Abstract
Hematopoietic stem cell (HSC) gene therapy offers promise for the development of new treatments for a variety of hematologic disorders. However, efficient in vivo modification of HSCs has proved challenging, thus imposing constraints on the therapeutic potential of this approach. Herein, we provide a gene-targeting strategy that allows site-specific in vivo gene modification in the HSCs of mice. Through conjugation of a triplex-forming peptide nucleic acid (PNA) to the transport peptide, antennapedia (Antp), we achieved successful in vivo chromosomal genomic modification of hematopoietic progenitor cells, while still retaining intact differentiation capabilities. Following systemic administration of PNA-Antp conjugates, sequence-specific gene modification was observed in multiple somatic tissues as well as within multiple compartments of the hematopoietic system, including erythroid, myeloid, and lymphoid cell lineages. As a true functional measure of gene targeting in a long-term renewable HSC, we also demonstrate preserved genomic modification in the bone marrow and spleen of primary recipient mice following transplantation of bone marrow from PNA-Antp-treated donor mice. Our approach offers a minimally invasive alternative to ex vivo gene therapy, by eliminating the need for the complex steps of stem cell mobilization and harvesting, ex vivo manipulation, and transplantation of stem cells. Therefore, our approach may provide new options for individualized therapies in the treatment of monogenic hematologic diseases such as sickle cell anemia and thalassemia.
Introduction
A substantial number of characterized genetic disorders can be attributed to changes in a single gene. Consequently, the concept of treating such diseases at the genetic point of origin has been appealing. While conventional gene therapy, which utilizes viral vectors for replacement gene delivery, has proven beneficial to some patients, its use is complex and is associated with some risks, including activation of an immune response and insertional oncogenesis. Allogeneic stem cell transplantation, a strategy aimed at replacing a patient's hematopoietic stem cells (HSCs) with those from a normal donor, addresses many of these factors, but is frequently limited by the availability of a compatible donor.1 To overcome this limitation, autologous transplantation of a patient's own HSCs that are genetically corrected ex vivo is an attractive therapeutic alternative. However, the ability to site-specifically modify the defective gene in a patient's HSCs in vivo would be even more advantageous.
To this end, we investigated the use of molecules such as triplex-forming peptide nucleic acids (PNAs) as tools for site-directed gene modification in vivo. These are achiral molecules comprised of purine and pyrimidine bases attached to a polyamide backbone that form a PNA:DNA:PNA triplex structure upon binding to target DNA sequences.2,3 PNA binding is sufficiently stable to block transcription initiation,4,5 suppress gene expression,6 and stimulate site-directed recombination of a co-transfected donor DNA, resulting in correction of a mutation in the target gene.7,8,9 Recent studies have further highlighted the potential utility of these gene-targeting molecules by identifying polypurine sites suitable for high-affinity triplex formation throughout the human genome.10
As a strategy for systemic administration of gene-targeting PNAs capable of modifying genes within selected hematopoietic compartments, we tested linkage of the PNAs to a cell-penetrating peptide (CPP) that is able to pass through cellular membranes and destabilize endosomal membranes.11,12 Cargoes that have been successfully internalized into cultured cells by CPPs range from small molecules to proteins.13,14 However, the use of such peptides to deliver PNA molecules to chromosomal sites as antigene agents in vivo has not been previously characterized. Because differences in intracellular localization have been reported among the various transport peptides, we focused on a defined fragment of the Drosophila antennapedia protein (Antp) that has been found to promote not only cellular uptake but also intranuclear targeting.15,16
We report here that triplex-forming PNAs covalently linked to the transport peptide Antp, can modify a target gene in the hematopoietic progenitor cells of mice following systemic administration via intraperitoneal (i.p.) injection. The treatment was found to produce genetic modification in multiple compartments within the hematopoietic system, including erythroid, myeloid, and lymphoid cell lineages. Furthermore, we demonstrate that the gene-modified HSCs retain their differentiation capabilities and were subsequently able to give rise to both erythroid and myeloid lineages. Identification of genomic modification in the whole bone marrow (wbm) and spleen of primary recipient mice following bone marrow transplantations from PNA-Antp-treated donor mice definitively provides evidence that systemic administration of these conjugates can be used to chromosomally modify HSCs in vivo. Moreover, the i.p. injection of the PNA-Antp conjugates also induced targeted gene modification in several somatic tissues, including liver and spleen. In addition, we were able to confirm increased bioavailability of the PNA-peptide conjugate following systemic administration through the use of fluorescence microscopy of tissue sections. Our work suggests PNA-peptide conjugates may provide an important step in the development of a gene-targeting strategy capable of inducing a heritable chromosomal change in tissue stem cells in vivo.
Results
Evaluation of PNA-Antp conjugate
In prior work, we developed a triplex-forming PNA (167-PNA) capable of modifying the chromosomally integrated supFG1 reporter gene in mouse cells.17 The supFG1 mutation reporter gene contains a homopurine/homopyrimidine target site to which the 167-PNA can bind to form a PNA:DNA:PNA triplex invasion complex,7 and prior work demonstrated that this complex triggers DNA repair as a means to catalyze genome modification.7 However, intracellular delivery of the PNAs in prior work was achieved by electroporation, a method unsuitable for in vivo administration.
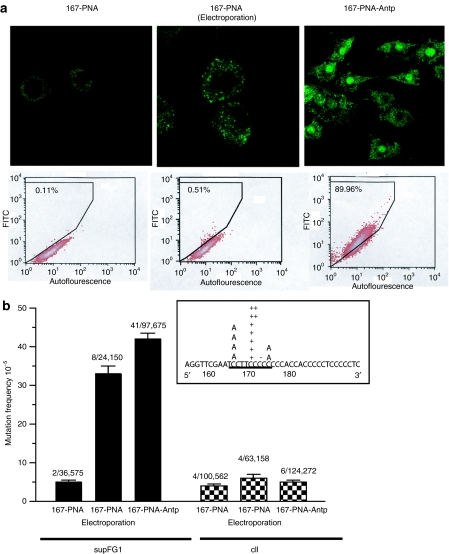
As an alternative delivery strategy that might ultimately be suitable for in vivo application, the supF-targeting PNA was synthesized in continuity with the Antp peptide. To evaluate cellular uptake in culture, fluorescein isothiocyanate (FITC)-labeled 167-PNA and 167-PNA-Antp were added to cells, followed by confocal microscopy and flow cytometry. To eliminate the possibility of artifactual uptake induced by cell fixation, microscopy was performed on live unfixed cells. As expected, there was minimal cellular uptake of FITC-167-PNA by passive diffusion (Figure 1a). The fluorescence detected following addition of 167-PNA to the cells was observed mainly in the cytoplasm. Cells were also transfected with FITC-167-PNA using electroporation, a method frequently used to deliver oligonucleotides. This method of transfection resulted in punctate distribution of fluorescence in the area surrounding the nucleus with negligible intranuclear uptake. However, treatment of the cells with FITC-167-PNA-Antp resulted in increased fluorescence throughout the cell. The majority of fluorescence was observed in the nucleus with lesser intensities detected in the cytoplasm. FACS analysis of cells treated with FITC-167-PNA and FITC-167-PNA-Antp support these results revealing 167-PNA-Antp to be the superior molecule for delivery (Figure 1a). These studies suggested that the CPP, Antp, was efficient at intracellular delivery and most importantly at mediating intranuclear distribution of the 167-PNA.
Figure 1.
Peptide nucleic acid (PNA)-mediated gene targeting in AV16 cells. (a) Cells were treated with FITC-167-PNA, electroporated FITC-167-PNA, or FITC-167-PNA-Antp, and examined by confocal microscopy and flow cytometry. (b) Targeted mutagenesis of the chromosomal supFG1 gene induced by 167-PNA and 167-PNA-Antp. Specificity of gene targeting was evaluated by analysis of mutation frequencies in the nontargeted control gene, cII. Sequence analysis of the supFG1 gene mutations induced after 167-PNA-Antp treatment. The PNA-binding site is underlined and (+) and (−) represent single base pair insertions and deletions. Antp, antennapedia; FITC, fluorescein isothiocyanate.
To further evaluate the effectiveness of PNA-Antp conjugation as a delivery strategy for chromosomal gene targeting, an assay for gene-targeted mutagenesis in mammalian cells was used. Because site-directed mutagenesis induced by PNAs at the supFG1 gene has been established in vitro,17 we utilized a mouse cell line (AV16), containing multiple copies of λsupFG1 shuttle vector DNA in a chromosomal locus.16 Using packaging extracts, the vector DNA can be isolated from genomic DNA into phage particles and subsequently analyzed for induced mutations. SupFG1 encodes an amber suppressor tRNA whose function can be scored in indicator bacteria. AV16 cells were treated with 167-PNA through addition to the media, electroporation or by addition to the medium via the Antp conjugate, 167-PNA-Antp. The cells were allowed 48 hours for mutation fixation before the vector DNA was isolated for genetic analysis. No gene modification above the background mutation frequency in the assay was observed when PNA was simply added to the cells (Figure 1b). This correlates with the negligible cellular uptake observed with the 167-PNA lacking the peptide conjugate in our microscopy studies. Because PNA is a neutrally charged molecule, and transfection by cationic lipids could not be used as a control without annealing to negatively charged DNA oligonucleotides,18 electroporation was used for comparison. PNA delivery via electroporation produced a mutation frequency (33 × 10−5) that was sevenfold higher than that obtained following direct addition of 167-PNA to the cells, but substantial cell death was observed following treatment. However, conjugation of 167-PNA to Antp resulted in a gene modification frequency (42 × 10−5) that was eightfold higher than observed in the 167-PNA-treated cells, with minimal cellular toxicity observed (Figure 1b). Hence, the Antp conjugation substantially enhances the bioactivity of the 167-PNA in cell culture.
DNA sequence analysis revealed that the majority of the mutations were located within the PNA-binding site and consisted of mostly single base pair insertions and deletions, consistent with previous studies.19 To provide additional evidence to support a mechanism of induced mutagenesis dependent on PNA clamp formation to a specific chromosomal target site and to confirm specificity of gene targeting, we screened the λ cII gene in the same vector for possible nonspecific off-target effects. We observed mutation frequencies at background levels in the cII gene, which does not contain the PNA-target site, in all of our treatment groups. Hence, one can conclude that in the absence of sequence-dependent PNA triplex clamp formation, the PNA-peptide conjugate does not induce detectable nonspecific mutations. Collectively, the analysis of the pattern of mutations and the evaluation of the nontargeted cII gene reveal that the increase in mutation frequency in supFG1 can be attributed to improved cellular uptake of the PNA-peptide conjugate and higher levels of sequence-specific binding of the PNA to the chromosomal target site.
In vivo biodistribution of PNA-Antp conjugate
The uptake and tissue distribution of systemically administered PNAs and PNA-Antp conjugates were then evaluated in mice using covalently linked rhodamine (Rhod) as a tag coupled with fluorescence microscopy. Because biodistribution studies have shown that oligonucleotides are rapidly eliminated from circulation following intravenous injection, we chose to administer the Rhod-labeled PNA compounds via i.p. injection and to collect tissue samples 2 hours and 24 hours following dosing. The tissues analyzed in the microscopy studies were fresh frozen and not fixed so as to avoid artifactual redistribution to the cytosol and nucleus.
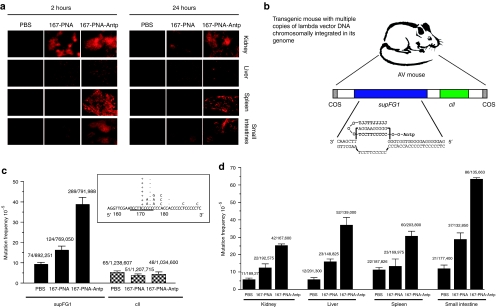
Rhod-167-PNA was administered to transgenic mice by i.p. injection. Except for the kidney, there was little uptake of Rhod-167-PNA in any of the tissue samples analyzed (Figure 2a). Also, the diffuse, intense fluorescence observed in the kidney at 2 hours was mostly eliminated by 24 hours, a phenomenon previously reported for unmodified PNAs.20 In contrast, conjugation of Rhod-167-PNA with Antp substantially improved PNA delivery to multiple tissues in vivo, as demonstrated by the higher levels of Rhod fluorescence observed in all tissues examined following i.p. injection of Rhod-167-PNA-Antp (Figure 2a). Rhod-167-PNA-Antp distribution in the kidney showed intense, diffuse fluorescent uptake throughout the tissue section. Although the intensity of fluorescence did decrease somewhat at 24 hours, the distribution of Rhod-167-PNA-Antp remained significantly greater at 24 hours than that of Rhod-167-PNA. This suggests that conjugation with Antp increases not only tissue uptake but also tissue retention. There was also substantial uptake in the small intestine and spleen at 2 hours and 24 hours for the Rhod-167-PNA-Antp molecule, with minimal fall off at 24 hours. In the liver, there was persistent uptake at 24 hours, but the fluorescence pattern in the liver samples was more punctate than observed in the other tissues. No visible toxicity to the mice was seen at the time of sacrifice as a result of PNA delivery. In the phosphate-buffered saline (PBS)-treated mice, all the tissues examined exhibited low background fluorescence, likely due to tissue autofluorescence.
Figure 2.
In vivo peptide nucleic acid (PNA)-Antp mediated gene targeting. (a) Mice were intraperitoneally (i.p.) injected with Rhod-167-PNA or Rhod-167-PNA-Antp. Tissues were harvested and visualized by fluorescence microscopy. (b) AV mice containing the chromosomally integrated supFG1 reporter gene and the control cII gene. The 167-PNA-Antp conjugate was designed to bind to the homopurine strand at positions 167–176 of the supFG1 gene. In this structure, one strand forms Watson–Crick hydrogen bonds with the purine-rich strand of the target duplex, while the other PNA strand forms Hoogsteen base pairs to the purine DNA strand in the PNA-DNA duplex. This strand invasion complex results in the displacement of the other DNA strand to form a D-loop, generating a PNA:DNA:PNA triple helix. (c) AV mice were treated with 167-PNA, 167-PNA-Antp, or phosphate-buffered saline (PBS). Tissue samples were harvested and analyzed by vector rescue from genomic DNA for analysis of the supFG1 reporter gene. Mutation frequencies were calculated and the combined totals from all tissues tested are displayed. Targeting specificity was evaluated by analysis of mutation frequencies in the nontargeted control gene, cII. Sequence analysis of the supFG1 gene mutations induced by treatment with 167-PNA-Antp further confirming site-specificity. The target site is underlined and (+) and (−) represent single base pair insertions and deletions. (d) Targeted mutagenesis of the supFG1 gene in selected somatic tissues of AV mice. Antp, antennapedia.
Targeted gene modification in mouse somatic tissue
To further evaluate the in vivo therapeutic applicability of the 167-PNA-Antp conjugate, we tested for targeted supF gene modification in the AV transgenic mouse model, which contains multiple copies of the λsupFG1 shuttle vector DNA chromosomally integrated into the genome (Figure 2b). As with the λ-containing cells, the vector DNA can be isolated from mouse genomic DNA and packaged into phage particles for subsequent analysis for induced mutations.21
I.p. injections of PBS, 167-PNA, or 167-PNA-Antp were administered on days 1 and 3 to AV mice at a concentration of 20 mg/kg for each dose with no signs of systemic toxicity being observed. Observations of the condition of the mice (behavior, grooming, feeding, and general appearance) throughout the study period revealed no difference between the three treatment groups. Fifteen days after treatment, tissue samples were harvested and analyzed for targeted mutations in the supFG1 gene. Necropsy findings for the mice at the time of euthanasia exposed no gross tissue toxicity or damage in any of the study mice. The combined mutation frequencies from a variety of tissues from the 167-PNA-treated mice (16 × 10−5) were found to be twofold higher than those obtained from control, PBS-treated mice (Figure 2c). This result implies that “naked” PNAs have some ability to enter cells in vivo, consistent with the low-level uptake observed in the microscopy experiments. However, combined mutation frequencies from 167-PNA-Antp-treated mice averaged 37 × 10−5, a substantial increase compared with the background seen in PBS-treated mice (Figure 2c) and significantly higher than those from the 167-PNA-treated mice, representing a more than fourfold increase in the observed mutation frequencies upon conjugation with Antp. In previous studies, we had tested the ability of “naked” DNA oligonucleotides (only end-capped with terminal amines to prevent degradation) to mediate gene targeting in mice via systemic administration, as done here.21 In that work, we observed average induced mutation frequencies of 27 × 10−5, somewhat higher than we have found here with “naked” PNAs.21 However, the targeting frequencies with “naked” DNA oligomers were lower than those observed with the PNA-peptide conjugate even though the DNA oligomers were administered in sixfold higher doses. Hence, it is likely that peptide conjugation would also serve to improve the in vivo bioactivity of triplex-forming DNA oligonucleotides, although this remains to be tested.
DNA sequence analysis of mutated supFG1 genes rescued from the 167-PNA-Antp-treated mice revealed that the majority of the mutations were located precisely within the PNA-binding site. The mutations consisted of mostly single base pair insertions and deletions (Figure 2c), consistent with the results from cell culture studies (Figure 1b). The majority of induced mutations were located within a stretch of eight consecutive G's within the PNA-binding site. Because this site has been previously shown to be prone to slippage events,22 it is likely that these mutations were produced by slippage errors during repair of the PNA-clamped structure. Our prior studies have shown that the ability of a PNA:DNA:PNA triple helix clamp to induce DNA repair is dependent upon the nucleotide excision repair pathway and is specifically XPA-dependent.7 As a result, it is essential to note that some of the bound PNA structures may have been repaired without mutation, so the supFG1 mutation read-out likely provides an underestimate of the true in vivo targeting frequency.
To provide additional evidence to support the specificity of the supFG1-targeted PNA, we screened the λ cII gene in the same vector for possible off-target mutagenesis by the PNA-peptide conjugate. We found that the cII gene, which does not contain the PNA-binding site, exhibited no induction of mutagenesis in the mice treated with either 167-PNA or 167-PNA-Antp compared with background levels (Figure 2c). Hence, there are no detectable off-target effects in the absence of the cognate target sequence for PNA clamp formation.
The tissue distribution of the PNA-induced mutagenesis was examined in detail by analysis of supFG1 genes rescued from the kidney, liver, spleen, and small intestine (Figure 2d). In some of the tissues examined, there was a small induction of mutations by 167-PNA. However, 167-PNA conjugation to Antp yielded significantly increased mutation frequencies in all tissues ranging from 25 × 10−5 in the kidney samples to 63 × 10−5 in the small intestine. These data indicate that PNA conjugation to Antp provides broad tissue distribution and intranuclear biological activity following administration via i.p. injections.
Sequence-specific gene targeting in wbm
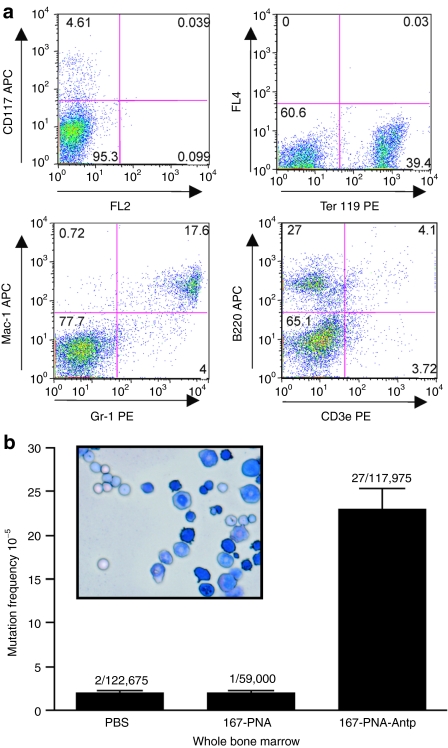
The results from these experiments prompted us to assess the feasibility of a strategy to utilize PNA-peptide conjugates as a noninvasive method to genetically modify cells within the hematopoietic system. Therefore, we investigated sequence-specific gene targeting in mouse wbm. WBM was harvested from AV mice 15 days after systemic administration of 167-PNA or 167-PNA-Antp. To determine composition of the wbm cell population harvested, expression markers for the major hematopoietic cell lineages were analyzed by flow cytometry and no significant difference in cell composition was detected between the treatment groups (Figure 3a). Morphologic analysis of the isolated wbm cells supports a heterogeneous population of hematopoietic cell lineages including erythroid, myeloid, and lymphoid. Genomic DNA was isolated from the wbm cells and analyzed for the induction of supFG1 mutations. Results indicated no induction of gene-targeted mutations in the 167-PNA-treated mice (2 × 10−5), with an identical mutation frequency as observed with the control PBS-treated mice (Figure 3b). However, 167-PNA conjugated to Antp significantly increased the chromosomal gene-targeting efficiency to 23 × 10−5, a 14-fold increase above background in mutation frequency.
Figure 3.
Sequence-specific gene targeting in whole bone marrow (wbm) cells. (a) FACS analysis for hematopoietic progenitor cell (CD117), erythroid (Ter119), lymphoid (B220 and CD3e), and myeloid (Mac-1 and Gr-1) markers of cells derived from the wbm of AV mice. Analysis is representative of both untreated and treated AV mice. (b) Morphology of wbm cells isolated from AV mice as analyzed on cytospins by hematologic staining. AV mice were treated with phosphate-buffered saline (PBS), 167-PNA, and 167-PNA-Antp. Samples analyzed for the induction of mutations 15 days post-treatment. Antp, antennapedia; PNA, peptide nucleic acid.
Induced genomic modification in multiple hematopoietic cell lineages
For clinical application in the treatment of monogenic hematopoietic diseases, the ultimate in vivo cellular target of the 167-PNA-Antp molecules would be hematopoietic stem and progenitor cells. Such an in vivo gene-targeting strategy (with PNAs designed to bind to disease-related genes, such as β-globin) would be expected to eliminate the requirement for autologous transplantation of HSCs genetically modified ex vivo. Therefore, we tested the ability of the PNA-Antp conjugate to induce site-specific mutations in hematopoietic progenitor cells and in multiple hematopoietic cell lineages following systemic administration. WBM cells were harvested from 167-PNA-Antp-treated mice and separated into three fractions using immunomagnetic techniques (Figure 4a): (i) erythroid lineage cells (Ter119+), (ii) myeloid lineage cells (Mac-1+, Gr-1+), and (iii) lineage marker-negative hematopoietic progenitor cells (CD117+). Lymphoid lineage cells (CD3e+, CD8a+, CD4+, CD45R/B220+) were isolated from splenic cells.
Figure 4.
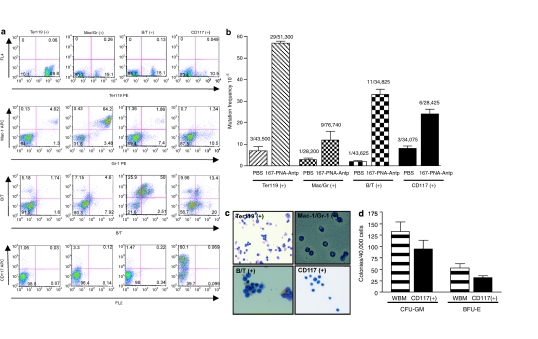
Targeted genome modification in multiple cell lineages. (a) FACS analysis for erythroid, myeloid, lymphoid, and hematopoietic progenitor markers following immunomagnetic separation of whole bone marrow (wbm) and splenic mouse cells. (b) Targeted mutagenesis of the supFG1 gene in multiple hematopoietic cell lineages: Ter119 (erythroid), Mac/Gr (myeloid), B/T (lymphoid), CD117 (hematopoietic progenitor cells). Mutation frequencies were calculated as previously described and standard errors were calculated as indicated by the error bars. (c) Morphology of Ter119, Mac/Gr, CD117, and B/T enriched cell lineages isolated from AV mouse wbm and spleen. (d) Colony formation ability of wbm and CD117+ cells isolated from 167-PNA-Antp-treated mice. Antp, antennapedia; BFU-E, burst-forming unit-erythroid; CFU-GM, colony-forming unit-granulocyte macrophage; PNA, peptide nucleic acid.
As shown in Figure 4b, targeted mutations were observed in all of the cell types analyzed. The highest mutation frequency was observed in the Ter119+ erythroid lineage cells, with a 167-PNA-Antp induced mutation frequency of 57 × 10−5. Although myeloid lineage cells had an induced mutation frequency above background (12 × 10−5), it was lower than that observed with the erythroid cells. B and T lymphocytic cells had an observed mutation frequency of 32 × 10−5. The uncommitted hematopoietic progenitor cells (CD117+) had an induced mutation frequency of 21 × 10−5. These results indicate that in vivo administration of the 167-PNA-Antp conjugate can produce targeted mutations not only in erythroid, myeloid, and lymphoid cells, but also in hematopoietic progenitor cells, which can subsequently give rise to genetically modified differentiated, lineage-specific cell populations.
In support of this, colony-forming assays indicate that CD117+ cells isolated from 167-PNA-Antp-treated mice were able to differentiate into granulocyte and monocyte/macrophage (colony-forming unit-granulocyte macrophage) and erythrocyte (burst-forming unit-erythroid) (Figure 4d). Detection of sequence-specific mutations in the Ter119+ population 15 days after 167-PNA-Antp treatment, further supports initial gene targeting within hematopoietic progenitor cells. During erythropoiesis, erythrocytes develop from proerythroblasts to mature anucleated red blood cells within ~7 days. In our studies, we analyzed induced mutations in the chromosomal DNA isolated from nucleated Ter119+ erythroid cells. In order to account for the presence of targeted mutations in the nucleated erythroid cells, gene targeting must have occurred in uncommitted HSCs. Detection of induced mutations up to 15 days after treatment also indicates a heritable change. While it is possible that separate erythroid, myeloid, and lymphoid progenitor cells may have been targeted by 167-PNA-Antp, these results clearly demonstrate that hematopoietic cells can be chromosomally modified in mice after only two doses of systemically administered 167-PNA-Antp. In fact, the self-renewal and differentiation capabilities of the hematopoietic progenitor CD117+ cells ensure that gene modification in a small population of cells will result in modification of a greater number of cells.
Targeted HSCs retain self-renewal and differentiation capabilities
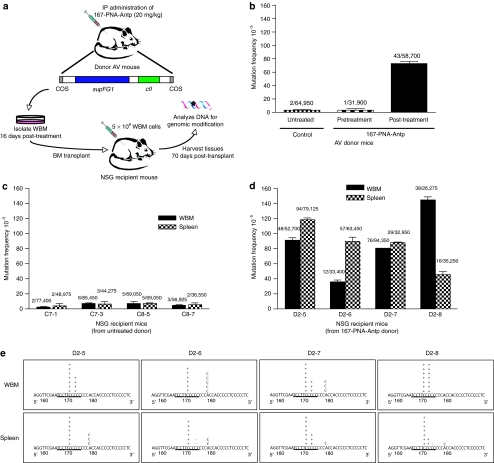
The true functional measure of a long-term renewable stem cell is the capability to engraft myeloablated recipients, repopulate their hematopoietic systems and sustain long-term multilineage hematopoiesis in vivo. In order to further confirm targeted genomic modification of HSCs in the 167-PNA-Antp-treated mice and provide definitive evidence of preservation of their self-renewal and differentiation capabilities, bone marrow transplantations were conducted using NOD-scid IL2rγ null (NSG) mice as recipients23 (Figure 5a). AV donor mice were treated (or not) as previously described and wbm was harvested 16 days post-treatment. Mutation frequencies of the AV donor mice were evaluated pre- and post-treatment. The 167-PNA-Antp-treated donor was determined to have a mutation frequency of 73 × 10−5 following administration of two systemic doses of 167-PNA-Antp by i.p. injection (Figure 5b). NSG recipient mice, which lack chromosomally integrated supFG1 reporter genes, were sublethally irradiated and intravenously injected with T-cell-depleted bm cells isolated from either untreated or 167-PNA-Antp-treated AV donor mice. Engraftment levels were ascertained 4 and 8 weeks following transplantation by detecting the presence of the supFG1 reporter gene in the peripheral blood. NSG recipient mice transplanted with bone marrow from untreated control AV donor mice showed a mutation frequency at background levels similar to that seen in the untreated donor mice. These results indicate that the process of transplantation itself is not mutagenic and lends support that any observation of induced mutations can be solely attributed to HSC gene targeting that had occurred in the donor mice (Figure 5c). Genomic DNA was isolated from the wbm and spleen of NSG recipient mice transplanted with bone marrow from 167-PNA-Antp-treated AV donor mice and then analyzed for the induction of supFG1 mutations. As shown in Figure 5d, targeted mutations were observed in the wbm and spleen of all of these NSG recipient mice, with mutation frequencies at or above that observed for the donor mice. In fact, one of the recipient mice (D2–8) had a mutation frequency of 145 × 10−5, which is twofold higher than that of the AV donor. These results emphasize that the self-renewal and differentiation characteristics of hematopoietic progenitor cells mean that modification of a small population of cells can result in modification of significantly greater numbers of genomically modified cells. DNA sequence analysis of mutated supFG1 genes rescued from the bm and spleen of the recipient mice reveal that the majority of the mutations were located precisely within the PNA-target site. As previously observed in the DNA of the directly PNA-treated mice, the mutations consisted of mostly single base pair insertions or deletions at the PNA target site. Identification of the targeted genomic modification in the wbm and spleen of recipient mice following bone marrow transplantation provides definitive evidence that systemic administration of PNA-Antp conjugates can successfully mediate gene targeting and modification in true HSCs.
Figure 5.
In vivo genomic modification of hematopoietic stem cells (HSCs) demonstrated by transplantation. (a) Scheme for bone marrow transplantations. (b) Targeted mutagenesis of the supFG1 gene in AV donor mice. Mutation frequencies were determined pre- and post-treatment with 167-PNA-Antp as previously described and standard errors were calculated as indicated by the error bars. (c) Analysis of the supFG1 gene for induced mutations in the whole bone marrow (wbm) and spleen of NSG recipient mice transplanted with bone marrow (bm) from untreated AV donor mice. (d) Identification of genomic modification in the wbm and spleen of primary NSG recipient mice following bm transplantations from AV donor mice treated by systemic administration of 167-PNA-Antp. (e) Sequence analysis of the supFG1 gene mutations induced by modified HSCs. The triplex-target site is underlined and (+) and (−) represent single base pair insertions and deletions. Antp, antennapedia; PNA, peptide nucleic acid.
Discussion
In this study, we have shown that conjugation of a PNA to the CPP, Antp, substantially increases the ability of the PNA to mediate targeted mutagenesis in multiple mouse tissues following systemic administration by i.p. injection in vivo compared with the “naked” PNA. Microscopy studies demonstrated cellular uptake in a number of mouse tissues following i.p. injection with Rhod-167-PNA-Antp, consistent with the gene-targeting data. Moreover, we demonstrate that this antigene agent is capable of targeted chromosomal gene modification in hematopoietic progenitor cells which are able to give rise to both erythroid and myeloid lineages, as indicated by colony-forming assays. In addition, we have shown that 167-PNA-Antp-induced gene modification can persist for at least 2 weeks after systemic administration and is present not only in hematopoietic progenitor cells but also in multiple hematopoietic cell lineages. The induced mutations were found to be site-specific as demonstrated by both sequence analysis and by the absence of induced mutations in a control gene, which contains a polypurine stretch but lacks the exact PNA target sequence. The cII gene was chosen as a control because it is conveniently located within the same construct as the target supFG1 gene and can be evaluated for the induction of mutations using a similar assay. However, we recognize that there is a possibility for PNA binding to occur at other polypurine sites elsewhere in the genome that were not evaluated in our present study.
Our study therefore suggests that this approach could provide the basis for new therapeutic alternatives for treatment of monogenetic hematopoietic diseases. Currently, allogeneic stem cell transplantation offers the only potential cure for hematologic disorders such as sickle cell anemia and thalassemia.24,25,26 However, the limited availability of histocompatible donors combined with the associated adverse effects such as graft-versus-host disease and infection limit the applicability of this therapeutic strategy. As a result, treatment of these diseases is usually aimed at avoiding crisis, relieving symptoms, and preventing complications.
As one alternative, efforts have focused on autologous stem cell transplantation of genetically modified HSCs. Modification strategies include gene correction using oligonucleotides8,9,27,28 or short DNA fragments29,30,31 or gene replacement via viral vectors.32,33,34 Such HSC-targeted gene therapy strategies offer potential methods to cure diseases in patients unable to find a compatible donor, and also alleviates some of the side effects associated with allogeneic transplants.
However, one obstacle for ex vivo HSC gene modification is the harvesting and ex vivo manipulation of sufficient numbers of hematopoietic progenitor cells necessary for sufficient re-engraftment of the corrected cells. The use of traditional methods, such as granulocyte colony-stimulating factor to mobilize HSCs, has limitations because it has been found that granulocyte colony-stimulating factor precipitates a severe pain crisis and acute chest syndrome in sickle patients.35,36 In vivo gene targeting of HSCs through the use of PNA-peptide conjugates eliminates the need for complex stem cell mobilization and harvesting, thus representing a more feasible therapeutic approach. Another aspect that has proven challenging is the retention of stem cell properties following ex vivo manipulation, in particular the self-renewal and proliferative capability of these cells. Our studies have demonstrated that PNA-peptide molecules are capable of introducing heritable changes in hematopoietic progenitor cells and that this change is detectable up to 15 days post-treatment in the treated mice themselves and up to 70 days in transplanted recipients. In addition, the PNA-Antp-treated HSCs maintained their pluripotency and were capable of differentiating into both erythroid and myeloid lineages. Primary bone marrow transplantations not only confirmed sequence-specific gene targeting of HSCs at the triplex-target site, but also provide unequivocal evidence that the targeted cells maintain their “stemness.” Indeed, the hematopoietic system in the recipients was repopulated with donor cells carrying the targeted mutation in the reporter gene, which, based on control experiments, could only have been produced in the donor mice. The higher mutation frequencies observed in the recipient mice also emphasize the clinical relevance of this gene-targeting strategy and signify that targeting of a few HSCs can guarantee long-term genomic modification of a larger population of cells. Although a single systemic administration of the PNA-peptide conjugates may be insufficient to achieve a clinically significant level of target gene modification, multiple doses, possible because of the ease of administration, can be given to attain the desired therapeutic threshold. Gene modification of HSCs following systemic administration of PNA-Antp conjugates therefore potentially addresses many of the complications encountered by autologous transplant strategies.
Although our current studies have focused on applying this technology to disorders that affect erythroid cells, this gene-targeting strategy has broader implications and could be implemented in the treatment of other monogenetic diseases. Chronic granulomatous disease is a primary immunodeficiency disorder that affects phagocytes of the innate immune system resulting in failure to kill a wide spectrum of bacteria and fungi.37 This leads to recurrent or persistent infections and hyperinflammation with widespread tissue granuloma formation. Currently the only curative option for patients with this disease is HSC transplantation. Although the overall success rate for patients fortunate to have a compatible donor is 81%, major risk factors are still associated, including graft-versus-host disease and severe inflammation at the time of neutrophil engraftment in response to ongoing inflammation and infection.38,39 Moreover, recent data has revealed that healthy carriers of X-linked chronic granulomatous disease have ≥10% normal neutrophils.40,41 This observation would suggest that functional correction of a small fraction of chronic granulomatous disease neutrophils might be sufficient to alleviate the symptoms of this disease. However, current ex vivo gene therapy strategies have been faced with problems achieving sufficient engraftment of gene-transduced HSCs with intact multipotency to provide long-term corrections.38,42 PNA-peptide-mediated gene targeting via systemic administration could address some of these limitations and so might be applicable to chronic granulomatous disease. In addition, the high levels of gene targeting observed in the liver in our experiments point to possible applications in liver-associated diseases such as hemophilia A, a severe X-linked bleeding disorder caused by a hereditary deficiency of factor VII. Targeting of factor VIII expression in liver sinusoid endothelial cells has been shown to result in phenotypic correction, and only a few corrected genes may be required to achieve therapeutic benefit. Taken together, our results demonstrate that systemically administered PNA-peptide conjugates can be effective reagents for in vivo gene targeting in multiple tissue compartments with potential application to the hematopoietic system and associated disorders.
Materials and Methods
PNA and peptide conjugates. PNA oligomers were purchased from BioSynthesis (Lewisville, TX). The 167-PNA oligomer, used for targeting the supFG1 gene, was synthesized with the sequence JJJJJTTJJT-O-O-O-TCCTCCCCCCKKK (O=8-amino-2,6,dioxaoctanoic acid), and designed to bind as a clamp to the homopurine strand at positions 167–176 of the gene, as previously described.17 The CPP, antennapedia, was covalently linked to the PNA at the C-terminal lysine with the following sequence, 167-PNA-O-O-KKKKKWKMRRNQFWIKIQR (167-PNA-Antp). FITC and Rhod were conjugated to the PNA at the N-terminus via two O linkers.
Cell culture and transfection. The mouse fibroblast line, AV16, containing ~100 chromosomally integrated copies of the λsupFG1 shuttle vector was used in the microscopy and mutagenesis assay.16 AV16 cells were treated with 167-PNA or 167-PNA-Antp and electroporation of 167-PNA was used as a positive control. Cells were collected for shuttle vector rescue and analysis 2–4 days following treatment. Genomic DNA was isolated from the cells and incubated with λ in vitro packing extracts for shuttle vector rescue and reporter gene analysis as previously described. Mutation frequency was calculated by dividing the number of colorless mutant plaques by the total number of plaques counted. Experiments were done in triplicate and standard errors were calculated for the mutation frequencies as indicated by the error bars.
Generation of transgenic mouse model. The AV supFG1 mice were derived from the CD1 background (Charles River Laboratories, Wilmington, MA) and were generated as described for the 3340 supFG1 mice.22 DNA dot blot analysis confirmed the AV founder mouse to carry ~50 copies of the λ shuttle vector DNA in their genomes. The mouse was bred with CD1 males and the presence or absence of the supFG1 reporter gene in the resulting pups was determined by PCR as described previously.22 Animal studies were approved and performed according to the guidelines of the Institutional Animal Care and Use Committee of Yale University.
Microscopy. AV16 cells were treated with 2 µmol/l of FITC-labeled 167-PNA and 167-PNA-Antp. For direct detection of uptake, live cells were analyzed 4 hours post-treatment on a Zeiss LSM 510 META confocal laser microscope as previously described.16 Transgenic AV mice were treated by i.p. with Rho-167-PNA or Rho-167-PNA-Antp (20 mg/kg). The mice were sacrificed 2 hours and 24 hours post-treatment, and tissue samples were harvested. Frozen 10 µm tissue sections were prepared, thaw mounted onto glass slides, and stored at −20 °C. Images of each tissue were taken by fluorescence microscopy using a Ziess Axiovert 200 microscope (Ziess, Thornwood, NY).
Targeted mutagenesis in the somatic tissues of mice. AV mice (14-days old) were treated i.p. with PBS, PNA, or PNA-Antp (20 mg/kg) on days 1 and 3. Mice were sacrificed 15 days after last treatment and tissue samples were collected. Genomic DNA was isolated and analyzed for induction of mutations as previously described.21 Sequence analysis was used to characterize the mutations induced by PNA treatment. Each treatment group consisted of three mice and standard errors were calculated for the mutation frequencies as indicated by the error bars.
Multiple cell lineage separation and isolation of hematopoietic progenitor cells. All biotinylated antibodies and magnetic nanoparticles used were purchased from BD Bioscience (San Jose, CA) and used according to manufacturer's instructions. Six AV mice (14 days old) were treated i.p. with PNA-Antp (20 mg/kg) on days 1 and 3. On day 15, wbm was harvested from the femurs and tibias of the mice by flushing the medullary cavities. A single cell suspension was incubated with anti-mouse Ter119 particles (50 µl/107 cells) and positive fraction was isolated. Fc receptors of the negative fraction were blocked with anti-mouse CD16 and CD32 antibodies (0.25 µg/106 cells; eBioscience, San Diego, CA) and myeloid lineage cells were positively selected by concurrently labeling with anti-mouse Ly-6G and Ly-6C(Gr-1) particles (50 µl/107 cells) and anti-mouse CD11b particles (50 µl/107 cells). The negative cell fraction was then enriched for HSCs using a biotinylated mouse lineage depletion cocktail which contains monoclonal antibodies to mouse CD3e, CD11b, CD45R/B220, Ly-6G, and Ly6C(Gr-1) and Ter119. Cells were also harvested from the spleens and Fc receptor blockade was performed as described above. A single-cell suspension was incubated with the following biotinylated anti-mouse antibodies: CD3e, CD4, CD8a, and CD45R/B220 (5 µl/106 cells). Splenic B 5 × 106 and T 5 × 106 cells were positively selected. All purified lineage populations were analyzed by FACS and genomic DNA was isolated and analyzed for the induction of mutations, as previously described.
Bone marrow transplantations. AV mice were treated with 167-PNA-Antp as previously described. WBM was harvested 16 days after last treatment from the femurs and tibias of the mice by flushing the medullary cavities. NOD-scid IL2rγnull (NSG) mice (female/4–6 weeks) were sublethally irradiated with 240 cGy using a 137Cs irradiator and then hematopoietically reconstituted 4 hours later with T-cell depleted wbm from the AV mice. BM cells from two independent 167-PNA-Antp-treated mice were transplanted into eight primary recipient NSG mice (four recipients/donor mouse) by i.v. injection at 5 × 106 cells/mouse. In addition, wbm cells originating from two independent untreated control AV mice were also transplanted into eight additional NSG recipient mice. The mice were given autoclaved food and maintained on sulfamethoxazole-trimethoprim-medicated water provided on alternative weeks. Tissue samples were collected 70 days post-transplantation. Genomic DNA was isolated and analyzed for induction of mutations as previously described.
Colony assays. A cell suspension of mouse wbm cells or uncommitted hematopoietic progenitor cells was mixed with MethoCult M3436 for burst-forming unit-erythroid and MethoCult M3434 (Stem Cell Technologies, Vancouver, British Columbia, Canada) for colony-forming unit-granulocyte macrophage following manufacturer's instructions. All assays were performed in duplicate. For the burst-forming unit-erythroid assay, colonies were scored on day 14. Myeloid colonies were assayed on day 12.
Flow cytometry. All monoclonal antibodies used were purchased from BD Bioscience. Cells were resuspended (1–2 × 107 cells/ml) in PBS with 2% fetal bovine serum and 2 mmol/l EDTA and Fc receptors were blocked as previously described. Cells were stained with the following antibodies: allophycocyanin CD117, Phycoerythrin (PE) Ter119, allophycocyanin Mac-1, PE Gr-1, allophycocyanin B220, PE CD3e, PE CD4, PE CD8a and the appropriate isotypes. The cells were analyzed on a FACSCalibur flow cytometer.
Acknowledgments
We are grateful to Jose Sangerman, Patrick Gallagher, Leonard Milstone, and Sara Rockwell for helpful commentary and Erica Schleifman for help in maintaining the AV mouse colony. In addition, we thank David Serreze, Leonard Shultz, and Dale Greiner for their expertise in mouse bone marrow transplantations. This work was supported by the National Institutes of Health (R01 HL082655 to P.M.G.; K22 CA120049 to F.A.R.) and the Yale Center of Excellence in Molecular Hematology (NIH DK072442). F.A.R. contributed to overall experimental design and specifically designed PNA-Antp, assisted in the generation of mouse model, and performed microscopy, mutagenesis, and wbm and hsc isolation experiments. S.S.L. performed FACS analysis, CFU experiments, and assisted in isolation of wbm and hsc cells. D.C.H. generated transgenic mouse model, and analyzed off-target effects. D.S.K. and P.M.G. contributed to experimental design and writing of the manuscript.
REFERENCES
- Biffi A., and, Cesani M. Human hematopoietic stem cells in gene therapy: pre-clinical and clinical issues. Curr Gene Ther. 2008;8:135–146. doi: 10.2174/156652308784049381. [DOI] [PubMed] [Google Scholar]
- Bukanov NO, Demidov VV, Nielsen PE., and, Frank-Kamenetskii MD. PD-loop: a complex of duplex DNA with an oligonucleotide. Proc Natl Acad Sci USA. 1998;95:5516–5520. doi: 10.1073/pnas.95.10.5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen PE, Egholm M., and, Buchardt O. Peptide nucleic acid (PNA). A DNA mimic with a peptide backbone. Bioconjug Chem. 1994;5:3–7. doi: 10.1021/bc00025a001. [DOI] [PubMed] [Google Scholar]
- Praseuth D, Grigoriev M, Guieysse AL, Pritchard LL, Harel-Bellan A, Nielsen PE.et al. (1996Peptide nucleic acids directed to the promoter of the alpha-chain of the interleukin-2 receptor Biochim Biophys Acta 1309226–238. [DOI] [PubMed] [Google Scholar]
- Janowski BA, Kaihatsu K, Huffman KE, Schwartz JC, Ram R, Hardy D.et al. (2005Inhibiting transcription of chromosomal DNA with antigene peptide nucleic acids Nat Chem Biol 1210–215. [DOI] [PubMed] [Google Scholar]
- Hu J., and, Corey DR. Inhibiting gene expression with peptide nucleic acid (PNA)–peptide conjugates that target chromosomal DNA. Biochemistry. 2007;46:7581–7589. doi: 10.1021/bi700230a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers FA, Vasquez KM, Egholm M., and, Glazer PM. Site-directed recombination via bifunctional PNA-DNA conjugates. Proc Natl Acad Sci USA. 2002;99:16695–16700. doi: 10.1073/pnas.262556899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonkar P, Kim KH, Kuan JY, Chin JY, Rogers FA, Knauert MP.et al. (2009Targeted correction of a thalassemia-associated beta-globin mutation induced by pseudo-complementary peptide nucleic acids Nucleic Acids Res 373635–3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin JY, Kuan JY, Lonkar PS, Krause DS, Seidman MM, Peterson KR.et al. (2008Correction of a splice-site mutation in the beta-globin gene stimulated by triplex-forming peptide nucleic acids Proc Natl Acad Sci USA 10513514–13519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Gaddis SS, MacLeod MC, Walborg EF, Thames HD, DiGiovanni J.et al. (2007High-affinity triplex-forming oligonucleotide target sequences in mammalian genomes Mol Carcinog 4615–23. [DOI] [PubMed] [Google Scholar]
- Vivès E, Brodin P., and, Lebleu B. A truncated HIV-1 Tat protein basic domain rapidly translocates through the plasma membrane and accumulates in the cell nucleus. J Biol Chem. 1997;272:16010–16017. doi: 10.1074/jbc.272.25.16010. [DOI] [PubMed] [Google Scholar]
- Bongartz JP, Aubertin AM, Milhaud PG., and, Lebleu B. Improved biological activity of antisense oligonucleotides conjugated to a fusogenic peptide. Nucleic Acids Res. 1994;22:4681–4688. doi: 10.1093/nar/22.22.4681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mäe M., and, Langel U. Cell-penetrating peptides as vectors for peptide, protein and oligonucleotide delivery. Curr Opin Pharmacol. 2006;6:509–514. doi: 10.1016/j.coph.2006.04.004. [DOI] [PubMed] [Google Scholar]
- Manceur A, Wu A., and, Audet J. Flow cytometric screening of cell-penetrating peptides for their uptake into embryonic and adult stem cells. Anal Biochem. 2007;364:51–59. doi: 10.1016/j.ab.2007.02.015. [DOI] [PubMed] [Google Scholar]
- Derossi D, Joliot AH, Chassaing G., and, Prochiantz A. The third helix of the Antennapedia homeodomain translocates through biological membranes. J Biol Chem. 1994;269:10444–10450. [PubMed] [Google Scholar]
- Rogers FA, Manoharan M, Rabinovitch P, Ward DC., and, Glazer PM. Peptide conjugates for chromosomal gene targeting by triplex-forming oligonucleotides. Nucleic Acids Res. 2004;32:6595–6604. doi: 10.1093/nar/gkh998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faruqi AF, Egholm M., and, Glazer PM. Peptide nucleic acid-targeted mutagenesis of a chromosomal gene in mouse cells. Proc Natl Acad Sci USA. 1998;95:1398–1403. doi: 10.1073/pnas.95.4.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton SE, Simmons CG, Kathiriya IS., and, Corey DR. Cellular delivery of peptide nucleic acids and inhibition of human telomerase. Chem Biol. 1999;6:343–351. doi: 10.1016/S1074-5521(99)80046-5. [DOI] [PubMed] [Google Scholar]
- Faruqi AF, Egholm M., and, Glazer PM. Peptide nucleic acid-targeted mutagenesis of a chromosomal gene in mouse cells. Proc Natl Acad Sci USA. 1998;95:1398–1403. doi: 10.1073/pnas.95.4.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon BM, Mays D, Lipsky J, Stewart JA, Fauq A., and, Richelson E. Pharmacokinetics and tissue distribution of a peptide nucleic acid after intravenous administration. Antisense Nucleic Acid Drug Dev. 2002;12:65–70. doi: 10.1089/108729002760070803. [DOI] [PubMed] [Google Scholar]
- Vasquez KM, Narayanan L., and, Glazer PM. Specific mutations induced by triplex-forming oligonucleotides in mice. Science. 2000;290:530–533. doi: 10.1126/science.290.5491.530. [DOI] [PubMed] [Google Scholar]
- Narayanan L, Fritzell JA, Baker SM, Liskay RM., and, Glazer PM. Elevated levels of mutation in multiple tissues of mice deficient in the DNA mismatch repair gene Pms2. Proc Natl Acad Sci USA. 1997;94:3122–3127. doi: 10.1073/pnas.94.7.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shultz LD, Lyons BL, Burzenski LM, Gott B, Chen X, Chaleff S.et al. (2005Human lymphoid and myeloid cell development in NOD/LtSz-scid IL2R gamma null mice engrafted with mobilized human hemopoietic stem cells J Immunol 1746477–6489. [DOI] [PubMed] [Google Scholar]
- Lucarelli G, Galimberti M, Giardini C, Polchi P, Angelucci E, Baronciani D.et al. (1998Bone marrow transplantation in thalassemia. The experience of Pesaro Ann N Y Acad Sci 850270–275. [DOI] [PubMed] [Google Scholar]
- Sodani P, Isgrò A, Gaziev J, Polchi P, Paciaroni K, Marziali M.et al. (2010Purified T-depleted, CD34+ peripheral blood and bone marrow cell transplantation from haploidentical mother to child with thalassemia Blood 1151296–1302. [DOI] [PubMed] [Google Scholar]
- Hsieh MM, Kang EM, Fitzhugh CD, Link MB, Bolan CD, Kurlander R.et al. (2009Allogeneic hematopoietic stem-cell transplantation for sickle cell disease N Engl J Med 3612309–2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu XS, Glazer PM., and, Wang G. Activation of human gamma-globin gene expression via triplex-forming oligonucleotide (TFO)-directed mutations in the gamma-globin gene 5' flanking region. Gene. 2000;242:219–228. doi: 10.1016/s0378-1119(99)00522-3. [DOI] [PubMed] [Google Scholar]
- Svasti S, Suwanmanee T, Fucharoen S, Moulton HM, Nelson MH, Maeda N.et al. (2009RNA repair restores hemoglobin expression in IVS2-654 thalassemic mice Proc Natl Acad Sci USA 1061205–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu LC, Sun CW, Ryan TM, Pawlik KM, Ren J., and, Townes TM. Correction of sickle cell disease by homologous recombination in embryonic stem cells. Blood. 2006;108:1183–1188. doi: 10.1182/blood-2006-02-004812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JC, Ye L., and, Kan YW. Correction of the sickle cell mutation in embryonic stem cells. Proc Natl Acad Sci USA. 2006;103:1036–1040. doi: 10.1073/pnas.0510177103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goncz KK, Prokopishyn NL, Abdolmohammadi A, Bedayat B, Maurisse R, Davis BR.et al. (2006Small fragment homologous replacement-mediated modification of genomic beta-globin sequences in human hematopoietic stem/progenitor cells Oligonucleotides 16213–224. [DOI] [PubMed] [Google Scholar]
- Miccio A, Cesari R, Lotti F, Rossi C, Sanvito F, Ponzoni M.et al. (2008In vivo selection of genetically modified erythroblastic progenitors leads to long-term correction of beta-thalassemia Proc Natl Acad Sci USA 10510547–10552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawliuk R, Westerman KA, Fabry ME, Payen E, Tighe R, Bouhassira EE.et al. (2001Correction of sickle cell disease in transgenic mouse models by gene therapy Science 2942368–2371. [DOI] [PubMed] [Google Scholar]
- Perumbeti A, Higashimoto T, Urbinati F, Franco R, Meiselman HJ, Witte D.et al. (2009A novel human gamma-globin gene vector for genetic correction of sickle cell anemia in a humanized sickle mouse model: critical determinants for successful correction Blood 1141174–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adler BK, Salzman DE, Carabasi MH, Vaughan WP, Reddy VV., and, Prchal JT. Fatal sickle cell crisis after granulocyte colony-stimulating factor administration. Blood. 2001;97:3313–3314. doi: 10.1182/blood.v97.10.3313. [DOI] [PubMed] [Google Scholar]
- Grigg AP. Granulocyte colony-stimulating factor-induced sickle cell crisis and multiorgan dysfunction in a patient with compound heterozygous sickle cell/beta+ thalassemia. Blood. 2001;97:3998–3999. doi: 10.1182/blood.v97.12.3998. [DOI] [PubMed] [Google Scholar]
- Segal BH, Leto TL, Gallin JI, Malech HL., and, Holland SM. Genetic, biochemical, and clinical features of chronic granulomatous disease. Medicine (Baltimore) 2000;79:170–200. doi: 10.1097/00005792-200005000-00004. [DOI] [PubMed] [Google Scholar]
- Seger RA, Gungor T, Belohradsky BH, Blanche S, Bordigoni P, Di Bartolomeo P.et al. (2002Treatment of chronic granulomatous disease with myeloablative conditioning and an unmodified hemopoietic allograft: a survey of the European experience, 1985-2000 Blood 1004344–4350. [DOI] [PubMed] [Google Scholar]
- Holland SM. Chronic granulomatous disease. Clin Rev Allergy Immunol. 2010;38:3–10. doi: 10.1007/s12016-009-8136-z. [DOI] [PubMed] [Google Scholar]
- Johnston RB, 3rd, Harbeck RJ., and, Johnston RB., Jr Recurrent severe infections in a girl with apparently variable expression of mosaicism for chronic granulomatous disease. J Pediatr. 1985;106:50–55. doi: 10.1016/s0022-3476(85)80463-7. [DOI] [PubMed] [Google Scholar]
- Woodman RC, Newburger PE, Anklesaria P, Erickson RW, Rae J, Cohen MS.et al. (1995A new X-linked variant of chronic granulomatous disease characterized by the existence of a normal clone of respiratory burst-competent phagocytic cells Blood 85231–241. [PubMed] [Google Scholar]
- Malech HL, Maples PB, Whiting-Theobald N, Linton GF, Sekhsaria S, Vowells SJ.et al. (1997Prolonged production of NADPH oxidase-corrected granulocytes after gene therapy of chronic granulomatous disease Proc Natl Acad Sci USA 9412133–12138. [DOI] [PMC free article] [PubMed] [Google Scholar]