Abstract
Formulations of antioxidant enzymes, superoxide dismutase 1 (SOD1, also known as Cu/Zn SOD) and catalase were prepared by electrostatic coupling of enzymes with cationic block copolymers, polyethyleneimine-poly(ethylene glycol) or poly(L-lysine)-poly(ethylene glycol), followed by covalent cross-linking to stabilize nanoparticles. Different cross-linking strategies (using glutaraldehyde, bis-(sulfosuccinimidyl)suberate sodium salt or 1-Ethyl-3-[3-dimethylaminopropyl]carbodiimide hydrochloride with N-hydroxysulfosuccinimide) and reaction conditions (pH and polycation/protein charge ratio) were investigated that allowed immobilizing active enzymes in cross-linked nanoparticles, termed “nanozymes”. Bi-enzyme nanoparticles, containing both SOD1 and catalase were also formulated. Formation of complexes was confirmed using denaturing gel electrophoresis and western blotting and physicochemical characterization was conducted using dynamic light scattering and atomic force microscopy. In vivo studies of 125I-labeled SOD1-containing nanozymes in mice demonstrated its increased stability in both blood and brain and increased accumulation in brain tissues, compared to non-cross-linked complexes and native SOD1. Future studies will evaluate potential of these formulations for delivery of antioxidant enzymes to central nervous system to attenuate oxidative stress associated with neurological diseases.
Keywords: polyion complexes, nanozymes, cross-linking, brain delivery
Background
Many central nervous system (CNS) diseases (i.e., HIV-associated dementia,1-3 multiple sclerosis, cerebral palsy, schizophrenia,4 Parkinson's and Alzheimer's diseases,5-7 and stroke8,9) have a common inflammatory component, which involves neurodegeneration10 and subsequent microglia activation accompanied by excessive production of reactive oxygen species (ROS). ROS damage cellular components and further contribute to neuronal cell death. Efficient transport of antioxidant therapeutics across the blood brain barrier (BBB) can considerably decrease the amount of ROS in the brain, and subsequently attenuate neuroinflammation and produce neuroprotection in patients with CNS disorders.11 Two antioxidant enzymes, SOD1 and catalase are known to be very effective scavengers of ROS. SOD1 catalyzes dismutation of superoxide radical to oxygen and hydrogen peroxide.12 Catalase catalyzes the decomposition of hydrogen peroxide to water and is one of the fastest known biocatalysts with highest turnover numbers of all known enzymes (40,000,000 molecules/second).13 Therefore, these enzymes alone or in combination could protect neuronal cells from ROS-induced damage and have a potential to improve the outcome of therapies for CNS disorders.
Synthetic SOD/catalase mimetics, EUK-189 and EUK-207, were shown to provide neuroprotection under acute ischemic conditions in the rat brain by elimination of free radicals.14 A few clinical studies evaluated efficacy of α-tocopherol on the rate of progression of Parkinson's disease.15 However, most studies failed to show significant improvements in part, perhaps, due to restricted transport of such agents across the BBB.16 Another limitation to the use of these enzymes as therapeutics is their rapid elimination from circulation and inactivation by proteases present in the body. Attempts to overcome these difficulties included modification with polyethylene glycol (PEG) (PEGylation)17 or incorporation into polymeric nanoparticles with PEG corona18-21 that resulted in prolonged circulation and increased stability of enzymes in the body.22 Unfortunately, encapsulation of enzymes into nanoparticles often resulted in their inactivation and/or unsatisfactory loading20,23 and moreover, PEGylation was shown to drastically decrease permeability of SOD1 across the brain microvessels.24
Here, we develop an alternate approach based on incorporation of these enzymes in polyion complexes with oppositely charged block copolymers. This approach was previously used by us and others to immobilize various biopolymers in micelle-like, core-shell particles with high colloidal stability.25-29 In particular, Kataoka and coworkers formulated positively-charged lysozyme with negatively-charged poly(ethylene glycol)-poly(α,β-aspartic acid) block copolymer. Noteworthy, the activity of lysozyme incorporated in such complex was higher compared to the native enzyme26,27 and it was hypothesized that the elevated enzymatic activity is a result of substrate concentration in the polyion complexes species26 and/or alterations in binding mode between lysozyme and substrate.27 Incorporation of an antioxidant enzyme, catalase, into a similar complex with polyethyleneimine-poly(ethylene glycol) (PEI-PEG) was previously described by us as a strategy for cell-mediated delivery of catalase to the brain.28
We now report preparation of polyion complexes formed by antioxidant enzymes (SOD1, catalase) and cationic block copolymers (PEI-PEG or poly(L-lysine)-poly(ethylene glycol) (pLL10-PEG and pLL50-PEG), followed by covalent cross-linking between polymer and enzyme. The resulting formulations contain a core of enzyme-polyelectrolyte complex/conjugate surrounded by a PEG shell (Figure 1). The block copolymer provides protection for the encapsulated enzyme, which nevertheless remains capable of ROS decomposition without the need for enzyme release from nanoparticles. In addition, bi-enzyme antioxidant system comprising of both SOD1 and catalase was encapsulated into the same polyion complex. Effect of various compositions of enzyme-polyion complexes on physicochemical characteristics (size, charge, and morphology), enzyme activity, in vitro accumulation and cytotoxicity, and in vivo behavior (stability and brain delivery) in mice were investigated. Based on the results we posit that incorporation of antioxidant enzymes into nanozymes may improve transport of active enzymes to the brain.
Figure 1. Schematic representation of polyion complexes.
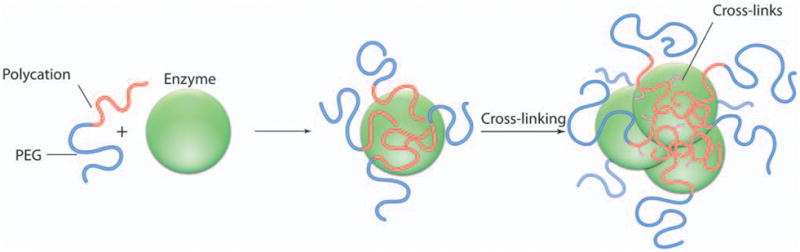
Complexes spontaneously form in aqueous solution as a result of electrostatic coupling of the enzyme and cationic block copolymer. Although only one protein globule is schematically shown here, the polyion complex may contain several protein globules. Cross-linker was added to pre-formed complexes that resulted in covalent stabilization.
Methods
Materials
Copper/Zinc superoxide dismutase (SOD1) from bovine erythrocytes, Bis-(sulfosuccinimidyl)suberate sodium salt (BS3), 1-Ethyl-3-[3-dimethylaminopropyl]carbodiimide Hydrochloride (EDC), N-hydroxysulfosuccinimide (S-NHS), glutaraldehyde (GA), pyrogallol, sodium dodecylsulfate (SDS), sodium borohydride (NaBH4), diethylene triamine pentaacetic acid (DTPA), Tris base, phosphate-buffered saline (PBS), dextran, trichloroacetic acid (TCA), bovine serum albumin (BSA), 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid) (HEPES), Triton X-100 and normal horse serum were purchased from Sigma-Aldrich (St. Louis, MO). Recombinant human SOD1 was kindly provided by Recsod, Russia. Catalase (bovine liver) was purchased from Sigma or Calbiochem (San Diego, CA). Polyethyleneimine-poly(ethylene glycol) (PEI2K-PEG10K) was synthesized as described previously.28 PLL-PEG with two different lengths of pLL (10 and 50 units – 1.6 and 8.2 kDa respectively); pLL10-PEG5K and pLL50-PEG5K were purchased from Alamanda Polymers Inc. (Madison, AL). The molecular masses and polydispersity indices (numbers in parentheses) of the copolymers were approximately 12 kDa (1.93), 6.6 kDa (1.13) and 13.2 kDa (1.12) for PEI-PEG, pLL10-PEG and pLL50-PEG, respectively. Na125I was purchased from PerkinElmer Life (Boston, MA). CATH.a neuronal cell line (CRL-11179™) was obtained from American Type Culture Collection (Manassas, VA). Fetal bovine serum (FBS) was purchased from Invitrogen (Carlsbad, CA). CellTiter® 96 aqueous cell proliferation assay was obtained from Promega (Madison, WI). All other chemicals were obtained from Fisher Scientific unless otherwise noted.
Cell culture
CATH.a cells were cultured in RPMI 1640 medium containing 4 mM L-alanyl-L-glutamine supplemented with 8% normal horse serum and 4% FBS at 37 °C in 5% CO2 atmosphere.
Preparation of nanozymes
Enzyme-polyelectrolyte complexes were prepared by mixing block copolymers (PEI-PEG or pLL-PEG) and enzymes (SOD1, catalase or both, as indicated). Briefly, pre-calculated amounts of the enzyme and the block copolymer were separately dissolved in 50 mM phosphate buffer, phosphate-buffered saline (PBS), 10 mM HEPES buffer or 10 mM HEPES-buffered saline (pH 7.4) at room temperature (RT). A solution of the block copolymer was added drop-by-drop to the enzyme solution and the mixture was gently vortexed during the entire addition. Complexes were allowed to sit at RT for 30 min before use in experiments. The charge ratio Z (+/-) was calculated as a ratio of concentration of amino groups in the block copolymer protonated at pH 7.4 (reported for PEI-PEG30 or as indicated by supplier for pLL10-PEG and pLL50-PEG) to the concentration of −COOH groups from glutamic acid and aspartic acid residues in the enzyme (estimated using Protein Calculator v3.3 software).
To further stabilize these complexes, we explored various cross-linking strategies to covalently link carboxyl- and/or amino groups of the enzyme to the amino groups of polycations. Cross-linking of pre-formed complexes was carried out using GA/NaBH4, BS3 or EDC/S-NHS. A list of nanozymes is presented in Table 1.
Table 1. Description of nanozymes.
| Sample ID | Sample | Block copolymer | Z | Cross-linker |
|---|---|---|---|---|
| nS | SOD1 (native enzyme) | - | - | - |
| S1 | non-cl* | PEI-PEG | 1 | - |
| S2 | cl† | PEI-PEG | 1 | GA |
| S3 | cl | PEI-PEG | 1 | BS3 |
| S4 | cl | PEI-PEG | 1 | EDC/S-NHS |
| S5 | cl | pLL50-PEG | 1 | EDC/S-NHS |
| S6 | non-cl | pLL10-PEG | 10 | EDC/S-NHS |
| S7 | cl | pLL10-PEG | 1 | EDC/S-NHS |
| S8 | cl | pLL10-PEG | 10 | EDC/S-NHS |
| nC | catalase (native enzyme) | none | - | - |
| C1 | non-cl | PEI-PEG | 1 | - |
| C2 | cl | PEI-PEG | 1 | GA |
| C3 | cl | PEI-PEG | 1 | BS3 |
| C4 | cl | PEI-PEG | 1 | EDC/S-NHS |
| C5 | cl | pLL50-PEG | 1 | EDC/S-NHS |
| C6 | cl | pLL50-PEG | 2 | EDC/S-NHS |
| C7 | cl | pLL50-PEG | 5 | EDC/S-NHS |
| C8 | non-cl | pLL10-PEG | 1 | - |
| C9 | cl | pLL10-PEG | 1 | EDC/S-NHS |
| SC1 | SOD1-catalase; non-cl | PEI-PEG | 1 | - |
| SC2 | SOD1-catalase; cl | PEI-PEG | 1 | GA |
| SC3 | SOD1-catalase; cl | PEI-PEG | 1 | BS3 |
| SC4 | SOD1-catalase; cl | PEI-PEG | 1 | EDC/S-NHS |
| SC5 | SOD1-catalase; non-cl | pLL10-PEG | 1 | - |
| SC6 | SOD1-catalase; cl | pLL10-PEG | 1 | BS3 |
| SC7 | SOD1-catalase; cl | pLL10-PEG | 1 | EDC/S-NHS |
| SC8 | SOD1-catalase; cl | pLL50-PEG | 1 | EDC/S-NHS |
non-cross-linked (non-cl)
cross-linked (cl)
In case of GA/NaBH4 and BS3, cross-linker excess was defined as the molar ratio of GA or BS3 to the total number of amino groups of polymer and enzyme (0.5-5 mM NaBH4 was added for 15-60 min after overnight incubation of the reaction mixture containing 2 to 100-fold GA excess). In case of EDC/S-NHS, it was defined as the molar ratio of EDC to the total number of carboxylic groups of the enzyme (5-10 mM S-NHS was added 5 min after EDC addition). Cross-linker excesses were varied from 2 to 100-fold as indicated. All mono enzyme samples were prepared in PBS, pH 7.4. In case of “bi-enzyme” samples, pH of the reaction buffer was varied from 5.2 to 7.4. Reaction mixtures were incubated overnight at 4 °C, and unreacted reagents were removed by desalting using NAP™-10 columns (GE Healthcare Bio-Sciences Corp. Piscataway, NJ). Wherever indicated, samples were filtered using a 20 nm (SOD1 nanozymes) or 100 nm (catalase nanozymes) pore size filter prior to use in experiments.
Electrophoretic Retention
The cross-linking of enzyme-polyion complexes was confirmed by polyacrylamide gel shift assay. Samples were loaded on a 10% polyacrylamide gel under denaturing conditions. Protein bands were visualized with sheep polyclonal anti-SOD (Calbiochem, San Diego, CA) for SOD1, and rabbit polyclonal anti-catalase (Ab 1877, Abcam Inc, Cambridge, MA) for catalase, and secondary horseradish peroxidase anti-rabbit Ig antibody (Amersham Life Sciences, Cleveland, OH).
Enzyme Activity
Enzyme activity in samples was measured using pyrogallol autoxidation followed by superoxide radical dismutation (for SOD1) and hydrogen peroxide decomposition (for catalase) methods, respectively (See SUPPLEMENTARY SECTION for complete details).
Dynamic Light Scattering (DLS)
Effective hydrodynamic diameter and zeta-potential (ζ-potential) of nanozymes was measured by photon correlation spectroscopy using Zetasizer Nano ZS (Malvern Instruments Ltd, UK) in a thermostatic cell at a scattering angle of 173° using previously described methods.31,32
Atomic Force Microscopy (AFM)
Five μL of an aqueous dispersion (ca. 0.01 mg/mL) was deposited onto positively-charged aminopropylytriethoxy silane (APS) mica surface33-35 for 2 min, washed with water and dried under argon atmosphere. AFM imaging was performed using a Multimode NanoScope IV system (Veeco, Santa Barbara, CA) operated in tapping mode. Particle widths and heights were estimated using Femtoscan software (Advanced Technologies Center, Russia).
Preparation of 125I-labeled SOD1
SOD1 was labeled with Na125I using IodoBEADS Iodination reagent (Pierce Biotechnology, Rockford, IL). Briefly, Na125I (500 μCi) was pre-incubated with IodoBEADS for 5 min, and then added to pre-calculated amount of SOD1 solution. The reaction mixture was incubated at RT for 15 min and purified on a NAP™-10 column. 125I-labeled-SOD1 nanozymes were prepared as described earlier.
Cellular Accumulation
Accumulation of various nanozymes in bovine brain microvessel endothelial cells (BBMEC) was evaluated as described earlier.36 Briefly, confluent cell monolayers grown on 24-well plates were pre-treated with assay buffer for 30 min at 37 °C and then incubated with 125I-labeled nanozymes for 1 h. Cells were then washed with ice-cold PBS, and solubilized in 1% Triton X-100 for subsequent determination of radioactivity counts (counts per minute; CPM) in a gamma counter. The cellular accumulation was normalized to cellular protein content determined by Pierce® BCA Protein Assay. Results are reported as average ± standard error mean (SEM) of quadruplicate samples.
Cytotoxicity Evaluation
Cytotoxicity of selected nanozymes was determined using the CellTiter® 96 Aqueous Cell Proliferation Assay (MTS). Twenty-thousand cells seeded in a 96-well plate 24 h before the experiment was incubated with increasing concentration of the respective samples in 100 μL of complete culture medium. The medium was removed after 24 h and replaced with a mixture of 100 μL of fresh RPMI and 20 μL of MTS reagent solution. The cells were incubated for 1 h at 37 °C in a CO2 incubator. The absorbance of each sample was then measured at 490 nm to determine cell viability. The results are expressed as the mean percentage cell viability relative to untreated cells ± SEM.
Animals
Male Balb/c mice (Charles River Laboratories, USA) 6-8 weeks old were used in the study. All procedures were approved by the Institutional Animal Care and Use Committee of the University of Nebraska Medical Center. Animals were housed and humanely handled in accordance with the Principles of Animal Care outlined by National Institutes of Health.
In Vivo Studies
Male Balb/c mice (8 weeks old) were anesthetized with a cocktail of ketamine (80 mg/kg) and xylazine (5 mg/kg) administered intraperitonially. Mice were injected with either native 125I-SOD1, non-cross-linked 125I-SOD1/pLL10-PEG nanozyme or cross-linked 125I-SOD1/pLL10-PEG. The nanozymes were prepared at Z = 10 and the cross-linked using 5-fold EDC excess (S-NHS: 5 mM) as described above. Nanozymes in saline were administered intravenously (i.v.) via tail vein in a 100 μL volume (1 mg SOD1/mL). One hour after injection, the abdomen and rib cage were opened and blood was collected from the heart. Twenty mL of lactated ringer's solution (LR) (B Braun Medical Inc., Irvine, CA) was then perfused through the left ventricle of the heart. Whole brains were dissected and weighed and the levels of radioactivity were counted in a gamma counter.
TCA Precipitation
Acid precipitation method was used to estimate stability of SOD1 by following 125I label degradation.37 Brain and serum samples were placed in an ice-cold glass homogenizer. The brain was homogenized by 10 vertical strokes in 1% w/v BSA acidified LR (pH 4.0). Homogenates were centrifuged at 4000 ×g for 25 min at 4 °C. The resulting supernatant (soluble fraction) and blood serum samples were precipitated with 30% v/v of TCA and centrifuged at 4000×g for 35 min. Radioactive counts (CPM) of resulting supernatant and precipitate (pellet) correspond to free 125I label and enzyme-bound 125I, respectively. Stability of nanozymes was assessed as a ratio of enzyme-bound 125I (pellet CPM) to overall 125I radioactivity (supernatant CPM + pellet CPM). The results obtained in these experiments represent average quantities of SOD1 in each sample/fraction without discrimination of its interstitial or intercellular localization in the organs.
Capillary Depletion
Male Balb/c mice were anesthetized as described earlier. Animals were treated and brain and serum samples were collected. The brain was homogenized by 10 vertical strokes in 0.8 mL of physiological buffer (10 mM HEPES, 141 mM NaCl, 4 mM KCl, 2.8 mM CaCl2, 1 mM MgSO4, 1 mM NaH2PO4, and 10 mM D-Glucose; pH 7.4). Dextran solution (1.6 mL of a 26% solution) was added to the homogenate, mixed, and homogenized with 3 additional vertical strokes. The homogenate was centrifuged at 4000×g for 25 min at 4 °C. The resulting supernatant (brain parenchymal fraction) and pellet (capillary fraction) were separated. The levels of radioactivity in the capillary and brain parenchymal fractions were counted in a gamma counter. The results are expressed as a compartment (parenchyma or capillaries)/serum ratio. Initial radioactivity values obtained in this experiment correspond to overall accumulation of complexes in the brain compartment that includes both 125I-labeled complexes as well as free label that dissociated from the nanoparticles. In order to quantify the presence of intact complexes that retain their structure in the brain compartments, initial radioactivity values obtained in capillary depletion experiment were multiplied by a ratio of enzyme-bound 125I (pellet CPM) to overall 125I radioactivity (supernatant CPM + pellet CPM) obtained in the acid precipitation experiment.
Statistical Analysis
All data are presented as the average ± SEM (n=4 or 5). Tests for significant differences between the groups were done using one-way ANOVA with multiple comparisons (Kruskal-Wallis) using GraphPad Prism 4.0 (GraphPad software, San Diego, CA). A minimum p value of 0.05 was set as the significance level in all cases.
Results
Preparation of cross-linked nanozymes
The polyion complexes were prepared by simple mixing of aqueous solutions of corresponding enzymes (SOD1 or catalase, or both) and block copolymers (PEI-PEG or pLL-PEG). Both SOD1 and catalase are negatively charged under physiological conditions (pI values are 4.95 and 5.8 for SOD1 and catalase, respectively). Unless stated otherwise, polyion complexes were prepared at pH 7.4 that favored electrostatic coupling of the enzyme and the block copolymers. Cross-linking of polyion complexes using GA/NaBH4, BS3, or EDC/S-NHS were done as described earlier.
Electrophoretic retention
The formation of cross-linked polyion complexes was confirmed by denaturing gel electrophoresis. Both enzymes are oligomeric proteins and dissociated under denaturing conditions into individual subunits of 16 kDa (SOD1) and 60 kDa (catalase) (Figure 2).
Figure 2. Gel retardation assay of enzyme/polyion complexes.
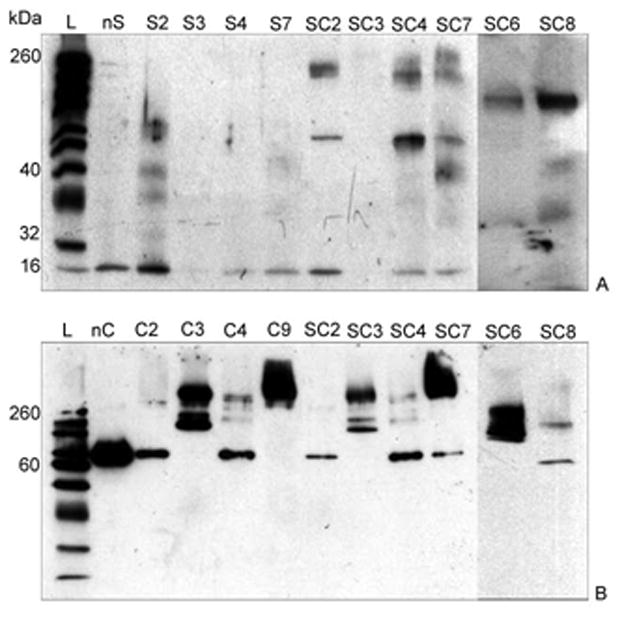
Five μg protein was loaded on a 10% polyacrylamide gel and electrophoresis was carried out as described earlier. Bands were visualized with A) anti-SOD1 and B) anti-catalase polyclonal antibodies. L refers to ladder. nS and nC refer to native SOD1 and catalase, respectively. The lanes correspond to the following samples of cross-linked nanozymes listed in Table 1: S2, S3 and S4 – SOD1/PEI-PEG (Z=1) cross-linked using GA, BS3 and EDC/S-NHS respectively; S7 – SOD1/pLL10-PEG (Z=10) cross-linked using EDC/S-NHS; C2, C3 and C4 - catalase/PEI-PEG (Z=1) cross-linked using GA, BS3 and EDC/S-NHS, respectively; SC2, SC3, SC4 – SOD1-catalase/PEI-PEG (Z=1) cross-linked using GA, BS3 and EDC/S-NHS, respectively; SC6, SC7, SC8 (all prepared at Z=1) indicate SOD1-catalase/pLL10-PEG (BS3), SOD1-catalase/pLL10-PEG (EDC/S-NHS) and SOD1-catalase/pLL50-PEG (EDC/S-NHS) respectively.
The migration pattern was different in case of cross-linked complexes. Usually, there was disappearance or retardation of the protein band, presumably due to inability of cross-linked complexes to migrate through the gel (lanes S3, S4, S7, C2 and SC3). Few conjugates did migrate through the gel, but their estimated molecular masses were greater than those of the respective oligomeric subunits (lanes S2, C3, C4, C9, SC4, SC7, SC6 and SC8). Complete disappearance of free subunits, suggesting nearly 100% conjugation was observed when PEI-PEG and BS3 (7 to 10-fold excess over NH2-groups) were used with both enzymes (lanes S3, C3, SC3, SC6) or pLL10-PEG and EDC/S-NHS (20 to 25-fold excess over −COOH groups) was used with catalase (lane C9). In contrast, use of GA/NaBH4 (2 to 5-fold excess over NH2 groups) and EDC/S-NHS (as earlier) resulted in partial conjugation under the experimental conditions (lanes S2, S4, S7, C2, C4, SC4, SC7 and SC8). In some cases, the extent of cross-linking could be improved by increasing the amount of the cross-linkers. For example, at 100-fold excess of GA the conjugation was nearly complete (not shown), however, the loss of enzyme activity (ca. 50%) prompted us to use lower excesses of GA (about 2 to 7-fold).
In order to obtain two enzymes in the same nanoparticle, we tried to enhance interactions of these enzymes with each other by rendering them oppositely charged using low pH of ∼5.2, which lies between pI values of 4.95 (SOD1) and 5.8 (catalase). However, this decreased the efficiency of cross-linking and led to a considerable loss of catalase activity. Therefore, in most cases, we carried out the cross-linking at relatively high pH ≥ 6.8 where both enzymes were negatively-charged. Formation of SOD1-catalase bi-enzyme nanoparticles was confirmed by visualizing both proteins within the bands of the same molecular mass using anti-SOD1 and anti-catalase antibodies (lanes SC4, SC7, SC6 and SC8). However, in few cases these antibodies stained bands of different molecular masses, suggesting that the enzymes segregated into particles of different type (lanes SC2 and SC3).
Characterization of nanozymes
Enzyme activity
Catalytic activity of SOD1 and catalase in nanozymes was measured as described earlier. Linear dependence of SOD1 and catalase catalytic activities on enzyme concentrations (Figure S1) was observed in the concentration range used; this allowed us to analyze changes in the activity of formulated enzymes using reaction rates that were normalized to the respective enzyme concentrations. As a quantitative analysis of catalase activity, the dependence of the reaction rate on substrate concentration was studied and kinetic parameters, maximal reaction rate (kinetic constant; kcat) and apparent Michaelis constant (Km) were obtained using double reciprocal coordinates (Figure S2). Apparent kinetic parameters (kcat and Km: 6.5×105 s-1 and 63 mM, respectively) calculated for catalase were found to be similar to those reported in published literature.38 For SOD1, the direct measurement of the superoxide dismutation is not possible. Therefore, enzyme activity was assayed indirectly by autoxidation of pyrogallol and was expressed as the amount of enzyme that resulted in 50% inhibition in rate of autoxidation. Two different substrate systems were also used for quantitative analysis of SOD1 activity, riboflavin/Nitro Blue Tetrazolium (Figure S3A) and luminol/xanthine/xanthine oxidase/oxygen systems (Figure S3B).
Before cross-linking, both enzymes retained 100% activity in the polyion complexes (Table 2). Significantly high levels of activity (at least 70%) were retained after cross-linking in most cases. Interestingly, the chemical structure of the block copolymer did not affect the activity after cross-linking. However, the type of cross-linker and reaction conditions showed a significant effect. For example, enzymes cross-linked with GA/NaBH4 retained 70-100% of activity (Table 2 and Figures S1B and S2) strongly depending not only on GA excess but also reduction time with NaBH4 (lesser time correlated with lower activity loss). In contrast, conjugation of nanozymes with EDC/S-NHS resulted in mild activation of the enzyme (residual activity 105-115%). Furthermore, as already mentioned low pH ∼5.2 resulted in irreversible inactivation of both enzymes (not shown).
Table 2. Residual enzyme activity of nanozymes.
| Sample | SOD1 residual activity, %* | Catalase residual activity, %* | pH | Cross-linker |
|---|---|---|---|---|
| SOD1 | 100 | - | 7.4 | - |
| SOD1 | 100 | - | 6.8 | - |
| SOD1/PEI-PEG; non-cl† | 100 | - | 7.4 | - |
| SOD1/PEI-PEG; cl‡ | 88 | - | 7.4 | GA/NaBH4 |
| SOD1/PEI-PEG; cl | 91 | - | 7.4 | BS3 |
| SOD1/PEI-PEG; cl | 115 | - | 7.4 | EDC/S-NHS |
| SOD1/pLL10-PEG; cl | 115 | - | 7.4 | EDC/S-NHS |
| SOD1/pLL50-PEG; cl | 109 | - | 7.4 | EDC/S-NHS |
| catalase (cat) | - | 100 | 7.4 | - |
| cat | - | 60 | 6.8 | - |
| cat/PEI-PEG; non-cl | - | 100 | 7.4 | - |
| cat/pLL10-PEG; non-cl | - | 100 | 7.4 | - |
| cat/PEI-PEG; cl | - | 80 | 7.4 | GA/NaBH4 |
| cat/PEI-PEG; cl | - | 100 | 7.4 | BS3 |
| cat/PEI-PEG; cl | - | 95 | 7.4 | EDC/S-NHS |
| cat/pLL10-PEG; cl | - | 100 | 7.4 | EDC/S-NHS |
| SOD1-cat/PEI-PEG; non-cl | 100 | 100 | 6.8 | - |
| SOD1-cat/pLL10-PEG; non-cl | 100 | 100 | 6.8 | - |
| SOD1-cat/PEI-PEG; cl | 80 | 71 | 6.8 | GA/NaBH4 |
| SOD1-cat/pLL10-PEG; cl | 95 | 78 | 6.8 | GA/NaBH4 |
| SOD1-cat/pLL10-PEG; cl | 85 | 88 | 7.4 | GA/NaBH4 |
| SOD1-cat/pLL10-PEG; cl | 70 | 98 | 6.8 | BS3 |
| SOD1-cat/PEI-PEG; cl | 100 | 105 | 6.8 | EDC/S-NHS |
| SOD1-cat/pLL10-PEG; cl | 97 | 82 | 6.8 | EDC/S-NHS |
| SOD1-cat/pLL10-PEG; cl | 110 | 115 | 6.8 | EDC/S-NHS |
Residual enzymatic activity of nanozymes (A/A0) was calculated based on native enzyme activity (A0) at the same pH. The initial activity values were 3780 U/mg protein for SOD1 and 46500 U/mg protein for catalase
non-cross-linked
cross-linked
Cross-linker excesses were 7 fold for GA and BS3 (over NH2 groups in enzyme and polymer), and 24-fold for EDC/S-NHS (over -COOH groups in enzyme).
Particle size
Hydrodynamic size (size) and surface charge (ζ-potential) of the nanozymes that showed considerable retention of enzyme activity was determined using DLS (Table 3A). Particles of native SOD1 showed a small size (4.6 nm), which was close to the theoretical diameter (4.5 nm) of a single protein globule estimated from molecular mass of the enzyme (32 kDa). In contrast, DLS analysis of native catalase revealed bimodal size distribution with a smaller size of 10 nm corresponding to a single globule and a larger size of 63 nm, which most likely represented aggregates. All complexes (cross-linked or non-cross-linked) were nearly electroneutral or weakly-charged. Compared to the native enzyme molecules, both SOD1- and catalase-based complexes displayed an increase in size and in most cases, their distribution was bimodal. In selected cases, we demonstrated that the particle uniformity can be improved by filtering nanozymes through a 20/100 nm pore size filter. This procedure resulted in SOD1 and catalase complexes of unimodal size distribution (Table 3B). It should be noted that there was no loss of enzyme activity (normalized to protein concentration) upon filtering.
Table 3. Physicochemical properties of selected nanozymes measured using DLS.
| Effective diameter, nm | ||||
|---|---|---|---|---|
| Sample | Linker | ζ-potential, mV | ||
| Peak 1 | Peak 2 | |||
| SOD1 | - | 4.6 | - | -2.6 |
| SOD1/PEI-PEG; non-cl* | - | 6.1 | 74.5 | -1.5 |
| SOD1/PEI-PEG; cl† | BS3 | - | 97.4 | 0.3 |
| SOD1/PEI-PEG; cl | EDC/S-NHS | 6.2 | 63.6 | -1.3 |
| SOD1/pLL10-PEG; cl | EDC/S-NHS | 6.3 | 64.2 | -2.4 |
| SOD1/pLL10-PEG‡; cl | EDC/S-NHS | 5.5 | 72.0 | 1.3 |
| SOD1/pLL50-PEG; cl | EDC/S-NHS | 5.8 | 81.0 | 2.6 |
| catalase (cat) | - | 10.0 | 63 | -5.0 |
| cat/PEI-PEG; non-cl | - | 12.5 | 68.4 | 0.5 |
| cat/PEI-PEG; cl | GA | 40.8 | 644.3 | n.d.§ |
| cat/PEI-PEG; cl | BS3 | 10.6 | 59.5 | -1.2 |
| cat/PEI-PEG; cl | EDC/S-NHS | 10.6 | 52.7 | -0.3 |
| cat/pLL10-PEG; non-cl | - | 12.0 | 71.0 | -2.4 |
| cat/pLL10-PEG; cl | EDC/S-NHS | 12.0 | 28.2; 504 | n.d.§ |
| SOD1-cat/PEI-PE; cl | GA | - | 30.0 | n.d.§ |
| SOD1-cat/PEI-PEG; cl | BS3 | - | 91.9 | -2.7 |
| SOD1-cat/PEI-PEG; cl | EDC/S-NHS | 11.7 | 91.0 | -2.9 |
| SOD1-cat/pLL10-PEG; cl EDC/S-NHS | 14.0 | 135.4 | -1.3 | |
| All nanozymes were prepared at Z=1 (unless indicated otherwise). | ||||
| *non-cross-linked, †cross-linked, ‡Z=10; §not determined | ||||
| Measurement error was typically ≤ 5% of reported values. | ||||
| Table 3B Diameter of filtered nanozymes measured using DLS | ||||
|---|---|---|---|---|
| Sample* | Linker | Dz, nm† | PdI‡ | ζ, mV§ |
| SOD1 | - | 5.2 ± 0.1¶ | 0.1± 0.03 | -2.3 ± 0.5 |
| SOD1/pLL50-PEG; non-cl# | - | 11.2 ± 0.5 | 0.2 ± 0.02 | 6.2 ± 0.5 |
| SOD1/pLL50-PEG; cl** | EDC/S-NHS | 9.7 ± 0.1 | 0.1 ± 0.02 | 0.6 ± 0.1 |
| catalase (cat) | - | 11.5 ± 0.1¶ | 0.2± 0.01 | n.d†† |
| cat/pLL50-PEG; non-cl | - | 16.1 ± 0.1 | 0.2 ± 0.01 | n.d. |
| cat/pLL50-PEG; cl | EDC/S-NHS | 19.8 ± 0.3 | 0.2 ± 0.01 | n.d. |
SOD1 and catalase formulations were prepared in 10 mM HEPES and 10 mM HEPES-buffered saline (pH 7.4), respectively.
All complexes are stoichiometric (Z=1)
Z-average diameter (Dz)
polydispersity index (PDI) and
ζ-potential. Data are mean ± SD of triplicate measurements.
Theoretical hydrodynamic diameters (Malvern Zetasizer Nano software) are 5.2 nm for SOD1 and 12.5 nm for catalase.
non-cross-linked;
cross-linked
ζ-potential not determined because of high ionic strength.
Particle morphology
Morphology and number-average sizes of nanozymes were studied by AFM. Typical image of cross-linked complexes is shown in Figure 3A. Analysis of the images revealed that the nanozymes displayed a broad range of sizes and appeared to be spherical particles.
Figure 3. AFM image of SOD1/pLL50-PEG complexes cross-linked using EDC/S-NHS.
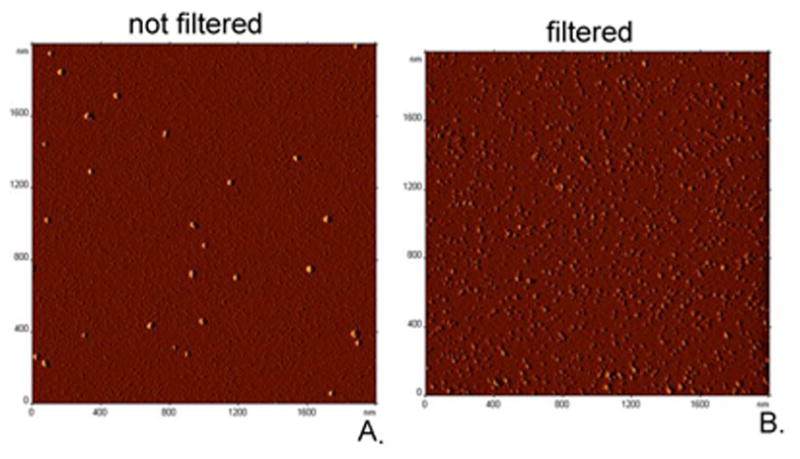
(A) not-filtered (B) filtered. Samples were prepared on an APS mica substrate, dried under argon and images were acquired on a Multimode NanoScope IV system. Scan size: 2 × 2 μm.
Their number-average diameters are presented in Table S1. For example, complexes that contained small particles having diameters ca. 25 nm also had bigger particles with diameters ca. 45-100 nm. These observations were in good agreement with the DLS data showing bimodal distribution. Native SOD1 showed smaller particles compared to SOD1 complexes, while native catalase showed aggregates in addition to small globular particles. It is worth mentioning that imaging in air usually provides lower numbers for the height as a result of the drying process, but higher numbers for the width, due to the tip convolution effect. In addition, interactions between the particles and positively charged mica might also affect size measured by AFM. AFM image of filtered cross-linked particles is shown in Figure 3B, demonstrating particles of spherical morphology and narrower distribution, in agreement with DLS data (Table 3B).
Accumulation of cross-linked nanozymes in BBMEC monolayers
Here, we used BBMEC monolayers as an in vitro model of BBB to evaluate cellular uptake of cross-linked nanozymes. We selected EDC/S-NHS as the cross-linking approach since the resulting nanozymes showed little if any loss in enzyme activity (Table 2). The following cross-linked nanozymes were prepared using pLL-PEG and EDC/S-NHS – cl SOD1/pLL10-PEG (Z=1), cl SOD1/pLL10-PEG (Z=10) and cl SOD1/pLL50-PEG (Z=1). BBMECs were incubated with 125I-labeled SOD1 nanozymes and accumulation was determined in cell lysates (Figure 4). Significant increase in cellular uptake was observed in case of nanozymes prepared using pLL10-PEG and pLL50-PEG block copolymers at Z=1. Interestingly, when a higher excess of the block copolymer was used (pLL10-PEG, Z=10) the uptake levels were not statistically different (P>0.05) from that of native SOD1, which may be due to a toxic effect of the excess polycation.
Figure 4. Accumulation of cross-linked SOD1 nanozymes and native SOD1 in BBMEC monolayers.
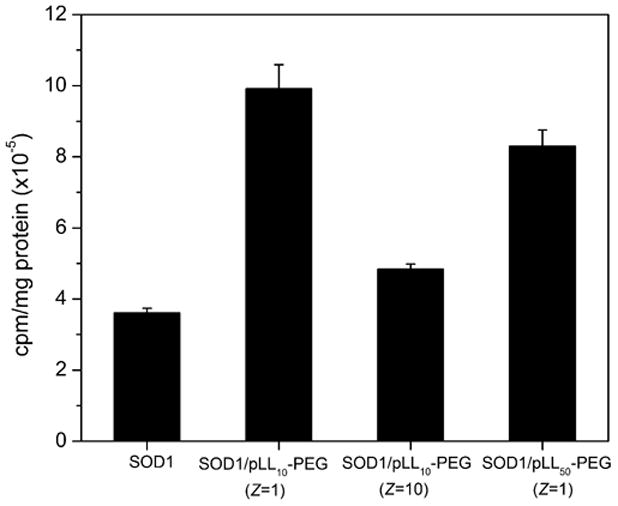
Cells were incubated with 125I-SOD1 formulations or native SOD1 for 1 hour, washed, lysed and 125I activity was measured in a gamma counter. SOD1 concentration in the incubation media was 1 mg/mL. The amount of cell-associated 125I was normalized to cellular protein content and values reported are mean ± SEM (n = 4).
Cytotoxicity of nanozymes in CATH.a neurons
Here, CATH.a neurons were used as an in vitro model to evaluate cytotoxicity of the following samples – native SOD1, non-cross-linked and EDC/S-NHS cross-linked SOD1/pLL10-PEG (Z=10) nanozymes (Figure 5). SOD1 concentration in each group ranged from 1 to 200 μg/mL (it should be noted that the SOD1 concentration used in following in vivo experiments is 50 μg/mL: 100 μg/mouse; assuming a blood volume of 2 mL). Almost no decrease in cell viability was observed in case of native SOD1 with an exception at 50 μg/mL concentration, but the viability was again >100% at higher concentrations. Cells treated with cross-linked nanozymes either showed the same/slightly higher viability (lower cytotoxicity) compared to those treated with non-cross-linked nanozymes; especially at concentrations ≥ 25 μg/mL. IC50 value could not be determined since the cell viability at the highest concentration used (200 μg/mL) was still ∼70-72%.
Figure 5. Cytotoxicity of non-cross-linked and cross-linked nanozymes in CATH.a neurons.
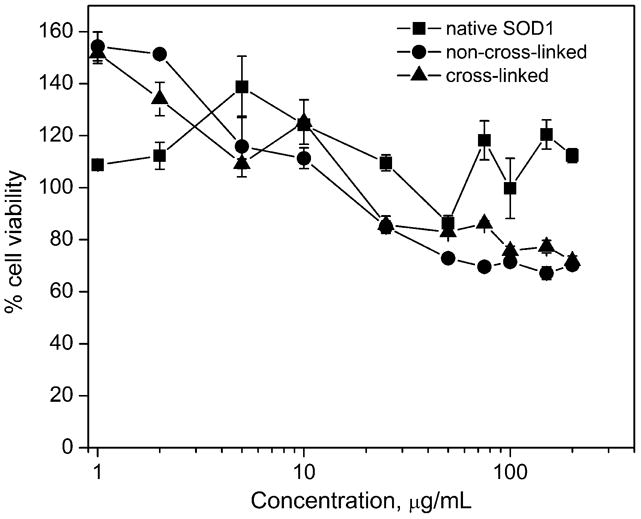
Cells were incubated for 24 h with either native SOD1, non-cross-linked SOD1/pLL10-PEG (Z=10), and SOD1/pLL10-PEG (Z=10) cross-linked using EDC/S-NHS in concentrations ranging from 1 to 200 μg/mL (SOD1). Cell viability was measured using a commercially available MTS assay kit and results are expressed as mean ± SEM (n = 3).
In vivo stability of SOD1 nanozymes
Further, we examined the stability of SOD1 in cross-linked and non-cross-linked nanozymes after i.v. administration in mice. The protein stability was determined using TCA precipitation in brain homogenates and blood serum 1 h after injection (Figure 6). At this time point, nearly 80% of native SOD1 in the brain and over 50% SOD1 in the serum were degraded. Interestingly, the non-cross-linked nanozyme did not show any improvement compared to native SOD1 despite the presence of a 10-fold excess of polycation in the formulation. However, once it was cross-linked, the stability of SOD1 was significantly increased both in the brain and blood. In this case, the amount of non-degraded protein was increased compared to native SOD1 or non-cross-linked nanozyme by ca. 1.8 and 1.2-fold in the brain and blood, respectively.
Figure 6.

In vivo stability of SOD1 in nanozyme formulations. The % 125I-SOD1 is the fraction of 125I precipitated by TCA in brain homogenates or serum. Samples include native SOD1, non-cross-linked SOD1/pLL10-PEG (Z=10), and SOD1/pLL10-PEG (Z=10) cross-linked using EDC/S-NHS. Hundred μL of sample (1 mg SOD1/mL) was injected i.v. into mice; blood and brain tissue were collected 1 hour post-injection and processed as described earlier. Values are mean ± SEM (n=4 or 5). Statistical comparisons were performed by Kruskal-Wallis ANOVA (*P<0.05).
Brain delivery of cross-linked nanozymes
Ability of nanozymes to deliver SOD1 to the brain was studied in a capillary depletion experiment (Figure 7). As earlier, samples were processed using acid precipitation technique to confirm that the radioactivity readout corresponded to enzyme-bound 125I. A 1.7- and 10-fold increase in compartment (parenchyma and capillaries, respectively) to serum ratios were observed in case of cross-linked nanozymes, compared to native SOD1 and non-cross-linked nanozymes. As earlier, we noted no significant differences between delivery of native SOD1 and non-cross-linked nanozymes.
Figure 7. Brain delivery of SOD1 formulations.
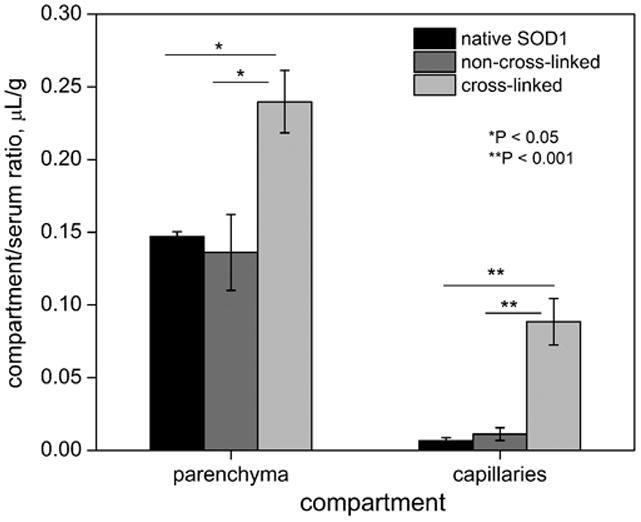
Hundred μg of each sample was injected i.v. into mice; brain tissues were collected 1 h post-injection, homogenized and fractionated in a dextran gradient. Values reported indicate average ± SEM (n=4 or 5) and statistical comparisons were performed by Kruskal-Wallis ANOVA (*P<0.05; **P<0.001).
Discussion
SOD1 and catalase are negatively-charged at physiological conditions. Physical mixing of enzymes with positively-charged block copolymers resulted in spontaneous formation of polyion complex nanoparticles, known as “block ionomer complex”39 with enzyme/polycation complex core and PEG shell. The reaction proceeded rapidly (minutes) at pH 7.4 due to the electrostatic coupling of negatively-charged carboxylic groups of enzymes and positively-charged amino groups of PEI or pLL in the block copolymers. Notably, both SOD1 and catalase retained nearly 100% of the initial catalytic activity after complexation. Compared to the DNA-based complexes studied earlier, nanozymes are less stable because of lower density of negative charges in the protein globule vs. DNA.40 Therefore, they can dissociate upon strong dilution or react with negatively charged competitor molecules, such as serum proteins, leading to premature release of the enzyme. To further stabilize these complexes in circulation, we explored various cross-linking strategies to covalently link carboxyl- and/or amino groups of the protein to the amino groups of polycations. We used water-soluble cross-linking reagents: 1) EDC/S-NHS; 2) GA/NaBH4; and 3) BS3.
We demonstrated that use of all three linkers resulted in cross-linked nanoparticles. Interestingly, use of BS3 resulted in uniform particles, which were larger compared to other nanozymes. We estimated that such particles may contain at least few thousands of SOD1 molecules. In contrast, GA and EDC/S-NHS resulted in formation of heterogeneous particle population. In most cases we obtained nanozymes with bimodal size distribution. The heterogeneity of nanozymes was also supported by AFM and gel electrophoresis data. Hydrodynamic size of particles can dramatically affect their in vivo behavior by influencing parameters such as blood half-life, hepatic filtration, renal clearance, and biodistribution.41-43 Aggregated particles were removed by filtration without any considerable loss in enzyme activity (normalized to protein content) and resulting nanozymes were of unimodal size distribution. Interestingly, with few exceptions, the sizes of the resulting nanoparticles and their charge did not depend on the chemical structure of the block copolymers, composition of the polyion complexes or cross-linker used. In all cases the ζ-potentials of the nanoparticles were either electroneutral, weakly-positive or weakly-negative. Altogether, our data were consistent with formation of stoichiometric polyion complexes that were almost electroneutral. They correspond to either complexes of single and/or several protein molecules as presented in Figure 1. Notably, even when a significant excess of block copolymer (pLL10-PEG, Z=10) was used the ζ-potential remained low, albeit weakly-positive and somewhat greater than the ζ-potential of stoichiometric complexes (Z=1). This suggests that excess of the polycation beyond stoichiometry cannot be incorporated in the block ionomer complex. In fact, a similar behavior is known for the block ionomer complexes formed between polynucleotides and cationic block copolymers. Such complexes are nearly electroneutral in contrast to polyion complexes formed by polynucleotides and homopolymer polycations that can incorporate excess of polycation and show an increase in positive charge at higher Z values. It appears that incorporation of excess of cationic block copolymers in block ionomer complexes is hindered by steric repulsion of the non-ionic PEG chains linked to the polycation molecules.44
The chemical structure of block copolymer in polyion complex affected neither size nor enzyme activity of the nanozymes. The most profound effect on activity was observed when different cross-linkers were used. We speculate that reactivity of the linker might play an important role in this effect. For instance, GA is known to be the most reactive agent45 among the cross-linkers used and therefore it might affect amino acids in proximity to the active site of the enzyme. Furthermore, high reactivity of GA leads to multiple byproducts45 that might affect activity of the enzyme. In addition, conjugation with GA requires the second step of Shiff-base reduction with NaBH4, which can contribute to enzyme inactivation (especially in the case of a heme-containing protein, catalase).
Although the idea to incorporate SOD1 and catalase in one particle might appear attractive from the standpoint of therapeutic applications, the technical difficulties of combining these two proteins together are significant. In particular, we observed segregation of two enzymes into different particles in some cases (Figure 2). An attempt to overcome this by increasing electrostatic interactions between SOD1 and catalase at low pH was offset by loss of enzyme activity and decreased cross-linking efficacy.
We further demonstrated that incorporation of SOD1 in nanozymes can increase its uptake in BBMEC monolayers, which were used as an in vitro model of BBB. Future studies will determine pathways of transport of these nanozymes in cells. We further tested cytotoxicity of selected formulations and demonstrated that CATH.a neurons treated with non-cross-linked or cross-linked nanozymes showed cell viabilities >70% even at the highest concentration tested, suggesting the promise of developing such systems for CNS delivery. Significant increase in the amount of 125I-SOD1 observed in case of cross-linked nanozymes in the acid precipitation study most-likely resulted from its improved stability against polyelectrolyte exchange reactions with serum proteins. However, we noted no improvement in stability of non-cross-linked nanozymes compared with native SOD1. Non-cross-linked nanozymes most-likely dissociated upon interactions with serum proteins and thus its behavior was similar to that of the native enzyme. Ongoing efforts in our lab demonstrate that fractionation using size exclusion chromatography under conditions that disassemble the non-cross-linked particles (mobile phase contained 0.15 M NaCl) result in a homogenous population exclusively comprised of cross-linked particles (manuscript in preparation). We believe that use of homogenous particles will further improve stability in vivo. Results from the capillary depletion study demonstrated increased delivery of 125I-SOD1 to both brain parenchyma and capillaries, suggesting that cross-linking improved metabolic stability of SOD1 across the BBB. Non-cross-linked particles behaved similar to native SOD1 most likely due to their dissociation as discussed above.
In summary, we prepared nanosized polyelectrolyte complexes of antioxidant enzymes by self-assembly of SOD1 or catalase with oppositely-charged block ionomers. Different synthetic strategies that allowed immobilizing active enzymes in cross-linked nanoparticles were evaluated. Preliminary evidence of utility of such nanozymes for the delivery of antioxidant enzymes in vivo was demonstrated. Overall, this study may lead to the development of a new technology for the delivery of antioxidant enzymes to CNS in order to attenuate pathology-associated oxidative stress.
Supplementary Material
Acknowledgments
We would like to thank Dr. William Banks (Professor, University of Washington and VA Puget Sound Health Care System - Seattle Division) for training and advice on iodine labeling, acid precipitation and capillary depletion experiments. We would like thank UNMC Nanoimaging Core Facility for help with AFM experiments.
This study was supported in parts by the United States National Institutes of Health (NIH) grants RR021937 (the Center for Biomedical Research Excellence (COBRE) Nebraska Center for Nanomedicine), NS057748, NS051334, United States National Science Foundation grant DMR 0513699, Russian Ministry of Science and Education (Contracts 02.740.11.5232 and 11.G34.31.0004), Atomic Force Microscopy facility was supported by grants from NIH (SIG program), UNMC Program of Excellence and Nebraska Research Initiative.
Footnotes
No competing interests are present.
References
- 1.Bachis A, Mocchetti I. Brain-Derived Neurotrophic Factor Is Neuroprotective against Human Immunodeficiency Virus-1 Envelope Proteins. Ann N Y Acad Sci. 2005;1053:247–57. doi: 10.1196/annals.1344.022. [DOI] [PubMed] [Google Scholar]
- 2.Ying Wang J, Peruzzi F, Lassak A, Del Valle L, Radhakrishnan S, Rappaport J, et al. Neuroprotective effects of IGF-I against TNFalpha-induced neuronal damage in HIV-associated dementia. Virology. 2003;305:66–76. doi: 10.1006/viro.2002.1690. [DOI] [PubMed] [Google Scholar]
- 3.Anderson E, Zink W, Xiong H, Gendelman HE. HIV-1-associated dementia: a metabolic encephalopathy perpetrated by virus-infected and immune-competent mononuclear phagocytes. J Acquir Immune Defic Syndr. 2002;31(Suppl 2):S43–54. doi: 10.1097/00126334-200210012-00004. [DOI] [PubMed] [Google Scholar]
- 4.Stolp H, Dziegielewska K. Role of developmental inflammation and blood-brain barrier dysfunction in neurodevelopmental and neurodegenerative diseases. Neuropathol Appl Neurobiol. 2008 doi: 10.1111/j.1365-2990.2008.01005.x. [DOI] [PubMed] [Google Scholar]
- 5.Brinton RD. A women's health issue: Alzheimer's disease and strategies for maintaining cognitive health. Int J Fertil Womens Med. 1999;44:174–85. [PubMed] [Google Scholar]
- 6.Gozes I. Neuroprotective peptide drug delivery and development: potential new therapeutics. Trends Neurosci. 2001;24:700–5. doi: 10.1016/s0166-2236(00)01931-7. [DOI] [PubMed] [Google Scholar]
- 7.Kroll RA, Neuwelt EA. Outwitting the blood-brain barrier for therapeutic purposes: osmotic opening and other means. Neurosurgery. 1998;42:1083–99. doi: 10.1097/00006123-199805000-00082. discussion 99-100. [DOI] [PubMed] [Google Scholar]
- 8.Koliatsos VE, Clatterbuck RE, Nauta HJ, Knusel B, Burton LE, Hefti FF, et al. Human nerve growth factor prevents degeneration of basal forebrain cholinergic neurons in primates. Ann Neurol. 1991;30:831–40. doi: 10.1002/ana.410300613. [DOI] [PubMed] [Google Scholar]
- 9.Dogrukol-Ak D, Banks WA, Tuncel N, Tuncel M. Passage of vasoactive intestinal peptide across the blood-brain barrier. Peptides. 2003;24:437–44. doi: 10.1016/s0196-9781(03)00059-7. [DOI] [PubMed] [Google Scholar]
- 10.Perry VH, Bell MD, Brown HC, Matyszak MK. Inflammation in the nervous system. Curr Opin Neurobiol. 1995;5:636–41. doi: 10.1016/0959-4388(95)80069-7. [DOI] [PubMed] [Google Scholar]
- 11.Prasad KN, Cole WC, Hovland AR, Prasad KC, Nahreini P, Kumar B, et al. Multiple antioxidants in the prevention and treatment of neurodegenerative disease: analysis of biologic rationale. Curr Opin Neurol. 1999;12:761–70. doi: 10.1097/00019052-199912000-00017. [DOI] [PubMed] [Google Scholar]
- 12.McCord JM, Fridovich I. Superoxide dismutase. An enzymic function for erythrocuprein (hemocuprein) J Biol Chem. 1969;244:6049–55. [PubMed] [Google Scholar]
- 13.Zamocky M. Phylogenetic relationships in class I of the superfamily of bacterial, fungal, and plant peroxidases. Eur J Biochem. 2004;271:3297–309. doi: 10.1111/j.1432-1033.2004.04262.x. [DOI] [PubMed] [Google Scholar]
- 14.Zhou M, Dominguez R, Baudry M. Superoxide dismutase/catalase mimetics but not MAP kinase inhibitors are neuroprotective against oxygen/glucose deprivation-induced neuronal death in hippocampus. J Neurochem. 2007;103:2212–23. doi: 10.1111/j.1471-4159.2007.04906.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Effects of tocopherol and deprenyl on the progression of disability in early Parkinson's disease. The Parkinson Study Group. N Engl J. 1993;328:176–83. doi: 10.1056/NEJM199301213280305. [DOI] [PubMed] [Google Scholar]
- 16.Pappert EJ, Tangney CC, Goetz CG, Ling ZD, Lipton JW, Stebbins GT, et al. Alpha-tocopherol in the ventricular cerebrospinal fluid of Parkinson's disease patients: dose-response study and correlations with plasma levels. Neurology. 1996;47:1037–42. doi: 10.1212/wnl.47.4.1037. [DOI] [PubMed] [Google Scholar]
- 17.Moghimi SM, Szebeni J. Stealth liposomes and long circulating nanoparticles: critical issues in pharmacokinetics, opsonization and protein-binding properties. Prog Lipid Res. 2003;42:463–78. doi: 10.1016/s0163-7827(03)00033-x. [DOI] [PubMed] [Google Scholar]
- 18.Prokop A, Kozlov E, Newman GW, Newman MJ. Water-based nanoparticulate polymeric system for protein delivery: permeability control and vaccine application. Biotechnol Bioeng. 2002;78:459–66. doi: 10.1002/bit.1200. [DOI] [PubMed] [Google Scholar]
- 19.Klyachko NL, Levashov AV. Bioorganic synthesis in reverse micelles and related systems. Curr Opin Colloid Int Sci. 2003;8:179–86. [Google Scholar]
- 20.Dziubla TD, Karim A, Muzykantov VR. Polymer nanocarriers protecting active enzyme cargo against proteolysis. J Control Release. 2005;102:427–39. doi: 10.1016/j.jconrel.2004.10.017. [DOI] [PubMed] [Google Scholar]
- 21.Simone EA, Dziubla TD, Colon-Gonzalez F, Discher DE, Muzykantov VR. Effect of polymer amphiphilicity on loading of a therapeutic enzyme into protective filamentous and spherical polymer nanocarriers. Biomacromolecules. 2007;8:3914–21. doi: 10.1021/bm700888h. [DOI] [PubMed] [Google Scholar]
- 22.Simone EA, Dziubla TD, Arguiri E, Vardon V, Shuvaev VV, Christofidou-Solomidou M, et al. Loading PEG-catalase into filamentous and spherical polymer nanocarriers. Pharm Res. 2009;26:250–60. doi: 10.1007/s11095-008-9744-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Langer R. Drug delivery and targeting. Nature. 1998;392:5–10. [PubMed] [Google Scholar]
- 24.Yoshida K, Burton GF, McKinney JS, Young H, Ellis EF. Brain and tissue distribution of polyethylene glycol-conjugated superoxide dismutase in rats. Stroke. 1992;23:865–9. doi: 10.1161/01.str.23.6.865. [DOI] [PubMed] [Google Scholar]
- 25.Kabanov A, Vinogradov S, Suzdaltseva Y, Alakhov V. Water-soluble block polycations as carriers for olygonucleotide delivery Bioconjugate Chemistry. 1995;6:639–43. doi: 10.1021/bc00036a001. [DOI] [PubMed] [Google Scholar]
- 26.Harada A, Kataoka K. Pronounced activity of enzymes through the incorporation into the core of polyion complex micelles made from charged block copolymers. J Control Release. 2001;72:85–91. doi: 10.1016/s0168-3659(01)00264-4. [DOI] [PubMed] [Google Scholar]
- 27.Harada A, Kataoka K. Switching by pulse electric field of the elevated enzymatic reaction in the core of polyion complex micelles. J Am Chem Soc. 2003;125:15306–7. doi: 10.1021/ja038572h. [DOI] [PubMed] [Google Scholar]
- 28.Batrakova EV, Li S, Reynolds AD, Mosley RL, Bronich TK, Kabanov AV, et al. A macrophage-nanozyme delivery system for Parkinson's disease. Bioconjug Chem. 2007;18:1498–506. doi: 10.1021/bc700184b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kawamura A, Harada A, Kono K, Kataoka K. Self-assembled nano-bioreactor from block ionomers with elevated and stabilized enzymatic function. Bioconjug Chem. 2007;18:1555–9. doi: 10.1021/bc070029t. [DOI] [PubMed] [Google Scholar]
- 30.Vinogradov S, Bronich T, Kabanov A. Self-assembly of polyamine-poly(ethylene glycol) copolymers with phosphorothioate oligonucleotides. Bioconjugate Chemistry. 1998;9:805–12. doi: 10.1021/bc980048q. [DOI] [PubMed] [Google Scholar]
- 31.Bronich T, Nguyen H, Eisenberg A, Kabanov A. Recognition of DNA topology in reactions between plasmid DNA and cationic copolymers. Journal of American Chemical Society. 2000;122:8339–43. [Google Scholar]
- 32.Vinogradov S, Batrakova E, Kabanov A. Poly(ethylene glycol)-polyethyleneimine NanoGel (TM) particles: novel drug delivery systems for antisense oligonucleotides Colloids and Surfaces B-Biointerfaces. 1999;16:291–304. [Google Scholar]
- 33.Lyubchenko YL, Shlyakhtenko LS. AFM for analysis of structure and dynamics of DNA and protein-DNA complexes. Methods. 2009;47:206–13. doi: 10.1016/j.ymeth.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lyubchenko YL, Shlyakhtenko LS, Gall AA. Atomic force microscopy imaging and probing of DNA, proteins, and protein DNA complexes: silatrane surface chemistry. Methods Mol Biol. 2009;543:337–51. doi: 10.1007/978-1-60327-015-1_21. [DOI] [PubMed] [Google Scholar]
- 35.Shlyakhtenko LS, Gall AA, Filonov A, Cerovac Z, Lushnikov A, Lyubchenko YL. Silatrane-based surface chemistry for immobilization of DNA, protein-DNA complexes and other biological materials. Ultramicroscopy. 2003;97:279–87. doi: 10.1016/S0304-3991(03)00053-6. [DOI] [PubMed] [Google Scholar]
- 36.Batrakova E, Miller D, Li S, Alakhov V, Kabanov A, Elmquist W. Pluronic P85 enhances the delivery of digoxin to the brain: in vitro and in vivo studies. J Pharmacol Exp Ther. 2001;296:551–57. [PubMed] [Google Scholar]
- 37.Price TO, Samson WK, Niehoff ML, Banks WA. Permeability of the blood-brain barrier to a novel satiety molecule nesfatin-1. Peptides. 2007;28:2372–81. doi: 10.1016/j.peptides.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 38.Jurgen-Lohmann DL, Legge RL. Immobilization of bovine catalase in sol-gels. Enzyme Microb Tech. 2006;39:626–33. [Google Scholar]
- 39.Kabanov A, Bronich T, Kabanov V, Yu K, Eisenberg A. Soluble stoichiometric complexes from poly(N-ethyl-4-vinylpyridinium) cations and poly(ethylene oxide)-block-poly(methacrylate) anions. Macromolecules. 1996;29:6797–802. [Google Scholar]
- 40.Vinogradov SV, Bronich TK, Kabanov AV. Self-assembly of polyamine-poly(ethylene glycol) copolymers with phosphorothioate oligonucleotides. Bioconjug Chem. 1998;9:805–12. doi: 10.1021/bc980048q. [DOI] [PubMed] [Google Scholar]
- 41.Alexis F, Pridgen E, Molnar LK, Farokhzad OC. Factors affecting the clearance and biodistribution of polymeric nanoparticles. Mol Pharm. 2008;5:505–15. doi: 10.1021/mp800051m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Choi HS, Ipe BI, Misra P, Lee JH, Bawendi MG, Frangioni JV. Tissue- and Organ-Selective Biodistribution of NIR Fluorescent Quantum Dots. Nano Lett. 2009;9:2354–9. doi: 10.1021/nl900872r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Choi HS, Liu W, Misra P, Tanaka E, Zimmer JP, Itty Ipe B, et al. Renal clearance of quantum dots. Nat Biotechnol. 2007;25:1165–70. doi: 10.1038/nbt1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seymour LW, Kataoka K, Kabanov AV. Cationic block copolymers as self-assembling vectors for gene delivery. John Wiley, Chichester; New York, Weinheim, Brisbane, Singapore, Toronto: 1998. [Google Scholar]
- 45.Hermanson GT. Bioconjugate techniques. Acad Press; San Diego, N.-Y., Boston, London, Sydney, Tokyo, Toronto: 1996. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


