Abstract
Knowledge of how the activity of enzymes is affected under in vivo conditions is essential for analyzing their regulation and constructing models that yield an integrated understanding of cell behavior. Current kinetic parameters for Lactococcus lactis are scattered through different studies and performed under different assay conditions. Furthermore, assay conditions often diverge from conditions prevailing in the intracellular environment. To establish uniform assay conditions that resemble intracellular conditions, we analyzed the intracellular composition of anaerobic glucose-limited chemostat cultures of L. lactis subsp. cremoris MG 1363. Based on this, we designed a new assay medium for enzyme activity measurements of growing cells of L. lactis, mimicking as closely as practically possible its intracellular environment. Procedures were optimized to be carried out in 96-well plates, and the reproducibility and dynamic range were checked for all enzyme activity measurements. The effects of freezing and the carryover of ammonium sulfate from the addition of coupling enzymes were also established. Activities of all 10 glycolytic and 4 fermentative enzymes were measured. Remarkably, most in vivo-like activities were lower than previously published data. Yet, the ratios of Vmax over measured in vivo fluxes were above 1. With this work, we have developed and extensively validated standard protocols for enzyme activity measurements for L. lactis.
INTRODUCTION
Lactococcus lactis is an industrially important lactic acid bacterium with prominence in the fermented dairy foods industry (3). It is known to convert nearly 90% of simple sugars like glucose into lactic acid at high growth rates (45). The genome of Lactococcus lactis subsp. cremoris MG 1363 has been characterized at the sequence level (27, 52), and recent attempts have been made to perform multilevel metabolomic, transcriptomic, and proteomic (multilevel-omics) analysis under different growth conditions (8, 10, 23). The integration of such multilevel-omics data sets in systems biology and bioinformatics studies relies crucially on well-validated standard protocols. Additionally the ability to measure many variables of the system at any given moment requires the development of sampling, storage, and measurement assay methods that optimally preserve the state of the organism.
One of the variables of increasing importance is the cellular enzyme activity, as it is an important target of many regulatory mechanisms, both through gene expression (acting on enzyme level) and through posttranslational modifications (acting on the catalytic efficiency of the enzyme). The impact of such regulation can be interpreted and predicted increasingly well with kinetic models. A few kinetic studies have been carried out on L. lactis (1, 18, 19, 32, 51). In some studies, metabolite data were generated by in vivo nuclear magnetic resonance (NMR) (32, 51), which is an elegant method but has strong limitations regarding measurements in growing microorganisms (30). In other studies (1, 18, 19), only limited or no kinetic data have been collected. Additionally, the sources of kinetic parameters span studies investigating different strains under a variety of growth conditions. For instance, the maximal enzyme catalytic rates (Vmax) are adopted from different studies (9, 28), using varying growth conditions, assay conditions, and sometimes even different strains. In fact, even in single studies, enzyme assay methods for L. lactis have been adopted from various studies of different organisms (Table 1). The assays are predominantly optimized to measure the maximal activity of a particular enzyme. Furthermore, different assays use different buffers and some also contain nonphysiological components like EDTA and arsenate, among others. Consequently, the dearth of well-established kinetic parameters under standardized conditions is a major setback for kinetic studies, which often limits the predictive power of models.
Table 1.
Enzyme assays for different organisms adopted for L. lactis (9)
In the era of systems biology, we require kinetic parameters under standardized conditions, ideally reflecting as closely as possible the conditions encountered by the enzymes in the cell. In a recent study, such an attempt to standardize the assay conditions for Saccharomyces cerevisiae has been made successfully (49). On similar lines, in the present study, we have developed an in vivo-like assay medium to standardize enzyme activity measurements in L. lactis. In addition, we established protocols for harvesting, storage, extract preparation, and specific enzyme assays of almost all enzymes in central energy metabolism in L. lactis. We apply these to measuring glycolytic and fermentative enzyme activities in batch and glucose-limited chemostat cultures of L. lactis.
MATERIALS AND METHODS
Strain and growth medium.
Lactococcus lactis subsp. cremoris MG 1363 (14) was grown on chemically defined medium for prolonged cultivation (CDMPC) as described elsewhere (F. Santos et al., unpublished data) with 25 mM glucose as the limiting nutrient and the following composition: (i) buffer (in g · liter−1), KH2PO4, 2.75; Na2HPO4, 2.85; NaCl, 2.9; (ii) vitamins (in mg · liter−1), dl-6,8-thioctic acid, 2; d-pantothenic acid hemicalcium salt, 0.5; biotin, 0.1; nicotinic acid, 1; pyridoxal hydrochloride, 1; pyridoxine hydrochloride, 1; thiamine hydrochloride, 1; (iii) metals (in mg · liter−1), ammonium molybdate tetrahydrate, 0.3; calcium chloride dihydrate, 3; cobalt(II) sulfate heptahydrate, 0.3; copper(II) sulfate pentahydrate, 0.3; iron(II) chloride tetrahydrate, 4; magnesium chloride hexahydrate, 200; manganese chloride tetrahydrate, 4; zinc sulfate heptahydrate, 0.3; and (iv) amino acids (in mg · liter−1), l-alanine, 130; l-arginine, 244; l-asparagine, 80; l-aspartic acid, 137; l-cysteine hydrochloride monohydrate, 61; l-glutamic acid, 97; l-glutamine, 96; glycine, 29; l-histidine, 24; l-isoleucine, 82; l-leucine, 117; l-lysine monohydrochloride, 187; l-methionine, 38; l-phenylalanine, 64; l-proline, 412; l-serine, 172; l-threonine, 68; l-tryptophan, 36; l-tyrosine, 50; l-valine, 86. The maximum specific growth rate of L. lactis in this growth medium under batch conditions is 0.7 h−1.
Culture conditions.
Glucose-limited chemostat cultures were grown in 2-liter bioreactors with a working volume of 1.2 liters at 30°C, under continuous stirring. The headspace was flushed at 5 headspace volume changes per hour, with a gas mixture of 95% N2 (99.998% pure) and 5% CO2 (99.7% pure) with oxygen impurity less than 34 volumes per million (vpm). A pH of 6.5 ± 0.05 was maintained by automatic titration with 5 M NaOH. Fermentors were inoculated with 4% (vol/vol) of standardized precultures consisting of 45 ml of CDMPC inoculated with 300 μl of a glycerol stock of L. lactis MG 1363 and incubated for 16 h at 30°C. After batch growth until an optical density at 600 nm (OD600) of around 1.8 was reached, medium was pumped at the appropriate dilution rate (D = 0.5, 0.2, or 0.15 h−1). The chemostats were harvested, assuming a steady state at 10 working volume changes (10). For assay standardization experiments, batch cultures were grown in static 50-ml cultures in the same medium at 30°C.
Analytical methods.
Cell density was measured spectrophotometrically at 600 nm and calibrated against cell dry weight measurements performed as follows. Four milliliters of culture was filtered through a predried, preweighed 0.2-μm cellulose nitrate filter (Whatman GmbH, Dassel, Germany), washed twice with deionized water, and dried to a constant weight. For one unit change of optical density, the change in dry weight was determined to be 0.31 ± 0.02 g · liter−1 · OD600−1.
Fermentation end product analysis.
Supernatant samples from chemostat fermentations were prepared by filtering the cultures through a 0.20 μm polyethersulfone (PES) filter (VWR International B.V., Amsterdam, The Netherlands) and storing the flowthrough at −20°C until further analysis. Extracellular concentrations of lactate, acetate, ethanol, formate, and glucose were determined by high-performance liquid chromatography (HPLC) on a Shimadzu LC-10AT liquid chromatograph equipped with a Shimadzu RID-10A refractive index detector for ethanol and glucose, and a Shimadzu SPD-10AVP UV-Vis absorbance detector set at 210 nm for the remaining metabolites. Separation was carried out on a Bio-Rad Aminex ion-exclusion HPX-87H column equilibrated at 55°C with an isocratic flow of 5 mM H2SO4 set to 0.5 ml · min−1. The injection volume used was 50 μl, and concentrations were estimated by comparison of peak areas to a calibration curve obtained with standards analyzed under the same conditions.
Element analysis.
Harvested samples from chemostats at 0.5 and 0.2 h−1 dilution rates were centrifuged (4°C, 5 min, 10,000 × g) and washed twice with 100 mM Tris-HCl buffer (pH 6.5). The supernatant was discarded, and the cell pellet, after snap-freezing in liquid nitrogen, was freeze-dried. The elemental composition of freeze-dried culture was determined by inductively coupled plasma atomic emission spectroscopy (ICP-AES) at the Energy Research Centre of The Netherlands (ECN Petten) (40). Values obtained were converted to intracellular concentrations by using a 1.67-μl intracellular volume per mg cell dry weight (47), assuming constant volume for both dilution rates (see the supplemental material).
Sampling and preparation of cell extracts.
An amount of cell culture containing 100 mg dry weight was centrifuged (4°C, 5 min, 8,000 rpm), washed once, and resuspended in 3 ml 50 mM HEPES [4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid]-KOH buffer at pH 7.5, containing 15% glycerol supplemented with Halt protease inhibitor single-use cocktail, EDTA free (Thermo Fischer Scientific, Rockford, IL). The rationale behind the choice of this buffer is addressed in the supplemental material. This suspension was divided into 0.5-ml aliquots added to 0.5-mg glass beads with 100-μm diameters (BioSpec Products, Bartlesville, OK) in screw-cap tubes and stored at −20°C until further analysis. Frozen samples were thawed, and MgCl2 was added to a final concentration of 2 mM. Cells were disrupted in a FastPrep FP120 homogenizer (BIO 101, Vista, CA) at a speed setting of 6, in 3 bursts of 20 s, with 120-s intermittent cooling periods (for details on optimization of this procedure, see Fig. S1 and S2 in the supplemental material). After centrifugation (4°C, 10 min, 10,000 × g), the supernatant was collected and a series of dilutions were prepared, which were used immediately for enzyme assays. Protein concentrations of whole cells and cell extracts used for enzyme assays were determined on the same day by the bicinchoninic acid (BCA) method (44) with a BCA protein assay kit (Pierce, Thermo Fisher Scientific) by using bovine serum albumin (BSA) (2 mg · ml−1 stock solution; Pierce), containing 2 mM MgCl2 and Halt protease inhibitor cocktail, as the standard.
Enzyme activity assays.
Enzyme activities were assayed at 30°C in freshly prepared cell extracts by coupling enzyme activity with the consumption or formation of NAD(P)H and monitoring its absorbance at 340 nm (A340) in Greiner flat-bottom polystyrene microplates in a Novostar spectrophotometer (BMG Labtech, Offenburg, Germany). In the P-transacetylase assay, the coenzyme A (CoA) formed in the reaction was oxidized by 5,5′-dithiobis-(2-nitrobenzoic) acid (DTNB). Reduction of DTNB was monitored at 405 nm. The enolase (ENO) assay was performed in two ways. One was by coupling with NADH formation as already described. The other was by measuring absorbance of its product, phosphoenol pyruvate (PEP), at 240 nm in Greiner flat-bottom UV-transparent (UV-star) microplates. Both assays resulted in the same activity (see Table S1 in the supplemental material). Absorbances of DTNB and PEP were monitored in a Spectramax spectrophotometer (Molecular Devices, Sunnyvale, CA). Extinction coefficients for 300-μl reagent mixtures with final composition as in the in vivo-like buffer in microplates were determined to be ε340 · L = 5.06 · 103 M−1 for NAD(P)H, ε405 · L = 10.494 · 103 M−1 for DTNB, and ε240 · L = 1.256 · 103 M−1 for PEP, where the path length (L) is 8.1 mm. For each assay, activities were corrected for background rates in controls, without the start reagent, without cell extract, and without both (see the supplemental material for the detailed protocol). Five dilutions in duplicate were used for all assays, and their proportionalities were checked. In all cases, activity values from at least two dilutions in duplicate were proportional with the amount of cell extract added and were used for calculation of mean activities. The values obtained from the assays yield the total activity of all isoenzymes in the cell extract and are expressed as the rate of substrate converted, relative to total protein in the extract. Where reported as fluxes, values were multiplied by the ratio of total protein content per dry weight estimated for the respective culture. All assay mixtures had a pH of 7.5 and contained the components of the in vivo-like buffer, i.e., 100 mM HEPES, 400 mM potassium glutamate, 50 mM NaCl, 1 mM potassium phosphate, and 1/10 of the concentration of metals present in CDMPC. Additional components are described in Table 2 for each assay. Acetaldehyde dehydrogenase (ALDH) was also assayed using a method for Escherichia coli (41). The reaction mixture contained 50 mM potassium phosphate buffer at pH 7, 0.1 mM CoA, 10 mM dithiothreitol (DTT), 0.5 mM NAD+, and initiator 40 mM acetaldehyde.
Table 2.
Composition of reagent mixtures for all enzyme assaysa
| Enzyme | Reaction mixture components |
|---|---|
| GLKb (EC 2.7.1.2) | 4 mM MgSO4, 2 mM ATP, 0.4 mM NADP+, 5 U · ml−1 G6PDH and initiator: 10 mM glucose |
| G6PDHb (EC 1.1.1.49) | 2 mM MgSO4, 0.4 mM NADP+ and initiator: 10 mM glucose-6P |
| PGI (EC 5.3.1.9; physiological direction) | 7 mM MgSO4, 5 mM ATP, 0.3 mM NADH, 1 U · ml−1 PFK, 1 U · ml−1 ALD, 2 U · ml−1 G3PD, 5 U · ml−1 TPI, and initiator: 10 mM glucose-6P |
| PGI (nonphysiological direction) | 2 mM MgSO4, 0.4 mM NADP+, 1.75 U · ml−1 G6PDH and initiator: 20 mM fructose-6P |
| PFK (EC 2.7.1.11) | 7 mM MgSO4, 5 mM ATP, 0.3 mM NADH, 1 U · ml−1 ALD, 2 U · ml−1 G3PD, 5 U · ml−1 TPI, and initiator: 20 mM fructose-6P |
| ALD (EC 4.1.2.13) | 2 mM MgSO4, 0.3 mM NADH, 2 U · ml−1 G3PD, 5 U · ml−1 TPI, and initiator: 30 mM fructose-1,6BP |
| TPI (EC 5.1.3.1) | 2 mM MgSO4, 0.3 mM NADH, 2 U · ml−1 G3PD, and initiator: 6 mM glyceraldehyde-3P |
| GAPDH (EC 1.2.1.12) | 5 mM MgSO4, 5 mM cysteine-HCl, 50 mM potassium phosphate, 3 mM ADP, 14.5 U · ml−1 PGK, 5 mM NAD+ and initiator: 10 mM glyceraldehyde-3P |
| PGK (EC 2.7.2.3; nonphysiological direction) | 7 mM MgSO4, 5 mM ATP, 0.3 mM NADH, 8 U · ml−1 GAPDH, and initiator: 10 mM 3-PGA |
| PGM (EC 5.4.2.1) | 5 mM MgSO4, 3 mM ADP, 0.1 mM 2,3BPG, 0.3 mM NADH, 2 U · ml−1 ENO, 5 U · ml−1 PYK, 10 U · ml−1 LDH, and initiator: 5 mM 3P-glycerate |
| ENO (EC 4.2.1.11; PEP absorbancec) | 5 mM MgSO4, 3 mM ADP, 0.3 mM NADH, 5 U · ml−1 PYK, 10 U · ml−1 LDH, and initiator: 5 mM 2P-glycerate; 2 mM MgSO4, and initiator: 5 mM 2P-glycerate |
| PYK (EC 2.7.1.40) | 5 mM MgSO4, 3 mM ADP, 5 mM fructose-1,6BP, 0.3 mM NADH, 10 U · ml−1 LDH, and initiator: 6 mM PEP |
| LDH (EC 1.1.1.27) | 2 mM MgSO4, 3 mM fructose-1,6BP, 0.3 mM NADH, and initiator: 6 mM PYR |
| ACK (EC 2.7.2.1) | 5 mM MgSO4, 3 mM ADP, 2 mM glucose, 0.4 mM NADP+, 8.5 U · ml−1 hexokinase, 12.7 U · ml−1 G6PDH, and initiator: 5 mM acetyl-P |
| PTA (EC 2.3.1.8; PTA control) | 2 mM MgSO4, 0.08 mM DTNB, and initiator: 0.4 mM acetyl-coenzyme A. 2 mM MgSO4, 0.008 mM DTNB, 2 mM acetyl-P, and initiator: 0.4 mM acetyl-CoA |
| ADH (EC 1.1.1.1) | 2 mM MgSO4, 0.3 NADH, and initiator: 20 mM acetaldehyde |
| ALDHd (EC 1.2.1.10; nonphysiological direction) | 2 mM MgSO4, 0.1 mM CoA, 1 mM DTT, 0.5 mM NAD+, and initiator: 40 mM acetaldehyde |
Desalting of coupling enzymes.
Enzymes obtained from commercial suppliers were desalted by centrifugal filtration in 3-kDa Microcon centrifugal filters (Millipore Corporation, Bedford, MA). Enzyme solutions were centrifuged (4°C, 60 min, 13,300 rpm), washed with an equal volume of demineralized water, centrifuged again, and suspended in the same volume of demineralized water. The filters were inverted and centrifuged again (4°C, 5 min, 9,000 rpm) to obtain desalted enzyme solutions.
RESULTS
The endeavor to establish an in vivo-like assay medium for L. lactis requires analysis of its cytosolic conditions regarding two major factors, namely, concentrations of ionic species and pH. We therefore started with analyzing chemostat cultures of L. lactis for concentrations of various elements.
Elemental analysis of L. lactis.
The intracellular concentrations of different elements in chemostat cultures of L. lactis for dilution rates of 0.5 and 0.2 h−1 are listed in Table 3. Two dilution rates were analyzed in order to test the possibility of using a single assay medium for analysis of cultures at dilution rates within this range. Among the dilution rates, all the elements had similar concentrations, except sodium, which differed by about 23 mM with a standard deviation of 13.6 mM. It is important to point out that these values denote total ion concentrations, and one cannot exclude the possibility of different proportions of liganded and free forms at various growth rates. Nevertheless, for practical purposes, an approach involving one single assay medium was chosen.
Table 3.
Intracellular concentrations of different elements in L. lactis at different dilution rates
| D (h−1) | Intracellular concna (mM) of: |
|||||
|---|---|---|---|---|---|---|
| Ca | K | Mg | Mn | Na | S | |
| 0.5b | 0.42 ± 0.24 | 559 ± 19 | 59 ± 2 | 0.68 ± 0.04 | 60 ± 4 | 59 ± 0.7 |
| 0.2c | 0.76 ± 0.44 | 570 ± 26 | 53 ± 2 | 0.67 ± 0.08 | 37 ± 13 | 63 ± 0.6 |
Calculated by dividing obtained quantities (in mg) of element per kg dry cells by 1.67 ml · kg dry cells−1 and converting mg element into moles (47).
Values represent the average ± standard deviation of three independent biological replicates.
Values represent the average ± standard deviation of two independent biological replicates.
pH of the assay medium.
pH has a strong influence on the kinetic parameters of enzymes (6), and therefore it is crucial that in all assays pH is kept constant and similar to the value in the cytoplasm. The pH of the assay medium was set to 7.5, the reported internal pH (34) at an external pH of 6.5 (the same as in CDMPC). Furthermore, it seems that the internal pH is kept relatively constant in vivo in the range of external pHs between 6 and 7 (29).
Element concentrations.
Based on the data in Table 3, the composition of the ions in the medium was rationalized. Calcium and manganese ions may bind to proteins and other components in cells (5, 12), and the values in Table 3 might not represent the free ion concentrations. Furthermore, due to their low concentrations (Table 3), it was decided not to add these ions to the standard buffer in separate solutions, but as a combined solution of many metal ions at low concentrations to improve reproducibility (see “Addition of other metal ions” below). Although this solution has a concentration of about 2 μM (each) Ca2+ and Mn2+, which is lower than that obtained by elemental analysis, it is sufficient to saturate enzymes in the assay. This is because the measured enzyme is only a fraction of the total protein content, which in the assay has a concentration of approximately 0.5 μM. For the sodium ion, an average concentration of 50 mM was selected, along with chloride as its counter ion.
The magnesium ion concentration was ∼55 mM, but inside the cell it is known to be bound to nucleic acids, adenine nucleotides (39), and cell wall teichoic acid (24). The cytosolic free concentration of Mg2+ in L. lactis has not been reported. However, for E. coli, it was reported to be the same as extracellular magnesium in a 1 μM to 10 mM range (20) and around 0.1 to 1 mM in Saccharomyces cerevisiae (2). The concentration of Mg2+ in the growth medium is ∼0.1 mM. For reproducibility of Mg2+ in the assay, an approximate free ion concentration of 2 mM was chosen. The ratio of ATP and ADP binding to Mg2+ is nearly 80% (43), which can consequently reduce the unbound Mg2+ concentration. Therefore, the addition of Mg2+ in assays was always 2 mM above the concentration of ATP or ADP added.
The equivalent elemental sulfur concentration in the cytoplasm was around 60 mM, but this value is likely to be representative of bound sulfur because free sulfur is not reported so far for L. lactis. However, sulfate was added as a counter ion for magnesium ions to a final concentration varying between 2 and 7 mM depending on the ATP or ADP concentration added.
The potassium ion concentration was by far the highest among all elements in L. lactis at approximately 550 mM. Similarly high potassium ion concentrations of 600 mM and 500 mM have been reported for L. lactis (36) and for another Gram-positive bacterium, Enterococcus faecalis, respectively (17). This value of 550 mM is representative of the free K+ concentration, because K+ does not generally interact with noncovalently interacting metabolite structures and thus may be evenly distributed in the cytoplasm (5). Hence, after finalizing Na+, Cl−, Mg2+, and K+, it was necessary to find a suitable counter ion for K+ in the assay medium.
Phosphate: an important effector.
Before proceeding further with the design, first, the workability of measuring enzymes in the standardized in vivo-like buffer designed for yeast was checked. It consists of 300 mM K+, 245 mM glutamate, and 50 mM phosphate (49), with phosphate offering the buffering action. The same buffer components are also present in L. lactis cytoplasm, albeit in different quantities. Assays of L. lactis were collected from the literature, and the buffers in each assay were replaced by the in vivo-like buffer of yeast at a pH of 7.5. These assays were then tested on commercially available enzymes and subsequently on cell extracts of L. lactis.
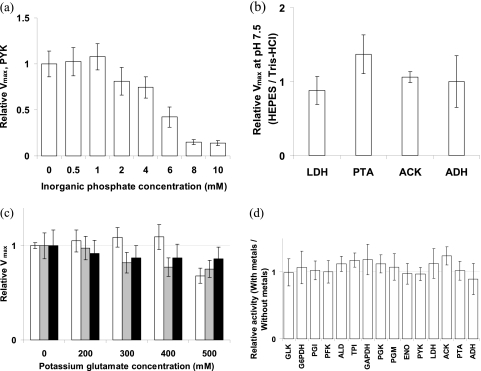
We observed no activity for pyruvate kinase (PYK) with the yeast buffer. We conceived that, for L. lactis, phosphate may not be the appropriate choice as a buffer, because it is a regulator of many glycolytic enzymes, pyruvate kinase in particular (4). Hence, the pyruvate kinase assay was tested with varying concentrations of phosphate to investigate its inhibitory effect in the presence of 5 mM fructose-1,6-bisphosphate (FBP) (required for activation of PYK). At a low concentration of 2 mM phosphate, pyruvate kinase activity is slightly inhibited (Fig. 1a). In view of the fact that the phosphate concentration in rapidly fermenting cells is substantially lower than 50 mM (<1 mM) (31), a final concentration of 1 mM free phosphate was chosen for the in vivo-like medium, with the realization that phosphate is variable and a potential effector of many enzymes.
Fig 1.
(a) Relative Vmax of PYK in the presence of 5 mM FBP and varying concentrations of inorganic phosphate, normalized to PYK Vmax in the absence of phosphate. (b) Relative Vmax at pH 7.5 in HEPES buffer, normalized to Vmax in Tris-HCl buffer. (c) Relative activities of LDH (white bars), PYK (gray bars), and GAPDH (black bars) with varying potassium glutamate concentrations normalized to activities in the absence of potassium glutamate. (d) Vmax values with 1/10 the metals present in CDMPC, relative to Vmax values without metals added to the assay buffer. Error bars represent standard deviations of a ratio (see the supplemental material) of average activities from at least three independent dilutions of a single cell extract. ACK, acetate kinase; G6PDH, glucose-6-phosphate dehydrogenase.
Buffering capacity.
With the lowering of phosphate, the buffering capacity of the assay medium was reduced considerably. Since a number of components are added to assays and pH changes due to reactant consumption or product formation can be expected, a buffer was needed in the assay medium. The cell has a number of salts and proteins, which together confer buffering capacity in the cytoplasm, but this is not practically reproducible in vitro. Hence, we searched for a buffer with a pKa closest to the pH of the in vivo-like assay medium, i.e., 7.5. The shortlisted candidates were DIPSO {3-[N,N-bis(2-hydroxyethyl)amino]-2-hydroxypropanesulfonic acid} and HEPES, with pKas of 7.52 and 7.48 and effective pH ranges of 7.0 to 8.2 and 6.8 to 8.2, respectively. Of these, we chose HEPES on account of its having a broader pH range and being one of the 12 Good's buffers with numerous desirable characteristics like low absorbance between 240 nm and 700 nm, minimal change with temperature, enzymatic stability, and limited effects due to solution composition, among others (15). We also compared the activities of a few enzymes in HEPES and Tris-HCl, the buffer used so far for many enzyme assays of L. lactis, as shown in Fig. 1b. Activities in the two buffers were similar in all cases, except for phosphotransacetylase (PTA), where the activity in HEPES was higher. On the preceding grounds, HEPES was chosen as the buffer for the in vivo-like assay. K+ was chosen as the counter ion to partially account for the high concentration required in the in vivo-like medium.
Counter ion for potassium.
In view of the fact that a high amount of K+ was present in L. lactis, the next step was to find a counter ion. Glutamate was found to be the best choice because it is the most abundant free amino acid in the cytoplasm of L. lactis, accounting for up to 50% of the total amino acid pool (37, 48). Furthermore, it is a better alternative to inorganic counter ions, which can significantly inhibit enzyme activities if present at high concentrations (35). Therefore, to examine the maximum noninhibiting concentration of glutamate, enzymes were tested with various concentrations of potassium glutamate. Up to a concentration of 400 mM glutamate, lactate dehydrogenase (LDH) was relatively insensitive, but at 500 mM, the activity dropped by 32% (Fig. 1c). For PYK, between 300 and 500 mM potassium glutamate, activity dropped 18 to 24%. For glyceraldehyde-3-phosphate dehydrogenase (GAPDH), above 300 mM glutamate, activity dropped around 13%. Keeping in mind that the cytoplasmic concentration of K+ in L. lactis is very high, it is important to have an ample amount of K+ in the assay medium. The consequence of this might be reduced enzyme activities. This can be supported by the fact that enzyme activities inside the cell might not actually function at the maximum rate as observed in conditions without K+. However, in order to avoid substantial inhibition, like that observed for LDH at 500 mM potassium glutamate, a final concentration of 400 mM was chosen for the in vivo-like medium.
Addition of other metal ions.
With the composition of the assay medium nearly complete, we thought of possible additions that would improve applicability and reproducibility. Chemically defined media for L. lactis cells always contain a concoction of metals necessary for its growth. Hence, it is possible that these metals are present in the cytoplasm. We tested the effect of adding 1/10 of the concentration of metals present in CDMPC (Table 4) to the in vivo-like medium. The activities in the presence of metals were slightly but not significantly higher and were not inhibited by metals (Fig. 1d). Since trace elements can easily enter the reaction mixture as lab contaminants, adding a known amount improves reproducibility without sacrificing the measurement. It also ensures that an enzyme activity will not be limited if it is dependent on some metal, making the assay suitable for enzymes other than those already tested. It was therefore decided to add to the in vivo-like assay medium 1/10 of the concentration of metals present in CDMPC.
Table 4.
Composition of the in vivo-like assay medium (version 1.0) for L. lactis
| Component | Concn |
|---|---|
| HEPES | 100 mM |
| K+ | 438 mM |
| Glutamate | 400 mM |
| Phosphate | 1 mM |
| NaCl | 50 mM |
| MgSO4 | 2 mM |
| (NH4)6Mo7O24 | 0.024 μM |
| CaCl2 | 2.04 μM |
| CoSO4 | 0.107 μM |
| CuSO4 | 0.120 μM |
| MgCl2 | 98.376 μM |
| MnCl2 | 2.021 μM |
| ZnSO4 | 0.104 μM |
| FeCl2 | 2.012 μM |
Proposed in vivo-like assay medium.
The composition of the proposed in vivo-like assay medium for L. lactis growing cultures was thus finalized and is described in Table 4.
Enzyme activities in the in vivo-like assay medium.
The activities of all enzymes involved in glycolysis and pyruvate metabolism were tested in the in vivo-like assay medium in cell extracts of L. lactis. At this point, we further standardized the procedure for reproducibility and the recognition of interfering reactions, enzyme effectors, the effect of incoming ammonium sulfate from coupling enzymes, and the effect of freezing on enzyme activities.
Proportionality and reproducibility.
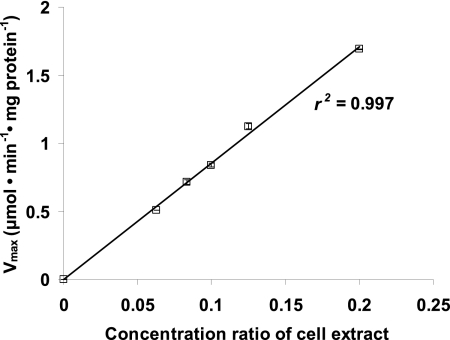
In order to ensure proportionality of activity with the amount of cell extract added, five serial dilutions in duplicate were used for all assays. In all cases, at least four slopes, i.e., two dilutions in duplicate, were linear in time as well as proportional with the amount of cell extract added, and thus at least four slopes were used for the calculation of activities. Figure 2 shows the proportionality between LDH activity values obtained for five dilutions of the cell extract with Pearson's correlation coefficient square (r2) of 0.9987. For some assays, less-diluted cell extract showed activities that were not proportional with the activities in more-diluted cell extracts. For the less-active glucokinase (GLK) and glucose-6-phosphate dehydrogenase (G6PDH), only undiluted and 2-fold-diluted cell extract could be used to detect activity. The ranges of activities around which measurements are linear in time and proportional with the amount of cell extract are listed in Table 5.
Fig 2.
Vmax of LDH at different dilutions of cell extract. The activity at each dilution is an average of duplicate measurements, and error bars represent deviations from the average.
Table 5.
Ranges of activity ratesa
| Enzyme | Range of Vmax (μmol · min−1 · ml−1) | r2 |
|---|---|---|
| GLK | 0.06–0.15 | 0.993 |
| G6PDH | 0.03–0.06 | 0.994 |
| PGI | 0.12–0.3 | 0.940 |
| PGIup | 0.02–0.05 | 0.998 |
| PFK | 0.05–0.2 | 0.981 |
| ALD | 0.15–0.3 | 0.997 |
| TPI | 0.2–0.5 | 0.972 |
| GAPDH | 0.08–0.18 | 0.999 |
| PGK | 0.3–0.5 | 0.998 |
| PGM | 0.1–0.5 | 0.983 |
| ENO | 0.1–0.4 | 0.998 |
| PYK | 0.15–0.4 | 0.999 |
| LDH | 0.15–0.5 | 0.997 |
| ACK | 0.1–0.2 | 0.982 |
| PTA | 0.03–0.08 | 0.985 |
| ADH | 0.03–0.06 | 0.992 |
Ranges of activity rates in reaction mixture for all enzymes, around which absorbance curves are linear (r2 ≥ 0.99). The listed r2 values indicate the extent to which these activity rates are proportional to dilutions of cell extract.
To check reproducibility, the assays were tested on cell extracts from three independent chemostat cultures grown at a dilution rate of 0.15 h−1. Every enzyme activity in the cell extract from each chemostat was measured at varying dilutions of cell extract, at least 4-fold and up to 12-fold, to test technical variation. An analysis of variance (ANOVA) on the data showed that an average technical variation of 5% was present and that any difference above this percentage can be attributed to biological variation. Based on this, activities of the replicates are in good agreement with each other (see Fig. S3 in the supplemental material).
Interfering reactions.
The present study uses cell extracts to determine enzyme activities. Due to this, there may be enzymatic activities present that could interfere with assays of interest. Among these are NADH-oxidoreductase (NOX) (EC 1.11.1.1) and l-glutamate N-acetyltransferase (EC 2.3.1.1). Apart from these, we did not find any other interfering enzyme reactions in the assay medium. The NOX activity was corrected for by subtracting background rates in controls with cell extract and without substrate for each enzyme. The acetyltransferase enzyme uses acetyl-CoA and glutamate as substrates to generate CoA and N-acetyl glutamate. It can therefore interfere with enzyme assays where acetyl-CoA is a substrate, for example, that with PTA. Therefore, in such assays it is essential to perform an additional control assay to correct for the activity of this enzyme. In this control, PTA is inhibited by adding its product, acetyl-P, in high concentrations (2 mM) above its half inhibition constant (Ki), which is 0.2 mM (19).
Enzyme effectors.
Certain enzymes need activation for exhibiting activity, and this was the case for GAPDH and phosphoglycerate mutase (PGM). For these enzymes, the assay as reported for the in vivo-like medium was obtained after a series of troubleshooting experiments which led to different versions of the assay. GAPDH was initially measured by a previous method, where 5 mM arsenate was used as a phosphate analogue to drive the reaction (13). All assays on the chemostat samples reported in this study were carried out with this nonoptimized method. However, later it was found that a higher concentration of arsenate (40 mM; from the method of Even et al. [9]) resulted in a higher GAPDH activity (results not shown). It was possible to measure GAPDH activity without arsenate but with a high concentration of phosphate (50 mM), provided 3-phosphoglycerate kinase (PGK) and ADP were added to the assay, to ensure consumption of the product, 1,3-bisphosphoglycerate (1,3BPG). This is the final assay that is reported here for GAPDH.
Only recently, it was found that L. lactis contains a PGM variant (dPGM), which requires 2,3BPG for activity (42). Based on this study it was decided to add 0.1 mM this metabolite to the PGM assay. Again, assays on chemostats in this paper report Vmax values obtained in the absence of 2,3BPG.
Effect of desalting ammonium sulfate from coupling enzymes.
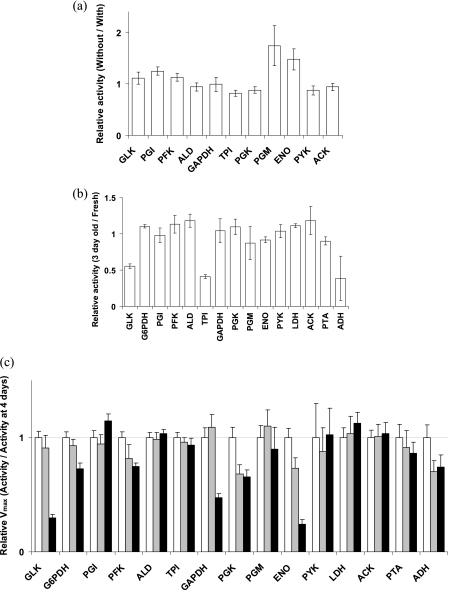
Many enzyme assay activities entail the usage of coupling activities of enzymes of interest to the formation of NAD(P)H. However, a majority of these coupling enzymes are commercially available in a suspension of ammonium sulfate with a concentration as high as 3.2 M. As a result, in some cases, such as the PGM assay, a concentration of ammonium sulfate of up to 60 mM would be present. Since our aim is to establish standard in vivo-like conditions for enzyme assays, we checked the effect of removing the ammonium sulfate in assays using coupling enzymes (Fig. 3a). The enzymes that were most sensitive were those which had assays with a high concentration of ammonium sulfate due a higher number of coupling enzymes. Activities of PGM (66 mM ammonium sulfate) and ENO (20 mM) were most affected, followed by those of glucose-6-phosphate isomerase (PGI) (41 mM) and phosphofructokinase (PFK) (41 mM). Activities of other enzymes did not differ greatly in the presence of small amounts (1 to 5 mM) of ammonium sulfate. For ENO, an alternative assay (26) measuring PEP absorbance at 240 nm is a more elegant way to solve the ammonium sulfate problem if measurements in the UV range are possible. In conclusion, ammonium sulfate could have an effect on enzyme activities, and it is better to work with desalted enzymes.
Fig 3.
(a) Vmax values of enzymes containing coupling enzymes in assays without ammonium sulfate relative to those with ammonium sulfate in the assay. (b) Vmax values measured in batch-grown cell samples frozen for 3 days relative to those obtained in fresh cells. (c) Relative Vmax values showing loss of enzyme activity during storage of cell samples at −20°C at 4 days (white bars), 12 days (gray bars), and 4 months (black bars) from the day of harvesting the chemostat at D of 0.15 h−1. Error bars represent standard deviations of a ratio of average activities from at least three independent dilutions from two independent batch cultures for panels a and b and from the cell extract of the chemostat culture for panel c.
Effect of freezing cells.
It is not always possible to measure all enzyme activities on the same day as cell harvesting. The cells thus have to be stored until they are taken for analysis. However, storage in a frozen state and thawing of L. lactis cells can affect the retention of enzyme activities (21) and can lead to conformational or catalytic changes of sensitive enzymes (38). It was therefore important to check whether cells could be frozen without change in enzyme activities. Enzyme activity measurements of fresh batch-grown cells and the same cells stored frozen for 3 days were compared (Fig. 3b). Furthermore, in order to see how long cell samples could be frozen without affecting enzyme activities, samples were analyzed for activities after 4 days, 12 days, and 4 months from the day the cells were harvested.
For all enzymes except GLK, triose phosphate isomerase (TPI), and alcohol dehydrogenase (ADH), freezing of cell samples for 3 days did not affect enzyme activities. However, PFK, PGK, and ENO activities declined between 4 to 12 days, and GAPDH declined after 4 months (Fig. 3c). Other enzymes were stable until the period checked, i.e., 4 months. Therefore, for accurate estimates of Vmax, GLK, TPI, and ADH should be measured on the same day as harvesting the cells, and GAPDH, PGK, and ENO must be measured within 3 days.
Comparison with measured fluxes.
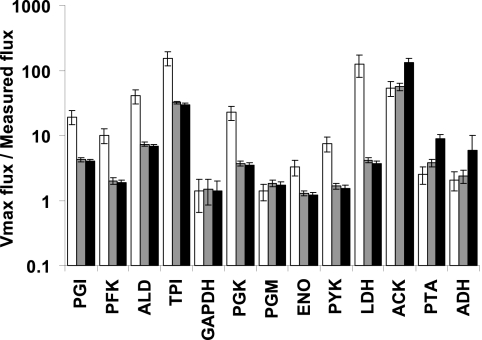
In order to check whether the in vivo-like Vmax values could support the actual flux in growing cells of L. lactis, they were compared with measured fluxes. In vivo-like maximal fluxes obtained from enzyme activities, corrected by the measured protein per gram cell dry weight, were compared with fluxes based on the organic acid concentrations, cell density, and set dilution rate in a chemostat. Additionally, maximal fluxes from batch cultures were compared with measured fluxes reported in the literature (11, 33). The ratios of the Vmax values to measured fluxes are above 1 for all enzymes in both culture conditions (Fig. 4), as expected. We can thus conclude that the in vivo-like maximal fluxes are high enough to sustain the glycolytic flux observed in L. lactis cells growing at maximal speed.
Fig 4.
Ratio of Vmax to measured fluxes for chemostat cultures at D = 0.15 h−1 (white bars) and for batch cultures with measured fluxes from the work of Even et al. (11) (gray bars) and from the work of Nordkvist et al. (33) (black bars). Error bars represent standard deviations of a ratio of average fluxes from three independent cultures.
DISCUSSION
The in vivo-like assay medium was tested for all enzymes involved in glycolysis and pyruvate metabolism in cell extracts of L. lactis. The only activity that could not be detected was the acetaldehyde dehydrogenase activity. It could not be detected in any sample in the in vivo-like assay in the nonphysiological direction or in the standard assay for E. coli from the literature (41). Notably, this activity has never been reported for L. lactis, and when it was assayed, it was undetectable (46). The gene coding for the enzyme catalyzing this reaction is adhE. This gene encodes a bifunctional acetaldehyde-CoA/ethanol dehydrogenase. It could be that this enzyme needs some additional factor to show acetaldehyde dehydrogenase activity, although the ethanol dehydrogenase activity is detectable under the current assay conditions.
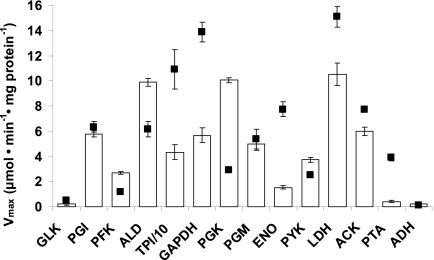
We compared the enzyme activities of all enzymes in batch culture with those reported in literature for L. lactis MG 1363 grown on glucose in a chemically defined medium (Fig. 5). Although enzyme activities can differ when cells are grown in different media, they can be compared if the cells are grown on the same sugar at similar growth rates. This was the case for growth conditions in this study and those of Even et al. (11), which differed only in some components in the defined medium. Therefore, Vmax values obtained in the in vivo-like assay medium were compared with those from Even et al. (11). About 9 of 14 enzymes show low activity under in vivo-like conditions compared to values reported in literature (Fig. 5). This is expected, since the assays for each enzyme reported in literature were optimized to observe maximal activity of that particular enzyme, even though under in vivo conditions the enzyme may not be maximally active. The fact that we do not see this for 5 enzymes demonstrates that the assays might not even have been optimal for enzymes in L. lactis in the first place. For instance, compared to the in vivo-like assays, in literature (i) the PYK assay lacked FBP and phosphate; (ii) PGM lacked 2,3BPG; (iii) PGK contained EDTA, which could scavenge an essential metal ion required for activity; and (iv) fructose-1,6-bisphosphate aldolase (ALD) assay had a high concentration of chloride ion which might have had an inhibitory effect. This is not surprising if one observes the various sources for enzyme assays in L. lactis (Table 1). Overall, the in vivo-like Vmax values differ from those in the literature, with some higher and some lower than those reported.
Fig 5.
Vmax values obtained in the in vivo-like assay medium (white bars) and those reported in the literature (11) (squares) for batch culture of L. lactis MG 1363. Error bars on the white bars represent standard deviations of average activities from two independent batch cultures.
Concluding remarks.
We developed a standardized in vivo-like assay medium to resemble the intracellular environment of growing cells of L. lactis as closely as is practically feasible. The design was based on the intracellular concentrations of ions measured from the biomass composition of chemostat cultures of L. lactis subsp. cremoris MG 1363. This newly developed assay medium was tested for the enzymes involved in glycolysis and pyruvate metabolism. This assay medium can also be used to assay other enzymes. In cases where enzyme levels of cells under starving or nongrowing conditions are being investigated, it will be important to adjust the concentration of phosphate in the medium, which significantly changes at different levels of glycolytic activity (31). We understand that changes in the assay medium might become necessary due to reasons that will become evident as the assay medium is used for further research studies. Hence for practical purposes, we propose that the composition described in this paper be referred to as “version 1.0” (refer to Table 4). Overall, we believe that the rational design process resulted in an assay medium that mimics the cytosolic conditions of L. lactis as much as practically possible. Together with our extensively validated protocols for sampling, storage, extract preparation, and measurement of individual enzymes, we hope that this endeavor should serve to unify the conditions of enzyme assays used in the systems biology initiatives for L. lactis. We also believe that it would be fruitful to apply this methodology to other microorganisms that are investigated with systems approaches in order to generate standard data sets that have a wider applicability.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Nakul Kumar Barfa (University of Amsterdam, The Netherlands) and Shannona Zimmerman (Vrije Universiteit, The Netherlands) for their proficient assistance in the laboratory.
This work is supported by the Technology Foundation STW (grant 08080) and by the Kluyver Centre for Genomics of Industrial Fermentation and the Netherlands Consortium for Systems Biology (NCSB), within the framework of the Netherlands Genomics Initiative (NGI).
Footnotes
Published ahead of print 21 October 2011
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Andersen AZ, et al. 2009. The metabolic pH response in Lactococcus lactis: an integrative experimental and modelling approach. Comput. Biol. Chem. 33:71–83 [DOI] [PubMed] [Google Scholar]
- 2. Beeler T, Bruce K, Dunn T. 1997. Regulation of cellular Mg2+ by Saccharomyces cerevisiae. BBA Biomembr. 1323:310–318 [DOI] [PubMed] [Google Scholar]
- 3. Cogan TM, Hill C. 1993. Cheese starter cultures, p 194–198. In Fox PF. (ed), Cheese: chemistry, physics and microbiology, 2nd ed, vol 1. Chapman & Hall, New York, NY [Google Scholar]
- 4. Collins LB, Thomas TD. 1974. Pyruvate kinase of Streptococcus lactis. J. Bacteriol. 120:52–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cossins BP, Jacobson MP, Guallar V. 2011. A new view of the bacterial cytosol environment. PLoS Comput. Biol. 7:e1002066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dixon M. 1953. The effect of pH on the affinities of enzymes for substrates and inhibitors. Biochem. J. 55:161–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dominguez H, et al. 1998. Carbon-flux distribution in the central metabolic pathways of Corynebacterium glutamicum during growth on fructose. Eur. J. Biochem. 254:96–102 [DOI] [PubMed] [Google Scholar]
- 8. Dressaire C, et al. 2009. Transcriptome and proteome exploration to model translation efficiency and protein stability in Lactococcus lactis. PLoS Comput. Biol. 5:e1000606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Even S, Lindley ND, Cocaign-Bousquet M. 2001. Molecular physiology of sugar catabolism in Lactococcus lactis IL1403. J. Bacteriol. 183:3817–3824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Even S, Lindley ND, Cocaign-bousquet M. 2003. Transcriptional, translational and metabolic regulation of glycolysis in Lactococcus lactis subsp. cremoris MG 1363 grown in continuous acidic cultures. Microbiology 149:1935–1944 [DOI] [PubMed] [Google Scholar]
- 11. Even S, Lindley ND, Loubiere P, Cocaign-Bousquet M. 2002. Dynamic response of catabolic pathways to autoacidification in Lactococcus lactis: transcript profiling and stability in relation to metabolic and energetic constraints. Mol. Microbiol. 45:1143–1152 [DOI] [PubMed] [Google Scholar]
- 12. Exterkate FA, Alting AC. 1999. Role of calcium in activity and stability of the Lactococcus lactis cell envelope proteinase. Appl. Environ. Microbiol. 65:1390–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Garrigues C, Loubiere P, Lindley ND, Cocaign-Bousquet M. 1997. Control of the shift from homolactic acid to mixed-acid fermentation in Lactococcus lactis: predominant role of the NADH/NAD+ ratio. J. Bacteriol. 179:5282–5287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gasson MJ. 1983. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J. Bacteriol. 154:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Good NE, et al. 1966. Hydrogen ion buffers for biological research. Biochemistry 5:467–477 [DOI] [PubMed] [Google Scholar]
- 16. Gracy RW, Tilley BE. 1975. Phosphoglucose isomerase of human erythrocytes and cardiac tissue. Methods Enzymol. 41:392–400 [DOI] [PubMed] [Google Scholar]
- 17. Harold FM, Kakinuma Y. 1985. Primary and secondary transport of cations in bacteria. Ann. N. Y. Acad. Sci. 456:375–383 [DOI] [PubMed] [Google Scholar]
- 18. Hoefnagel MHN, van Der Burgt A, Martens DE, Hugenholtz J, Snoep JL. 2002. Time dependent responses of glycolytic intermediates in a detailed glycolytic model of Lactococcus lactis during glucose run-out experiments. Mol. Biol. Rep. 29:157–161 [DOI] [PubMed] [Google Scholar]
- 19. Hoefnagel MHN, et al. 2002. Metabolic engineering of lactic acid bacteria, the combined approach: kinetic modelling, metabolic control and experimental analysis. Microbiology 148:1003–1013 [DOI] [PubMed] [Google Scholar]
- 20. Hurwitz C, Rosano CL. 1967. The intracellular concentration of bound and unbound magnesium ions in Escherichia coli. J. Biol. Chem. 242:3719–3722 [PubMed] [Google Scholar]
- 21. Kamaly KM, Marth EH. 1989. Enzyme activities of cell-free extracts from mutant strains of lactic streptococci subjected to sublethal heating or freeze-thawing. Cryobiology 26:496–507 [DOI] [PubMed] [Google Scholar]
- 22. Kulbe KD, Foellmer H, Fuchs J. 1982. Simultaneous purification of glyceraldehyde-3-phosphate dehydrogenase, 3-phosphoglycerate kinase, and phosphoglycerate mutase from pig liver and muscle. Methods Enzymol. 90:498–511 [DOI] [PubMed] [Google Scholar]
- 23. Lahtvee P-J, et al. 2011. Multi-omics approach to study the growth efficiency and amino acid metabolism in Lactococcus lactis at various specific growth rates. Microb. Cell Fact. 10:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lambert PA, Hancock IC, Baddiley J. 1975. The interaction of magnesium ions with teichoic acid. Biochem. J. 149:519–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Le Bloas P, Guilbert N, Loubiere P, Lindley ND. 1993. Growth inhibition and pyruvate overflow during glucose metabolism of Eubacterium limosum are related to a limited capacity to reassimilate CO2 by the acetyl-CoA pathway. Microbiology 139:1861–1868 [Google Scholar]
- 26. Lee BH, Nowak T. 1992. Influence of pH on the Mn2+ activation of and binding to yeast enolase: a functional study. Biochemistry 31:2165–2171 [DOI] [PubMed] [Google Scholar]
- 27. Linares DM, Kok J, Poolman B. 2010. Genome sequences of Lactococcus lactis MG1363 (revised) and NZ9000 and comparative physiological studies. J. Bacteriol. 192:5806–5812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lopez de Felipe F, Gaudu P. 2009. Multiple control of the acetate pathway in Lactococcus lactis under aeration by catabolite repression and metabolites. Appl. Environ. Microbiol. 82:1115–1122 [DOI] [PubMed] [Google Scholar]
- 29. Molina-Gutierrez A, Stippl V, Delgado A, Ganzle MG, Vogel RF. 2002. In situ determination of the intracellular pH of Lactococcus lactis and Lactobacillus plantarum during pressure treatment. Appl. Environ. Microbiol. 68:4399–4406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Neves AR, Pool WA, Kok J, Kuipers OP, Santos H. 2005. Overview on sugar metabolism and its control in Lactococcus lactis — the input from in vivo NMR. FEMS Microbiol. Rev. 29:531–554 [DOI] [PubMed] [Google Scholar]
- 31. Neves AR, et al. 2002. Effect of different NADH oxidase levels on glucose metabolism by Lactococcus lactis: kinetics of intracellular metabolite pools determined by in vivo nuclear magnetic resonance. Appl. Environ. Microbiol. 68:6332–6342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Neves AR, et al. 1999. In vivo nuclear magnetic resonance studies of glycolytic kinetics in Lactococcus lactis. Biotechnol. Bioeng. 64:200–212 [DOI] [PubMed] [Google Scholar]
- 33. Nordkvist M, Jensen NBS, Villadsen J. 2003. Glucose metabolism in Lactococcus lactis MG1363 under different aeration conditions: requirement of acetate to sustain growth under microaerobic conditions. Appl. Environ. Microbiol. 69:3462–3468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Olsen KN, et al. 2002. Noninvasive measurement of bacterial intracellular pH on a single-cell level with green fluorescent protein and fluorescence ratio imaging microscopy. Appl. Environ. Microbiol. 68:4145–4147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pollard A, Jones RGW. 1979. Enzyme activities in concentrated solutions of glycinebetaine and other solutes. Planta 144:291–298 [DOI] [PubMed] [Google Scholar]
- 36. Poolman B, Hellingwerf KJ, Konings WN. 1987. Regulation of the glutamate-glutamine transport system by intracellular pH in Streptococcus lactis. J. Bacteriol. 169:2272–2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Poolman B, Smid EJ, Veldkamp H, Konings WN. 1987. Bioenergetic consequences of lactose starvation for continuously cultured Streptococcus cremoris. J. Bacteriol. 169:1460–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ray B, Speck ML. 1973. Freeze-injury in bacteria. Crit. Rev. Clin. Lab. Sci. 4:161–213 [DOI] [PubMed] [Google Scholar]
- 39. Romani A, Scarpa A. 1992. Regulation of cell magnesium. Arch. Biochem. Biophys. 298:1–12 [DOI] [PubMed] [Google Scholar]
- 40. Rouf MA. 1964. Spectrochemical analysis of inorganic elements in bacteria. J. Bacteriol. 88:1545–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rudolph FB, Purich DL, Fromm HJ. 1968. Coenzyme A-linked aldehyde dehydrogenase from Escherichia coli. J. Biol. Chem. 243:5539–5545 [PubMed] [Google Scholar]
- 42. Solem C, Petranovic D, Koebmann B, Mijakovic I, Jensen PR. 2010. Phosphoglycerate mutase is a highly efficient enzyme without flux control in Lactococcus lactis. J. Mol. Microbiol. Biotechnol. 18:174–180 [DOI] [PubMed] [Google Scholar]
- 43. Storer AC, Cornish-Bowden A. 1976. Concentration of MgATP2- and other ions in solution. Calculation of the true concentrations of species present in mixtures of associating ions. Biochem. J. 159:1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Stoscheck CM. 1990. Quantitation of protein. Methods Enzymol. 182:50–68 [DOI] [PubMed] [Google Scholar]
- 45. Thomas TD, Ellwood DC, Longyear VM. 1979. Change from homo- to heterolactic fermentation by Streptococcus lactis resulting from glucose limitation in anaerobic chemostat cultures. J. Bacteriol. 138:109–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Thomas TD, Turner KW, Crow VL. 1980. Galactose fermentation by Streptococcus lactis and Streptococcus cremoris: pathways, products, and regulation. J. Bacteriol. 144:672–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Thompson J. 1976. Characteristics and energy requirements of an alpha-aminoisobutyric acid transport system in Streptococcus lactis. J. Bacteriol. 127:719–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Thompson J, Curtis MA, Miller SP. 1986. N5-(1-carboxyethyl)-ornithine, a new amino acid from the intracellular pool of Streptococcus lactis. J. Bacteriol. 167:522–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. van Eunen K, et al. 2010. Measuring enzyme activities under standardized in vivo-like conditions for systems biology. FEBS J. 277:749–760 [DOI] [PubMed] [Google Scholar]
- 50. Vasconcelos I, Girbal L, Soucaille P. 1994. Regulation of carbon and electron flow in Clostridium acetobutylicum grown in chemostat culture at neutral pH on mixtures of glucose and glycerol. J. Bacteriol. 176:1443–1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Voit E, Neves AR, Santos H. 2006. The intricate side of systems biology. Proc. Natl. Acad. Sci. U. S. A. 103:9452–9457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wegmann U, et al. 2007. Complete genome sequence of the prototype lactic acid bacterium Lactococcus lactis subsp. cremoris MG1363. J. Bacteriol. 189:3256–3270 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.