Abstract
We have engineered Escherichia coli to overproduce saturated and monounsaturated aliphatic methyl ketones in the C11 to C15 (diesel) range; this group of methyl ketones includes 2-undecanone and 2-tridecanone, which are of importance to the flavor and fragrance industry and also have favorable cetane numbers (as we report here). We describe specific improvements that resulted in a 700-fold enhancement in methyl ketone titer relative to that of a fatty acid-overproducing E. coli strain, including the following: (i) overproduction of β-ketoacyl coenzyme A (CoA) thioesters achieved by modification of the β-oxidation pathway (specifically, overexpression of a heterologous acyl-CoA oxidase and native FadB and chromosomal deletion of fadA) and (ii) overexpression of a native thioesterase (FadM). FadM was previously associated with oleic acid degradation, not methyl ketone synthesis, but outperformed a recently identified methyl ketone synthase (Solanum habrochaites MKS2 [ShMKS2], a thioesterase from wild tomato) in β-ketoacyl-CoA-overproducing strains tested. Whole-genome transcriptional (microarray) studies led to the discovery that FadM is a valuable catalyst for enhancing methyl ketone production. The use of a two-phase system with decane enhanced methyl ketone production by 4- to 7-fold in addition to increases from genetic modifications.
INTRODUCTION
Aliphatic methyl ketones are naturally occurring compounds that were first discovered in rue (Ruta graveolens) more than a century ago (30) and have since been commonly found in microorganisms, plants, insects, and mammalian cells (10). These compounds have a variety of important natural and commercial roles, including acting as pheromones and natural insecticides in plants (1) or providing scents in essential oils and flavoring in cheese and other dairy products (10). Biosynthesis of methyl ketones has been hypothesized to derive from a variety of different biological pathways such as fatty acid β-oxidation or aerobic alkene/alkane degradation (10, 21). However, studies to elucidate the genes and biochemical pathways involved in the synthesis of these compounds have been quite rare until recently. One research group in particular has carried out extensive biochemical and genetic studies in a wild tomato species (Solanum habrochaites) and identified two key genes, methyl ketone synthase I (ShMKS1) and methyl ketone synthase II (ShMKS2), that are essential for methyl ketone synthesis from fatty acid intermediates in this plant (6, 11, 31). ShMKS2, which belongs to the 4-hydroxybenzoyl coenzyme A (CoA) thioesterase (4-HBT) family, is hypothesized to hydrolyze a β-ketoacyl-acyl carrier protein (ACP) thioester intermediate to generate a β-keto acid; ShMKS1, an enzyme that belongs to the α/β-hydrolase superfamily, apparently decarboxylates the β-keto acid released by ShMKS2 to yield a methyl ketone (31).
Despite the commercial relevance of methyl ketones and their prevalence in nature, no genes other than ShMKS1, ShMKS2, and At1g68260 (a ShMKS2 homolog from Arabidopsis thaliana) have been recombinantly expressed and shown to be associated with methyl ketone biosynthesis (31). Metabolic engineering of microbes to overproduce methyl ketones may merit additional attention, as these compounds could be relevant to the biofuel industry as well as the flavor and fragrance industry by virtue of their highly reduced, aliphatic character. Indeed, several other fatty acid-derived compounds have already been successfully synthesized from metabolically engineered microbes for use as biofuels, such as fatty acid ethyl esters (26), alkanes (24), alkenes (5, 18, 22, 28), and n-alcohols (9).
In this article, we report on engineering of Escherichia coli to overproduce saturated and monounsaturated methyl ketones in the C11 to C15 (diesel) range for potential application to biofuel production. We describe specific improvements that resulted in more than 4,500-fold enhancement in methyl ketone titer relative to that of a fatty acid-overproducing E. coli strain, including reengineering of β-oxidation, overexpression of a thioesterase native to E. coli (FadM), and use of decane overlays. Methyl ketone titers in the best producing strains were in the range of 380 mg/liter.
MATERIALS AND METHODS
Bacterial strains, plasmids, oligonucleotides, and reagents.
Bacterial strains and plasmids used in this study are listed in Table 1. Plasmid extractions were carried out using the Qiagen (Valencia, CA) miniprep and midiprep kits. Oligonucleotide primers were designed using the web-based PrimerBlast program (http://www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi?LINK_LOC=BlastHomeAd) and synthesized by Bioneer (Alameda, CA). Primer sequences for amplification of E. coli DH1 and Micrococcus luteus open reading frames (ORFs) are listed in Table 2. The coding sequences (CDS) corresponding to the enzymes ShMKS1 (GenBank accession no. AAV87156) (11) and ShMKS2 (GenBank accession no. ADK38536) (31) from S. habrochaites and FatB1 from Umbellularia californica (UcFatB1; GenBank accession no. Q41635) (32) were synthesized and codon optimized for expression in E. coli by GenScript (Piscataway, NJ). Codon-optimized sequences are listed in Table S1 in the supplemental material.
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic | Source or reference |
|---|---|---|
| Strains | ||
| E. coli strains | ||
| BL21(DE3) | F−ompT gal dcm lon hsdSB(rB− mB−) λ(DE3) | 27 |
| DH1 | endA1 recA1 gyrA96 thi-1 glnV44 relA1 hsdR17(rK− mK+) λ− | 19 |
| LT-ΔfadE | DH1 ΔfadE with pKS1 | 26 |
| EGS084 | LT-ΔfadE with pEC-XK99E | 5 |
| EGS212 | LT-ΔfadE with pEG205 | 5 |
| EGS514 | BL21(DE3) with pEG513 | This study |
| EGS517 | BL21(DE3) with pEG516 | This study |
| EGS522 | DH1 ΔfadE ΔfadA | This study |
| EGS560 | EGS522 with pEG530 and pEC-XK99E | This study |
| EGS700 | EGS522 with pEG530 and pEG205 | This study |
| EGS735 | LT-ΔfadE with pEG705 | This study |
| EGS790 | LT-ΔfadE with pEG775 | This study |
| EGS860 | LT-ΔfadE with pEG855 | This study |
| EGS895 | EGS522 with pEG530 and pEG855 | This study |
| EGS975 | EGS522 with pEG955 and pEG855 | This study |
| EGS1015 | EGS522 with pEG530 and pEG990 | This study |
| EGS1080 | EGS522 with pEG530 and pEG1065 | This study |
| EGS1085 | EGS522 with pEG530 and pEG1070 | This study |
| EGS1090 | EGS522 with pEG530 and pEG1075 | This study |
| EGS1115 | EGS522 with pEG530 and pEG1101 | This study |
| EGS1120 | EGS522 with pEG530 and pEG1106 | This study |
| EGS1135 | DH1 with pEG1065 | This study |
| EGS1140 | DH1 with pEG1075 | This study |
| EGS1150 | LT-ΔfadE with pEG1065 | This study |
| EGS1155 | LT-ΔfadE with pEG1075 | This study |
| M. luteus strain ATCC 4698 | Wild type | ATCC |
| Plasmids | ||
| pEC-XK99E | Kmr; E. coli-Corynebacterium glutamicum shuttle expression vectors based on the medium-copy-no. plasmid pGA1 and containing the trc promoter | 14 |
| pKS1 | Cmr; p15a derivative containing ′tesA under the lacUV5 promoter | 26 |
| pKS104 | Ampr, ColE1 derivative with fadD (M335I), atfA under the lacUV5 promoter | 26 |
| pSKB3 | Kmr; a derivative of the expression vector pET-28a with the thrombin protease site replaced by a TEVb protease site | Burleya |
| pEG205 | Kmr; ∼1-kb fragment of Mlut_09310 (MlfabH) cloned into pEC-XK99E at EcoRI and XbaI sites | 5 |
| pEG513 | Kmr; ∼2.2-kb fragment of fadB (EcDH1_4135) cloned into pSKB3 at NdeI and SalI sites | This study |
| pEG516 | Kmr; ∼2.1-kb fragment of Mlut_11700 cloned into pSKB3 at NdeI and SalI sites | This study |
| pEG530 | Cmr; ∼2.1-kb fragment of Mlut_11700 and ∼2.2-kb fragment of fadB (EcDH1_4135) cloned downstream of the ′tesA gene in pKS1 by SLIC | This study |
| pEG705 | Kmr; QuikChange mutagenesis of MlfabH in pEG205 to the following residues: C123S, H275A, and N306A | This study |
| pEG775 | Kmr; ∼0.4-kb fragment of paaI (EcDH1_2249) cloned into pEC-XK99E at EcoRI and XbaI sites | This study |
| pEG855 | Kmr; ∼0.4-kb fragment of fadM (EcDH1_3166) cloned into pEC-XK99E at EcoRI and XbaI sites | This study |
| pEG955 | Cmr; ∼2.2-kb translational fusion of a leaderless maltose-binding protein gene (l-mbp) and the UcfatB1 gene, ∼2.1-kb fragment of Mlut_11700 and 2.2-kb fragment of fadB cloned into pKS1 (digested with MfeI and SalI to remove ′tesA) by SLIC | This study |
| pEG990 | Kmr; ∼1.7-kb of fadD M335I allele from pKS104 cloned downstream of fadM in pEG855 by SLIC | This study |
| pEG1065 | Kmr; ∼0.8-kb fragment of ShMKS1 and ∼0.6-kb fragment of ShMKS2 cloned into pEC-XK99E at BamHI and SalI sites by SLIC | This study |
| pEG1070 | Kmr; ∼0.8-kb fragment of ShMKS1 cloned into pEC-XK99E at BamHI and XbaI sites | This study |
| pEG1075 | Kmr; ∼0.6-kb fragment of ShMKS2 cloned into pEC-XK99E at BamHI and XbaI sites | This study |
| pEG1101 | Kmr; ∼ 0.9-kb fragment of Ptrc-ShMKS1 cloned downstream of fadM in pEG855 by SLIC | This study |
| pEG1106 | Kmr; ∼ 0.8-kb fragment of ShMKS1 cloned downstream of fadM in pEG855 by SLIC | This study |
| pEG1145 | Kmr; ∼1.2-kb fragment of hcaT (EcDH1_1132) into pEC-XK99E at EcoRI and XbaI sites | This study |
Stephen K. Burley.
TEV, tobacco etch virus.
Table 2.
Primers used in this study
| Primer use and target gene | Primer name | Primer sequencea (5′ → 3′) |
|---|---|---|
| Target gene amplification | ||
| fadB | DH1_fadB_SLIC_F1 | GCGAAGCAGTTGCAGCCTTTAGTAAATCATGACTCATAAGAGCTCGGTACGACCAGATCACCTTGCGG |
| DH1_fadB_SLIC_R1 | TGGACGGTCATGACGATGCTCCTGTTCGTGAGTGGGGGCGTTCGAACGGCCCATCGGGGT | |
| DH1_fadB_F1 | CTGCCATATGCTTTACAAAGGCGACACCCTGT | |
| DH1_fadB_R1 | TACAGAATTCGAACGGCCCATCGGGGTG | |
| fadM | DH1_fadM_F1 | CGCTGAATTCACAACGTAAGGTTATTGCGCTATGC |
| DH1_fadM_R1 | ATGTTCTAGACTTGAGCATCCGGCACCACAAAAC | |
| hcaT | DH1_hcaT_F1 | TACTGAATTCCCTGACGGGAGGGACTCATGGT |
| DH1_hcaT_R1 | GCTATCTAGAGGAGCAGATCCGCAAAATGCTCG | |
| l-mbp | l-mbp_SLIC_F1 | TGTGGAATTGTGAGCGGATAACAATTGCACCAACAAGGACCATAGCATATGAAAATCGAAGAAGGTAAACTGGT |
| l-mbp_SLIC_R1 | AAGGCGCTTGCCAGGCTCGTCGTTGCCATCCCGAGGTTGTTGTTATTGTTATTGTTG | |
| paaI | DH1_paaI_F1 | AGTGGAATTCGGGCGCTTCTGGAGAGCGGTTA |
| DH1_paaI_R1 | TTATTCTAGAGGCTTCACGCATCAGGCTTCTCC | |
| ptrc | Ptrc_SLIC_F1 | GTTTTGTGGTGCCGGATGCTCAAGTCTAGATATCATCGACTGCACGGTGC |
| Ptrc_SLIC_R1 | TTCCATGTTTCCTCCTGCGCAGGGAATTCCATGGTCTGTTTCCTGTGTGA | |
| ShMKS1 | MKS1_SLIC_F1(MKS2) | CGTCCAGCATCATCTGTAATCTAGACCTGCGCAGGAGGAAACATGGAA |
| MKS1_SLIC_F2 (fadM) | TTTTGTGGTGCCGGATGCTCAAGTCTAGACCTGCGCAGGAGGAAACATGGAA | |
| MKS1_SLIC_F3 (ptrc) | TCACACAGGAAACAGACCATGGAATTCCCTGCGCAGGAGGAAACATGGAA | |
| MKS1_SLIC_R1 | GCCAAGCTTGCATGCCTGCAGGTCGACTCATTTGTATTTATTAGCGATGG | |
| ShMKS2 | MKS2_SLIC_F1 | TCACACAGGAAACAGACCATGGGATCCCCTGCGCAGGAGGAAACATGTCAC |
| MKS2_SLIC_R1 | TTCCATGTTTCCTCCTGCGCAGGTCTAGATTACAGATGATGCTGGACG | |
| Mlut_09310 | Mlut_09310_C123S_F1 | TCTCCGCCGCGAGCGCCGGCTAC |
| Mlut_09310_C123S_R1 | GTAGCCGGCGCTCGCGGCGGAGA | |
| Mlut_09310_H275A_F1 | CCGCGTTCATCCCGGCCCAGGCCAACATGC | |
| Mlut_09310_H275A_R1 | GCATGTTGGCCTGGGCCGGGATGAACGCGG | |
| Mlut_09310_N306A_F1 | GCGGACGCCGGCGCCACGTCGGCCGC | |
| Mlut_09310_N306A_R1 | GCGGCCGACGTGGCGCCGGCGTCCGC | |
| Mlut_11700 | Mlut_11700_SLIC F1 | GTCATTGTCGATGCAATTCGCACCCCGATGGGCCGTTCGAACGCCCCCACTCACGAACAGG |
| Mlut_11700_SLIC R1 | TGCCTCTAGCACGCGTCTCACTATAGGGCGAATTGGAGCTCCACCGCGAGGTGACGGGG | |
| Mlut_11700_F2 | GATTCATATGACCGTCCACGAGAAGCTCGC | |
| Mlut_11700_R2 | GATTGAATTCACCGCGAGGTGACGGGGG | |
| UcfatB1 | UcfatB1_SLIC_F1 | CAACAATAACAATAACAACAACCTCGGGATGGCAACGACGAGCCTGGCAAGCGCCTT |
| UcfatB1_SLIC_R1 | ATCCGCAAGGTGATCTGGTCGTACGAGCTCTCACACACGCGGTTCAGCCGGAAT | |
| Real-time PCR | ||
| fadM | fadM_qPCR_ F1 | CCGCTACCTTGAATTTCTCG |
| fadM_qPCR_ R1 | ACGACGAAGGCGATGTTATG | |
| hcaT | hcaT_qPCR_ F1 | GCTGATGCTGGTGATGATTG |
| hcaT_qPCR_R1 | AGTCGCACTTTGCCGTAATC |
Underlined sequences indicate restriction sites or homology regions used for cloning purposes. Bold sequences indicate nucleotide changes from wild-type gene to generate site-directed mutations.
Media and bacterial growth.
E. coli was propagated as previously described (23). For studies of heterologous gene expression in E. coli strains, cells were grown in 15 ml of tryptic soy broth (containing 0.2% glucose) in 30-ml glass tubes with 200-rpm agitation at 37°C, unless indicated otherwise, for up to 72 h before being harvested for analysis. Frozen glycerol stocks were used as inocula for the studies described here, unless noted otherwise. When required, antibiotics were added to the growth medium at the following final concentrations: chloramphenicol, 25 μg/ml; kanamycin, 50 μg/ml. A final concentration of 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) was added to cultures after 6 h when induction of genes was required.
Plasmid and strain construction for heterologous expression in E. coli.
Cloning of M. luteus and E. coli genes into expression plasmids was carried out as previously described (5). All primers used to amplify target genes are listed in Table 2. PCR products and plasmid DNA were digested with the appropriate restriction enzymes and purified with QIAquick gel extraction and/or PCR purification kits (Qiagen) before being ligated and transformed into E. coli. When no appropriate restriction sites were available for generating cohesive ends for ligation, sequence and ligation independent cloning (SLIC) was performed as described by Li and Elledge (17). Proper clone construction was confirmed by DNA sequencing, which was performed by Quintara Biosciences (Berkeley, CA). Expression of heterologous genes in constructs was confirmed by extraction of proteins, tryptic digestion, and analysis of the resulting peptides by electrospray ionization liquid chromatography-tandem mass spectrometry (LC/MS/MS; QSTAR Elite Hybrid Quadrupole TOF; Applied Biosystems). Mutations of genes were performed as described for the QuikChange site-directed mutagenesis kit (Agilent) using primers designed with nucleotide changes that corresponded to the desired amino acid substitutions. To knock out E. coli genes, in-frame chromosomal deletion of E. coli genes was carried using the method of Datsenko and coworkers (2, 7).
Extraction of methyl ketones and related metabolites from bacterial cultures.
For most samples, methyl ketones and other metabolites were extracted from cultures using a decane overlay. For overlay extractions, 1 ml of decane (≥ 99% purity; ReagentPlus; Sigma) amended with perdeuterated decane (C10D22) and tetracosane (C24D50) internal standards was added to 15-ml cultures in 30-ml glass tubes following induction with IPTG. Fifty microliters of decane overlay was removed at specified time points, up to 72 h, for direct gas chromatography-mass spectrometry (GC/MS) analysis. For low-concentration samples in which methyl ketones were not detectable using decane overlays, extractions of cell pellets were performed as previously described (5). For all extractions, culture tubes were precleaned with high-purity acetone before being autoclaved. All other glass and polytetrafluoroethylene (PTFE) surfaces were also rigorously cleaned with high-purity acetone, and an effort was made to ensure that solvent extracts contacted only glass or PTFE surfaces, whenever possible. Metabolite data described in Results are from 72-h overlays unless indicated otherwise. For fatty acid analysis, 50-μl aliquots of extracts were derivatized with ethereal diazomethane to generate fatty acid methyl esters (FAMEs), as previously described (5).
Analysis by GC/MS.
For electron ionization (EI) GC/MS analyses with a quadrupole mass spectrometer, studies were performed with a model 7890A GC (Agilent) with a DB-5 fused silica capillary column (30-m length, 0.25-mm inner diameter, 0.25-μm film thickness; J & W Scientific) coupled to an HP 5975C series mass selective detector; 1-μl injections were performed by a model 7683B autosampler. The GC oven was typically programmed from 40°C (held for 3 min) to 300°C at 15°C/min and then held for 5 min; the injection port temperature was 250°C, and the transfer line temperature was 280°C. The carrier gas, ultra-high-purity helium, flowed at a constant rate of 1 ml/min. Injections were splitless, with the split turned on after 0.5 min. For full-scan data acquisition, the MS typically scanned from 50 to 600 atomic mass units at a rate of 2.7 scans per s. For saturated methyl ketones (C11, C13, C15), external standard quantification (m/z 58 areas) was performed with authentic standards. For monounsaturated ketones, no authentic standards were available, so external standard quantification relied on total ion chromatogram (TIC) areas and saturated methyl ketone standards with the appropriate chain length. Thus, in the absence of authentic standards, unsaturated methyl ketone data should be considered estimates.
Analysis by liquid chromatography-atmospheric pressure chemical ionization-time of flight (LC-APCI-TOF) mass spectrometry.
Liquid chromatographic separation of methyl ketones was conducted at 55°C with an Inertsil ODS-3 reverse-phase column (250-mm length, 2.1-mm internal diameter, 3-μm particle size; GL Sciences, Inc., Torrance, CA) using a 1200 series high-performance liquid chromatography (HPLC) system (Agilent Technologies, CA). The injection volume for each measurement was 2 μl. The mobile phase was composed of water (solvent A) and methanol (solvent B) (HPLC grade; Honeywell Burdick & Jackson, CA). Methyl ketones were separated with the following gradient: 60% to 98% B for 10 min, held at 98% B for 15 min, 98% to 60% B for 17 min, held at 60% B for 8 min. A flow rate of 0.19 ml/min was used throughout.
The HPLC system was coupled to an Agilent Technologies 6210 time-of-flight mass spectrometer (TOF MS). Nitrogen gas was used as both the nebulizing and drying gas to facilitate the production of gas-phase ions. The drying and nebulizing gases were set to 10 liters/min and 30 lb/in2, respectively, and a drying gas temperature of 325°C was used throughout. The vaporizer and corona were set to 350°C and 4 μA, respectively. APCI was conducted in the positive-ion mode with a capillary voltage of 3 kV. MS experiments were carried out in the full-scan mode (m/z 102 to 1,000) at 0.86 spectra per s for the detection of [M + H]+ ions. The instrument was tuned for a range of m/z 50 to 1,700. Prior to LC-APCI-TOF MS analysis, the TOF MS was calibrated with the Agilent APCI TOF tuning mix. Data acquisition and processing were performed by the MassHunter software package (Agilent Technologies).
In vitro assay to generate pentadecenone.
His-tagged acyl-CoA oxidase (Mlut_11700) and His-tagged E. coli FadB were purified as previously described (5). A 1-ml acyl-CoA oxidase assay was conducted in a screw-cap glass vial containing 1.5 mM palmitoleoyl-CoA (Sigma), 400 μg of acyl-CoA oxidase, 150 μg/ml bovine serum albumin (BSA), 0.1 mM FAD, and 0.1 M potassium phosphate buffer (pH 7.5). The reaction mixture was incubated on a rotary shaker at 30°C for 3 h, 4 U of catalase (Sigma) was added to the mixture, and the mixture was incubated as before for another 30 min at 37°C to remove the H2O2 generated by the acyl-CoA oxidase. The acyl-CoA oxidase reaction mixture (250 μl) was added to a 4-ml screw-cap glass vial with a polytetrafluoroethylene (PTFE)-lined septum for the 1-ml FadB assay, which also contained 400 μg/ml of BSA, 300 mM NAD, 600 μg of FadB, and 0.1 M potassium phosphate buffer (pH 7.5). Controls included assay mixtures without FadB. Reaction mixtures were incubated on the rotary shaker overnight (∼18 h) at 37°C. For extraction of assay products, 1 ml hexane (amended with C10D22 internal standard) was added to the assay solution, mixed well, and allowed to sit for 20 min, and the solvent layer was transferred to a 10-ml conical glass vial. The extraction step was repeated, and the two 1-ml aliquots of hexane were combined and then concentrated to 50 μl under a gentle stream of ultra-high-purity N2 for subsequent analysis by GC/MS.
Transcriptional studies of E. coli with RT-qPCR and microarray analyses.
For transcriptional studies, E. coli cultures were grown in 15 ml of tryptic soy broth in a 30-ml glass tube as described above, induced with IPTG after 6 h, and harvested at 8 h into 2 ml of ethanol solution containing 5% phenol to stop further transcription and preserve RNA integrity. Cell cultures were spun down, and the pellets were immediately frozen in liquid nitrogen and stored at −80°C until RNA extraction. Extraction and purification of RNA were carried out with the Qiagen RNeasy Minikit and treated on-column with RNase-free DNase I (Qiagen). Concentration and integrity of RNA were determined with a Thermo Scientific Nanodrop ND-1000 spectrophotometer and an Agilent 2100 BioAnalyzer, respectively.
Synthesis of cDNA for reverse transcription-quantitative PCR (RT-qPCR) analysis was carried out using 1 μg of total RNA primed with 60 μM random hexamers and reverse transcribed using a Transcriptor First Strand cDNA synthesis kit (Roche, Germany). qPCR analyses were then conducted on an Applied Biosystems StepOne system using 1 μl of the reverse transcription reaction mixture, gene-specific primers (Table 2), and the PerfeCTa SYBR green FastMix (Quanta Biosciences, Gaithersburg, MD). Quantitative PCR cycle parameters were as follows: initial denaturation at 95°C for 5 min, followed by 40 cycles of 1 s denaturation at 95°C and 30 s annealing and extension at 60°C. Fluorescence measurements were taken between each cycle. At the conclusion of the qPCR cycle, melting curve analysis was conducted by denaturing the PCR products from 60°C to 95°C and making fluorescence measurements at 0.3°C increments. All reactions were performed in triplicate. Transcripts were quantified with a standard curve generated by serial dilution of pEG855 (from 105 to 1010 copies/reaction) and normalized to the internal reference gene, hcaT (34).
To perform microarray analyses, 10 μg of total RNA primed with 5 μg of random hexamers (Roche, Germany) was reverse transcribed using the SuperScript Indirect cDNA labeling kit (Invitrogen). Alexa Fluor 555 dyes (Invitrogen) were then incorporated into amino-allyl-dUTP-labeled cDNA, and the fluorescently labeled cDNA was purified with the QIAquick PCR purification kit (Qiagen) and dried under vacuum (Vacufuge Speed Vac; Eppendorf). Labeled cDNA was hybridized to the four-plex NimbleGen E. coli K-12 (Roche) expression microarray chip (catalog no. A6697-00-01), which contains duplicates of 8 different 60-mer probes for each of the 4,254 genes in the E. coli K-12 genome, at 42°C for 20 to 24 h as recommended by the manufacturer. After hybridization, microarray chips were scanned with a GenePix 4000B scanner and data were extracted using NimbleScan software. Array normalization was performed using the robust multiarray average (RMA) technique as described by Irizarry et al. (13). The normalized expression values generated in RMA pair files were imported into Excel software, and statistical analyses were performed with the Significance Analysis of Microarray (SAM) add-on (29).
CN determination.
Cetane number (CN) determinations of selected methyl ketones (Sigma) were performed by the Southwest Research Institute (San Antonio, TX) according to ASTM (American Society for Testing and Materials) method D613, with no modifications.
Microarray data accession number.
Microarray data have been deposited in the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo) under accession number GPL14649.
RESULTS
Detection of methyl ketones in E. coli fatty acid-overproducing strains.
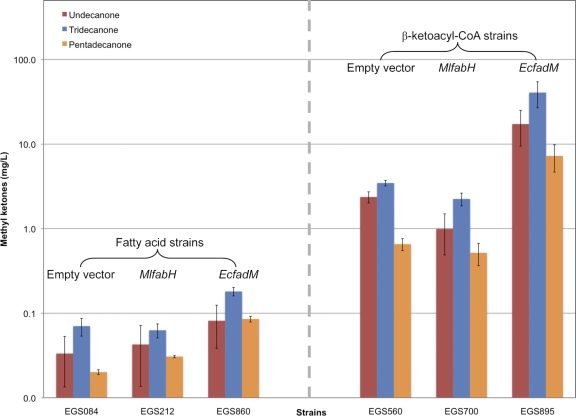
Previous studies of alkene biosynthesis in Micrococcus luteus (5) in which M. luteus condensing enzymes (e.g., FabH [β-ketoacyl-ACP synthase III] and FabF [β-ketoacyl-ACP synthase II]) were heterologously expressed in a fatty acid-overproducing strain of E. coli DH1 resulted in unexpected GC/MS detection of methyl ketones. Authentic standards were used to confirm that these compounds were 2-undecanone (C11), 2-tridecanone (C13; the predominant methyl ketone), and 2-pentadecanone (C15). Furthermore, we observed that overexpression of the M. luteus fabH (MlfabH; Mlut_09310) resulted in an increase in methyl ketone concentration relative to the fatty acid-overproducing control strain, particularly on an optical density (OD)-normalized basis (Fig. 1; see Fig. S1 in the supplemental material).
Fig 1.
Methyl ketone production in fatty acid- and β-ketoacyl-CoA-overproducing strains. Bar heights represent the averages of at least three biological replicates, and error bars represent 1 standard deviation. EcfadM, E. coli fadM.
Enhancement of methyl ketone generation by overproduction of β-ketoacyl-CoAs.
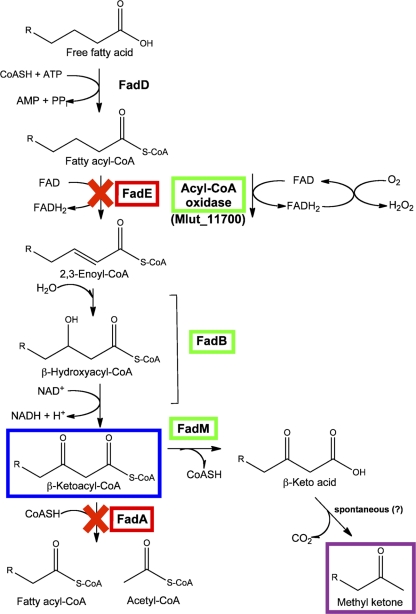
Several factors led us to hypothesize that increasing the production of β-ketoacyl-CoAs would lead to better production of methyl ketones: (i) the long-held hypothesis that, in fungi, methyl ketones arise from incomplete β-oxidation of fatty acids and decarboxylation of β-keto acids (10), (ii) methyl ketones were observed at higher concentration in fatty acid-overproducing DH1 strains than in wild-type DH1 (data not shown), and (iii) the carbon-chain lengths of the observed methyl ketones were consistent with decarboxylation of prominent fatty acids in DH1 (i.e., C12, C14, and C16). To test this hypothesis and increase levels of β-ketoacyl-CoAs, we constructed a modified, truncated fatty acid β-oxidation pathway in DH1 (Fig. 2).
Fig 2.
Summary of engineered pathway to convert fatty acids to methyl ketones in E. coli DH1. Green boxes indicate overexpressed genes, and red boxes indicate chromosomal deletions. The blue box indicates the putative substrate for FadM (producing free β-keto acids), and the purple box indicates the final methyl ketone product (putatively generated by spontaneous decarboxylation of β-keto acids). The ′TesA thioesterase used for fatty acid overproduction is not depicted in this figure.
The native fatty acid β-oxidation pathway in E. coli strain DH1 begins with the conversion of free fatty acids into acyl-CoAs by an acyl-CoA synthetase (FadD). The acyl-CoA is then oxidized to a trans-2-enoyl-CoA by a FAD-dependent acyl-CoA dehydrogenase (FadE). Next, FadB catalyzes a hydratase reaction to form a β-hydroxyacyl-CoA, which is then oxidized to a β-ketoacyl-CoA (also catalyzed by the bifunctional FadB). The cycle is completed by CoA-mediated thiolytic cleavage of a β-ketoacyl-CoA to acetyl-CoA and a shorter (n − 2) acyl-CoA, a reaction catalyzed by FadA. Our strategy to increase levels of β-ketoacyl-CoAs involved the following steps: (i) overexpression of a heterologous acyl-CoA oxidase used in lieu of FadE, (ii) overexpression of the native FadB, and (iii) deletion of fadA from the chromosome to truncate the β-oxidation cycle at β-ketoacyl-CoA. We chose to replace FadE with an acyl-CoA oxidase because the latter enzyme is a highly soluble protein (FadE is membrane associated) and has much higher specific activity than FadE (3, 4). Based upon reports of a high-activity acyl-CoA oxidase from Arthrobacter ureafaciens (3), we selected an apparent homolog (Mlut_11700; 63% protein sequence identity) from a related actinobacterium, M. luteus. Both Mlut_11700 and E. coli fadB were cloned into the low-copy pKS1 vector downstream of the ′tesA (thioesterase) gene (Table 1). The chromosomal deletion of fadA in E. coli DH1 was performed as described in Materials and Methods.
GC/MS analyses of extracts of β-ketoacyl-CoA-overproducing strains indicated dramatic increases in methyl ketone production relative to fatty acid-overproducing strains (e.g., an ∼75-fold increase for strain EGS560 versus strain EGS084) (Table 3; Fig. 1). Concentration trends were similar on an OD-normalized basis (compare Fig. 1 and Fig. S1 in the supplemental material). 2-Tridecanone was the predominant methyl ketone observed in β-ketoacyl-CoA-overproducing strains, as it was in fatty acid-overproducing strains (Fig. 1).
Table 3.
Fold improvements in total methyl ketone productiona resulting from genetic modifications and the presence of a decane overlay
| Methyl ketone source and strain | Fold improvement for overlay |
||
|---|---|---|---|
| EGS895b | EGS560c | EGS084d | |
| Overlay | |||
| EGS084 | 700 | 76 | NAe |
| EGS560 | 9.0 | NA | NA |
| Pelletf | |||
| EGS084 | 4,600 | 500 | 6.6 |
| EGS560 | 61 | 6.6 | NA |
| EGS895 | 4.7 | NA | NA |
Ratios of total methyl ketone concentrations at 39 h. Individual and total methyl ketone concentrations in these strains are presented in Table S3 in the supplemental material.
Strain EGS895, β-ketoacyl-CoA overproducing, FadM overexpressing (full description in Table 1).
Strain EGS560, β-ketoacyl-CoA-overproducing control without FadM (full description in Table 1).
Strain EGS084, fatty acid-overproducing control without FadM (full description in Table 1).
NA, not applicable.
Cell pellet extracted after incubation; no decane overlay used.
Identification of candidate E. coli thioesterase genes involved in methyl ketone production.
We demonstrated that overproduction of β-ketoacyl-CoAs increased methyl ketone production; however, it was unclear whether native E. coli proteins were facilitating conversion of the β-ketoacyl-CoAs to methyl ketones (e.g., by hydrolysis of the CoA thioester bond to generate a free β-keto acid and/or decarboxylation of the β-keto acid; Fig. 2). Further investigation of the enhancement of methyl ketone production in the presence of MlfabH suggested that indeed native E. coli proteins were facilitating conversion of β-ketoacyl-CoAs to methyl ketones. More specifically, when we mutated the conserved, well-characterized catalytic triad residues (C123S-H275A-N306A) of MlFabH (strain EGS735; Table 1), which should have rendered FabH enzymatically inactive (8), enhancement of methyl ketones was comparable to that observed in the strain expressing wild-type MlFabH (EGS212) (within 10%). This suggested that MlfabH expression had an epigenetic rather than catalytic effect, potentially upregulating native genes whose products facilitated methyl ketone production.
To explore the possibility that native E. coli DH1 proteins that could facilitate methyl ketone synthesis were being upregulated in the presence of MlfabH, we performed whole-genome transcriptional (microarray) analysis of strains EGS212 (MlfabH; Table 1) and EGS084 (control; empty vector). Using the Significance Analysis of Microarray (SAM) software package, we were able to narrow down to 55 the number of significantly upregulated genes that had a false discovery rate (FDR) of 0.6% or less (see Fig. S2 and Table S2 in the supplemental material). Of these significantly upregulated genes, only 7 were annotated to be associated with metabolism, and two thioesterases (paaI and fadM) were the most upregulated genes in this group (Table 4). RT-qPCR analyses confirmed that fadM was upregulated approximately 2-fold in strain EGS212 compared to strain EGS084.
Table 4.
Metabolic genes that were significantly upregulated during heterologous expression of MlFabHa
| Gene identifier | Gene name | Fold change | Annotation |
|---|---|---|---|
| b1396 | paaI | 3.4 | Predicted thioesteraseb |
| b0443 | fadM | 2.3 | Long-chain acyl-CoA thioesterase IIIb |
| b0459 | maa | 2.1 | Maltose O-acetyltransferase |
| b4040 | ubiA | 2.0 | p-Hydroxybenzoate octaprenyltransferase |
| b3769 | ilvM | 2.0 | Acetolactate synthase II, small subunit |
| b4039 | ubiC | 1.9 | Chorismate pyruvate-lyase |
| b1400 | paaY | 1.7 | Predicted hexapeptide repeat acetyltransferase |
Based upon whole-genome microarray analysis of strain EGS212 and control strain EGS084.
The two thioesterase genes used for further characterization are indicated in bold.
Overexpression of the E. coli fadM thioesterase enhances methyl ketone production.
The two thioesterase genes observed to be upregulated in the presence of MlFabH were overexpressed in a fatty acid-overproducing host (fadM in strain EGS860 and paaI in strain EGS790; Table 1), and the effect on methyl ketone production was assessed. Overexpression of paaI slightly decreased methyl ketone production (∼ 30%; data not shown), but overexpression of fadM resulted in an approximately 2-fold increase in 2-tridecanone (relative to the empty-vector control, strain EGS084) (Fig. 1). Furthermore, overexpression of fadM in a β-ketoacyl-CoA-overproducing strain (strain EGS895; Table 1) resulted in a 9-fold increase in methyl ketone production (relative to the empty-vector control, strain EGS560) (Table 3; Fig. 1).
A broader range of methyl ketones (including monounsaturates) is produced in β-ketoacyl-CoA-overproducing strains expressing FadM.
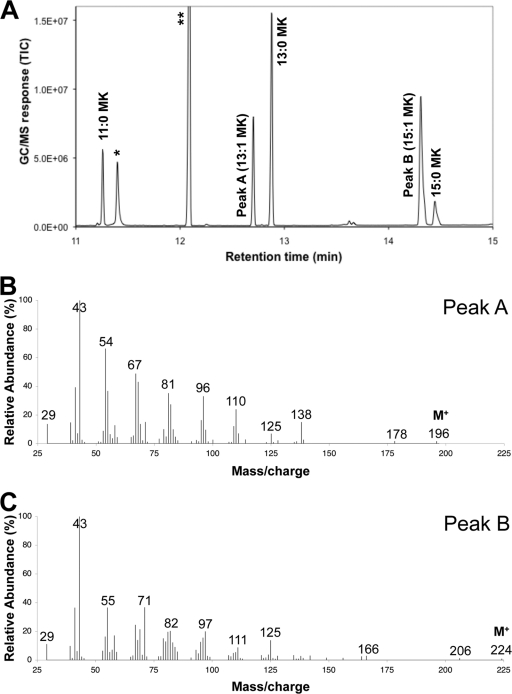
In addition to producing higher concentrations of 2-undecanone, 2-tridecanone, and 2-pentadecanone relative to fatty acid-overproducing strains and/or strains without fadM overexpression (Fig. 1), strain EGS895 also produced a wider range of detectable methyl ketones. This included 2-nonanone (C9) and 2-heptadecanone (C17) at low relative concentration (<1% of 2-tridecanone levels) and prominent peaks that are identified as monounsaturated methyl ketones. A representative GC/MS chromatogram of a diluted decane overlay of strain EGS895 is presented in Fig. 3A. Peaks A and B (Fig. 3A) are identified as tridecenone (C13H24O) and pentadecenone (C15H28O), respectively, based upon electron ionization GC/MS spectra (Fig. 3B and C), LC-APCI-TOF MS analysis, and comparison to a pentadecenone standard synthesized in vitro. Although authentic standards are not commercially available for tridecenone and pentadecenone, the TOF-determined accurate masses of the molecular ions representing peaks A and B agreed extremely well (within 0.5 ppm relative error) with the calculated masses for C13H24O and C15H28O. Furthermore, the base peak at m/z 43 in both EI spectra (Fig. 3B and C) is consistent with the [CH3-CO+] fragment characteristic of methyl ketones. Finally, an in vitro assay containing the CoA thioester of palmitoleic acid [(Z)-9-hexadecenoic acid], acyl-CoA oxidase (from M. luteus), E. coli DH1 FadB, and appropriate cofactors resulted in the formation of a compound with an identical GC/MS retention time and mass spectrum as peak B; this compound was not observed in an assay lacking FadB. Notably, an analogous assay using tetradecanoyl-CoA rather than palmitoleoyl-CoA resulted in the formation of 2-tridecanone. This strongly suggests that peak B is (Z)-8-pentadecen-2-one (15:1 methyl ketone), which was derived from palmitoleic acid (16:1 fatty acid). By analogy to peak B, it is logical to conclude that peak A is (Z)-8-tridecen-2-one derived from myristoleic acid (14:1 fatty acid). However, the mass spectral fragmentation patterns of peaks A and B differ somewhat in the region between m/z 50 and 120, so the position of the double bond in the tridecenone is less certain.
Fig 3.
GC/MS chromatogram of methyl ketone mixture generated by the best producing strain (strain EGS895) and mass spectra of prominent monounsaturated methyl ketones. (A) GC/MS total ion chromatogram (TIC) of diluted decane overlay featuring region with C11 to C15 saturated and monounsaturated methyl ketones (MK). X:Y notation is described in Table 5. ∗, component of growth medium; ∗∗, hydrocarbon contaminant in decane. (B) The 70-eV electron ionization mass spectrum of peak A, which was identified as tridecenone (see text). (C) The 70-eV electron ionization mass spectrum of peak B, which was identified as pentadecenone (see text).
A summary of the quantitative relationships between methyl ketones (both saturated and unsaturated) and their presumed fatty acid precursors is presented in Table 5. Among the trends apparent from Table 5 is that ratios of fatty acid precursors to the daughter methyl ketones are much greater in fatty acid-overproducing strains (EGS084 and EGS860) than in β-ketoacyl-CoA-overproducing strains (EGS560 and EGS895), suggesting that overall conversion of fatty acids to methyl ketones is far more efficient in the β-ketoacyl-CoA-overproducing strains. In addition, ratios of fatty acid precursors to the daughter methyl ketones are typically lower in strains with overexpressed FadM (EGS860 and EGS895) than in those without (EGS084 and EGS560, respectively), further suggesting that FadM improves the conversion of fatty acids to methyl ketones.
Table 5.
Molar ratios of precursor fatty acids to their daughter methyl ketones in fatty acid- and β-ketoacyl-CoA-overproducing strains of E. coli DH1 with and without fadM overexpression
| Straina | Molar ratio |
||||
|---|---|---|---|---|---|
| C12 fatty acid/C11 methyl ketoneb | C14 fatty acid/C13 methyl ketone | C14:1 fatty acid/C13:1 methyl ketonec | C16 fatty acid/C15 methyl ketone | C16:1 fatty acid/C15:1 methyl ketone | |
| EGS084 | 35 | 112 | NAd | 220 | NA |
| EGS860 | 33 | 30 | 6.2 | 41 | 68 |
| EGS560 | 0.50 | 0.40 | 0.13 | 0.97 | 0e |
| EGS895 | 0.078 | 0.041 | 0.018 | 0.17 | 0.0052 |
Strains are listed in order of increasing methyl ketone production.
Fatty acids were determined as methyl esters.
X:Y notation represents number of carbon atoms:number of carbon-carbon double bonds.
NA, not applicable; unsaturated methyl ketone was not detected.
Fatty acid (16:1) not detected.
Further characterization of the best methyl ketone-producing strain (EGS895).
The relative distribution of methyl ketones produced by strain EGS895 (the best producing strain in this study) is as follows (expressed as percentage of total methyl ketones): 2-undecanone (15%), 2-tridecenone (16%), 2-tridecanone (36%), 2-pentadecenone (26%), 2-pentadecanone (6%). The total concentration of methyl ketones produced by strain EGS895 was 380 ± 38 mg/liter for freshly transformed cells (pEG855) and 110 ± 32 mg/liter in cells grown from frozen glycerol stocks. A time series of methyl ketone production by strain EGS895 over 72 h (see Fig. S3 in the supplemental material) indicates that production begins in postexponential phase and that the production rate decreases between 48 and 72 h.
Strategies to modify methyl ketone composition.
Degree of unsaturation and chain length are important factors that mediate key properties of diesel fuels (e.g., low-temperature properties, represented here by melting point, and CN). Three modifications to the genotype or cultivation of strain EGS895 were examined to determine their impact on overall methyl ketone composition and production.
The first strategy involved changing the cultivation temperature of EGS895 to increase degree of unsaturation and thereby decrease melting point. We found that indeed the ratios of the dominant unsaturated methyl ketones (C13 and C15) to their saturated analogs increased considerably when strain EGS895 was cultivated at lower temperature. To illustrate, at 37°C, the ratio of tridecenone/tridecanone was 0.45, but at 15°C it increased to 0.93. Similarly, at 37°C, the ratio of pentadecenone/pentadecanone was 4, but at 15°C it increased to 8.5.
The second strategy was to replace the native ′TesA acyl-ACP thioesterase with UcFatB1 (strain EGS975; Table 1), a plant-derived thioesterase that has a stronger preference toward C12:0 acyl-ACP than does ′TesA (32). Based on the substrate preference of UcFatB1, we anticipated an increase in the proportion of undecanone (derived from C12 fatty acid) and a corresponding decrease in melting point. As expected, the ratio of undecanone to tridecanone increased from 0.1 in strain EGS895 to 0.4 in strain EGS975, but unexpectedly, the pentadecanone-to-tridecanone ratio increased from 0.24 in strain EGS895 to 0.82 in strain EGS975.
Although both strategies achieved the intended objective of altering methyl ketone composition, they also resulted in lower total methyl ketone production (from 2- to 5-fold lower) than in strain EGS895 cultivated at 37°C. Finally, an attempt was made to increase methyl ketone production by increasing the flux of free fatty acids into the β-oxidation pathway. To accomplish this, E. coli FadD (fatty acyl-CoA synthetase; Fig. 2) was overexpressed in strain EGS895. However, this modification also resulted in a 2-fold decrease rather than an increase in methyl ketone production.
Methyl ketone production in strains containing fadM compared to production in strains containing known methyl ketone synthases.
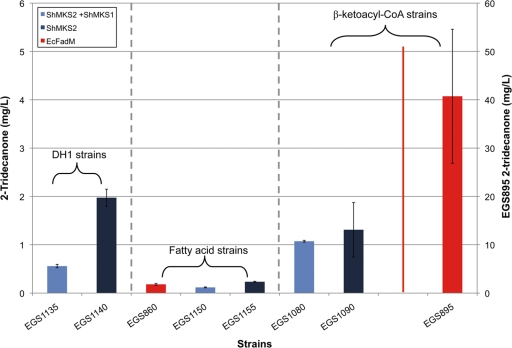
To date, the only proteins that have been experimentally verified to be methyl ketone synthases are ShMKS1 and ShMKS2 from S. habrochaites and homologous proteins in other plants (6, 31). ShMKS2 has been described as a “hot-dog”-fold-family thioesterase that hydrolyzes β-ketoacyl-ACPs (intermediates of fatty acid biosynthesis), and ShMKS1 is a decarboxylase that acts on β-keto acids (such as those produced by ShMKS2) (31). Since FadM, like ShMKS2, is a thioesterase belonging to the “hot-dog”-fold protein family (in this case hydrolyzing long-chain acyl-CoAs) (20), we were curious about the relative effects of overexpression of these proteins on methyl ketone production. Comparisons were made of methyl ketone (2-tridecanone) production in wild-type, fatty acid-overproducing, and β-ketoacyl-CoA-overproducing DH1 strains overexpressing fadM, ShMKS2, or ShMKS1 plus ShMKS2 (Fig. 4). Proteomics analyses confirmed ample expression of ShMKS1 and ShMKS2 in these studies. In all strains tested, constructs overexpressing ShMKS2 or ShMKS1 plus ShMKS2 never produced a 2-tridecanone concentration exceeding 5% that of strain EGS895 (a β-ketoacyl-CoA-overproducing, FadM-overexpressing strain). Two aspects of the data in Fig. 4 were unexpected: (i) the best methyl ketone production in a strain containing ShMKS2 was in the wild-type host (strain EGS1140) rather than in a fatty acid- or β-ketoacyl-CoA-overproducing host, and (ii) overexpression of ShMKS1 with ShMKS2 never improved methyl ketone production, and in some cases it detracted considerably from methyl ketone production. Regarding the latter point, overexpression of ShMKS1 also detracted from methyl ketone production in strains overexpressing FadM. To illustrate, in β-ketoacyl-CoA-overproducing DH1 strains overexpressing fadM plus ShMKS1 (with or without its own Ptrc promoter; strains EGS1115 and EGS1120, respectively), 2-tridecanone concentrations were approximately 5-fold lower than in strain EGS895, which did not contain ShMKS1 (data not shown). The reason that ShMKS1 decreased methyl ketone production is unknown.
Fig 4.
2-Tridecanone concentration in DH1 wild-type, fatty acid-overproducing, or β-ketoacyl-CoA-overproducing strains expressing various methyl ketone synthases. Note that the scale for 2-tridecanone concentration in strain EGS895 is on the right-hand y axis. Bar heights represent averages, and error bars represent 1 standard deviation.
Effect of decane overlay on production.
In strains with very low methyl ketone production (primarily wild-type E. coli DH1), an exhaustive extraction of the cell pellet (using methods described previously [5]) was necessary. However, decane overlays were usable for all other strains. Methyl ketone production was considerably higher when fatty acid- or β-ketoacyl-CoA-overproducing strains were incubated with a decane overlay than when they were sacrificed and the cell pellet was exhaustively extracted. To illustrate, for the best producing strain (EGS895; Table 1), the methyl ketone concentration was more than 4-fold greater in the overlay than in the pellet extract at 39 h (Table 3). This may be explained by one or more of several factors, including the following: (i) removal of the methyl ketone products provides a thermodynamic driving force for production, (ii) the overlay efficiently sequesters methyl ketones that might otherwise be volatilized during cultivation, and (iii) removal of methyl ketones (or other metabolites) from the medium may alleviate potentially inhibitory or toxic effects from their accumulation. A comparison between the results of overlay and pellet extractions supports both points (i) and (ii). First, the ratio of C14:0 fatty acid to C13:0 methyl ketone for strain EGS895 was 30-fold lower in overlays than in pellet extractions; this lower ratio in overlays could be explained by more efficient flux of fatty acids to methyl ketones in the presence of the overlay. Second, the ratio of C11:0 methyl ketone to C15:0 methyl ketone is 2-fold higher for the overlay than for the pellet. Since the C11:0 methyl ketone is more volatile than the C15:0 methyl ketone, the higher ratio in overlays supports the notion that the decane overlay facilitates capture of volatile compounds that would be lost without an overlay. Regarding the final explanation (toxicity mitigation), this seems unlikely because OD values for strain EGS895 were similar in the presence and absence of an overlay, suggesting that methyl ketones are not particularly toxic (at least, not at these concentrations).
Cetane number determination of selected methyl ketones.
Cetane number (CN) is a key index indicating overall diesel fuel quality, much as octane number is a widely used indicator of gasoline fuel quality. More specifically, CN is a measure of ignition delay during compression ignition; a higher CN indicates a shorter ignition delay period and is more favorable than a lower CN (up to a CN of 55 to 60). In the United States, diesel fuel must have a minimum CN of 40, in accordance with ASTM standard D975. The CN for 2-undecanone (Sigma) was 56.6 and for a 50/50 (wt/wt) mixture of 2-undecanone and 2-tridecanone was 58.4.
DISCUSSION
We have engineered a small number of modifications into E. coli DH1 that resulted in a 700-fold increase in methyl ketone concentration relative to a fatty acid-overproducing strain. Accounting for the use of decane overlays, the overall increase was more than 4,500-fold (Table 3). The modifications included overproduction of β-ketoacyl-CoAs (by overexpression of an acyl-CoA oxidase from M. luteus and native FadB, as well as chromosomal deletion of fadA) and overexpression of the native thioesterase, FadM. In all host strains tested (wild-type, fatty acid-overproducing, β-ketoacyl-CoA-overproducing DH1), overexpression of the methyl ketone synthase ShMKS2 never produced methyl ketones at concentrations that were more than 5% of those observed for the best producing FadM-overexpressing strain.
To some extent, the difference in behavior of the two thioesterases, FadM and ShMKS2, can be explained by their known substrates. FadM has relatively high activity on acyl-CoA substrates between C12 and C18 (particularly 3,5-cis-tetradecadienoyl-CoA) (20), whereas ShMKS2 appears to be well suited to β-ketoacyl-ACPs (31). It follows that a thioesterase that hydrolyzes CoA thioesters (FadM) would be more amenable to acting on β-oxidation intermediates, whereas a thioesterase that hydrolyzes ACP thioesters would be more effective at hydrolyzing fatty acid biosynthetic intermediates (β-ketoacyl-ACPs in particular). That said, a limited amount of information is available on the substrate ranges of these two thioesterases (particularly ShMKS2), so the extent to which each favors CoA versus ACP thioesters is unknown (25). Although FadM apparently hydrolyzes β-ketoacyl-CoAs sufficiently to markedly increase methyl ketone yields, it is reported to have considerably (at least 10-fold) higher activity on C16 acyl-CoA than on C16 β-ketoacyl-CoA (20).
The best methyl ketone producer studied here (strain EGS895) did not have an added decarboxylase to convert free β-keto acids to methyl ketones. Either a native enzyme catalyzed this reaction or it occurred abiotically, as β-keto acids are well-known to be inherently unstable and prone to spontaneous decarboxylation (16). Spontaneous decarboxylation would not be surprising, as we observed substantial methyl ketone yields from in vitro reaction mixtures that produced β-ketoacyl-CoAs from acyl-CoAs; these reaction mixtures lacked both decarboxylases and thioesterases (the only enzymes that they contained were acyl-CoA oxidase and FadB). For unknown reasons, overexpression of the ShMKS1 decarboxylase, which is reported to play a role in methyl ketone synthesis in S. habrochaites, markedly decreased methyl ketone synthesis in this study (including strains EGS1115 and EGS1120, which were simply ShMKS1-amended versions of EGS895).
As is the case for other fatty acid-derived biofuels, such as fatty acid ethyl esters, saturated, medium-chain methyl ketones addressed in this article have favorable cetane numbers (CNs). A less favorable property of the saturated methyl ketones addressed in this article is relatively high melting point (e.g., 30.5°C for 2-tridecanone [12]), which is related to cold-temperature diesel fuel properties such as cloud point. This disadvantage could be significantly mitigated by the prominent monounsaturated methyl ketones observed in the best producing strains (monounsaturated methyl ketones account for ∼40% of total methyl ketones in strain EGS895). Melting point depression caused by monounsaturation in fatty acid methyl esters illustrates this point. For example, for C16 and C18 fatty acid methyl esters, the cis-Δ9 monounsaturated homologs have melting points approximately 60°C lower than those of their saturated counterparts (the melting point of methyl palmitoleate [16:1] is −33.9°C, whereas that of methyl palmitate [16:0] is 30°C; the melting point of methyl oleate [18:1] is −19.5°C, whereas that of methyl stearate [18:0] is 39°C) (15). However, unsaturation can also be expected to decrease CN (e.g., a decrease of ∼30 in CN applies to C16 fatty acid methyl esters [15]). In addition to degree of unsaturation, chain length will also affect fuel properties (increasing chain length increases CN and melting point). The ensemble of saturated and unsaturated methyl ketones generated by strain EGS895 (and related strains) may have sufficiently favorable collective fuel properties to be appropriate for blending with petroleum-based diesel. Nonetheless, future efforts will be directed at enhancing methyl ketone production (e.g., by enhancing intracellular malonyl-CoA levels [33]) and modulating the methyl ketone composition to optimize diesel fuel properties.
Supplementary Material
ACKNOWLEDGMENTS
We thank Tanveer Batth and Christopher Petzold (Functional Genomics Department, Technology Division, JBEI) for mass spectrometric analysis of protein samples, Kenneth Childress (Southwest Research Institute) for cetane number analysis, and Taek Soon Lee (JBEI) for helpful comments on the manuscript.
J.D.K. has a financial interest in LS9, Amyris, and Lygos.
This work conducted by the Joint BioEnergy Institute was supported by the Office of Science, Office of Biological and Environmental Research, of the U.S. Department of Energy under contract no. DE-AC02-05CH11231.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Antonious GF, Dahlman DL, Hawkins LM. 2003. Insecticidal and acaricidal performance of methyl ketones in wild tomato leaves. Bull. Environ. Contam. Toxicol. 71:400–407 [DOI] [PubMed] [Google Scholar]
- 2. Baba T, et al. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bakke M, Setoyama C, Miura R, Kajiyama N. 2007. N-Ethylmaleimide-resistant acyl-coenzyme A oxidase from Arthrobacter ureafaciens NBRC 12140: molecular cloning, gene expression and characterization of the recombinant enzyme. Biochim. Biophys. Acta 1774:65–71 [DOI] [PubMed] [Google Scholar]
- 4. Baltazar MF, Dickinson FM, Ratledge C. 1999. Oxidation of medium-chain acyl-CoA esters by extracts of Aspergillus niger: enzymology and characterization of intermediates by HPLC. Microbiology 145:271–278 [DOI] [PubMed] [Google Scholar]
- 5. Beller HR, Goh EB, Keasling JD. 2010. Genes involved in long-chain alkene biosynthesis in Micrococcus luteus. Appl. Environ. Microbiol. 76:1212–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ben-Israel I, et al. 2009. Multiple biochemical and morphological factors underlie the production of methylketones in tomato trichomes. Plant Physiol. 151:1952–1964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Davies C, Heath RJ, White SW, Rock CO. 2000. The 1.8 Å crystal structure and active-site architecture of β-ketoacyl-acyl carrier protein synthase III (FabH) from Escherichia coli. Structure 8:185–195 [DOI] [PubMed] [Google Scholar]
- 9. Dellomonaco C, Clomburg JM, Miller EN, Gonzalez R. 2011. Engineered reversal of the beta-oxidation cycle for the synthesis of fuels and chemicals. Nature 476:355–359 [DOI] [PubMed] [Google Scholar]
- 10. Forney FW, Markovetz AJ. 1971. The biology of methyl ketones. J. Lipid Res. 12:383–395 [PubMed] [Google Scholar]
- 11. Fridman E, et al. 2005. Metabolic, genomic, and biochemical analyses of glandular trichomes from the wild tomato species Lycopersicon hirsutum identify a key enzyme in the biosynthesis of methylketones. Plant Cell 17:1252–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Haynes WM. 2010-2011. CRC handbook of chemistry and physics, 91st ed. CRC Press, Boca Raton, FL [Google Scholar]
- 13. Irizarry RA, et al. 2003. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4:249–264 [DOI] [PubMed] [Google Scholar]
- 14. Kirchner O, Tauch A. 2003. Tools for genetic engineering in the amino acid-producing bacterium Corynebacterium glutamicum. J. Biotechnol. 104:287–299 [DOI] [PubMed] [Google Scholar]
- 15. Knothe G. 2008. “Designer” biodiesel: optimizing fatty ester composition to improve fuel properties. Energy Fuels 22:1358–1364 [Google Scholar]
- 16. Kornberg A, Ochoa S, Mehler AH. 1947. Spectrophotometric studies on the decarboxylation of β-keto acids. Fed. Proc. 6:268. [PubMed] [Google Scholar]
- 17. Li MZ, Elledge SJ. 2007. Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC. Nat. Methods 4:251–256 [DOI] [PubMed] [Google Scholar]
- 18. Mendez-Perez D, Begemann MB, Pfleger BF. 2011. Modular synthase-encoding gene involved in α-olefin biosynthesis in Synechococcus sp. strain PCC 7002. Appl. Environ. Microbiol. 77:4264–4267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Meselson M, Yuan R. 1968. DNA restriction enzyme from E. coli. Nature 217:1110–1114 [DOI] [PubMed] [Google Scholar]
- 20. Nie L, Ren Y, Schulz H. 2008. Identification and characterization of Escherichia coli thioesterase III that functions in fatty acid β-oxidation. Biochemistry 47:7744–7751 [DOI] [PubMed] [Google Scholar]
- 21. Patel RN, Hou CT, Laskin AI, Felix A, Derelanko P. 1980. Microbial oxidation of gaseous hydrocarbons: production of methylketones from corresponding n-alkanes by methane-utilizing bacteria. Appl. Environ. Microbiol. 39:727–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rude MA, et al. 2011. Terminal olefin (1-alkene) biosynthesis by a novel P450 fatty acid decarboxylase from Jeotgalicoccus species. Appl. Environ. Microbiol. 77:1718–1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 24. Schirmer A, Rude MA, Li X, Popova E, del Cardayre SB. 2010. Microbial biosynthesis of alkanes. Science 329:559–562 [DOI] [PubMed] [Google Scholar]
- 25. Spencer AK, Greenspan AD, Cronan JE., Jr 1978. Thioesterases I and II of Escherichia coli. Hydrolysis of native acyl-acyl carrier protein thioesters. J. Biol. Chem. 253:5922–5926 [PubMed] [Google Scholar]
- 26. Steen EJ, et al. 2010. Microbial production of fatty-acid-derived fuels and chemicals from plant biomass. Nature 463:559–562 [DOI] [PubMed] [Google Scholar]
- 27. Studier FW, Moffatt BA. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189:113–130 [DOI] [PubMed] [Google Scholar]
- 28. Sukovich DJ, Seffernick JL, Richman JE, Gralnick JA, Wackett LP. 2010. Widespread head-to-head hydrocarbon biosynthesis in bacteria and role of OleA. Appl. Environ. Microbiol. 76:3850–3862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tusher VG, Tibshirani R, Chu G. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. U. S. A. 98:5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Williams CG. 1858. On the constitution of the essential oil of rue. Philos. Trans. R. Soc. Lond. 148:199–204 [Google Scholar]
- 31. Yu G, et al. 2010. Enzymatic functions of wild tomato methylketone synthases 1 and 2. Plant Physiol. 154:67–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yuan L, Voelker TA, Hawkins DJ. 1995. Modification of the substrate specificity of an acyl-acyl carrier protein thioesterase by protein engineering. Proc. Natl. Acad. Sci. U. S. A. 92:10639–10643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zha W, Rubin-Pitel SB, Shao Z, Zhao H. 2009. Improving cellular malonyl-CoA level in Escherichia coli via metabolic engineering. Metab. Eng. 11:192–198 [DOI] [PubMed] [Google Scholar]
- 34. Zhou K, et al. 2011. Novel reference genes for quantifying transcriptional responses of Escherichia coli to protein overexpression by quantitative PCR. BMC Mol. Biol. 12:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.