Abstract
The aging heart undergoes well characterized structural changes associated with functional decline, though the underlying mechanisms are not understood. The aim of this study was to determine to what extent ventricular myocardial protein expression was altered with age and which proteins underwent protein nitration. Fischer 344 X Brown Norway F1 hybrid (FBN) rats of four age groups were used, 4, 12, 24, and 34 months. Differential protein expression was determined by 2-DE and proteins were identified by peptide mass fingerprinting. Altered protein nitration with age was assessed by immunoblotting. Over 1,000 protein spots per sample were detected, and 255 were found to be differentially expressed when all aged groups were compared to young rats (4 months) (p ≤ 0.05). A strong positive correlation between differential protein expression and increasing age (p=0.03, R2=0.997) indicated a progressive, rather than abrupt, change with age. Of 46 differentially expressed proteins identified, 17 have roles in apoptosis, 10 in hypertrophy, 7 in fibrosis, and 3 in diastolic dysfunction, aging-associated processes previously reported in both human and FBN rat heart. Protein expression alterations detected here could have beneficial effects on cardiac function; thus, our data indicate a largely adaptive change in protein expression during aging. In contrast, differential protein nitration increased abruptly, rather than progressively, at 24 months of age. Altogether, the results suggest that differential myocardial protein expression occurs in a progressive manner during aging, and that a proteomic-based approach is an effective method for the identification of potential therapeutic targets to mitigate aging-related myocardial dysfunction.
Keywords: aging, F344 X BN hybrid, heart, MALDI-TOF, nitrosylation, peptide mass fingerprinting, proteomics, two-dimensional electrophoresis
1. Introduction
Aging is a major risk factor for developing disease, including cardiovascular disease and cancer (Lakatta and Levy, 2003). Thus, as the elderly percentage of the population rapidly increases in the near future, there will be a resultant rise in age-related cardiovascular disease. A major aspect of the aging process is a gradual decline in biological functions, and the human heart undergoes structural and functional changes with age, resulting in a decline in cardiac performance (Gates et al., 2003; Groban, 2005; Oxenham and Sharpe, 2003). While these systemic changes are well documented, the changes that occur at the cellular and molecular level are less well understood. Growing evidence implicates oxidative stress such as that caused by reactive nitrogen species (RNS) as a causal factor for cellular dysfunction and it is well accepted that oxidative stress increases with age (Barja, 2002; Sohal and Weindruch, 1996).
To provide a better understanding of the protein molecular machinery underlying the aging process, we employed a proteomic platform capable of characterizing changes in protein expression and post-translational modifications. Although many theories of aging have been proposed, such as cellular senescence (Hayflick, 1965), gene expression order (Helfand and Rogina, 2000), and free radical generation (Harman, 1981), the mechanism that accounts for biological aging is currently viewed as a complex, multifactorial process. Alterations in proteins also have been proposed to contribute to aging-related impairments (Levine and Stadtman, 2001), and tools of proteomics such as the field’s cornerstone, two-dimensional electrophoresis (2-DE), are well suited to analyze complex quantitative and qualitative changes with age. This analysis of differential expression and modification yields a semi-global view of the underlying molecular changes accompanying physiological processes such as aging, which can assist in elucidating its mechanisms. Furthermore, these exploratory experiments are necessary to increase the likelihood of discovering novel proteins regulating aging phenomena and potential therapeutic targets for aging related dysfunction.
The majority of aging studies have compared only very young to extremely old animals. A primary goal of this study was to determine whether global and individual changes in ventricular myocardial protein expression during aging occurred in a progressive or in an abrupt manner. We also examined post-translational protein nitration (3-nitrotyrosine) of specific proteins as an index of age-related oxidative stress. To this end, hearts from Fischer 344 X Brown Norway hybrid (FNB) rats, ages 4, 12, 24, and 34 months were obtained from the NIA for analysis of differential protein expression and nitrotyrosine modification.
2. Materials and Methods
2.1 Materials
IPG strips and acrylamide for slab gels were purchased from Bio-Rad (Richmond, CA). Other ultrapure electrophoretic reagents were obtained from Bio-Rad (Richmond, CA), Sigma Chemical Co. (St. Louis, MO), or BDH (Poole, UK). Sequence grade trypsin was obtained from Promega (Madison, WI). Ammonium bicarbonate was purchased from Mallinckrodt Chemicals Paris, KY). Formic acid, iodoethanol, and triethylphosphine were obtained from Sigma-Aldrich Co. (St.Louis, MO). Acetonitrile and hydrochloric acid solution N/10 were obtained from Fisher Scientific (Fair Lawn, NJ). PVDF for western blotting was obtained from Millipore Co. (Billerica, MA). All other chemicals used were of the highest grade obtainable.
2.2 Tissues
Male Fischer 344 X Brown Norway hybrid (FBN) rat hearts (ages 4, 12, 24, and 34 months; n=5) were purchased from the National Institute of Aging (NIA). The rodent colonies were barrier maintained and kept Specific Pathogen Free (SPF). The rats were euthanized by CO2 asphyxiation and their hearts were removed, flash frozen in liquid N2, and stored at −80°C. The hearts were shipped on dry ice and stored at −45°C immediately upon arrival.
2.3 Preparation of Ventricular Myocardium (VM)
On ice, 150 mg of tissue was cut from the apex from each heart (VM) and massaged in ice cold filtered saline to remove as much blood as possible. The VM was then thoroughly minced with surgical scissors in a 50 ml beaker along with 8 volumes of a solution containing 9M urea, 4% Igepal CA-630 ([octylphenoxy] polyethoxyethanol), 1% DTT and 2% carrier ampholytes (pH 3–10). The minced samples were then placed in 3 ml DUALL® ground-glass tissue grinders and manually homogenized. To complete the solubilization, the minced/ground samples were sonicated with 3 × 2 second bursts at instrument setting 3 using a Microson Ultrasonic Cell Disruptor (Misonix, Inc.). This was repeated every 15 min for 1 hr at room temperature. After complete solubilization at room temperature for 120 min, samples were centrifuged at 50,000 rpm at 15°C for 20 min using a Beckman TL-100 ultracentrifuge to remove nucleic acid and insoluble materials, and the supernates stored at −45°C until 2-DE separation.
2.4 Two-Dimensional Electrophoresis and Image Analysis
Reagent Compatible Detergent Compatible (RC DC) protein concentration assays were performed for each of the 20 samples using an RC DC Protein Assay Kit according to the manufacturer’s protocol (BioRad). Aliquots (~20–50 µl each) containing 400 µg of protein from each sample were diluted with ~450–480 µl rehydration buffer (8M urea, 2% CHAPS, 15 mM DTT, 0.2% ampholytes pH 3–10, and 0.001% orange G). The resulting 500 µl protein dilutions were loaded onto IPG strips (24 cm, linear pH 3–10) by overnight, passive rehydration at room temperature. Isoelectric focusing was performed simultaneously on all IPG strips using two Protean IEF Cells (BioRad), by a program of progressively increasing voltage (150 V for 2 h, 300 V for 4 h, 1500 V for 1 h, 5000 V for 5 h, 7000 V for 6 h, and 10,000V for 3 h) for a total of 100,000 Vh. A computer-controlled gradient casting system was used to prepare the second-dimension SDS gradient slab gels (20 × 25 × 0.15 cm) in which the acrylamide concentration varied linearly from 11% to 17% T. First-dimension IPG strips were loaded directly onto the slab gels following equilibration for 10 minutes in Equilibration Buffer I and 10 minutes in Equilibration Buffer II (Equilibration Buffer I: 6M urea, 2% SDS, 0.375M Tris-HCl pH 8.8, 20% Glycerol, 130mM DTT; Equilibration Buffer II: 6M urea, 2% SDS, 0.375M Tris-HCl pH 8.8, 20% Glycerol, 135mM iodoacetamide) for alkylation and reduction of proteins. Second-dimension slab gels were run in parallel at 8°C for 18 h at 160V. Slab gels were stained using a colloidal Coomassie Blue G-250 procedure (Candiano et al., 2004). Gels were fixed in 1.5 L/10 gels of 50% ethanol/2% phosphoric acid overnight followed by three 30 min washes in 2 L/10 gels of deionized water. Gels were transferred to 1.5 L/10 gels of 30% methanol/17% ammonium sulfate/3% phosphoric acid for 1 h followed by addition of 1.2 g/L of powdered Coomassie Blue G-250 stain. After 96 h, gels were washed several times with water and scanned at 95.3 µm/pixel resolution using a GS-800 Calibrated Imaging Densitometer (Bio-Rad, Hercules CA). The resulting 12 bit images were analyzed using PDQuest™ software (Bio-Rad, v.7.1). Background was subtracted and peaks for the protein spots located and counted. Each gel image was normalized against the total gel density. PDQuest™ performs a preliminary statistical analysis using Student’s t-test to compare groups and determine differential expression candidates. Sigma Stat 3.0 (Systat Software, Inc., San Jose, CA) was used to calculate the regression coefficient when comparing changes in the number of these candidates with age. PDQuest™ data was exported to Sigma Stat 3.0 for 1-way analysis of variance ANOVA). Proteins were selected for MS identification based on these calculations.
2.5 Post-Translational Modification Analysis via Western blotting
Immediately upon completion of a second, replicate 2-DE run, gels from all four groups (n=3) were carefully removed and equilibrated in transfer buffer (49 mM TRIS, 39mM Glycine, 10% SDS, 20% MeOH, pH 9.2) for 1 h at room temperature. The PVDF membrane was immersed briefly in 100% methanol and then transfer buffer before being placed on the transfer soaked filter paper in the Transblot® SD Semi-Dry Transfer Cell (BioRad). Equilibrated gels were placed directly on top of the PVDF membrane. A maximum of 2 gels were stacked in the Semi-Dry Transfer Cells and each Cell was connected to a separate Power Pac HC (BioRad). Power supplies were set at 25 V and protein spots were transferred to PVDF paper over the course of 2 hrs. Following transfer, the gels were removed and stained as described above to evaluate transfer efficiency. The PVDF membranes were removed from the Transfer Cells and allowed to dry completely. Each membrane was then blocked with PBS-0.3% Tween20 for 2 hrs using 3–5 washes at room temperature and then incubated overnight with a primary anti-3-nitrotyrosine antibody (clone 1A6, Upstate/Millipore, Billerica, MA), diluted 1:10,000 in PBS-0.1% Tween20. The unbound antibody was rinsed off with 5–6 changes of PBS-0.1% Tween20 in 30–45 min. The secondary antibody (Goat Anti-mouse IgG, HRP conjugate, Upstate/Millipore, Billerica, MA) was diluted 1:10,000 in PBS-0.1% Tween20 and incubated for 2 hrs at room temperature. The unbound antibody was rinsed off with 5–6 changes of PBS-0.1% Tween20 in 30–45 min. The membranes are then developed in 10–15mls of tetramethylbenzidine (TMB) for exactly 5 min at room temperature. The membranes were scanned at 95.3 µm/pixel resolution using a GS-800 Calibrated Densitometer. The resulting images were analyzed using PDQuest™ software and the resultant data exported to Sigma Stat 3.0 (Jandel Scientific, CA) for 1-way analysis of variance (ANOVA) after PDQuest™ performed a preliminary statistical analysis using Student’s t-test to determine differential expression candidates.
2.6 In-gel Tryptic Digestion and Peptide Mass Fingerprinting
Each pre-selected spot that could be detected against a light box was cut from the gel by hand using a 1.5 mm gel cutting tool. Each cut spot was placed in one well of a 96-well plate, as was grp78 (standard) and one gel blank, and was processed using the Multiprobe II (Perkin-Elmer, Boston MA). In this automated system, the excised protein spots were first destained with 50 mM ammonium bicarbonate-50% acetonitrile followed by 100% acetonitrile. Reduction with 10 mM DTT and alkylation with 55 mM iodoacetamide was carried out prior to overnight tryptic digestion using modified trypsin (6 ng/µl). The grp78 (StressGen, Victoria, BC Canada) calibrant and a gel blank were digested using identical conditions. The resulting peptides were extracted by addition of 25 µl 0.2% formic acid (aqueous) and 7 µl of acetonitrile solution to each well, and shaking plates at 37°C for one hour. The resulting solution was placed in a separate 96 well plate and dried via a speedvac. The dehydrated peptides were then reconstituted in 5 µL of 0.2% formic acid and 1 µL of acetonitrile with continuous shaking of the plate for 5 minutes. Aliquots from peptide extracts (in 3 µL volumes) were then placed onto a MALDI target plate, air dried, and the application repeated until all the extraction solution was used. Just before the peptides finished drying, 0.8 µl of matrix (2 mg/ml W-cyano-4-hydroxycinnamic acid in 50% acetonitrile) was added to each peptide spot and allowed to dry completely.
Peptide masses were analyzed by MALDI-TOF-MS using a Waters Micromass M@LDI SYSTEM (Micromass, Milford, MA) and the prOTOF™ 2000 MALDI Orthogonal Time of Flight Mass Spectrometer (PerkinElmer/SCIEX, Concord, ON). Prior to data collection, the instrument was calibrated externally using a mixture of peptide standards, digested standard (grp78), and experiment artifact peaks based on tryptic autolysis. Proteins were identified by manually searching the Rattus NCBInr database using ProFound (The Rockefeller University) software and the mass lists obtained from exported MALDI spectra of the excised proteins. An “Expectation” value less than 0.001 was the threshold for what was considered a positive identification.
2.7 Bioinformatic Analysis of Identified Proteins
Functions, processes, and components (sub-cellular locations) of identified proteins were analyzed by the Gene Ontology tools available from Princeton University (http://go.princeton.edu/) (Harris et al., 2004). A list of the altered proteins’ Rat Genome Database Accession numbers (RGD number) was generated and entered into the Princeton University Generic Gene Ontology (GO) Term Mapper (http://go.princeton.edu/cgi-bin/GOTermMapper). The output files were analyzed and transformed into histograms.
3. Results
3.1 Differential Protein Expression
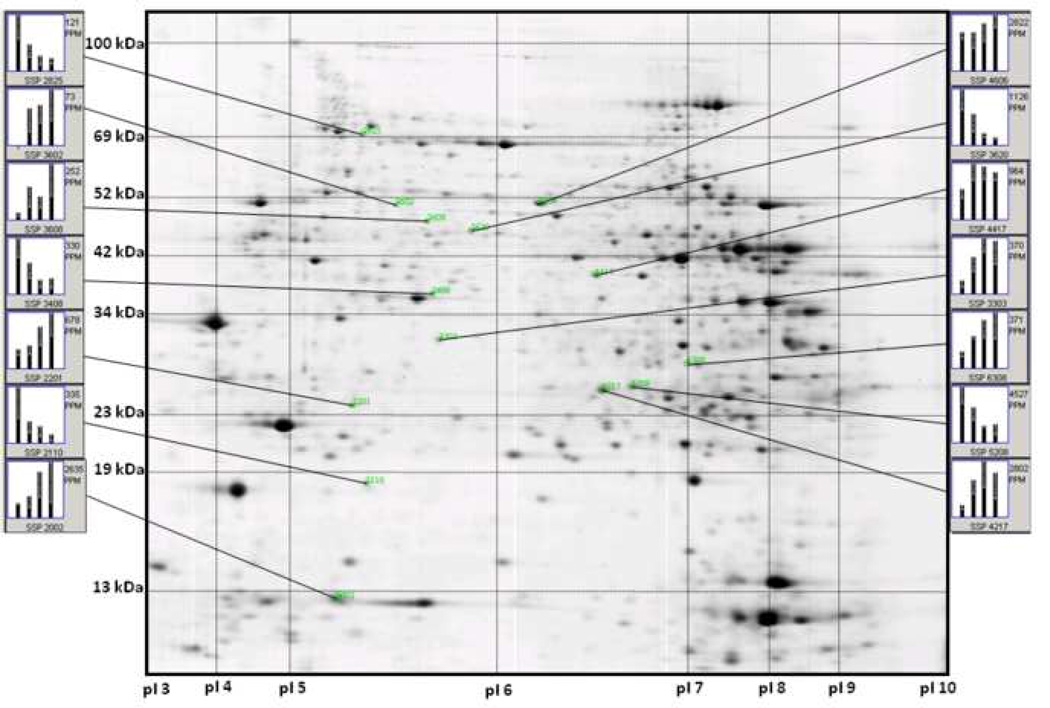
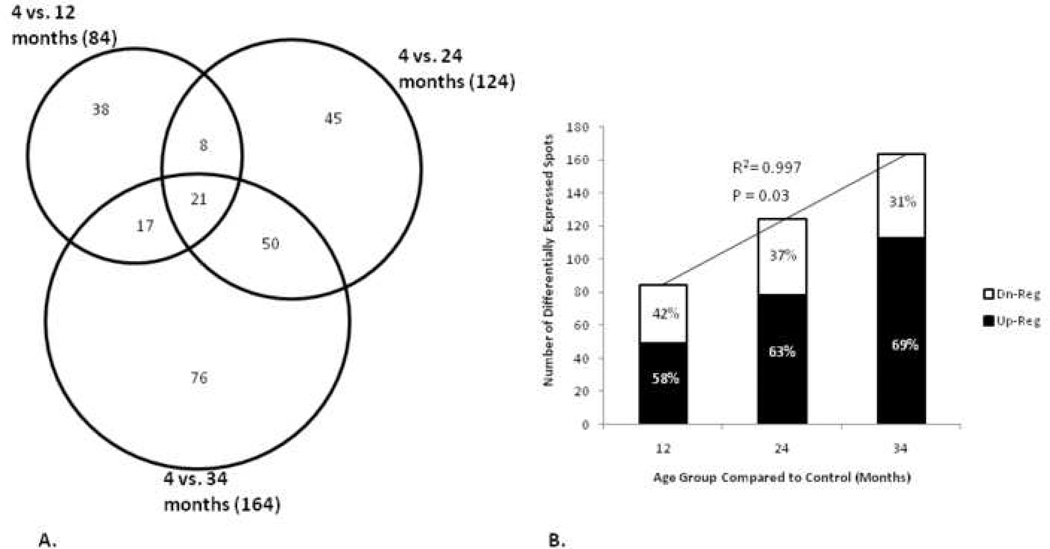
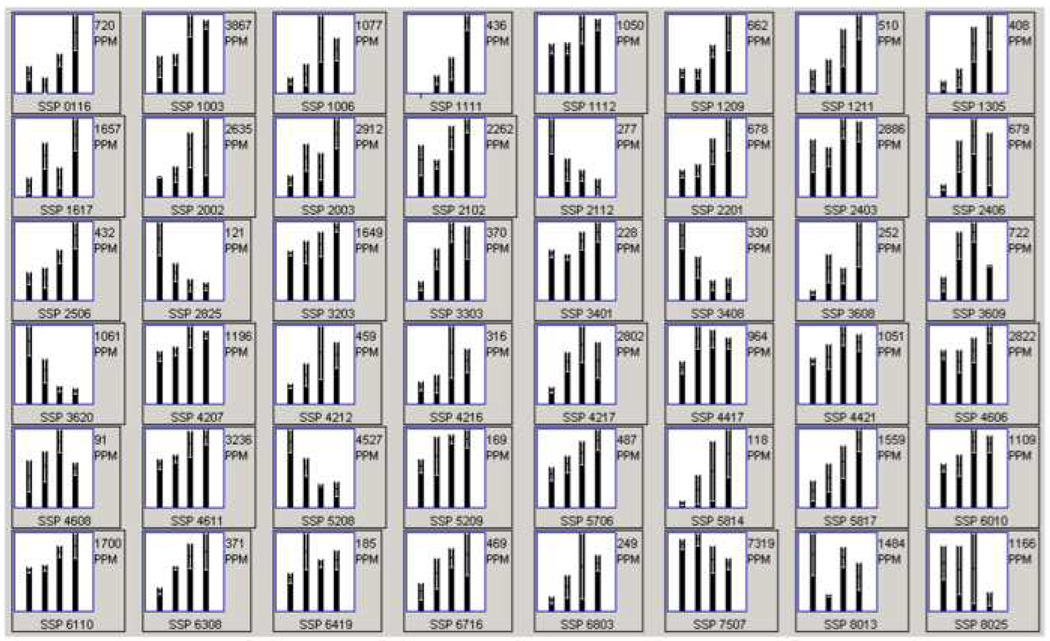
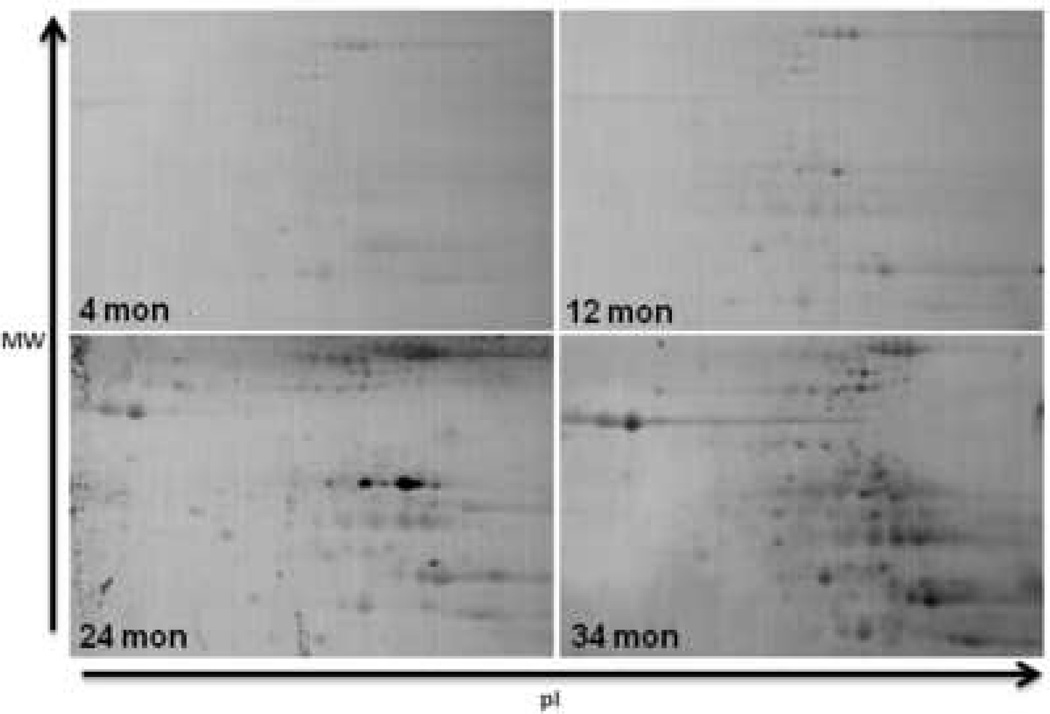
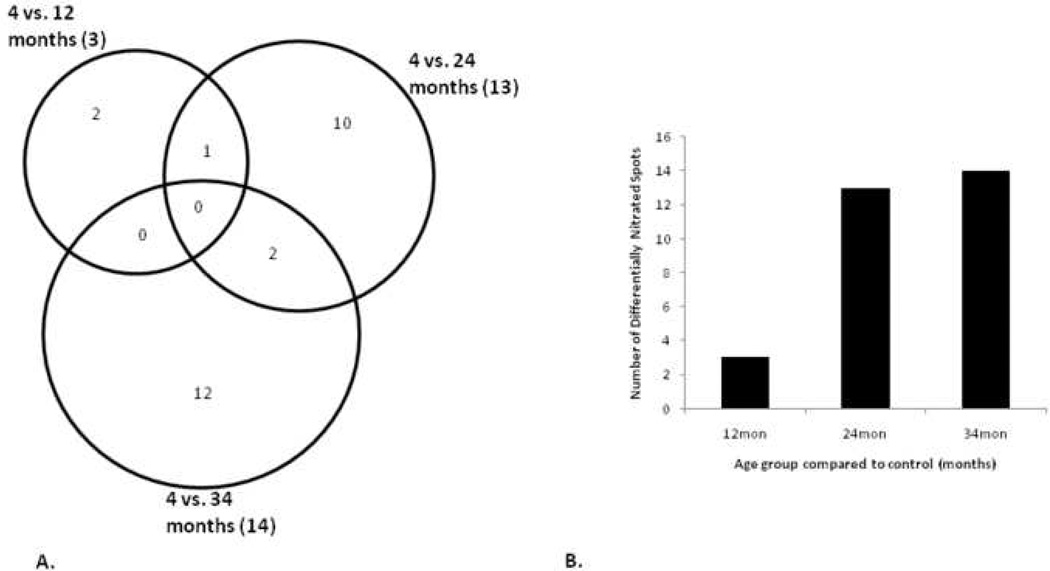
Rat hearts from four different age groups, 4, 12, 24, and 34 month were solubilized and proteins separated by 2-DE. A representative image of these 2D gels is shown in Figure 1. Axes were calibrated based on calculated pI and MW from identified proteins, using the Compute pI/Mw Tool (http://us.expasy.org/tools/pi_tool.html) and from known masses of homologous proteins in a rat ventricular myocardium 2D gel available from the WORLD-2DPAGE List (http://us.expasy.org). On average, 1,010 protein spots were detected, and 73.4% were matched to the master pattern on average. Student’s T-test was used as a preliminary statistical test to determine differential expression candidates via comparisons with control. The results are shown in Figure 2A in a Venn diagram. The union of all three comparisons is 255 protein spots while the intersection contains 21 spots (p ≤ 0.05), 12 of which have been identified by MALDI-TOF PMF and are listed in bold in Table 1. Figure 2B illustrates the strong, significant correlation of differential expression and aging. There was an approximate 2-fold increase in the number of differentially expressed proteins from 12 to 34 months when compared to the 4-month-old controls. The proportion of those proteins that were up-regulated increased with age while the proportion of down-regulated proteins decreased with age. The candidate proteins were then analyzed by ANOVA using Sigma Stat, and those with sufficient staining intensity (>200 PPM) were cut and identified by PMF. Those proteins identified with a sufficient expectation score are listed in Table 1, and their corresponding bar graphs are shown in Figure 3. Each bar represents the mean spot abundance in PPM of that group, where the groups are 4, 12, 24, and 34 months illustrated from left to right, each with standard error bars. Each protein’s average abundance is listed to the right in PPM, and the SSP number, a unique identification number given to each spot by PDQuest, is listed at the bottom of each bar graph and in the same row as the corresponding protein identity in Table 1. Some of the proteins demonstrating progressively increasing or decreasing trends over the rats’ lifespan, such as several of the aforementioned 21 in Figure 2A, are also shown in Figure 1 with their respective bar graphs in the margins. Note that not every SSP number shown in Figure 1 is listed in Table 1 because some proteins could not be identified with a sufficient expectation score.
Figure 1.
Detection of differential protein expression. Shown is a representative Coomassie stained 2D gel of ventricular myocardial proteins. The gel was calibrated for isoelectric point (pI) in the first dimension (X-axis) and molecular weight (kDa) in the second dimension (Y-axis). Individual bars in each graph represent mean spot abundance per group which are 4, 12, 24, and 34 months from left to right. The overall mean spot abundance is listed to the right (PPM) and the spot’s SSP number is listed below each graph.
Figure 2.
Analysis of differential protein expression. A) Venn diagram illustrating the number of differentially expressed protein spots in each age group compared to control (p ≤ 0.05) using Student’s T-test as a preliminary statistical analysis. The totals in each group are listed in parentheses after the two group comparison label while the numbers of individual components are listed separately. B) Histogram where each bar represents the total number of differentially expressed protein spots per two group comparison and the percent that are either up- or down-regulated. The linear regression trend line with respective R2 and p-value illustrates the correlation of differential expression and aging.
Table 1.
Differentially expressed proteins identified by MALDI-TOF PMF. Results of the ANOVA analysis are indicated by arrows in the far right 3 columns representing up- or down-regulation of protein abundance when compared to the young (4-month-old) control group.
| Swiss Prot ID | SSP# | Annotation | pI | Mass | 4→12 | 4→24 | 4→34 |
|---|---|---|---|---|---|---|---|
| Q9Z1N4 | 3401 | 3(2),5-bisphosphate nucleotidase | 5.6 | 33.5 | - | ↑ | ↑ |
| P07953 | 2112 | 6-Phosphofructo-2-KinaseFRUCTOSE-2,6-Bisphosphatase | 5.7 | 22.34 | ↓ | ↓ | ↓ |
| P38918 | 5209 | Aflatoxin Aldehyde Reductase | 6.6 | 37.08 | - | ↑ | ↑ |
| Q5M890 | 1211 | Apolipoprotein N | 5.3 | 28.5 | - | ↑ | ↑ |
| Q80UA1 | 1111 | Beta-1 adducin | 4.9 | 20.9 | - | ↑ | ↑ |
| P02564 | 3620 | Cardiac myosin heavy chain 5 (fragment) | 5.5 | 47.89 | ↓ | ↓ | ↓ |
| Q7TP19 | 5208 | Cb1–812 | 6.8 | 30.53 | ↓ | ↓ | ↓ |
| Q68FW9 | 3608 | COP9 subunit 2 | 5.4 | 51.86 | ↑ | - | ↑ |
| Q6AY36 | 6716 | Coronin, actin binding protein 2A | 7 | 61.59 | - | ↑ | ↑ |
| P09605 | 7507 | Creatine kinase, sarcomeric mitochondrial precursor | 9.2 | 48.87 | - | ↓ | ↓ |
| P48675 | 1617 | Desmin | 5.2 | 53.46 | ↑ | - | ↑ |
| O95173 | 3303 | Dimethylglycine dehydrogenase-like protein | 5.9 | 30.09 | ↑ | ↑ | ↑ |
| P04764 | 4611 | Enolase 1 | 6.7 | 53.3 | - | ↑ | ↑ |
| Q9Z1E1 | 4608 | Flotillin-1 | 6.7 | 47.77 | - | ↑ | - |
| P54311 | 2406 | G protein beta 1 subunit | 5.5 | 38.59 | ↑ | ↑ | - |
| P18645 | 6419 | Galactose-4-epimerase, UDP | 7.9 | 38.55 | ↑ | - | - |
| P04905 | 4216 | Glutathione S-transferase Y-b subunit | 7.9 | 22.04 | - | ↑ | - |
| P26772 | 8013 | Heat Shock Protein 10 | 8.9 | 10.88 | ↓ | - | ↓ |
| P42930 | 2201 | Heat Shock Protein 27 | 6.1 | 22.86 | - | ↑ | ↑ |
| Q641X3 | 5706 | Hexosaminidase A | 6.9 | 61.09 | - | ↑ | ↑ |
| Q9UL88 | 8025 | Immunoglobulin heavy chain variable region | 8.9 | 11.11 | - | - | ↑ |
| Q6IE13 | 4207 | Kallikrein 1 precursor | 6.3 | 26.13 | - | ↑ | ↑ |
| Q64361 | 4212 | Latexin | 5.8 | 26.36 | - | ↑ | ↑ |
| P11884 | 4606 | Mitochondrial aldehyde dehydrogenase precursor | 6.4 | 56.07 | - | - | ↑ |
| RGD:62029 | 3408 | Myosin H (fragment) | 5.4 | 49.48 | ↓ | ↓ | ↓ |
| Q6LEH0 | 1112 | Myosin heavy chain (fragment) | 5.3 | 20.16 | - | ↑ | ↑ |
| P02563 | 2506 | Myosin heavy chain (fragment) | 5.2 | 44.9 | - | ↑ | ↑ |
| P02601 | 116 | Myosin Light Chain 3-f | 4.6 | 16.72 | - | - | ↑ |
| P08733 | 1003 | Myosin regulatory light chain 2 | 4.2 | 12.57 | - | ↑ | ↑ |
| Q63862 | 2825 | Myosin-11 (fragment) | 5.1 | 71.76 | ↓ | ↓ | ↓ |
| P04462 | 4217 | Myosin-8 (fragment) | 6.4 | 29.83 | ↑ | ↑ | ↑ |
| P02625 | 1006 | Parvalbumin | 5 | 12.58 | - | ↑ | ↑ |
| O35244 | 3203 | Peroxiredoxin 6 | 5.6 | 25.61 | - | - | ↑ |
| P31044 | 2102 | Phosphatidylethanolamine binding protein | 5.5 | 20.9 | - | ↑ | ↑ |
| Q63321 | 5814 | Procollagen-lysine, 2-oxoglutarate 5-dioxygenase 1 | 6.3 | 84.16 | - | - | ↑ |
| Q6AYD3 | 4417 | Proliferation-associated 2G4 | 6.4 | 45.75 | ↑ | ↑ | ↑ |
| P09216 | 6803 | Protein kinase C epsilon | 6.6 | 84.93 | - | - | ↑ |
| O35331 | 4421 | Pyridoxal kinase | 6.3 | 35.83 | - | ↑ | ↑ |
| P49432 | 2403 | Pyruvate dehydrogenase (lipoamide) beta | 6.2 | 40.11 | - | ↑ | ↑ |
| P62824 | 1209 | RAB3C | 5.2 | 26.11 | - | ↑ | ↑ |
| Q6PAG9 | 6110 | RBL-NDP kinase 18kDa subunit | 6.9 | 17.88 | - | ↑ | ↑ |
| P60711 | 3609 | Similar to cytoplasmic beta-actin | 5.3 | 43.04 | ↑ | ↑ | - |
| Q6AXM9 | 6308 | Similar to RIKEN cDNA C430008C19 | 6.5 | 23.9 | ↑ | ↑ | ↑ |
| Q5I0J4 | 2002 | Spindle and kinetochore-associated protein 2 | 5.7 | 16.67 | - | ↑ | ↑ |
| Q920L4 | 2003 | TAFIIB | 5.5 | 14.16 | - | - | ↑ |
| P12346 | 5817 | Transferrin | 7 | 78.57 | - | ↑ | ↑ |
| Q5FVG5 | 1305 | Tropomyosin 1 | 4.6 | 33 | - | ↑ | ↑ |
| P62989 | 6010 | Ubiquitin | 6.6 | 8.55 | - | ↑ | ↑ |
Figure 3.
Relative quantitation of differential protein expression. Bar graphs were generated in PDQuest for the differentially expressed proteins listed in Table 1 (ANOVA). The SSP numbers correspond to the protein ID listed in Table 1. Each bar represents the relative mean abundance for one group, which are represented as follows: 4, 12, 24, and 34 months from left to right. The overall mean spot abundance is listed to the right (PPM) and the spot’s unique SSP number is listed below each graph.
3.2 Ontological Analysis
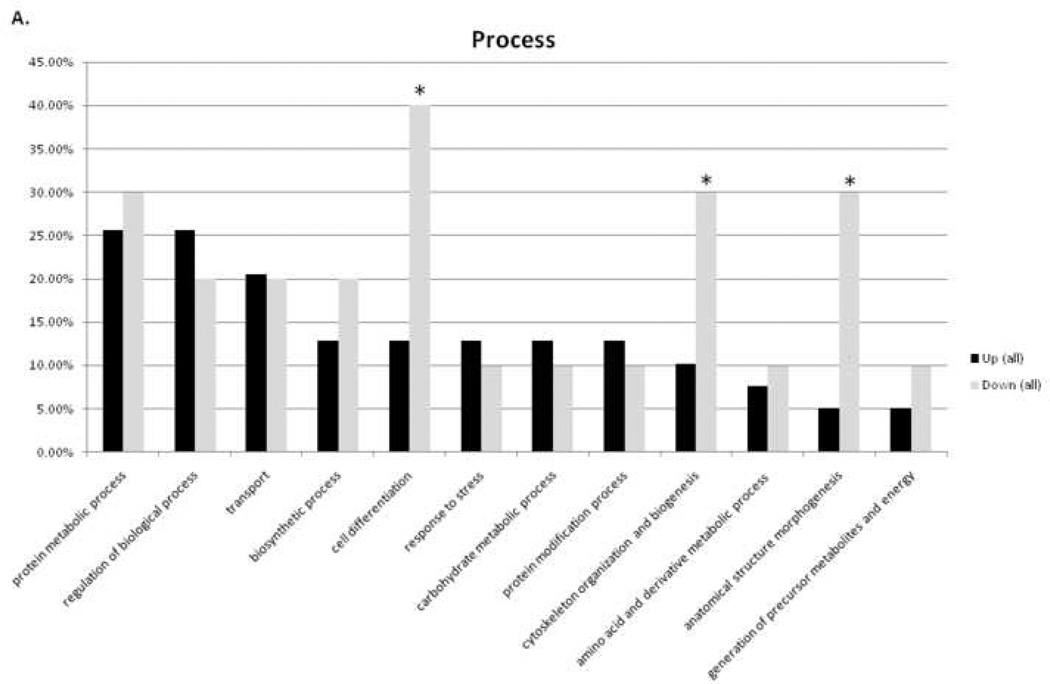
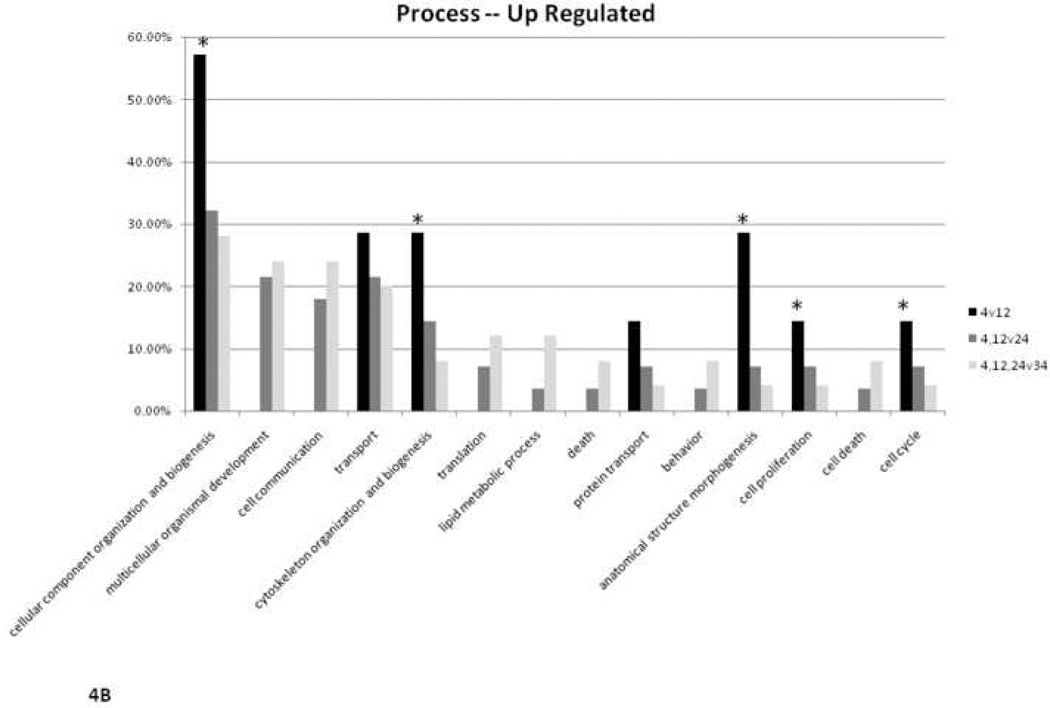
Altered proteins were organized using the Gene Ontology (GO) Term Mapper (http://go.princeton.edu/cgi-bin/GOTermMapper), into groups based on the cellular processes in which they are involved. Figure 4A illustrates the percent of up- and down-regulated proteins that belong to a particular process. The asterisks in the figure highlight the observation that a large portion of the down-regulated proteins are involved in cell differentiation, biogenesis, and morphogenesis. Decreased protein expression in these categories is consistent with an expected smaller role for these processes in an aging, terminally differentiated tissue bed with limited proliferative capacity. Figure 4B illustrates the analysis of up-regulated proteins by age group comparison. Again, the asterisks highlight the expected finding that fewer biogenesis proteins are highly expressed with age. Some 4v12 bars are not present indicating there were no differentially expressed proteins in that two group comparison involved in that process marked below.
Figure 4.
Ontological analysis of cellular processes. A) Percent of up- or down-regulated proteins belonging to particular categories of cellular processes. B) Percent of up-regulated proteins in 12-, 24-, or 34-month-old comparisons belonging to particular categories of cellular processes. * highlights result expected for aging heart (see Discussion section for further explanation).
3.3 Differential Nitrosylation
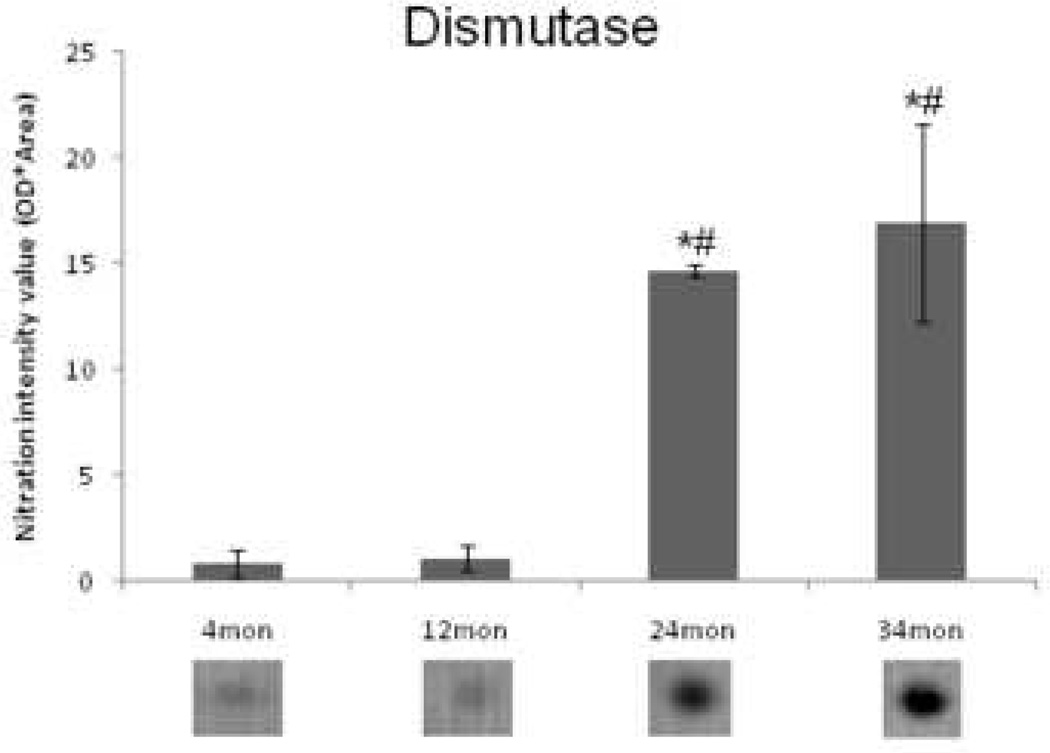
Nitrotyrosine immunoblots from all four age groups were analyzed to determine which proteins had undergone changes in nitration with age. Images of representative blots from all four groups are shown in Figure 5. Most of the proteins that were nitrated were resolved near the basic end of the gel (~pH 6–9) and at a midrange of molecular weight (~30–80kDa). Student’s T-test was used as a preliminary statistical analysis to determine differential modification candidates in pairwise comparisons with control. The results are illustrated in Figure 6A in a Venn diagram. The number of differentially nitrosylated proteins increased with age (Figure 6B), but in an abrupt rather than in a progressive manner as seen with protein expression. All of these proteins exhibited increased nitrosylation compared to controls (data not shown). Corresponding protein spots were located in the Coomassie-stained 2D gel by pattern matching and homologous position and were cut out and identified. Those identified with sufficient expectation scores are listed in Table 2, with upward arrows indicating the protein spots that demonstrated a significant increase (ANOVA) in nitrosylation with age. The spot identified as dismutase was chosen for illustration with an enlarged view of a representative nitrated spot for each group under the corresponding bar in the graph in Figure 7. Dismutase exhibited a significant increase in nitration at 24 and 34 months compared to 4 and 12 months.
Figure 5.
Detection of protein nitration. Representative Western blots from all four groups, 4,12, 24, and 34 months, are shown from rat ventricular myocardium 2D gels that were probed with an anti-3-nitrotyrosine primary antibody. Indicated by arrows are the isoelectric point (pI) in the first dimension (X-axis) and molecular weight (kDa) in the second dimension (Y-axis).
Figure 6.
Analysis of protein nitration. A) Venn diagram illustrating the number of differentially expressed nitrated proteins in each age group compared to control (p ≤ 0.05) using Student’s T-test as a preliminary statistical analysis. The totals in each group are listed in parentheses after the two group comparison label while the numbers of individual components are listed separately. B) Histogram where each bar represents the total number of differentially expressed proteins per two groups illustrating the relationship between aging and nitration in these animals.
Table 2.
Differentially nitrotyrosinylated proteins identified by MALDI-TOF PMF. Each protein in the list demonstrated increases in the mean value of tyrosine nitration when compared to the young (4-month-old) control group, but only those significant by ANOVA analysis are indicated by arrows in the far right 3 columns.
| Swiss Prot ID | SSP# | Annotation | pI | Mass | 4→12 | 4→24 | 4→34 |
|---|---|---|---|---|---|---|---|
| Q9ER34 | 6506 | Aconitase 2, mitochondrial | 8.2 | 88.39 | - | - | - |
| Q5TF02 | 4602 | Adenosine monophosphate deaminase 1 | 6.5 | 89.24 | - | - | - |
| Q64057 | 6318 | Aldehyde dehydrogenase family 6 | 8.9 | 59.59 | - | - | - |
| Q5M7U6 | 4312 | ARP2 actin-related protein 2 homolog | 6.3 | 46.05 | - | ↑ | ↑ |
| O88501 | 4501 | Calpain 6 | 6.9 | 77.18 | - | - | - |
| P07632 | 3006 | Dismutase | 5.9 | 16.11 | - | ↑ | ↑ |
| Q6I7R1 | 4503 | Down-regulated in nephrectomized rat kidney | 6.2 | 68.99 | - | - | - |
| P04764 | 3306 | Enolase 1 protein | 6.7 | 53.26 | - | - | - |
| P04797 | 6202 | Glyceraldehyde-3-phosphate dehydrogenase | 8.7 | 37.24 | - | - | - |
| P83581 | 2503 | Heat Shock Protein 70 | 5.5 | 72.29 | ↑ | ↑ | - |
| Q9ESM0 | 6314 | Inositol hexaphosphate kinase 1 | 6.8 | 51.03 | - | ↑ | ↑ |
| Q80YG1 | 6208 | Phosphoserine aminotransferase | 8.5 | 41.92 | - | - | - |
| Q04631 | 2303 | Protein geranylgeranyltransferase type I, beta subunit | 6.2 | 43.97 | - | ↑ | - |
| Q63477 | 6508 | Protein tyrosine phosphatase epsilon C | 7.2 | 78.91 | - | - | - |
| P52847 | 6209 | Sulfotransferase family cytosolic 1B member 1 | 8.4 | 36.39 | - | ↑ | ↑ |
| P80204 | 3411 | TGF-beta receptor type-1 precursor | 7.3 | 58.22 | - | ↑ | - |
| P81155 | 6210 | Voltage-dependent anion channel 2 | 7.7 | 33.32 | - | - | - |
Figure 7.
Relative quantitation of dismutase nitration. Mean nitration intensity, as measured by PDQuest™ following immunoblot, of dismutase increases significantly in 24 and 34 month old FBN rat hearts compared to 4 and 12 month old rats (1-way ANOVA). * denotes a significant increase from 4 months; # denotes a significant increase from 12 months. Shown below each bar is an enlarged view of a representative immunoblot dismutase protein spot for each group.
4. Discussion
4.1 Differential Protein Expression
The total number of differentially expressed protein spots increased linearly with age compared to the 4-month-old controls, demonstrating that differential expression increases progressively, rather than abruptly, during aging, (Fig. 2). These results are consistent with the well-described functional and structural changes that occur in the aging heart. Furthermore, progressive increases and decreases in individual protein abundance with age, as shown in Figure 3, strongly suggest they are causally related. The relevance of these individual changes with aging is better understood by addressing individual protein function, as detailed categorically below, based on the ontological analyses.
4.2 Apoptosis
Aging is characterized by a loss of cardiomyocytes in both humans (Olivetti et al., 1991) and rats (Anversa et al., 1986; Anversa et al., 1990), a process due, at least in part, to apoptosis (Hacker et al., 2006). We found 12 up-regulated and 2 down-regulated proteins with antiapoptotic properties, and only 1 up-regulated pro-apoptotic protein. The majority of changes therefore appear to be adaptive rather than to facilitate apoptosis. For example, parvalbumin increased at 24 and 34 months, and over-expressing parvalbumin has been reported to delay apoptosis (Tombal et al., 2002). Desmin, a type III intermediate filament protein, also increased with age, and studies have shown that decreased desmin expression results in apoptosis and cardiomyopathy (Kumarapeli and Wang, 2004), implying desmin may facilitate myocyte survival. HSP27 expression increased by 3-fold from 4 to 34 months. HSP27 is known to attenuate certain models of myocardial dysfunction (Liu et al., 2007) and interfere with apoptosis, which is dependent on its ability to control reactive oxygen species (ROS) (Mehlen et al., 1997).
We found that peroxiredoxin 6 (Prx6) increased linearly with age. Overexpressing Prx6 has been shown to protect cells against oxidative stress (Manevich and Fisher, 2005) and reduce the number of apoptotic cells after UV irradiation in keratinocytes (Kumin et al., 2006). Furthermore, certain peroxiredoxins have been shown to have an inhibitory effect on cellular senescence (Han et al., 2005; Han et al., 2006). We also found that the protein kallikrein, a subfamily of serine proteases, increased with age (4 vs. 34 month), and kallikrein gene delivery has been reported to reduce myocardial apoptosis (Agata et al., 2002; Yin et al., 2005). We observed a consistent upregulation of glutathione s-transferase (GST) with age. GST overexpression has been found to reduce both ROS and apoptosis in the H9C2 rat heart tissue cell line (L'Ecuyer et al., 2004). The anti-apoptotic effect of Pyridoxal kinase is inferred because downregulation of the protein induces apoptosis (Schlieper et al., 2007). Thus the upregulation of pyridoxal kinase, as well as others such as mitochondrial aldehyde dehydrogenase, ubiquitin, and protein kinase C epsilon (PKCe), clearly favor cell survival (Chen et al., 2006; Inagaki et al., 2006; Powell, 2006). It is possible that age related susceptibility to oxidative stress may result in increased signals to initiate programmed cell death, and therefore, a global upregulation of cell survival proteins would be favorable to counter these signals and maintain the life of the organism.
4.3 Hypertrophy
Though the heart loses cells with age, the remaining cells enlarge as the heart hypertrophies during aging. In the FBN rat, an increase in the left ventricle mass-to-body-weight ratio is accompanied by a decrease in systolic function (Hacker et al., 2006). Hypertrophic growth is consistent with our finding that anti-apoptotic proteins increase with age, leaving the cells in a pro-growth state. For example, the up-regulated anti-apoptotic protein HSP27 is thought to play a regulatory role in growth of certain tissues through its actin cytoskeleton stabilizing properties (Pantos et al., 2004). Protein Kinase C epsilon (PKCe) increased stepwise with age in our study. Increased PKCe expression and activity is associated with hypertrophy, and the generally accepted notion is that PKCe mediates pathological hypertrophy. However, Wu et al. found that increasing PKCe translocation/activation improved cardiac contractile function in a hypertrophic mouse model indicating that PKCe signaling is a compensatory rather than pathological event (Wu et al., 2000). Desmin previously has been found to be associated with hypertrophy (Wang et al., 2002), but upregulation, even up to 12 fold of its normal mRNA levels, of desmin is not detrimental (Wang et al., 2001). We observed a maximum 6 fold upregulation in protein expression (4 vs. 34 month). Also, proteins such as peroxiredoxin 6, kallikrein, and ubiquitin, were up-regulated with age in our study and have been implicated as potentially important proteins in preventing myocardial hypertrophy (Agata et al., 2002; Matsushima et al., 2006; Okada et al., 2004).
4.4 Fibrosis
Hypertrophy is commonly associated with fibrosis in aging humans and animal models, including the FBN rat (Hacker et al., 2006). Fibrosis is an abnormal accumulation of connective tissue involving structural remodeling of the fibrillar collagen matrix, which leads to compromised elasticity and therefore contributes to diastolic function (Burlew and Weber, 2002). Interestingly, the aging associated increase in collagen is not due to up-regulation of the collagen gene but a decrease in its degradation and increase in cross-linking of mature collagen (Razza, 2005). We did not identify collagen in our 2D gels because collagen is approximately 130 kDa, which is typically too large to enter an 11% acrylamide gel. We did find, however, a stepwise increase, up to 15 fold that of control, in procollagen-lysine,2-oxoglutarate 5-dioxygenase 1 (lysyl hydroxylase 1, or LH1) with age. LH1 forms hydroxylysine sites that are essential for the stability of the intermolecular collagen cross-links. It has been reported that in Fischer 344 rat hearts cross-linking increases approximately 5 fold from 5 to 23 months of age (Thomas et al., 2001). This upregulation may promote stabilization of collagen leading to a decrease in its degradation and contribute to the increase in fibrosis observed in aging. To our knowledge, this is the first report of LH1 upregulation in the aging heart.
Consistent with other observations in this study, we found a number of alterations in protein abundance that may be compensatory for dysfunction. For example, desmin, peroxiredoxin, and kallikrein were all found to be upregulated. Mice null for desmin develop myocardial fibrosis (Fountoulakis et al., 2005), indicating desmin may protect against fibrosis. Peroxiredoxin and kallikrein also have been shown to have protective effects against fibrosis (Agata et al., 2002; Matsushima et al., 2006), which is consistent with their reported protective effects against hypertrophy.
4.5 Diastolic Dysfunction
Diastolic dysfunction is a hallmark of aging in the human heart, occurs independently of disease, but may contribute to the increased incidence of congestive heart failure in the elderly (Lakatta, 1993). Diastolic dysfunction occurs in the FBN rat model of aging as well (Brenner et al., 2001; Hacker et al., 2006; Walker et al., 2006; Wanagat et al., 2002). We identified two proteins with possible roles in diastolic dysfunction, both of which increased in abundance in stepwise fashion from 4 to 34 months: myosin regulatory light chain 2 (MLC-2) and parvalbumin. It has been reported that MLC-2 overexpression causes diastolic dysfunction (Palermo et al., 1995; Swynghedauw, 1996), thus the upregulation of MLC-2 we observed may play some role in impairing relaxation with age. The upregulation of parvalbumin, however, is most likely adaptive to diastolic dysfunction. It has been reported that parvalbumin gene transfer into the myocardium improves diastolic dysfunction in senescent FBN myocytes (Huq et al., 2004). Reuptake of Ca2+ into the sarcoplasmic reticulum (SR) is one of the major determinants of the rate of relaxation in the heart, and it has been reported that aging is accompanied by a prolonged increase in intracytosolic Ca2+ after depolarization which prolongs relaxation (Muscari et al., 1992). Parvalbumin is a calcium binding protein that can function as a Ca2+ sink to facilitate myocardial relaxation (Schmidt et al., 2005). To our knowledge, this is the first report of parvalbumin and MLC-2 upregulation in the heart with aging.
4.6 Nitration
Increases in oxidative stress are associated with an age dependent decline in function of the heart (Sohal, 2002) and have been implicated as a cause of aging (Golden et al., 2002; Sohal, 2002; Sohal and Weindruch, 1996). Tyrosine nitration is a biomarker of oxidative stress, increases in certain cardiac proteins with age (Kanski et al., 2005), and is known to cause protein dysfunction (Beckman, 1996). We confirmed a previously observed increase in the number of proteins modified with age (Fig. 6B); however, this trend appeared more abrupt with most of the changes occurring at 24 and 34 months rather than a gradual increase as seen in the differential expression data (Fig. 2B). This disconnect indicates that nitration may not be the oxidative stress signal that is potentially triggering differential expression. It is possible that protein nitration does not reflect total protein oxidation, which is expected to increase in a gradual manner.
We identified 17 nitrated myocardial proteins, 7 of which exhibited a significant increase in nitration, or nitrosylation, with aging (ANOVA) indicated by upward arrows in Table 2; however it should be noted that a higher degree of certainty awaits confirmation of both identity and post-translational modification of specific peptides by tandem mass spectrometry. We observed that nitration of dismutase, a protein that destroys oxygen radicals produced within the cells, increases at 24 and 34 months (Figure 7). Manganese superoxide dismutase (mnSOD) has been found to be nitrated with age (Kanski et al., 2005) and a single tyrosine residue in the active catalytic sites of mnSOD can inactivate it (MacMillan-Crow et al., 1998; Xu et al., 2006). We also observed an increase in nitration of heat shock protein 70 (HSP70). It has been shown that removing HSP70 might induce cardiac dysfunction and hypertrophy (Kim et al., 2006), thus nitration related HSP70 dysfunction may partially explain age related myocardial dysfunction.
5 Final Remarks
Our results show that differential protein expression increased progressively with aging. This progressive trend was observed not only on a group basis but, with few exceptions, in individual proteins as well. The results of this investigation reveal an adaptive or compensatory response to aging at the protein level. Of those identified altered proteins with known functions in apoptosis, fibrosis, hypertrophy, and diastolic dysfunction, 68% have properties that would compensate rather than mediate dysfunction. Rather than serving as an explanation as to why the aging animals have compromised cardiac function and increased senescence, the data seem to account for their longevity. Animals without these proteomic characteristics may not have survived as long. In fact, the 36 month mean survival age of the FBN rat is longer than most other rat strains, and they have been described as having fewer age-related pathologies (Lipman et al., 1996). Differential nitrosylation also increased with age, a corroboration of previous studies with similar findings. The trend we observed was abrupt as opposed to progressive, indicating nitration may not be the causal factor behind the increase in differential expression. Future work must confirm some of the most prominent age-related changes in protein expression, using Western bloting, immunohistochemistry, and definitive tandem mass spectrometric-based protein identification. In conclusion, proteomic analysis continues to be an extremely useful approach for characterizing changes in protein levels during aging, generating new hypotheses regarding mechanisms that might govern aging, and discovering potential therapeutic targets to mitigate aging-related myocardial dysfunction.
Supplementary Material
Acknowledgements
The authors thank Heather Ringham for her expert technical assistance and Robert Snyder for his critical reading of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Lakatta EG, Levy D. Arterial and cardiac aging: major shareholders in cardiovascular disease enterprises: Part I: aging arteries: a "set up" for vascular disease. Circulation. 2003;107:139–146. doi: 10.1161/01.cir.0000048892.83521.58. [DOI] [PubMed] [Google Scholar]
- Gates PE, Tanaka H, Graves J, Seals DR. Left ventricular structure and diastolic function with human ageing. Relation to habitual exercise and arterial stiffness. Eur Heart J. 2003;24:2213–2220. doi: 10.1016/j.ehj.2003.09.026. [DOI] [PubMed] [Google Scholar]
- Groban L. Diastolic dysfunction in the older heart. J Cardiothorac Vasc Anesth. 2005;19:228–236. doi: 10.1053/j.jvca.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Oxenham H, Sharpe N. Cardiovascular aging and heart failure. Eur J Heart Fail. 2003;5:427–434. doi: 10.1016/s1388-9842(03)00011-4. [DOI] [PubMed] [Google Scholar]
- Barja G. Endogenous oxidative stress: relationship to aging, longevity and caloric restriction. Ageing Res Rev. 2002;1:397–411. doi: 10.1016/s1568-1637(02)00008-9. [DOI] [PubMed] [Google Scholar]
- Sohal RS, Weindruch R. Oxidative stress, caloric restriction, and aging. Science. 1996;273:59–63. doi: 10.1126/science.273.5271.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayflick L. The Limited in Vitro Lifetime of Human Diploid Cell Strains. Exp Cell Res. 1965;37:614–636. doi: 10.1016/0014-4827(65)90211-9. [DOI] [PubMed] [Google Scholar]
- Helfand SL, Rogina B. Regulation of gene expression during aging. Results Probl Cell Differ. 2000;29:67–80. doi: 10.1007/978-3-540-48003-7_4. [DOI] [PubMed] [Google Scholar]
- Harman D. The aging process. Proc Natl Acad Sci U S A. 1981;78:7124–7128. doi: 10.1073/pnas.78.11.7124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine RL, Stadtman ER. Oxidative modification of proteins during aging. Exp Gerontol. 2001;36:1495–1502. doi: 10.1016/s0531-5565(01)00135-8. [DOI] [PubMed] [Google Scholar]
- Candiano G, Bruschi M, Musante L, Santucci L, Ghiggeri GM, Carnemolla B, Orecchia P, Zardi L, Righetti PG. Blue silver: a very sensitive colloidal Coomassie G-250 staining for proteome analysis. Electrophoresis. 2004;25:1327–1333. doi: 10.1002/elps.200305844. [DOI] [PubMed] [Google Scholar]
- Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, Foulger R, Eilbeck K, Lewis S, Marshall B, Mungall C, Richter J, Rubin GM, Blake JA, Bult C, Dolan M, Drabkin H, Eppig JT, Hill DP, Ni L, Ringwald M, Balakrishnan R, Cherry JM, Christie KR, Costanzo MC, Dwight SS, Engel S, Fisk DG, Hirschman JE, Hong EL, Nash RS, Sethuraman A, Theesfeld CL, Botstein D, Dolinski K, Feierbach B, Berardini T, Mundodi S, Rhee SY, Apweiler R, Barrell D, Camon E, Dimmer E, Lee V, Chisholm R, Gaudet P, Kibbe W, Kishore R, Schwarz EM, Sternberg P, Gwinn M, Hannick L, Wortman J, Berriman M, Wood V, de la Cruz N, Tonellato P, Jaiswal P, Seigfried T, White R. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32:D258–D261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivetti G, Melissari M, Capasso JM, Anversa P. Cardiomyopathy of the aging human heart. Myocyte loss and reactive cellular hypertrophy. 1991;68:1560–1568. doi: 10.1161/01.res.68.6.1560. [DOI] [PubMed] [Google Scholar]
- Anversa P, Hiler B, Ricci R, Guideri G, Olivetti G. Myocyte cell loss and myocyte hypertrophy in the aging rat heart. J Am Coll Cardiol. 1986;8:1441–1448. doi: 10.1016/s0735-1097(86)80321-7. [DOI] [PubMed] [Google Scholar]
- Anversa P, Palackal T, Sonnenblick EH, Olivetti G, Meggs LG, Capasso JM. Myocyte cell loss and myocyte cellular hyperplasia in the hypertrophied aging rat heart. Circ Res. 1990;67:871–885. doi: 10.1161/01.res.67.4.871. [DOI] [PubMed] [Google Scholar]
- Hacker TA, McKiernan SH, Douglas PS, Wanagat J, Aiken JM. Age-related changes in cardiac structure and function in Fischer 344 x Brown Norway hybrid rats. Am J Physiol Heart Circ Physiol. 2006;290:H304–H311. doi: 10.1152/ajpheart.00290.2005. [DOI] [PubMed] [Google Scholar]
- Tombal B, Denmeade SR, Gillis JM, Isaacs JT. A supramicromolar elevation of intracellular free calcium ([Ca(2+)](i)) is consistently required to induce the execution phase of apoptosis. Cell Death Differ. 2002;9:561–573. doi: 10.1038/sj.cdd.4400999. [DOI] [PubMed] [Google Scholar]
- Kumarapeli AR, Wang X. Genetic modification of the heart: chaperones and the cytoskeleton. J Mol Cell Cardiol. 2004;37:1097–1109. doi: 10.1016/j.yjmcc.2004.07.004. [DOI] [PubMed] [Google Scholar]
- Liu L, Zhang X, Qian B, Min X, Gao X, Li C, Cheng Y, Huang J. Over-expression of heat shock protein 27 attenuates doxorubicin-induced cardiac dysfunction in mice. Eur J Heart Fail. 2007;9:762–769. doi: 10.1016/j.ejheart.2007.03.007. [DOI] [PubMed] [Google Scholar]
- Mehlen P, Hickey E, Weber LA, Arrigo AP. Large unphosphorylated aggregates as the active form of hsp27 which controls intracellular reactive oxygen species and glutathione levels and generates a protection against TNFalpha in NIH-3T3-ras cells. Biochem Biophys Res Commun. 1997;241:187–192. doi: 10.1006/bbrc.1997.7635. [DOI] [PubMed] [Google Scholar]
- Manevich Y, Fisher AB. Peroxiredoxin 6, a 1-Cys peroxiredoxin, functions in antioxidant defense and lung phospholipid metabolism. Free Radic Biol Med. 2005;38:1422–1432. doi: 10.1016/j.freeradbiomed.2005.02.011. [DOI] [PubMed] [Google Scholar]
- Kumin A, Huber C, Rulicke T, Wolf E, Werner S. Peroxiredoxin 6 is a potent cytoprotective enzyme in the epidermis. Am J Pathol. 2006;169:1194–1205. doi: 10.2353/ajpath.2006.060119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han YH, Kim HS, Kim JM, Kim SK, Yu DY, Moon EY. Inhibitory role of peroxiredoxin II (Prx II) on cellular senescence. FEBS Lett. 2005;579:4897–4902. doi: 10.1016/j.febslet.2005.07.049. [DOI] [PubMed] [Google Scholar]
- Han YH, Kwon JH, Yu DY, Moon EY. Inhibitory effect of peroxiredoxin II (Prx II) on Ras-ERK-NFkappaB pathway in mouse embryonic fibroblast (MEF) senescence. Free Radic Res. 2006;40:1182–1189. doi: 10.1080/10715760600868552. [DOI] [PubMed] [Google Scholar]
- Agata J, Chao L, Chao J. Kallikrein gene delivery improves cardiac reserve and attenuates remodeling after myocardial infarction. Hypertension. 2002;40:653–659. doi: 10.1161/01.hyp.0000036035.41122.99. [DOI] [PubMed] [Google Scholar]
- Yin H, Chao L, Chao J. Kallikrein/kinin protects against myocardial apoptosis after ischemia/reperfusion via Akt-glycogen synthase kinase-3 and Akt-Bad.14-3-3 signaling pathways. J Biol Chem. 2005;280:8022–8030. doi: 10.1074/jbc.M407179200. [DOI] [PubMed] [Google Scholar]
- L'Ecuyer T, Allebban Z, Thomas R, Vander Heide R. Glutathione S-transferase overexpression protects against anthracycline-induced H9C2 cell death. Am J Physiol Heart Circ Physiol. 2004;286:H2057–H2064. doi: 10.1152/ajpheart.00778.2003. [DOI] [PubMed] [Google Scholar]
- Schlieper A, Anwar M, Heger J, Piper HM, Euler G. Repression of anti-apoptotic genes via AP-1 as a mechanism of apoptosis induction in ventricular cardiomyocytes. Pflugers Arch. 2007;454:53–61. doi: 10.1007/s00424-006-0180-4. [DOI] [PubMed] [Google Scholar]
- Chen T, Cui J, Liang Y, Xin X, Owen Young D, Chen C, Shen P. Identification of human liver mitochondrial aldehyde dehydrogenase as a potential target for microcystin-LR. Toxicology. 2006;220:71–80. doi: 10.1016/j.tox.2005.12.001. [DOI] [PubMed] [Google Scholar]
- Inagaki K, Churchill E, Mochly-Rosen D. Epsilon protein kinase C as a potential therapeutic target for the ischemic heart. Cardiovasc Res. 2006;70:222–230. doi: 10.1016/j.cardiores.2006.02.015. [DOI] [PubMed] [Google Scholar]
- Powell SR. The ubiquitin-proteasome system in cardiac physiology and pathology. Am J Physiol Heart Circ Physiol. 2006;291:H1–H19. doi: 10.1152/ajpheart.00062.2006. [DOI] [PubMed] [Google Scholar]
- Pantos C, Malliopoulou V, Varonos DD, Cokkinos DV. Thyroid hormone and phenotypes of cardioprotection. Basic Res Cardiol. 2004;99:101–120. doi: 10.1007/s00395-003-0449-0. [DOI] [PubMed] [Google Scholar]
- Wu G, Toyokawa T, Hahn H, Dorn GW., 2nd Epsilon protein kinase C in pathological myocardial hypertrophy. Analysis by combined transgenic expression of translocation modifiers and Galphaq. J Biol Chem. 2000;275:29927–29930. doi: 10.1074/jbc.C000380200. [DOI] [PubMed] [Google Scholar]
- Wang X, Osinska H, Gerdes AM, Robbins J. Desmin filaments and cardiac disease: establishing causality. J Card Fail. 2002;8:S287–S292. doi: 10.1054/jcaf.2002.129279. [DOI] [PubMed] [Google Scholar]
- Wang X, Osinska H, Dorn GW, 2nd, Nieman M, Lorenz JN, Gerdes AM, Witt S, Kimball T, Gulick J, Robbins J. Mouse model of desmin-related cardiomyopathy. Circulation. 2001;103:2402–2407. doi: 10.1161/01.cir.103.19.2402. [DOI] [PubMed] [Google Scholar]
- Matsushima S, Ide T, Yamato M, Matsusaka H, Hattori F, Ikeuchi M, Kubota T, Sunagawa K, Hasegawa Y, Kurihara T, Oikawa S, Kinugawa S, Tsutsui H. Overexpression of mitochondrial peroxiredoxin-3 prevents left ventricular remodeling and failure after myocardial infarction in mice. Circulation. 2006;113:1779–1786. doi: 10.1161/CIRCULATIONAHA.105.582239. [DOI] [PubMed] [Google Scholar]
- Okada K, Minamino T, Tsukamoto Y, Liao Y, Tsukamoto O, Takashima S, Hirata A, Fujita M, Nagamachi Y, Nakatani T, Yutani C, Ozawa K, Ogawa S, Tomoike H, Hori M, Kitakaze M. Prolonged endoplasmic reticulum stress in hypertrophic and failing heart after aortic constriction: possible contribution of endoplasmic reticulum stress to cardiac myocyte apoptosis. Circulation. 2004;110:705–712. doi: 10.1161/01.CIR.0000137836.95625.D4. [DOI] [PubMed] [Google Scholar]
- Burlew BS, Weber KT. Cardiac fibrosis as a cause of diastolic dysfunction. Herz. 2002;27:92–98. doi: 10.1007/s00059-002-2354-y. [DOI] [PubMed] [Google Scholar]
- Razza MS, editor. Fibrogenesis: Cellular and Molecular Basis. Plenum Publishers; 2005. [Google Scholar]
- Thomas DP, Cotter TA, Li X, McCormick RJ, Gosselin LE. Exercise training attenuates aging-associated increases in collagen and collagen crosslinking of the left but not the right ventricle in the rat. Eur J Appl Physiol. 2001;85:164–169. doi: 10.1007/s004210100447. [DOI] [PubMed] [Google Scholar]
- Fountoulakis M, Soumaka E, Rapti K, Mavroidis M, Tsangaris G, Maris A, Weisleder N, Capetanaki Y. Alterations in the heart mitochondrial proteome in a desmin null heart failure model. J Mol Cell Cardiol. 2005;38:461–474. doi: 10.1016/j.yjmcc.2004.12.008. [DOI] [PubMed] [Google Scholar]
- Lakatta EG. Cardiovascular regulatory mechanisms in advanced age. Physiol Rev. 1993;73:413–467. doi: 10.1152/physrev.1993.73.2.413. [DOI] [PubMed] [Google Scholar]
- Brenner DA, Apstein CS, Saupe KW. Exercise training attenuates age-associated diastolic dysfunction in rats. Circulation. 2001;104:221–226. doi: 10.1161/01.cir.104.2.221. [DOI] [PubMed] [Google Scholar]
- Walker EM, Jr, Nillas MS, Mangiarua EI, Cansino S, Morrison RG, Perdue RR, Triest WE, Wright GL, Studeny M, Wehner P, Rice KM, Blough ER. Age-associated changes in hearts of male Fischer 344/Brown Norway F1 rats. Ann Clin Lab Sci. 2006;36:427–438. [PubMed] [Google Scholar]
- Wanagat J, Wolff MR, Aiken JM. Age-associated changes in function, structure and mitochondrial genetic and enzymatic abnormalities in the Fischer 344 x Brown Norway F(1) hybrid rat heart. J Mol Cell Cardiol. 2002;34:17–28. doi: 10.1006/jmcc.2001.1483. [DOI] [PubMed] [Google Scholar]
- Palermo J, Gulick J, Ng W, Grupp IL, Grupp G, Robbins J. Remodeling the mammalian heart using transgenesis. Cell Mol Biol Res. 1995;41:501–509. [PubMed] [Google Scholar]
- Swynghedauw B. Transgenic Models of Myocardial Dysfunction. Heart Failure Reviews. 1996;1:277–290. [Google Scholar]
- Huq F, Lebeche D, Iyer V, Liao R, Hajjar RJ. Gene transfer of parvalbumin improves diastolic dysfunction in senescent myocytes. Circulation. 2004;109:2780–2785. doi: 10.1161/01.CIR.0000131764.62242.96. [DOI] [PubMed] [Google Scholar]
- Muscari C, Caldarera I, Rapezzi C, Branzi A, Caldarera CM. Biochemical correlates with myocardial aging. Cardioscience. 1992;3:67–78. [PubMed] [Google Scholar]
- Schmidt U, Zhu X, Lebeche D, Huq F, Guerrero JL, Hajjar RJ. In vivo gene transfer of parvalbumin improves diastolic function in aged rat hearts. Cardiovasc Res. 2005;66:318–323. doi: 10.1016/j.cardiores.2004.06.028. [DOI] [PubMed] [Google Scholar]
- Sohal RS. Role of oxidative stress and protein oxidation in the aging process. Free Radic Biol Med. 2002;33:37–44. doi: 10.1016/s0891-5849(02)00856-0. [DOI] [PubMed] [Google Scholar]
- Golden TR, Hinerfeld DA, Melov S. Oxidative stress and aging: beyond correlation. Aging Cell. 2002;1:117–123. doi: 10.1046/j.1474-9728.2002.00015.x. [DOI] [PubMed] [Google Scholar]
- Kanski J, Behring A, Pelling J, Schoneich C. Proteomic identification of 3-nitrotyrosine-containing rat cardiac proteins: effects of biological aging. Am J Physiol Heart Circ Physiol. 2005;288:H371–H381. doi: 10.1152/ajpheart.01030.2003. [DOI] [PubMed] [Google Scholar]
- Beckman JS. Oxidative damage and tyrosine nitration from peroxynitrite. Chem Res Toxicol. 1996;9:836–844. doi: 10.1021/tx9501445. [DOI] [PubMed] [Google Scholar]
- MacMillan-Crow LA, Crow JP, Thompson JA. Peroxynitrite-mediated inactivation of manganese superoxide dismutase involves nitration and oxidation of critical tyrosine residues. Biochemistry. 1998;37:1613–1622. doi: 10.1021/bi971894b. [DOI] [PubMed] [Google Scholar]
- Xu S, Ying J, Jiang B, Guo W, Adachi T, Sharov V, Lazar H, Menzoian J, Knyushko TV, Bigelow D, Schoneich C, Cohen RA. Detection of sequence-specific tyrosine nitration of manganese SOD and SERCA in cardiovascular disease and aging. Am J Physiol Heart Circ Physiol. 2006;290:H2220–H2227. doi: 10.1152/ajpheart.01293.2005. [DOI] [PubMed] [Google Scholar]
- Kim YK, Suarez J, Hu Y, McDonough PM, Boer C, Dix DJ, Dillmann WH. Deletion of the inducible 70-kDa heat shock protein genes in mice impairs cardiac contractile function and calcium handling associated with hypertrophy. Circulation. 2006;113:2589–2597. doi: 10.1161/CIRCULATIONAHA.105.598409. [DOI] [PubMed] [Google Scholar]
- Lipman RD, Chrisp CE, Hazzard DG, Bronson RT. Pathologic characterization of brown Norway, brown Norway x Fischer 344, and Fischer 344 x brown Norway rats with relation to age. J Gerontol A Biol Sci Med Sci. 1996;51:B54–B59. doi: 10.1093/gerona/51A.1.B54. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.