Abstract
Spinal muscular atrophy (SMA) is a neurodegenerative disease caused by loss of survival motor neuron-1 (SMN1). A nearly identical copy gene, SMN2, is present in all SMA patients. Although the SMN2 coding sequence has the potential to produce full-length SMN, nearly 90% of SMN2-derived transcripts are alternatively spliced and encode a truncated protein. SMN2, however, is an excellent therapeutic target. Previously, we developed antisense-based oligonucleotides (bifunctional RNAs) that specifically recruit SR/SR-like splicing factors and target a negative regulator of SMN2 exon-7 inclusion within intron-6. As a means to optimize the antisense sequence of the bifunctional RNAs, we chose to target a potent intronic repressor downstream of SMN2 exon 7, called intronic splicing silencer N1 (ISS-N1). We developed RNAs that specifically target ISS-N1 and concurrently recruit the modular SR proteins SF2/ASF or hTra2β1. RNAs were directly injected in the brains of SMA mice. Bifunctional RNA injections were able to elicit robust induction of SMN protein in the brain and spinal column of neonatal SMA mice. Importantly, hTra2β1-ISS-N1 and SF2/ASF-ISS-N1 bifunctional RNAs significantly extended lifespan and increased weight in the SMNΔ7 mice. This technology has direct implications for SMA therapy and provides similar therapeutic strategies for other diseases caused by aberrant splicing.
Introduction
Spinal muscular atrophies (SMA) (MIM 253300; 253550; 253400; 271150) are a heterogeneous group of neuromuscular disorders characterized by degeneration of lower motor neurons in the anterior horn of the spinal cord. The progressive loss of innervating α-motor neurons causes denervation of voluntary muscles, leading to muscle weakness, and muscle atrophy of the limbs and trunk. SMA is the second most common autosomal recessive disorder after cystic fibrosis. SMA is the leading genetic cause of infantile death worldwide with an incidence of ~1 in 6,000 live births1 and a carrier frequency of ~1 in 35.2 There are two nearly identical gene copies of SMN located on chromosome 5q13. The telomeric copy of the gene is SMN1. In humans, a genomic duplication and inversion event has resulted in a second copy of the gene-termed SMN2. The disease is caused by the loss of the survival motor neuron-1 (SMN1) gene, whereas each patient retains at least one SMN2 copy. Despite the ubiquitous expression of SMN, motor neurons are particularly sensitive to low levels of SMN, resulting in the development of SMA.
A variety of molecular strategies and therapeutic approaches have been developed to attenuate the SMA disease severity, some of which are SMN-dependent, while others are SMN-independent, including: histone deacetylase inhibition;3,4 central nervous system-targeted gene therapy utilizing an adeno-associated virus vector;5,6,7,8,9 lentiviral SMN gene transfer;10 SMN2 modulating oligonucleotides;11,12,13,14,15,16,17 read-through of an SMN2 stop codon;18,19 induction of muscle growth;20 and altering actin cytoskeletal dynamics.21 The presence of SMN2 represents a unique therapeutic target for SMA because the SMN2 gene has the capacity to encode a fully functional protein, and all SMA patients have retained at least one copy of SMN2.
Alternative splicing plays an important role in gene expression and abnormal splicing has been recognized as a cause of an increasing number of diseases.22 With regard to SMA, SMN1 and SMN2 encode identical proteins, however, in contrast to SMN1, which primarily produces full-length mRNA transcripts; SMN2-derived transcripts undergo alternative splicing resulting in low levels of functional protein. This is due to a silent C to T transition found at position 6 of exon 7.23 Nearly 90% of SMN2-derived transcripts encode a truncated protein that lacks the final coding exon (exon 7).24 Pre-mRNA splicing of SMN exon 7 is regulated by a variety of cis- and trans-acting factors and is catalyzed by a complex macromolecular machine termed the spliceosome. Regulation of the SMN2 exon 7 alternative splicing event in SMA depends on several non-spliceosomal factors that bind to pre-mRNA sequences called exonic or intronic splicing enhancers and silencers [exonic splicing silencers or intronic splicing silencer (ISSs)]. The role of these enhancer and silencer motifs is to promote or repress the splice-site decision. It has been previously shown that there are a multitude of alternative splice signals within and flanking SMN exon 7.3,25
Antisense oligonucleotides (ASOs) targeting regulatory elements have become powerful tools to modulate alternative splicing. We and other labs have previously developed bifunctional RNAs as a means to modulate SMN2 splicing to increase exon-7 inclusion.11,26,27,28,29,30 Bifunctional RNAs derive their name from the presence of two functional domains: an RNA sequence that is complementary to a specific cellular RNA (e.g., SMN exon 7, intron-6); and an untethered RNA segment that serves as a sequence-specific binding platform for cellular splicing factors, such as SR proteins. In the initial design, the 5′ end of exon 7 was targeted with the antisense element; however, it was possible that an antisense sequence within exon 7 does not allow for proper recognition of the necessary splicing signals. To enhance the activity of the SMN bifunctional RNAs, we developed a set of RNAs that targeted an intronic repressor within intron-6, Element 1. By targeting a repressor sequence with the antisense sequence, there was a twofold mechanism of SMN induction: inhibition of the intronic repressor and recruitment of SR proteins via the SR recruitment sequence of the bifunctional RNA. Since the SMN2 exon 7 region is highly regulated and additional negative regulatory elements exist, in the present study, we have targeted a potent intronic silencer of SMN2 exon 7 inclusion with a new generation of bifunctional RNAs. SMN2 exon 7 has a weak 5′ splice-site likely due to a nonconsensus U1 snRNA binding site found within that region.31 Previously, it has been determined that there is a unique silencer adjacent to the U1-binding site within intron 7 that can modulate exon 7 splicing termed intronic splicing silencer N1 (ISS-N1).32 Therefore, we developed bifunctional RNAs that specifically target the ISS-N1 region located in SMN intron 7. The 2′-O-methyl bifunctional RNAs were delivered directly into the central nervous system of SMA mice via intracerebroventricular (ICV) injections.11,26,27,33,34 RNA injections were able to elicit a strong induction of SMN protein in the brain and throughout the spinal column of neonatal SMA mice. The mean life span as well as the percent weight gained after the injections was extended following the delivery of bifunctional RNAs. Additionally, the motor skills of the treated animals were significantly improved. These results demonstrate that bifunctional RNAs are a means to increase SMN expression and reduce disease severity in an important model of disease. This type of treatment has direct implications not only for the development of a SMA therapy, but demonstrates the utility of antisense RNAs for a number of diseases caused by abnormal pre-mRNA splicing.
Results
Development of bifunctional RNAs targeting ISS-N1
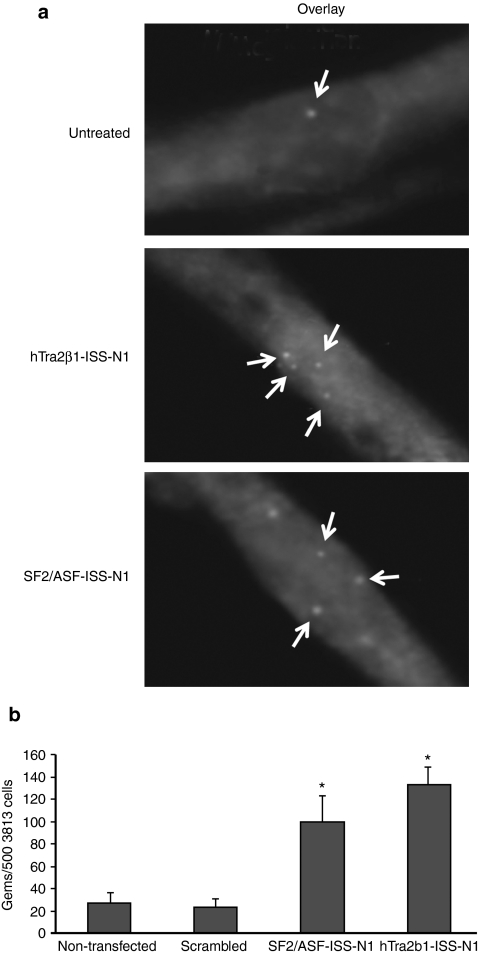
To increase full-length SMN expression from SMN2, we targeted the potent ISS-N1 repressor with a series of bifunctional RNAs that contained three tandem repeats of high-affinity exonic splicing enhancers for hTra2β1 or SF2/ASF. Both of these SR proteins have been previously shown to mediate high levels of SMN exon 7 inclusion35,36 (Figure 1). As a means to assess activity of the RNA molecules in a cell-based model of disease, the bifunctional RNAs and control RNAs were transfected into primary fibroblasts derived from a type I SMA patient. SMN localizes to discrete nuclear bodies termed Gemini-of-coiled-bodies or gems; the frequency of gems is considered a surrogate marker for total SMN protein levels.37 Following transfection of the bifunctional RNAs into SMA fibroblasts, SMN-positive gems were markedly increased (Figure 2a). A quantitative analysis of the transfected cell populations demonstrated that the hTra2β1 or SF2/ASF bifunctional RNAs significantly elevated gem numbers several fold above cells transfected with a negative control RNA of similar size or in mock transfected cells (Figure 2b). Based upon these results in a relatively straightforward disease context, we next examined the ISS-N1 bifunctional RNAs in a more complex disease context, a SMA mouse model.
Figure 1.
Schematic representation of specific bifunctional RNAs targeting intronic splicing silencer N1 (ISS-N1). The organization of the bifunctional RNA is illustrated with the antisense domain targeting the sequences of the ISS-N1 repressor; with high-affinity exonic splice enhancer sequences for either transformer-2 protein homolog β (hTra2β1) or splicing factor2/alternative splicing factor (SF2/ASF). The ISS-N1 bifunctional RNAs have an uninterrupted antisense specific for ISS-N1 with an additional six nucleotides downstream to ensure coverage of the negative region. In addition the RNA has a linked domain that contains triplicate exonic splicing enhancer (ESE) recruitment domains (data not shown).
Figure 2.
2′-O-Methyl bifunctional RNAs increase survival motor neuron (SMN) protein and gem number in patient fibroblasts. (a) Spinal muscular atrophy (SMA) patient fibroblasts (3,813 cells) were transiently transfected with 100 ng of hTra2β1-ISS-N1 and SF2/ASF-ISS-N1 2′-O-methyl bifunctional RNAs for 48 hours. Representative cells after immunofluorescence (IMF) staining. 3,813 cells are shown in the top row as positive control staining (b) 500 3,813 cells were randomly counted (n = 3) and total gem number determined. ISS-N1, intronic splicing silencer N1.
Bifunctional RNAs targeting the ISS-N1 repressor increase SMN protein levels in the SMNΔ7 mouse model
To determine whether the bifunctional RNAs specifically designed to target the ISS-N1 repressor could elevate SMN levels in vivo, we performed ICV injections on 1-day-old SMA mice (PND1). The advantages of ICV injections are the direct delivery of RNAs into the central nervous system as the injection bypasses the blood–brain barrier, the rapid distribution throughout the spinal column, and the ability to deliver relatively high concentrations of the modified RNAs. To assay in vivo activity, we selected the SMNΔ7 model. These animals lack the murine Smn, but have two copies of human SMN2 and expresses the SMNΔ7 complementary DNA from a separate transgene (Smn−/−; SMN2+/+; SMNΔ7+/+).38 This animal model is widely used for translational studies based upon several factors including: representation of a relatively severe form of SMA with loss of lumbar motor neurons and progressive muscle weakness; the rapid attaining of peak body weight at approximately PND10; the quantifiable nature of gross motor deficits such as inability to walk and right themselves; and the presence of SMN2 allowing the analysis of SMN2 splicing modulators. In order to deliver the RNAs to the central nervous system, we used ICV injections of the ISS-N1 antisense-alone, the SF2/ASF-ISS-N1 and the hTra2β1-ISS-N1 bifunctional RNAs on PND1, PND3, and PND5. Two hours following the last injection on PND5, brain and spinal cord tissues were harvested and protein extracts were generated to determine SMN levels by a minimum of five western blots per sample group. Consistent with the cell-based analysis, the delivery of either the SF2/ASF-ISS-N1 or the hTra2β1-ISS-N1 bifunctional RNAs significantly increased SMN protein levels in the brain and spinal cord (Figure 3a,b). Lower levels of SMN were detected when an antisense oligonucleotide was injected that lacked the exonic splicing enhancer-recruitment motifs (ISS-N1). As expected, very low levels of SMN were detected in untreated SMA pups and SMN levels in aged-matched heterozygous/unaffected littermates were much higher. Densitometric analysis of the blots revealed a significant increase in SMN protein in brain and spinal cord tissues from animals injected with either of the bifunctional RNAs (Figure 3). We observed a trend toward increased SMN protein levels in the ISS-N1 antisense samples compared to the noninjected controls, although statistical significance was not achieved (Figure 3a,b).
Figure 3.
Injection of 2′-O-methyl bifunctional RNAs targeting the intronic splicing silencer N1 (ISS-N1) repressor increase total survival motor neuron (SMN) protein in the SMNΔ7 mouse model. Intracerebroventricular (ICV) injections of hTra2β1-ISS-N1 and SF2/ASF-ISS-N1 2′-O-methyl bifunctional RNAs increase SMN protein levels in (a) brain and (b) spinal cord. Western blots (n = 5) for each treatment group were performed on brain and spinal cord at PND6 and the fold increase in SMN protein induction was compared to the nontreated group (n = 5).
Delivery of 2′-O-methyl bifunctional RNA increases weight gain in the SMNΔ7 model
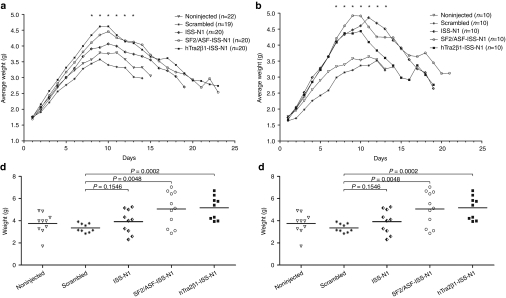
To determine whether increases in SMN led to a lessening of the SMA phenotype, we next analyzed body weight of treated and untreated SMNΔ7 mice. While this is a relatively severe model, SMNΔ7 animals gain weight during the first 10 days of life, although not at the same rate as unaffected animals. On PND1, PND3, and PND5, an ICV injection was administered that delivered 6 µg/day of the respective RNAs (scrambled; ISS-N1 antisense alone; SF2/ASF-ISS-N1 or hTra2β1-ISS-N1) and animals were weighed daily. Animals treated with the bifunctional RNAs (SF2/ASF-ISS-N1 and hTra2β1-ISS-N1) reached a higher average peak weight on PND8-13 and continued to maintain significant weight compared to the animals that received the negative control injection (scrambled) or noninjected groups. These data are consistent with the previous western blot results showing that the bifunctional RNAs elevated SMN levels. Interestingly, the animals injected with the RNA targeting only the ISS-N1 repressor region (ISS-N1) also showed increased weight compared to the untreated and scrambled ASO-treated animals, although the weight gain was not as significant compared to the bifunctional RNAs (Figure 4a).
Figure 4.
Bifunctional RNAs injected animals were heavier than untreated spinal muscular atrophy (SMA) controls starting at PND6. Intracerebroventricular (ICV) injections of 2′-O-methyl intronic splicing silencer N1 (ISS-N1) bifunctional RNAs into SMNΔ7 mice on PND1, PND3 and PND5. (a) Total body weight was measured daily for noninjected, scrambled RNA, ISS-N1, SF2/ASF-ISS-N1, and hTra2β1-ISS-N1 injected mice with oligo concentration of 1 µg/µl. (b) Spinal muscular atrophy (SMA) animals injected with the same bifunctional RNAs and controls with an oligo concentration of 10 µg/µl. Mice were weighed daily after injection. Days with significant weight gain are indicated with (*). (c) Individual weights on PND11 for all animals treated with 1 µg/µl of 2′-O-methyl ISS-N1 bifunctional RNAs. Student's t-test was used to compare each group against the scrambled control: ISS-N1 P = 0.0262; SF2/ASF P = 0.005; hTra2β1-ISS-N1 P = 0.0021. (d) Individual weights on PND11 for animals treated with 10 µg/µl of 2′-O-methyl ISS-N1 bifunctional RNAs. Student's t-test comparing each group against the scrambled control: ISS-N1 P = 0.1546; SF2/ASF P = 0.0048; hTra2β1-ISS-N1 P = 0.0002. SMN, survival motor neuron
To determine whether an increased dose would lead to a greater increase in weight, we increased the concentration of the oligonucleotide injections on PND1, PND3, and PND5 by tenfold compared to the initial concentration. Again, the treated animals showed significant weight gain compared to the control groups, and bifunctional-treated animals were able to achieve nearly 5 g (Figure 4b). As before, weight increase was observed as early as PND6, and some of the treated animals gained ~5–15% more weight than the animals injected with the lower concentration of the same bifunctional RNAs (Figure 4b). In side by side comparison of both treatment concentrations, the mice injected with hTra2β1-ISS-N1 and SF2/ASF-ISS-N1 show a significant increase in weight gained, and although the effect was modest, the 10× concentration resulted in greater weight gain for all bifunctional RNA-treated animals. Injection of the bifunctional RNAs into unaffected animals did not alter weight gain (data not shown).
For a more thorough picture of the differences in weight gain between the experimental groups, we plotted the individual weights for each animal on PND11. We selected PND11 since this was the day when the untreated SMNΔ7 mice reached their peak weights. The low- and high concentration-treated animals significantly outperformed their littermates from noninjected and scramble-treated groups (Figure 4c,d). The scatter plots highlight that several animals treated with the high concentration of the bifunctional RNAs reached weights between 6 and 7 g, significantly higher than the typical weight for the SMNΔ7 at this age (~3.5 g). Collectively, these results demonstrate that bifunctional RNAs lessen the severity of the disease phenotype and result in increases in weight not achieved by the antisense alone (Figure 4c,d).
Significant improvement in gross motor function observed in animals treated with bifunctional RNA
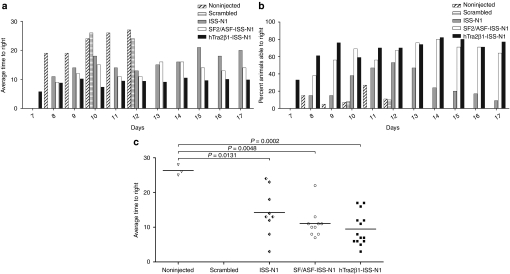
To determine whether the bifunctional RNA injections impacted the gross motor function of the treated animals, we measured time-to-right for all experimental groups. Time-to-right was previously shown to be a sensitive measurement of gross motor function for SMA animals.20,39,40 Animals were placed on their backs and the time required to turn upright was measured. The test was terminated at 30 seconds and if an animal had not turned by this time, it was recorded as “30 seconds.” Time-to-right success rate and speed tests were initiated on PND7 because unaffected animals start to turn over at this time; trials concluded on PND17. A significant improvement (smaller bars) in the motor functions of all groups injected with bifunctional RNAs was observed. Animals from the SF2/ASF-ISS-N1-injected group were able to right themselves on average within 15 seconds and the mice injected with hTra2β1-ISS-N1 were able to right themselves within 10 seconds on each day (Figure 5a). Mice injected with the ISS-N1 antisense also showed a significant improvement in motor performance compared to both negative control groups (Figure 5a–d). A substantially greater percentage of the pups treated with bifunctional RNA were able to right when compared to either the antisense alone or the negative controls (Figure 5b). The individual righting measurements for PND11 are emphasized for the individual animals from each treatment group and the controls. Notably, only a small number (n = 3) of the nontreated animals can right themselves and no animals (n = 0) from the scrambled RNA injected group were able to right. In contrast, the bifunctional RNA treated animals, on average, could perform the time-to-right test within ~10 seconds and all of the animals could right themselves (Figure 5c). The antisense-alone treated animals were also improved compared to the negative controls, although the average time-to-right was slower than the bifunctional-treated groups (Figure 5c).
Figure 5.
Overall fitness was assessed by the ability of the mice to right themselves from a prone position. (a) Graph representing raw data of the average time to right from PND7 to PND17. Animals injected with the hTra2β1-ISS-N1 and SF2/ASF-ISS-N1 bifunctional RNAs were able to right themselves within 10 and 15 seconds, respectively. (b) Graph representing the fraction of animals able to right themselves within 30 seconds measured from PND7 to PND17. Treated spinal muscular atrophy (SMA) mice performed considerably better than negative control and untreated SMA mice. (c) Scatter plot of time-to-right (TTR) performance of mice injected with hTra2β1-ISS-N1 bifunctional RNA. To highlight the performance of individual mice, TTR values are shown for PND11 All three treatment groups outperformed the noninjected controls (Student's t-test ISS-N1 P = 0.0131; SF2/ASF P = 0.0001; hTra2β1-ISS-N1 P = 0.0001). ISS-N1 intronic splicing silencer N1.
Bifunctional RNA treatment increases survival in the SMNΔ7 mouse model
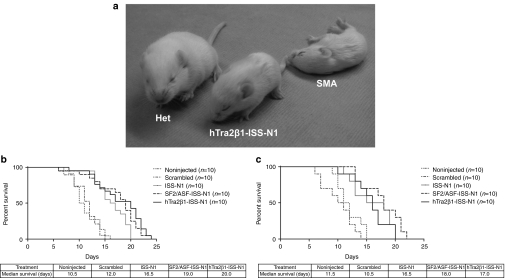
The SMNΔ7 mice die prematurely following progressive muscle weakness, having an average lifespan of <2 weeks.38 To determine whether ICV injection of the bifunctional RNAs extended survival of the SMA animals, lifespan for each treatment group was analyzed by Kaplan–Meier survival curves and compared to the lifespan of the animals from the control groups. ICV delivery for the ISS-N1 antisense or the ISS-N1 bifunctional RNAs significantly increased the average lifespan of the SMA mice, compared to either noninjected or scramble-treated animals (Figure 6a,b). The median survival for the SF2/ASF-ISS-N1- and hTra2β1-ISS-N1-treated animals increased to 19 and 20 days, respectively (Figure 6b). The antisense-alone RNA targeting the ISS-N1 repressor also increased the median survival of the tested animals to 16.5 days (Figure 6c). Additionally, the extension of the lifespan of the treated animals was similar for the low and high concentrations of the RNAs administered (Figure 6b,c). Collectively, these results demonstrate that bifunctional RNAs can be identified that increase SMN in cell-based assays that can then be used to reduce disease severity in more complex experimental models, the severe SMNΔ7 mice.
Figure 6.
SMNΔ7 mice showed significant improvement in survival and motor function tests after intracerebroventricular (ICV) injections with bifunctional RNA oligonucleotides. (a) The treated spinal muscular atrophy (SMA) animal (labeled “Tra2-ISS-N1”) and the untreated heterozygous littermate (labeled “Het”) were able to right themselves within 10 seconds and to consistently ambulate compared to the untreated SMA pup (labeled “SMA”) at PND10. (b, c) Bifunctional RNA treatment increased longevity of SMNΔ7 mice. Kaplan–Meier survival curves were constructed from the various treatment groups as indicated. Log-rank (Mantel–Cox) statistics were applied for comparisons between groups. [(b) (1 µg/µl) For all tested groups survival of noninjected versus intronic splicing silencer N1 (ISS-N1), SF2/ASF-ISS-N1, and hTra2β1-ISS-N1 P < 0.0001; Survival of: ISS-N1 versus SF2/ASF-ISS-N1 P = 0.06; ISS-N1 versus hTra2β1-ISS-N1 P = 0.01; (c) (10 µg/µl) Survival of noninjected versus ISS-N1 P < 0.006; noninjected versus SF2/ASF-ISS-N1 P < 0.0005; Noninjected versus hTra2β1-ISS-N1 P < 0.002; ISS-N1 versus SF2/ASF-ISS-N1 P = 0.16; ISS-N1 versus hTra2β1-ISS-N1 P = 0.85]. The survival curves depict a significant increase in life expectancy for SF2/ASF-ISS-N1 and hTra2β1-ISS-N1 injected animals with increases in median survival for both treatment concentrations [(b) 1 µg/µl or (c) 10 µg/µl]. SMN, survival motor neuron.
Discussion
RNA therapeutics, and antisense strategies specifically, have been the subject of considerable investigation in recent years. Antisense molecules are being investigated in a variety of clinical trials, including Duchenne muscular dystrophy and amyotrophic lateral sclerosis. In the SMA field, our lab has provided further evidence that the use of antisense RNA and bifunctional RNAs can increase SMN protein levels in an important preclinical model of the disease. The results presented here demonstrate that administration of bifunctional RNAs that target ISS-N1 are beneficial in a relatively severe model of SMA. Our strategy in designing these bifunctional RNAs was two-pronged: inhibit ISS-N1 through the antisense component, and at the same time recruit known activators of SMN exon-7 inclusion: SF2/ASF or hTra2-β1. Through mutagenesis and antisense approaches, several labs have highlighted the importance of ISS-N1 in cultured cells and more recently in SMA animal models region.5,12,16,32,41 Using the 2′-O-methyl chemistry, we demonstrate that the antisense alone molecule that targets ISS-N1 is sufficient to increase SMN as well as extend the life span of the SMNΔ7 model. The bifunctional RNAs, which along with the ISS-N1 antisense also contain a recruiting platform splicing factors, provide an additional benefit above the antisense alone.
Recent work using the 2′-O-(2-methoxyethyl) phosphorothioate-modified antisense chemistry has demonstrated that additional backbone chemistries can be used to alter the activity of antisense RNAs, likely by extending the half-life and/or altering cellular uptake. Remarkably, an antisense molecule targeting ISS-N1, called ASO-10-27, can nearly completely reverse the SMN2 splicing pattern for several months following the final administration of ASO-10-27.13 In this work, a transgenic model was examined that does not present a detectable SMA-like phenotype, however, the endogenous Smn gene is knocked-out and four copies of SMN2 are present. Although postnatal delivery of the RNA does not rescue the tail and ear necrosis common to this model, a single embryonic administration does ameliorate nearly all of the necrosis as well as correcting the SMN2 splicing defect. When ASO-10-27 was delivered via ICV injection to SMNΔ7 mice, this resulted in a significant extension in median survival from 16 days to 17–25 days, depending upon the concentration injected at P0.34 In our current work, the bifunctional RNAs result in an extension of medial survival to 18–20 days, although the longest lived animals in our experiments lived ~25 days, compared to the 50+ days observed in the ASO-10-27-treated animals. The antisense sequence used in our current studies overlaps ASO-10-27 in its entirety and extends for an additional three nucleotides down the 3′ end of the SMN transcript (e.g., ASO-10-30). In the present experiments, the bifunctional RNAs perform better in many of the experiments compared to the ASO-alone counterpart, suggesting that the recruiting platforms of the bifunctional RNAs confers even greater levels of SMN protein production. However, the direct comparison between our bifunctional RNAs and ASO-10-27 is not possible in this report as the chemistries of the compounds are significantly different. Clearly, however, our antisense alone RNA is not nearly as effective as the comparable 2′-O-(2-methoxyethyl) chemistry and the ideal comparison would be to synthesize the bifunctional RNAs with a variety of chemistries and compared to the antisense-alone molecule. It is important to state that these experiments were not designed to compare and contrast a variety of backbone chemistries, rather they were designed to determine the utility of bifunctional RNAs and if bifunctional RNAs conferred a greater level of activity in a SMA mouse model compared to the antisense alone. Using the 2′-O-Methyl chemistry presented in the current experiments, the bifunctional RNA conferred greater activity in many of the assays we examined.
Alternative splicing is a complex event where many cis- and trans-elements work in a coordinated and dynamic manner leading to gene expression control that can be modulated temporally, developmentally, and spatially. This work and that of others highlight the importance of understanding the molecular constituents that regulate SMN splicing as a means to identify potential targets for therapeutics. SMN1 exon 7 is a constitutively included in nearly all transcripts, whereas SMN2 exon 7 is largely excised and is regulated by a complex and dynamic suite of cis- and trans-factors. The region immediately downstream from exon 7 is an intriguing target for therapeutics as it appears to function as a nucleation point for hnRNP-A1 which in turn functions as a potent repressor of exon-7 inclusion. A variety of antisense molecules, including those reported here, have been developed that are designed to block the formation of the secondary structure of the ISS-N1 region and/or the ISS-N1/hnRNP-A1 complex. Recently, an additional factor has been identified that interacts near the ISS-N1 site, slightly downstream in intron 7, called TIA1.42
A number of translational programs are accelerating in SMA research, including small molecules, gene therapy, and antisense RNAs. The most striking results have been obtained using self-complementary adeno-associated virus vectors to deliver full-length SMN complementary DNA.5,6,8,9 Rather remarkably, four independent laboratories have recently reported exceptionally similar dramatic extensions in survival and near reversals of the SMA phenotype. Small molecules have a relatively well-defined pathway for US Food and Drug Administration approval; however, it is likely that biologics such as gene therapy, ASOs, and cellular therapies will be greatly enhanced once delivery and sustainability issues can be examined in a larger animal model of disease, such as swine. Recently, the initial characterization of the swine SMN gene has been reported and the development of a pSMN heterozygous animal (pSMN+/−) has been completed.43 An important distinction between gene replacement and RNA-based therapeutics is that ASO-based applications alter expression of the native endogenous SMN2 transcript, and therefore, spatial and temporal expression is dictated by the natural promoter. It will be of particular interest to determine whether this regulatory nuance is advantageous for RNA-based therapeutics compared to gene replacement.
Materials and Methods
Animal procedures. All animals were housed and treated in accordance with Animal Care and Use Committee guidelines of the University of Missouri, Columbia, MO. Smn−/−; SMN2+/+; SMNΔ7+/+ mice38 were genotyped on the day of birth, and injected on PND1. ICV injections were performed on PND1, PND3, and PND5 as previously described.26,33,44 Briefly, mice were immobilized via cryoanesthesia and injected using microliter calibrated sterilized glass micropipettes. The injection site was ~0.25-mm lateral to the sagittal suture and 0.50–0.75 mm rostral to the neonatal coronary suture. The needles were inserted perpendicular to the skull surface using a fiber-optic light (Boyce Scientific, Gray Summit, MO) to aid in illuminating pertinent anatomical structures. Needles were removed after 15 seconds of discontinuation of plunger movement to prevent backflow. Mice recovered for 5–10 minutes in a warmed container until movement was restored. Single injections of 6 µl (1 µg/1 µl and 10 µg/1 µl, respectively) of each 2′-O-methyl oligonucleotides were delivered ICV as described above for all mice.
2′-O-Methyl RNAs. The following oligos were modified at every base with 2′-O-methyl groups (IDT, Coralville, IA); hTra2β-ISS-N1, 5′-GAA GGA GGG AAG GAG GGA AGG AGG GAU UCA CUU UCA UAA UGC UGG-3′, SF2/ASF-ISS-N1, 5′-CAC ACG ACA CAC GAC ACA CGA GAU UCA CUU UCA UAA UGC UGG-3′, and negative control RNA, scrambled 5′-CCU UCC CUG AAG GUU CCU CC-3′.
Immunofluorescence imaging. For all immunofluorescence staining, subconfluent type I patient fibroblasts cells (3,813 cells; Coriell Cell Repositories, Camden, NJ) were transfected in eight chambered slides (BD Biosciences, Bedford, MA) with 2′-O-methyl RNA oligonucleotides, incubated for 48 hours in Dulbecco's modified eagle medium supplemented with 10% fetal bovine serum and antibiotics. Transfected cells were fixed with a solution of acetone/methanol (1/1 vol/vol) and then washed with phosphate-buffered saline (PBS) (Gibco Cell Culture; Invitrogen, Carlsbad, CA). Cells were then blocked in PBS + 5% bovine serum albumin and then washed again in PBS. A pooled group of three previously described anti-SMN monoclonal antibodies was added, diluted 1:10 in PBS + 1.5% bovine serum albumin.32 Cells were washed again in PBS and a secondary monoclonal antibody, an anti-mouse conjugated to Texas red (The Jackson Laboratory, Bar Harbor, ME), or conjugated to Fluorescein isothiocyanate (Sigma-Aldrich, St Louis, MO) for either the plasmid or 2′-O-methyl transfected cells, respectively, diluted 1:200 in PBS + 1.5% bovine serum albumin. After washing in PBS, DAPI was added to each chamber for 5 minutes, and samples were washed again. Chambers were then fitted with cover slips using mounting media (DABCO Thermo Fisher Scientific, Pittsburgh, PA, 2.3% (wt/vol), 10% PBS, 87.7% glycerol) and sealed with nail polish. Microscope images were captured using Nikon Eclipse E1000 using Meta-Morph software (Nikon, Melville, NY). Cells for gem counting were chosen at random. Each randomly chosen nucleus was examined for nuclear gem accumulation and gems were counted.
Western blots. For the SMNΔ7 mouse western blots, indicated tissues were collected at selected time points and immediately frozen in liquid nitrogen. Tissue samples were placed at −80 °C until ready for analysis. Roughly 100 mg of tissue was homogenized in JLB buffer (50 mmol/l Tris−HCl pH 7.5, 150 mmol/l NaCl, 20 mmol/l NaH2(PO4), 25 mmol/l NaF, 2 mmol/l EDTA, 10% glycerol, 1% Triton X-100, and protease inhibitors (Roche, Indianapolis, IN)). Equal amounts of protein were separated on 12% sodium dodecyl sulphate-polyacrylamide gel electrophoresis gels. SMN immunoblots were performed using a mouse SMN-specific monoclonal antibody (BD Biosciences, San Jose, CA) diluted 1:3,000 in TBST (Tris-buffered saline Tween-20 (10 mmol/l Tris–HCl, pH 7.5, 150 mmol/l NaCl, 0.2% Tween-20)) in 1.5% dry milk. Then blots were visualized by chemiluminescence on a Fujifilm imager LAS-3000 and the corresponding software. To verify equal loading the Westerns were then stripped using H2O2 for 30 minutes at room temperature and reprobed with anti-β-actin rabbit and anti-rabbit horseradish peroxidase secondary antibody. Western blots were performed in quadruplicate or more and representative blots are shown.
Acknowledgments
We thank Travis D. Baughan (Department of Neurology, University of Washington, Seattle, WA) for early work on this project; Hansjörg Rindt (Department of Veterinary Pathobiology, University of Missouri, Columbia, MO) for assistance with the statistical analysis and John Marston (Department of Veterinary Pathobiology, University of Missouri, Columbia, MO) for expert technical assistance in animal husbandry. This work was supported by a grant from The Eunice Kennedy Shriver National Institute of Child Health and Human Development to C.L.L. (R01HD054413). The authors declared no conflict of interest.
REFERENCES
- Crawford TO., and, Pardo CA. The neurobiology of childhood spinal muscular atrophy. Neurobiol Dis. 1996;3:97–110. doi: 10.1006/nbdi.1996.0010. [DOI] [PubMed] [Google Scholar]
- Feldkötter M, Schwarzer V, Wirth R, Wienker TF., and, Wirth B. Quantitative analyses of SMN1 and SMN2 based on real-time lightCycler PCR: fast and highly reliable carrier testing and prediction of severity of spinal muscular atrophy. Am J Hum Genet. 2002;70:358–368. doi: 10.1086/338627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorson CL, Rindt H., and, Shababi M. Spinal muscular atrophy: mechanisms and therapeutic strategies. Hum Mol Genet. 2010;19 R1:R111–R118. doi: 10.1093/hmg/ddq147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kernochan LE, Russo ML, Woodling NS, Huynh TN, Avila AM, Fischbeck KH.et al. (2005The role of histone acetylation in SMN gene expression Hum Mol Genet 141171–1182. [DOI] [PubMed] [Google Scholar]
- Passini MA, Bu J, Roskelley EM, Richards AM, Sardi SP, O'Riordan CR.et al. (2010CNS-targeted gene therapy improves survival and motor function in a mouse model of spinal muscular atrophy J Clin Invest 1201253–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foust KD, Wang X, McGovern VL, Braun L, Bevan AK, Haidet AM.et al. (2010Rescue of the spinal muscular atrophy phenotype in a mouse model by early postnatal delivery of SMN Nat Biotechnol 28271–274. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Bevan AK, Hutchinson KR, Foust KD, Braun L, McGovern VL, Schmelzer L.et al. (2010Early heart failure in the SMNDelta7 model of spinal muscular atrophy and correction by postnatal scAAV9-SMN delivery Hum Mol Genet 193895–3905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez E, Marais T, Chatauret N, Benkhelifa-Ziyyat S, Duque S, Ravassard P.et al. (2011Intravenous scAAV9 delivery of a codon-optimized SMN1 sequence rescues SMA mice Hum Mol Genet 20681–693. [DOI] [PubMed] [Google Scholar]
- Valori CF, Ning K, Wyles M, Mead RJ, Grierson AJ, Shaw PJ.et al. (2010Systemic delivery of scAAV9 expressing SMN prolongs survival in a model of spinal muscular atrophy Sci Transl Med 235ra42. [DOI] [PubMed] [Google Scholar]
- Azzouz M, Le T, Ralph GS, Walmsley L, Monani UR, Lee DC.et al. (2004Lentivector-mediated SMN replacement in a mouse model of spinal muscular atrophy J Clin Invest 1141726–1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baughan TD, Dickson A, Osman EY., and, Lorson CL. Delivery of bifunctional RNAs that target an intronic repressor and increase SMN levels in an animal model of spinal muscular atrophy. Hum Mol Genet. 2009;18:1600–1611. doi: 10.1093/hmg/ddp076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JH, Schray RC, Patterson CA, Ayitey SO, Tallent MK., and, Lutz GJ. Oligonucleotide-mediated survival of motor neuron protein expression in CNS improves phenotype in a mouse model of spinal muscular atrophy. J Neurosci. 2009;29:7633–7638. doi: 10.1523/JNEUROSCI.0950-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Y, Sahashi K, Hung G, Rigo F, Passini MA, Bennett CF.et al. (2010Antisense correction of SMN2 splicing in the CNS rescues necrosis in a type III SMA mouse model Genes Dev 241634–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geib T., and, Hertel KJ. Restoration of full-length SMN promoted by adenoviral vectors expressing RNA antisense oligonucleotides embedded in U7 snRNAs. PLoS ONE. 2009;4:e8204. doi: 10.1371/journal.pone.0008204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burghes AH., and, McGovern VL. Antisense oligonucleotides and spinal muscular atrophy: skipping along. Genes Dev. 2010;24:1574–1579. doi: 10.1101/gad.1961710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh NN, Shishimorova M, Cao LC, Gangwani L., and, Singh RN. A short antisense oligonucleotide masking a unique intronic motif prevents skipping of a critical exon in spinal muscular atrophy. RNA Biol. 2009;6:341–350. doi: 10.4161/rna.6.3.8723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Y, Vickers TA, Okunola HL, Bennett CF., and, Krainer AR. Antisense masking of an hnRNP A1/A2 intronic splicing silencer corrects SMN2 splicing in transgenic mice. Am J Hum Genet. 2008;82:834–848. doi: 10.1016/j.ajhg.2008.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattis VB, Ebert AD, Fosso MY, Chang CW., and, Lorson CL. Delivery of a read-through inducing compound, TC007, lessens the severity of a spinal muscular atrophy animal model. Hum Mol Genet. 2009;18:3906–3913. doi: 10.1093/hmg/ddp333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heier CR., and, DiDonato CJ. Translational readthrough by the aminoglycoside geneticin (G418) modulates SMN stability in vitro and improves motor function in SMA mice in vivo. Hum Mol Genet. 2009;18:1310–1322. doi: 10.1093/hmg/ddp030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose FF, Jr, Mattis VB, Rindt H., and, Lorson CL. Delivery of recombinant follistatin lessens disease severity in a mouse model of spinal muscular atrophy. Hum Mol Genet. 2009;18:997–1005. doi: 10.1093/hmg/ddn426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowerman M, Anderson CL, Beauvais A, Boyl PP, Witke W., and, Kothary R. SMN, profilin IIa and plastin 3: a link between the deregulation of actin dynamics and SMA pathogenesis. Mol Cell Neurosci. 2009;42:66–74. doi: 10.1016/j.mcn.2009.05.009. [DOI] [PubMed] [Google Scholar]
- Faustino NA., and, Cooper TA. Pre-mRNA splicing and human disease. Genes Dev. 2003;17:419–437. doi: 10.1101/gad.1048803. [DOI] [PubMed] [Google Scholar]
- Lorson CL, Hahnen E, Androphy EJ., and, Wirth B. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc Natl Acad Sci USA. 1999;96:6307–6311. doi: 10.1073/pnas.96.11.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorson CL., and, Androphy EJ. An exonic enhancer is required for inclusion of an essential exon in the SMA-determining gene SMN. Hum Mol Genet. 2000;9:259–265. doi: 10.1093/hmg/9.2.259. [DOI] [PubMed] [Google Scholar]
- Bebee TW, Gladman JT., and, Chandler DS. Splicing regulation of the survival motor neuron genes and implications for treatment of spinal muscular atrophy. Front Biosci. 2010;15:1191–1204. doi: 10.2741/3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson A, Osman E., and, Lorson CL. A negatively acting bifunctional RNA increases survival motor neuron both in vitro and in vivo. Hum Gene Ther. 2008;19:1307–1315. doi: 10.1089/hum.2008.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baughan T, Shababi M, Coady TH, Dickson AM, Tullis GE., and, Lorson CL. Stimulating full-length SMN2 expression by delivering bifunctional RNAs via a viral vector. Mol Ther. 2006;14:54–62. doi: 10.1016/j.ymthe.2006.01.012. [DOI] [PubMed] [Google Scholar]
- Owen N, Zhou H, Malygin AA, Sangha J, Smith LD, Muntoni F.et al. (2011Design principles for bifunctional targeted oligonucleotide enhancers of splicing Nucleic Acids Res 397194–7208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skordis LA, Dunckley MG, Yue B, Eperon IC., and, Muntoni F. Bifunctional antisense oligonucleotides provide a trans-acting splicing enhancer that stimulates SMN2 gene expression in patient fibroblasts. Proc Natl Acad Sci USA. 2003;100:4114–4119. doi: 10.1073/pnas.0633863100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skordis LA, Dunckley MG, Burglen L, Campbell L, Talbot K, Patel S.et al. (2001Characterisation of novel point mutations in the survival motor neuron gene SMN, in three patients with SMA Hum Genet 108356–357. [DOI] [PubMed] [Google Scholar]
- Singh NN, Androphy EJ., and, Singh RN. In vivo selection reveals combinatorial controls that define a critical exon in the spinal muscular atrophy genes. RNA. 2004;10:1291–1305. doi: 10.1261/rna.7580704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh NK, Singh NN, Androphy EJ., and, Singh RN. Splicing of a critical exon of human Survival Motor Neuron is regulated by a unique silencer element located in the last intron. Mol Cell Biol. 2006;26:1333–1346. doi: 10.1128/MCB.26.4.1333-1346.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coady TH, Baughan TD, Shababi M, Passini MA., and, Lorson CL. Development of a single vector system that enhances trans-splicing of SMN2 transcripts. PLoS ONE. 2008;3:e3468. doi: 10.1371/journal.pone.0003468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passini MA, Bu J, Richards AM, Kinnecom C, Sardi SP, Stanek LM.et al. (2011Antisense oligonucleotides delivered to the mouse CNS ameliorate symptoms of severe spinal muscular atrophy Sci Transl Med 372ra18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann Y, Lorson CL, Stamm S, Androphy EJ., and, Wirth B. Htra2-β1 stimulates an exonic splicing enhancer and can restore full-length SMN expression to survival motor neuron 2 (SMN2) Proc Natl Acad Sci USA. 2000;97:9618–9623. doi: 10.1073/pnas.160181697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartegni L., and, Krainer AR. Disruption of an SF2/ASF-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN1. Nat Genet. 2002;30:377–384. doi: 10.1038/ng854. [DOI] [PubMed] [Google Scholar]
- Liu Q., and, Dreyfuss G. A novel nuclear structure containing the survival of motor neurons protein. EMBO J. 1996;15:3555–3565. [PMC free article] [PubMed] [Google Scholar]
- Le TT, Pham LT, Butchbach ME, Zhang HL, Monani UR, Coovert DD.et al. (2005SMNDelta7, the major product of the centromeric survival motor neuron (SMN2) gene, extends survival in mice with spinal muscular atrophy and associates with full-length SMN Hum Mol Genet 14845–857. [DOI] [PubMed] [Google Scholar]
- Avila AM, Burnett BG, Taye AA, Gabanella F, Knight MA, Hartenstein P.et al. (2007Trichostatin A increases SMN expression and survival in a mouse model of spinal muscular atrophy J Clin Invest 117659–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butchbach ME, Edwards JD., and, Burghes AH. Abnormal motor phenotype in the SMNDelta7 mouse model of spinal muscular atrophy. Neurobiol Dis. 2007;27:207–219. doi: 10.1016/j.nbd.2007.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh RN. Evolving concepts on human SMN pre-mRNA splicing. RNA Biol. 2007;4:7–10. doi: 10.4161/rna.4.1.4535. [DOI] [PubMed] [Google Scholar]
- Singh NN, Seo J, Ottesen EW, Shishimorova M, Bhattacharya D., and, Singh RN. TIA1 prevents skipping of a critical exon associated with spinal muscular atrophy. Mol Cell Biol. 2011;31:935–954. doi: 10.1128/MCB.00945-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorson MA, Spate LD, Samuel MS, Murphy CN, Lorson CL, Prather RS.et al. (2011Disruption of the Survival Motor Neuron (SMN) gene in pigs using ssDNA Transgenic Res epub ahead of print; [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passini MA., and, Wolfe JH. Widespread gene delivery and structure-specific patterns of expression in the brain after intraventricular injections of neonatal mice with an adeno-associated virus vector. J Virol. 2001;75:12382–12392. doi: 10.1128/JVI.75.24.12382-12392.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]