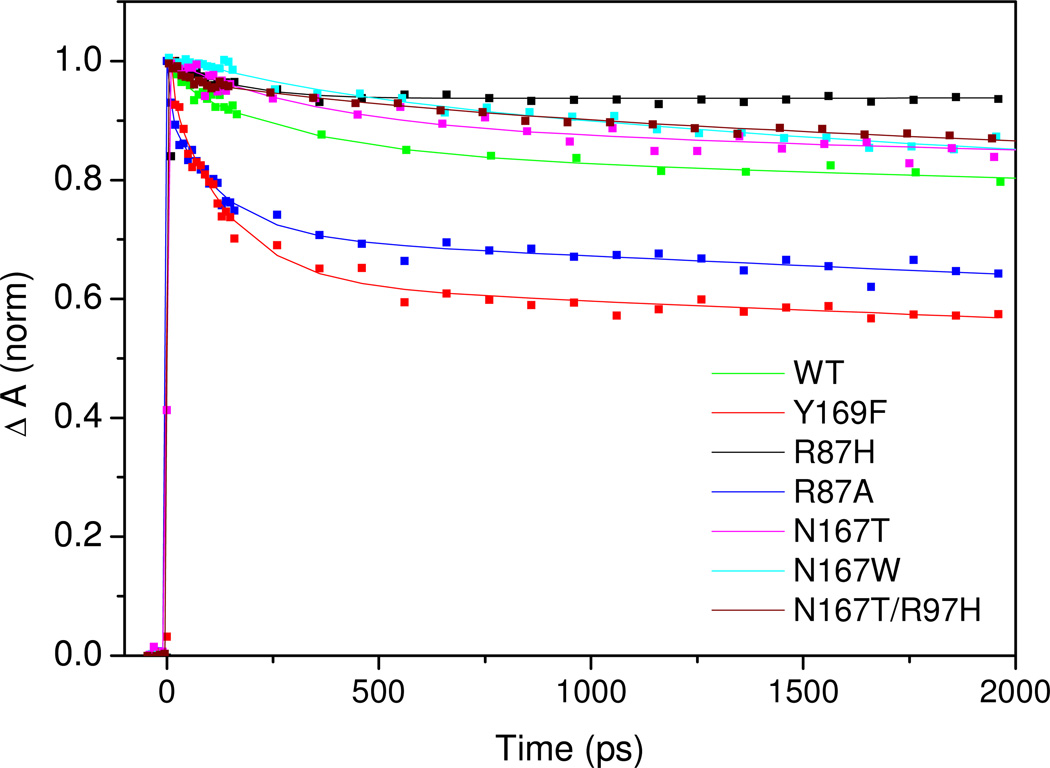
To further investigate the origin of the relatively low yields of ligand escape in DosT, we performed experiments on the CO and O2 complexes of mutant DosT GAF A domains. A distal tyrosine (Tyr169 in DosT) is in close interaction with heme-bound O2 via its OH group (15) (Inset Fig. 1B); this residue is part of a ligand-binding GAF motif and also conserved in DosS, where it has been shown to play an important role in ligand discrimination (16). Non-conserved residues in close contact with Tyr169 include Arg87 and Asp167, which are respectively His and Thr in DosS; both residues are thought to align a ligand exchange pathway (15). Consequently we constructed mutants Y169F, R87H, N167T and the R87H/N167T double mutant and, in order to investigate steric effects, the single mutants R87A and N167W.
Fig. 5 shows the CO rebinding data for these mutant sensor domains; fit results are collected in Table 1. All proteins show significant and multiexponential geminate rebinding. In the R87H, N167T and R87H/N167T mutant proteins that are altered towards the corresponding residues in DosS, recombination is somewhat less extensive than in WT DosT. Interestingly, only the R87H single mutation suppresses all longer (> 1 ns) recombination phases; its CO rebinding properties are similar to those of WT DosS. This finding suggests that His in place of the more flexible Arg at position 87 allows the creation of a CO docking site where, after initial relaxation phases, recombination is strongly hindered. The much stronger and continuing recombination observed in the R87A mutant is consistent with a steric effect of the residue at position 87. In contrast, the influence of the residue at position 167 appears more subtle, as replacement of Asp167 by the bulky Trp only has a modest effect. Finally, in the Y169F mutant protein, CO rebinding is markedly enhanced, in general agreement with the close proximity of Tyr169 with the heme-bound ligand.
Fig. 5.
Kinetics of CO rebinding to the WT and mutant GAF domains of DosT, measured at 435 nm.
The characteristics of oxygen photodissociation and rebinding are summarized in Table 3. With the exception of the N167W substitution, the photodissociation QY for the mutations involving Arg87 and Asp167 is similar as for WT, indicating that the amino acid differences at positions 87 and 167 between DosS and DosT are not at the origin of the differences in QY. The higher QY (0.09) observed for N167W points at a steric effect disrupting the interaction of bound oxygen with its environment.
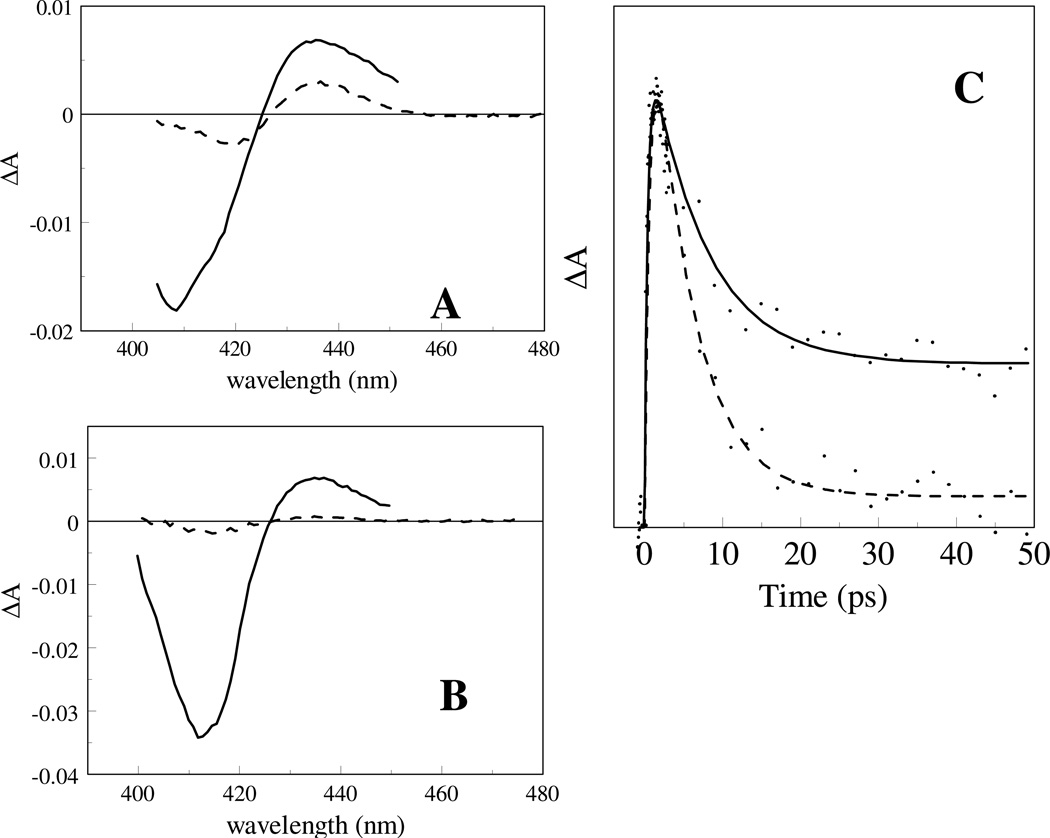
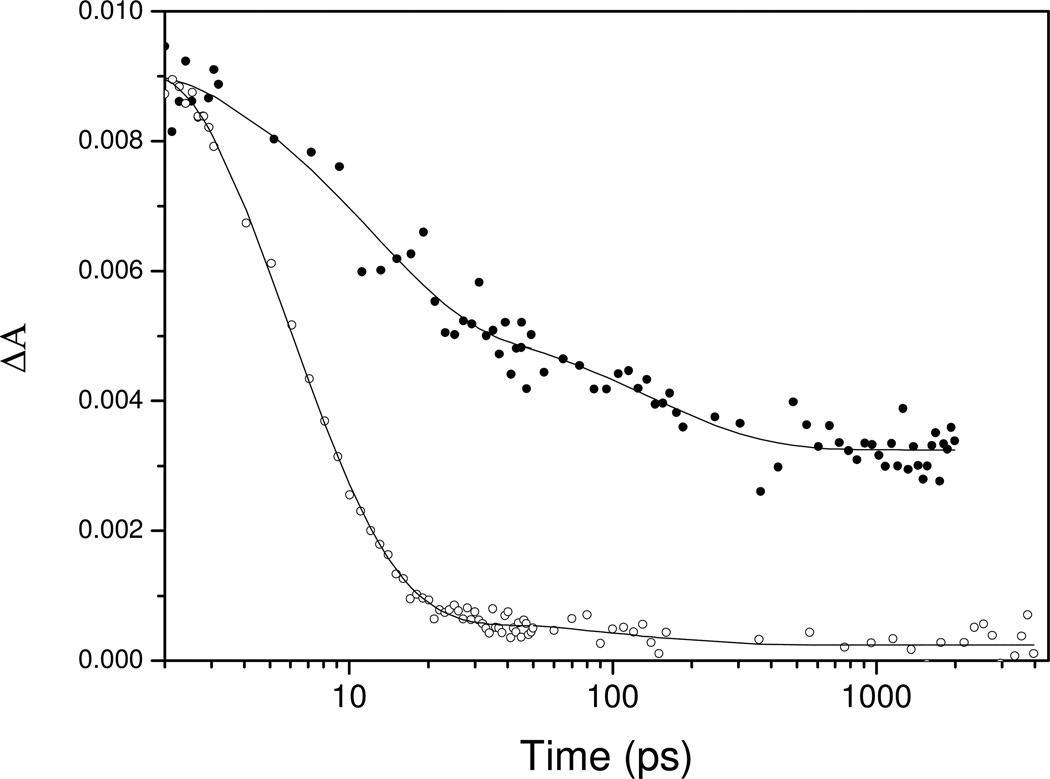
The most dramatic effect, an increase in the dissociation QY of nearly an order of magnitude compared to WT, was observed in the DosT Y169F substitution (Fig. 6, Table 3). This finding points to a crucial role of Tyr169 in maintaining O2 in a favourable position for binding. The rebinding kinetics of the dissociated O2, on the timescale > 5 ps, is very similar in Y169F and WT DosT. Altogether, remarkably, the overall effect of the Y169F mutation is an inversion of the ligand escape properties for CO and O2 in this sensor.
Fig. 6.
Kinetics at 435 nm after excitation of the O2 complex of WT (open circles) and Y169F (closed circles) DosT. The decay after ~20 ps reflects O2 recombination.
Molecular dynamics simulations
To better understand the role of Tyr169 in the low QY of O2 dissociation, molecular dynamics simulations were performed on the WT and Y169F GAF domain models. In these classical simulations, impulsive Fe-O2 bond dissociation was imposed in independent trajectories, but bond reformation was not taken into account. Therefore rather than monitoring directly the yield of dissociated O2 on the picosecond timescale (as measured in our experiments), information on the positioning of O2 immediately after dissociation is obtained.
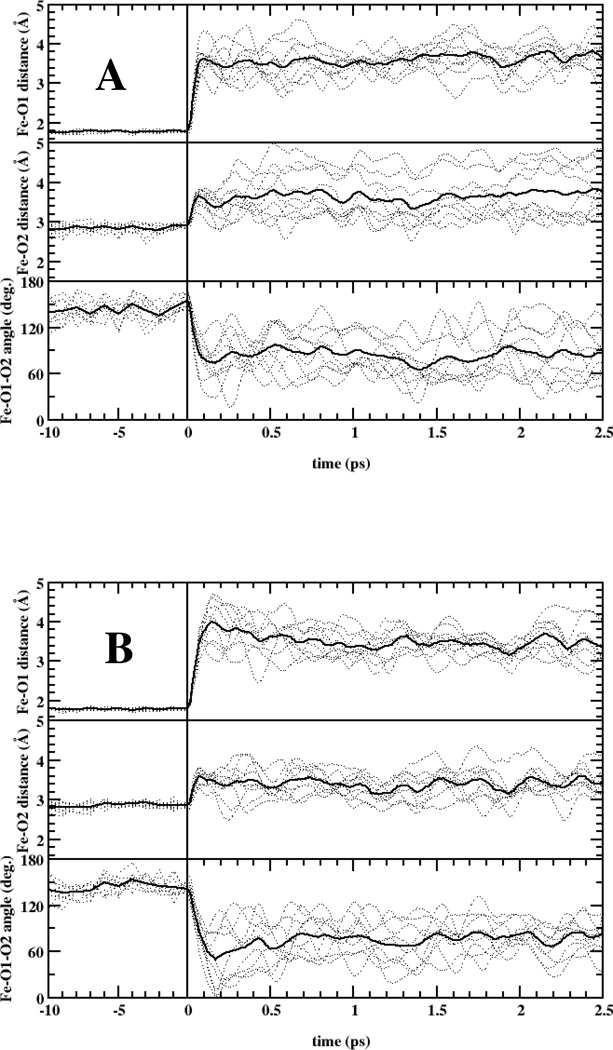
Fig. 7 shows the evolution of characteristic parameters in the Fe-O2 interaction following dissociation. The evolution appears generally similar, and in particular in both models within the first ~100 fs the heme-binding oxygen atom (O1) moves away from the heme iron and the O2 molecule rotates towards the heme plane. However, close comparison shows that in the Y169F substitution, both the Fe-O1 distancing and the O2 rotational motions can be significantly further extended than in WT. These observations indicate that the H-bonding interaction between Tyr169 and the O2 molecule keeps the dissociated ligand closer to the heme-bound configuration in the WT model. These features may allow O2 to more efficiently reform a bond with the heme on the timescale of 100 fs or less, and are therefore in qualitative agreement with the experimental QY findings described above.
Fig. 7.
Molecular dynamics simulations of O2 dissociation in WT (A) and Y169F (B) DosT. O2 was dissociated at t=0 ps. Dotted curves represent independent trajectories and bold solid curves averaged trajectories.
Discussion
Like in other sensor proteins (19, 23–25), rebinding of NO and CO with heme was found to be efficient, indicating that the heme pocket acts as an effective ligand trap. Ultrafast recombination of NO and heme is often used to probe the heme environment (17). For DosS and DosT, picosecond heme-NO rebinding is very efficient and the kinetic properties are very similar, both in the ferrous and ferric state (Fig. 3), although the overall kinetics are somewhat slower in DosS and in the ferric state the escape probability is also higher for DosS, pointing at a higher ligand mobility.
Picosecond CO rebinding in DosS is sizeable but modest (7%) (in many heme proteins, geminate CO rebinding does not occur (17)), and can be described by a single exponential (280 ps). Here, in the simplest model, upon CO dissociation competition occurs between occupation (in ~300 ps) of a site from which rebinding is very inefficient and rebinding with heme (in ~4 ns). In contrast, in DosT, more extensive and multiexponential heme-CO rebinding is observed, suggesting transiently a heterogeneous distribution of configurations from which rebinding is possible. Investigation of the DosT mutants (Fig. 5) indicates that the R87H mutation alters the CO rebinding kinetics towards those observed for DosS. This indicates that the flexibility of Arg87 (His in DosS) in particular allows dissociated CO to occupy a range of conformations in the distal heme cavity, including those from which rebinding to the heme can occur in competition with escape through a ligand entry and exit tunnel, which has been proposed to start out near this residue (15). In DosS and in the DosT R87H substitution, the rigid His residue may block access of dissociated CO to the heme and/or facilitate access to the ligand tunnel. The opposite effect of the R87A mutation is in agreement with the proposed steric role of this residue at position 87. We remark that the difference in CO binding properties between DosT and DosS must involve the subtle interplay between a number of residues and not uniquely the His residue, as in the R87H/N167T double mutant CO rebinding is more extensive than in the R87H single substitution and the WT DosS protein. Furthermore the effects of the R87H and N167T mutations are not additive.
The most remarkable differences between DosT and DosS that we observed concern O2 dissociation patterns. DosT displays a very low O2 photodissociation yield, similar to what has been observed in the PAS-domain sensor proteins FixL and EcDos (23), implying that the heme domain acts as an effective O2 trap. By contrast, in DosS the yield is much higher and approaches that of the oxygen storage protein myoglobin (Fig. 4, Table 3). As previously shown for FixL (21), these differences probably arise from very early dynamic processes occurring within the 100 fs timescale and involving interactions with distal residues that can keep O2 close to the heme after dissociation, thus favoring ultrafast rebinding. In FixL, a hydrogen bond between a distal arginine and O2 plays an important role in this interaction (20, 21). In DosT, Tyr169, that forms a strong hydrogen bond with the heme-ligated O2 (15), is likely implicated in such interactions. Indeed, mutation of this tyrosine to phenylalanine, which eliminates this H-bond, results in a much higher O2 escape yield (Fig. 7, Table 3). Our MD simulations indicate that this occurs by releasing the motional restrictions on the 100 fs timescale imposed by the H-bond (Fig. 7). Yet, this finding does not explain the difference between DosT and DosS in this respect, as DosS also carries Tyr at the corresponding position. Mutation of the nearby non-conserved residues Arg87 and Asp167 in DosT did not lead to significantly higher escape yields, indicating that yet to be identified further differences between DosT and DosS are involved. However, the ensemble of our results does strongly suggest that the hydrogen bond between the distal tyrosine and O2, although clearly present (16), is weaker in DosS than in DosT. The observed insensitivity of the oxygen binding and autoxidation properties of DosS towards mutation of the distal tyrosine (11) is in agreement with this proposal. Determination of the crystal structure of the DosS sensor domain oxycomplex may further clarify this issue.
The rate of O2 escape from the protein due to thermal dissociation of the heme Fe-O2 bond (the oxygen off rate) has been suggested to be similar for DosT and DosS, although a directly determined O2 off rate for DosT is not available (7). If this is correct, and assuming that the intrinsic rate for thermal dissociation of the Fe-O2 bond is similar for DosT and DosS, our data, showing a much higher escape yield for DosS than for DosT on the time scale of 4 ns, suggests that in DosS additional O2 rebinding phases take place in the time window of 4 ns to ~1 ms (that has not been investigated so far). This would imply that in DosS rebinding may take place simultaneously while the ligand switching process is already in progress, a feature that may make fine tuning of the sensor efficiency more sensitive to environmental factors. By contrast, the DosT sensor functions with a mechanism where the switch would fully proceed once the oxygen has escaped the heme pocket (a relatively rare event), as we previously proposed for FixL and EcDos (23). In this view, the initially operating DosT sensor acts more like other heme-based gas sensors (see Introduction).
The most prominent effect of our mutation studies is observed for the Y169F mutant, both on CO and O2 dynamics. This is not surprising, as Tyr169 is in closest contact with the ligand in the available X-ray structures and is proposed to be involved in the switching pathway (15). However it is interesting to note that the effect is inverted for CO and O2: whereas the hydrogen bond with Tyr169 clearly constrains O2 in the heme pocket as discussed above, allowing much more O2 escape from the heme pocket in the mutant, the same Y169F mutation leads to a much lower escape yield for CO. Assuming this latter effect is due to the difference in hydrogen bonding capacity, it may reflect suppression of a hydrogen bond of Tyr169 with CO or, alternatively, a different site. The first possibility is unlikely as in this case hydrogen bonding would be expected to lead to less rather than more CO escape, as for instance observed in the R220H mutation in FixL (20). Furthermore, at least in DosS, hydrogen bonding of the distal tyrosine to heme-bound CO hardly occurs (16). Regarding the second possibility, Tyr169 has been invoked to play a key role in the initial signalling pathway. One possible mechanism involves locking the residue in a different configuration in the presence of other ligands than O2 via interaction with another hydrogen-bond partner, either a residue or a water molecule. Such a mechanism has been invoked to explain the ligand discrimination effect of the corresponding tyrosine in DosS (16). In this mechanism, mutation to Phe would unlock the distal residue so that it can accommodate a position similar to that in the oxygen bound form and favour CO maintenance close to the heme iron after dissociation and thus enhance fast geminate recombination and decrease the escape yield. Our results suggest that such a hydrogen bonding “switch” takes place in DosT signal transmission and also that Tyr169 plays an important role in ligand discrimination, as it does in DosS (16). Further detailing of the molecular mechanism, and in particular possible hydrogen bond partners in the active form, must await determination of the crystal structure of the CO-bound form or application of appropriate time-resolved vibrational spectroscopic techniques.
Abbreviations
- Mb
myoglobin
- QY
quantum yield
- WT
wild type
Footnotes
This work was supported by National Institutes of Health grant AI074824 to P.R.O.d M. and by a New Zealand Lottery Health Research Grant to J.J. E.-R.
References
- 1.World Health Organization. Global Tuberculosis Control. 2010
- 2.Wayne LG, Sohaskey CD. Nonreplicating persistence of Mycobaterium tuberculosis. Ann. Rev. Microbiol. 2001;55:139–163. doi: 10.1146/annurev.micro.55.1.139. [DOI] [PubMed] [Google Scholar]
- 3.Nathan C, Shiloh MU. Reactive oxygen and nitrogen intermediates in the relationship between mammalian hosts and microbial pathogens. Proc. Natl. Acad. Sci. U.S.A. 2000;97:8841–8848. doi: 10.1073/pnas.97.16.8841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sherman DR, Voskuil M, Schnappinger D, Liao R, Harrell MI, Schoolnik GK. Regulation of the Mycobacterium tuberculosis hypoxic response gene encoding α-crystallin. Proc. Natl. Acad. Sci. U.S.A. 2001;98:7534–7539. doi: 10.1073/pnas.121172498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roberts DM, Liao RP, Wisedchaisri G, Hol WGJ, Sherman DR. Two Sensor Kinases Contribute to the Hypoxic Response of Mycobacterium tuberculosis. J. Biol. Chem. 2004;279:23082–23087. doi: 10.1074/jbc.M401230200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sardiwal S, Kendall SL, Movahedzadeh F, Rison SCG, Stoker NG, Djordjevic S. A GAF Domain in the Hypoxia/NO-inducible Mycobacterium tuberculosis DosS Protein Binds Haem. J. Mol. Biol. 2005;353:929–936. doi: 10.1016/j.jmb.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 7.Sousa EHS, Tuckerman JR, Gonzalez G, Gilles-Gonzalez MA. DosT and DevS are oxygen-switched kinases in Mycobacterium tuberculosis. Protein Sci. 2007;16:1708–1719. doi: 10.1110/ps.072897707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Honaker RW, Leistikow RL, Bartek IL, Voskuil MI. Unique Roles of DosT and DosS in DosR Regulon Induction and Mycobacterium tuberculosis Dormancy. Infect. Immun. 2009;77:3258–3263. doi: 10.1128/IAI.01449-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim M-J, Park K-J, Ko I-J, Kim YM, Oh J-I. Different Roles of DosS and DosT in the Hypoxic Adaptation of Mycobacteria. J. Bacteriol. 2010;192:4868–4875. doi: 10.1128/JB.00550-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ioanoviciu A, Yukl ET, Moënne-Loccoz P, Ortiz de Montellano PR. DevS, a Heme-Containing Two-Component Oxygen Sensor of Mycobacterium tuberculosis. Biochemistry. 2007;46:4250–4260. doi: 10.1021/bi7008695. [DOI] [PubMed] [Google Scholar]
- 11.Ioanoviciu A, Meharenna YT, Poulos TL, Ortiz de Montellano PR. DevS Oxy Complex Stability Identifies This Heme Protein as a Gas Sensor in Mycobacterium tuberculosis Dormancy. Biochemistry. 2009;48:5839–5848. doi: 10.1021/bi802309y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar A, Toledo JC, Patel RP, Lancaster JR, Steyn AJC. Mycobacterium tuberculosis DosS is a redox sensor and DosT is a hypoxia sensor. Proc. Natl. Acad. Sci. U.S.A. 2007;104:11568–11573. doi: 10.1073/pnas.0705054104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yukl ET, Ioanoviciu A, Sivaramakrishnan S, Nakano MM, Ortiz de Montellano PR, Moënne-Loccoz P. Nitric Oxide Dioxygenation Reaction in DevS and the Initial Response to Nitric Oxide in Mycobacterium tuberculosis. Biochemistry. 2011;50:1023–1028. doi: 10.1021/bi1015315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cho HY, Cho HJ, Kim YM, Oh JI, Kang BS. Structural Insight into the Heme-based Redox Sensing by DosS from Mycobacterium tuberculosis. J. Biol. Chem. 2009;284:13057–13067. doi: 10.1074/jbc.M808905200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Podust LM, Ioanoviciu A, Ortiz de Montellano PR. 2.3 Ã X-ray Structure of the Heme-Bound GAF Domain of Sensory Histidine Kinase DosT of Mycobacterium tuberculosis. Biochemistry. 2008;47:12523–12531. doi: 10.1021/bi8012356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yukl ET, Ioanoviciu A, Nakano MM, Ortiz de Montellano PR, Moënne-Loccoz P. A Distal Tyrosine Residue Is Required for Ligand Discrimination in DevS from Mycobacterium tuberculosis. Biochemistry. 2008;47:12532–12539. doi: 10.1021/bi801234w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vos MH. Ultrafast dynamics of ligands within heme proteins. Biochim. Biophys. Acta. 2008;1777:15–31. doi: 10.1016/j.bbabio.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 18.Négrerie M, Bouzhir-Sima L, Martin J-L, Liebl U. Control of nitric oxide dynamics by guanylate cyclase in its activated state. J. Biol. Chem. 2001;276:46815–46821. doi: 10.1074/jbc.M102224200. [DOI] [PubMed] [Google Scholar]
- 19.Kumazaki S, Nakajima H, Sakaguchi T, Nakagawa E, Shinahara H, Yoshihara K, Aono S. Dissociation and recombination between ligands and heme in a CO-sensing transcriptional activator CooA. J. Biol. Chem. 2000;275:38378–38383. doi: 10.1074/jbc.M005533200. [DOI] [PubMed] [Google Scholar]
- 20.Jasaitis A, Hola K, Bouzhir-Sima L, Lambry J-C, Balland V, Vos MH, Liebl U. Role of distal arginine in early sensing intermediates in the heme domain of the oxygen sensor FixL. Biochemistry. 2006;45:6018–6026. doi: 10.1021/bi060012i. [DOI] [PubMed] [Google Scholar]
- 21.Kruglik SG, Jasaitis A, Hola K, Yamashita T, Liebl U, Martin J-L, Vos MH. Subpicosecond Oxygen Trapping in the Heme Pocket of the Oxygen Sensor FixL Observed by Time-Resolved Resonance Raman Spectroscopy. Proc. Natl. Acad. Sci. U.S.A. 2007;104:7408–7413. doi: 10.1073/pnas.0700445104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lechauve C, Bouzhir-Sima L, Yamashita T, Marden MC, Vos MH, Liebl U, Kiger L. Heme Ligand Binding Properties and Intradimer Interactions in the Full-length Sensor Protein Dos from Escherichia coli and Its Isolated Heme Domain. J. Biol. Chem. 2009;284:36146–36159. doi: 10.1074/jbc.M109.066811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liebl U, Bouzhir-Sima L, Négrerie M, Martin J-L, Vos MH. Ultrafast ligand rebinding in the heme domain of the oxygen sensors FixL and Dos: general regulatory implications for heme-based sensors. Proc. Natl. Acad. Sci. U.S.A. 2002;99:12771–12776. doi: 10.1073/pnas.192311699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liebl U, Bouzhir-Sima L, Kiger L, Marden MC, Lambry J-C, Négrerie M, Vos MH. Ligand binding dynamics to the heme domain of the oxygen sensor Dos from Escherichia coli. Biochemistry. 2003;42:6527–6535. doi: 10.1021/bi027359f. [DOI] [PubMed] [Google Scholar]
- 25.Yamashita T, Bouzhir-Sima L, Lambry J-C, Liebl U, Vos MH. Ligand dynamics and early signalling events in the heme domain of the sensor protein Dos from Escherichia coli. J. Biol. Chem. 2008;283:2344–2352. doi: 10.1074/jbc.M708123200. [DOI] [PubMed] [Google Scholar]
- 26.Silkstone G, Jasaitis A, Wilson MT, Vos MH. Ligand dynamics in an electron-transfer protein: picosecond geminate recombination of carbon monoxide to heme in mutant forms of cytochrome c. J. Biol. Chem. 2007;282:1638–1649. doi: 10.1074/jbc.M605760200. [DOI] [PubMed] [Google Scholar]
- 27.Brooks BR, Bruccoleri RE, Olafson BD, Swaminathan S, Karplus M. CHARMM: A Program for Macromolecular Energy, Minimization, and Dynamics Calculations. J. Comput. Chem. 1983;4:187–212. [Google Scholar]
- 28.Petrich JW, Poyart C, Martin J-L. Photophysics and reactivity of heme proteins: a femtosecond absorption study of hemoglobin, myoglobin and protoheme. Biochemistry. 1988;27:4049–4060. doi: 10.1021/bi00411a022. [DOI] [PubMed] [Google Scholar]
- 29.Gilles-Gonzalez MA, Gonzalez G, Perutz MF, Kiger L, Marden MC, Poyart C. Heme-based sensors, exemplified by the kinase FixL, are a new class of heme protein with distinctive ligand binding and autoxidation. Biochemistry. 1994;33:8067–8073. doi: 10.1021/bi00192a011. [DOI] [PubMed] [Google Scholar]
- 30.Ye X, Yu A, Champion PM. Dynamics of Nitric Oxide Rebinding and Escape in Horseradish Peroxidase. J. Am. Chem. Soc. 2006;128:1444–1445. doi: 10.1021/ja057172m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ye X, Demidov A, Champion PM. Measurements of the Photodissociation Quantum Yields of MbNO and MbO2 and the Vibrational Relaxation of the Six-Coordinate Heme Species. J. Am. Chem. Soc. 2002;124:5914–5924. doi: 10.1021/ja017359n. [DOI] [PubMed] [Google Scholar]
- 32.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comp. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]