Abstract
Microorganisms have developed mechanisms to combat reactive nitrogen species (RNS); however, only a few of the fungal genes involved have been characterized. Here we screened RNS-resistant Aspergillus nidulans strains from fungal transformants obtained by introducing a genomic DNA library constructed in a multicopy vector. We found that the AN0121.3 gene (hemC) encodes a protein similar to the heme biosynthesis enzyme porphobilinogen deaminase (PBG-D) and facilitates RNS-tolerant fungal growth. The overproduction of PBG-D in A. nidulans promoted RNS tolerance, whereas PBG-D repression caused growth that was hypersensitive to RNS. PBG-D levels were comparable to those of cellular protoheme synthesis as well as flavohemoglobin (FHb; encoded by fhbA and fhbB) and nitrite reductase (NiR; encoded by niiA) activities. Both FHb and NiR are hemoproteins that consume nitric oxide and nitrite, respectively, and we found that they are required for maximal growth in the presence of RNS. The transcription of hemC was upregulated by RNS. These results demonstrated that PBG-D is a novel NO-tolerant protein that modulates the reduction of environmental NO and nitrite levels by FHb and NiR.
INTRODUCTION
Nitric oxide (NO) is the best-characterized of the reactive nitrogen species (RNS), which react to and damage DNA, lipids, and enzymes, as do reactive oxygen species (31, 39). Both enzymatic and chemical reactions can generate NO under physiological conditions. Mammalian NO synthase oxidizes arginine to produce NO, which transduces signals for neuronal communication, inflammation, and smooth-muscle relaxation (24). Macrophages produce NO by inducible NO synthase to fight infective microorganisms and parasites (5). Microorganisms generate NO as an intermediate of denitrification (10). Bacterial nitrification and denitrification processes involve nitrite (NO2−) as an intermediate, and improper aeration often results in the accumulation of NO2−, which is concentration-dependently dismutated to NO, especially in an acidic environment (43). Thus, NO is ubiquitous in the microbial milieu, and exposure to NO2− and NO should induce microbial mechanisms to survive RNS toxicity.
Microorganisms have developed constitutive and inducible strategies for combating RNS. Flavohemoglobin (FHb) is the best known enzyme that is involved in the RNS response, and it is widespread in bacteria, yeast, and filamentous fungi (29). It comprises protoheme-containing hemoglobin-like and flavin-containing ferredoxin-NAD(P)H reductase-like domains that are located in the amino- and carboxy-terminal regions, respectively (16). Flavohemoglobin functions as a nitric oxide dioxygenase (NOD), converts oxygen and NO to less-toxic nitrate, and facilitates NO tolerance (9). Besides FHb, bacteria and fungi produce cytochrome b- and cytochrome P450-type NO reductases (Nor), which reduce NO to nitrous oxide and consequently minimize NO toxicity (34, 42). Another bacterial flavorubredoxin-type Nor is NorVW, which contains a di-iron catalytic center and reduces and detoxifies NO (7). The primary function of cytochrome c NO2− reductases (NiRs) is the unambiguously dissimilatory reduction of NO2−. Nevertheless, reports indicate that the Escherichia coli isozyme reduces NO to less-harmful ammonium both in vitro (35) and in vivo (28) and facilitates RNS tolerance. S-Nitrosoglutathione (GSNO) reductase, which is conserved from bacteria to humans, reduces GSNO to ammonia and glutathione disulfide and consequently enhances cellular resistance to nitrosative stress (20). The genome of Aspergillus nidulans encodes FHb and GSNO reductase orthologs, and no other RNS-detoxifying enzyme has been identified in the fungus.
Transcriptome analyses have shown that more than 100 genes of Saccharomyces cerevisiae, Candida albicans, and Histoplasma capsulatum fungi are regulated by NO (14, 17, 25). The functions of a large number of these genes are unknown, indicating that the fungal mechanisms of the RNS response are not fully understood. FHb-deficient Cryptococcus neoformans and C. albicans strains still show considerable RNS-tolerant growth (4, 37), indicating that a non-FHb-dependent mechanism(s) confers RNS tolerance on these fungi.
The genus Aspergillus contains strains of industrial, pharmaceutical, and medical importance, such as Aspergillus niger, A. oryzae, A. flavus, and A. fumigatus. Recent reports indicate that A. oryzae produces two FHb isozymes that function as NODs and that they are located in the cytosol and mitochondria (40, 41). Aspergillus nidulans is a powerful model organism of the genus Aspergillus that produces FHb in order to survive under nitrosative stress (32). We previously reported that A. nidulans dissimilated nitrate to generate ammonium via NO2− under hypoxic conditions (36). We also reported that hypoxic A. nidulans cells accumulate NO2−, which damages purine residues of DNA (33). A recent report has suggested that A. nidulans NAD(P)H-NO2− reductase (NiR, the product of the niiA gene) plays a potential role in NO detoxification by sequestering NO-generating NO2− (32). These findings indicate that the fungal mechanism(s) involved in the RNS response is more complex than previously imagined.
We adopted a systematic approach to understanding the mechanism of fungal RNS tolerance based on the phenotypic screening of RNS tolerance genes using a genomic DNA library constructed in a multicopy vector of A. nidulans. One gene that was identified encoded porphobilinogen deaminase (PBG-D), which is involved in the heme biosynthetic pathway. Porphobilinogen deaminase modulated cellular protoheme biosynthesis and the activities of heme-containing FHb and NiR, thus generating NO-tolerant growth. These results demonstrated that PBG-D is a novel NO-tolerant protein through which the fungus increases protoheme synthesis and RNS tolerance.
MATERIALS AND METHODS
Strains, culture, and media.
Aspergillus nidulans strain FGSC713 (yA2; pyroA4; niiA4) was obtained from the Fungal Genetic Stock Center (FGSC; University of Missouri, Kansas City). Strain ABPU1 (biA1 pyrG89; wA3; argB2; pyroA4) was a gift from Hiroyuki Horiuchi (University of Tokyo, Tokyo, Japan). Strains YMT (yA2 pyrG89; pyroA4) and MST5 (the same genotype as YMT) were progenies of a meiotic cross between strains FGSC713 and ABPU1. All fungi were grown at 37°C in minimal medium (MM) (1% glucose, 10 mm NaNO3, 10 mM KH2PO4, 7 mM KCl, 2 mM MgSO4, 2 ml liter−1 Hunter's trace metals) (36) supplemented appropriately (0.55 g liter−1 uracil, 0.6 g liter−1 uridine, 0.4 mg liter−1 pyridoxine, 0.4 mg liter−1 biotin, 10 mM proline, 10 mM methionine, and/or 10 μg ml−1 ergosterol).
Screening of genes for RNS tolerance.
Strain MST5 was transformed with the pRG3-AMA1-NotI wild-type (WT) library provided by the FGSC as described previously (36). After a 3-day incubation, transformants were replicated in MM containing 25 mM NaNO2 (pH 5.5), and growth was monitored at 37°C for 3 days. Total DNA prepared from transformants cultured in MM as described by Takasaki et al. (36) was introduced into Escherichia coli DH5α to generate E. coli transformants. Plasmids were recovered from the bacterial transformants, and the nucleotide sequences of the insert fragments were determined.
Assaying RNS-tolerant growth.
Serial dilutions of 2.5-day-old conidia of the A. nidulans strains were spotted onto agar plates of MM containing 25 mM NaNO2 (pH 5.5) or 10 mM NOC-18 (Dojindo, Kumamoto, Japan). After incubation at 37°C for 2 days, the morphology of the colonies was examined to determine whether growth was RNS tolerant.
Quantitative PCR.
Total RNA and first-strand cDNA were prepared from fungal cells cultured at 37°C in MM for 12 h as described previously (33). Quantitative PCR proceeded using iQ SYBR Green Supermix (Bio-Rad Laboratories, Hercules, CA) and MiniOpticon, version 3.1 (Bio-Rad) according to the manufacturer's instructions. Gene expression was normalized against that of actA, and the results are presented as relative expression rates. The primers are listed in Table S1 in the supplemental material.
Disruption of the hemC and fhb genes.
The pyrG gene was amplified by PCR using A. nidulans total DNA with primers pyrG-F and pyrG-R (see Table S1 in the supplemental material), digested with SmaI, and inserted into the SmaI site of pBluescript KS+ to construct pBSpyrG. To construct a hemC disruption cassette, the 5′ untranslated region of hemC was amplified by PCR using primers ΔhemC-1 and ΔhemC-2, digested with PstI and EcoRI, and inserted into pBSpyrG, which had been cut beforehand with the same enzymes. The resulting plasmid was digested with SacII and NotI and was ligated to the 3′ region of hemC, which was amplified with primers ΔhemC-3 and ΔhemC-4 and was pretreated with the same enzymes, to generate pBSpyrGhemC. Plasmids pBSpyrGfhbA and pBSargBfhbB, for the disruption of fhbA and fhbB, were constructed as described above using the corresponding primers (see Table S1 in the supplemental material), pBSpyrG and pSSH1 (harboring the A. nidulans argB gene) (36), respectively. Aspergillus nidulans YMT was transformed using pBSpyrGhemC, and A. nidulans ABPU1 was transformed using pBSpyrGfhbA. We confirmed fhbA gene disruption and then introduced pBSargBfhbB into the strain to obtain fhbA fhbB double knockout strains. Southern blot analysis confirmed gene disruption as described below.
Construction of the hemC conditional mutant.
The 0.3-kb DNA fragment corresponding to the alcA gene promoter was amplified using primers alcA-F and alcA-R (see Table S1 in the supplemental material). The DNA fragment containing the hemC open reading frame (ORF) was amplified using primers hemC-F and hemC-R. These DNA fragments were fused by overlap extension PCR using primers alcA-F and hemC-R. The fused product was first digested with SacII and NotI and then inserted into the same restriction sites of pBSpyrG to construct pBSpyrG-alcPhemC. The 5′ region of hemC was amplified using primers ΔhemC-1 and ΔhemC-2, digested with PstI and EcoRV, and inserted into pBSpyrG-alcPhemC spliced with the same restriction enzymes. The resulting plasmid was introduced into YMT, and transformants in which the chromosomal gene promoter for hemC was replaced with that for alcA were selected by Southern blotting as described below.
Southern blot analysis.
Total DNAs of the fungal strains were digested with the appropriate restriction enzymes, resolved by agarose gel electrophoresis, and transferred to Hybond N+ membranes (GE Healthcare, Buckinghamshire, England). Hybridization probes were amplified with primer sets (see Table S1 in the supplemental material) and were labeled using a digoxigenin (DIG) PCR labeling kit (Roche Diagnostics, Basel, Switzerland). Hybridization and signal detection proceeded using the DIG system according to the instructions supplied. Signals were detected using a luminescent image analyzer (ImageQuant LAS 4000 mini; GE Healthcare, Sweden).
Preparation of cell extracts and cell fractions.
Fungal cells were collected by filtration, washed twice with 0.7% NaCl, suspended in buffer A (20 mM potassium phosphate [pH 7.5], 0.25 M sucrose, 0.3 mM N-tosyl-l-phenylalanyl chloromethylketone, 0.3 mM phenylmethylsulfonyl fluoride), and homogenized as described previously (36).
Enzyme assays.
Porphobilinogen deaminase activity was determined by measuring the absorbance of uroporphyrin formed by the light-induced oxidation of uroporphyrinogen, the immediate product of enzymatic deamination, as described by Ickowicz Schwartz et al. (15), with the following modification. The cell extract (2 mg of proteins) was incubated at 37°C for 30 min in the dark with 1 ml of 1 mM porphobilinogen and 250 mM sodium phosphate-citric acid buffer (pH 7.5). After the addition of 2 ml of ethyl acetate-acetic acid (3:1, vol/vol), the reaction mixture was first separated by centrifugation at 1,000 × g and then exposed to ambient light at room temperature for 15 min. The upper layer (1.6 ml) containing porphyrin was mixed with 1 ml of 0.5 M HCl, and the mixture was separated by centrifugation at 2,500 × g for 10 min. The lower layer was then transferred to a fluorescence microplate reader (DTX 880 multimode detector; Beckman Coulter, Brea, CA) and was analyzed at excitation and emission filter settings of 409 and 595 nm.
Aconitase activity was measured in a reaction mixture containing 100 mM Tris-HCl (pH 8.0), 30 mM sodium citrate, 0.6 mM MnCl2, 0.2 mM NADP+, 1 U ml−1 isocitrate dehydrogenase, and cell extracts. Nitrite reductase activity was measured in a reaction mixture containing 50 mM potassium phosphate (pH 7.5), 0.2 mM sodium nitrite, 1 mM NADPH, 10 μM flavin adenine dinucleotide (FAD), and 10 μg of cell extracts. Both reactions were monitored by following absorbance changes in NADPH at 25°C over a 5-min period. The molar extinction coefficient used for calculating NADPH was 6.22 × 103 mM−1 cm−1.
The activity of NOD was measured using a nitric oxide electrode (ISO-NO Mark II; World Precision Instruments, Sarasota, FL). The reaction mixture contained 2 ml of 50 mM citrate-NaOH buffer (pH 5.5), 0.1 mM NADH, 10 μM FAD, and 3 μM NOC-18. The reaction was initiated by adding cell extracts (20 μg).
Incubation with a proteasome inhibitor.
Aspergillus nidulans WT cells were precultured in MM for 20 h and were then incubated in 10 ml MM containing 0.003% sodium dodecyl sulfate (SDS) for 3 h, followed by 120 μM MG132 (Merck KGaA, Darmstadt, Germany) or dimethyl sulfoxide (the solvent for MG132) for 1 h. Subsequently, the cells were collected, and intracellular PBG-D activity was measured as described above.
Measurement of respiration rates.
Fungal cells were homogenized as described above; cellular debris was removed by centrifugation at 1,500 × g for 10 min; and mitochondrial fractions were pelleted by centrifugation at 10,000 × g for 15 min. Respiration rates were monitored using an oxygen electrode connected to a biological oxygen monitor (model 5300; YSI Inc., Yellow Springs, OH) at 25°C. The reaction mixture contained 20 mM Tris-HCl (pH 7.4), 10 mM KH2PO4, 5 mM MgCl2, 20 mM KCl, and the mitochondrial fractions, and the reaction was initiated by the addition of 0.5 mM NADH.
Determination of protoheme levels.
Fungal cells (wet weight, 4 g) were disrupted by grinding in liquid nitrogen and were then suspended in 25 ml of 80% acetone saturated with CaCO3. The suspension was filtered, and the resulting pellet was thoroughly rinsed with 80% acetone. Heme was extracted from suspensions of the residue in 20 ml of 2% HCl in acetone, filtered, and mixed with 30 ml of diethyl ether. Deionized water (100 ml) was added; the ether layer was evaporated in darkness to a final volume of 1 ml; and the residue was analyzed by high-performance liquid chromatography (HPLC) with an HP-1100 system (Hewlett-Packard, Palo Alto, CA) equipped with a TSKgel ODS-80 column (4.6 by 150 mm; Tosoh, Tokyo, Japan). The mobile phase was 0.1 M ammonium phosphate (pH 3.5), with a methanol gradient (60% to 100%) in 15 min. Absorbance at 405 nm was monitored.
RESULTS
Screening of a novel fungal nitrosative stress resistance gene.
To isolate A. nidulans genes involved in the mechanism of RNS tolerance, we transformed A. nidulans strain MST5 with a fungal genomic DNA library constructed using a high-copy-number vector (pRG3) that contained an autonomously replicating sequence of this fungus (AMA1) (26). A total of 26,000 transformants were replicated in minimal medium (MM) containing 40 mM acidified NO2− (pH 5.5) as an RNS donor, and viable transformants were selected. Their total DNAs were extracted and introduced into E. coli to recover plasmids. Nucleotide sequencing showed that four plasmids prepared from independent fungal transformants carried inserted DNA fragments with overlapping regions harboring AN0120.3 and AN0121.3 (see Fig. S1 in the supplemental material). Plasmids from another two fungal transformants contained genes that differed from AN0120.3 and AN0121.3, suggesting that our screening was not saturated. The truncated DNA fragments in the four plasmids were subcloned into pRG3 and were introduced into A. nidulans YMT (the wild-type [WT] strain), and the growth of the resulting transformants was then examined using a medium containing acidified NO2−. The transformant harboring the 1.7-kb DNA fragment containing AN0121.3 (strain PD1) grew at a rate similar to that of the WT strain harboring pRG3 in the absence of acidified NO2− (Fig. 1A). The addition of 25 mM acidified NO2− to the culture medium obviously suppressed the growth of the WT more than that of PD1 (Fig. 1A). The transformants harboring the DNA fragment containing AN0120.3 were as sensitive to 25 mM acidified NO2− as the WT (data not shown). These results indicated that AN0120.3 does not allow growth in the presence of acidified NO2− and that AN0121.3 conferred RNS tolerance on the fungus.
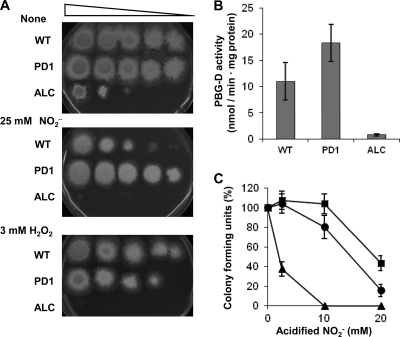
Fig 1.
Growth, PBG-D activities, and colony formation for the WT strain, PD1, and ALC. (A) Colonies appeared on MM agar without NaNO2 or containing 25 mM NaNO2 (pH 5.5) or 3 mM H2O2 after incubation with serial dilutions of conidia from the WT strain, PD1, or ALC at 37°C for 48 h. (B) Mycelia (1 g) were incubated for 12 h at 37°C in 100 ml of MM containing 15 mM NaNO2 (pH 5.5), and cellular PBG-D activities were determined. (C) Conidia of the strains were spread on MM plates containing 25 mM NaNO2 (pH 5.5). Colonies were counted after a 48-h incubation, and CFU are expressed as percentages of the CFU for strains incubated without NaNO2. Circles, WT; squares, PD1; triangles, ALC. Data are means for three experiments, and error bars represent standard errors. P, <0.01.
Characterization of AN0121.3.
The AN0121.3 gene encodes a protein comprising 348 amino acid residues. The amino acid sequence was similar to those of the porphobilinogen deaminases (PBG-Ds) of S. cerevisiae (Hem3p) (44% identity) and E. coli (HemC) (35% identity) (see Fig. S2 in the supplemental material). These PBG-Ds condense four porphobilinogen molecules in a head-to-tail fashion to form hydroxymethylbilane (2), which is critical for synthesizing heme. To understand the function of AN0121.3 (referred to below as hemC) in RNS tolerance, we attempted to disrupt the hemC gene. After introducing a plasmid designed for double crossover at both the 5′ and 3′ regions of chromosomal hemC, we obtained slowly growing transformants of heterokaryons. None of their mononucleate conidia formed colonies on selective medium, indicating that hemC is essential for growth (data not shown). We replaced the chromosomal hemC gene promoter with the alcA gene promoter, the expression of which is induced by ethanol or threonine and repressed by glucose (1), and designated the strain ALC (see Fig. S3 in the supplemental material). After the incubation of fungal mycelia in MM containing glucose as the sole source of carbon, more PBG-D activity was detected in PD1 than in the WT, whereas little activity was detected in ALC (Fig. 1B). This indicated that hemC expression was limited in strain ALC and that strains PD1 and ALC overproduced and repressed PBG-D activity, respectively.
The growth of strain ALC, cultured using glucose as the sole source of carbon, was severely retarded (Fig. 1A), supporting our notion that hemC is essential for growth. The slight growth observed suggested that the low background expression of hemC due to the presence of the alcA gene promoter is sufficient to support minimal growth. Acidified NO2− repressed the growth of strain ALC more than that of the WT strain (Fig. 1A), indicating that hemC contributes to growth under RNS stress. The rates at which conidia formed colonies on a medium containing glucose plus acidified NO2− were highest for PD1, intermediate for the WT, and lowest for ALC (Fig. 1C). This indicated that high PBG-D levels are associated with colony formation in the presence of acidified NO2−.
We cultured A. nidulans using nitrate and sulfate as the sole nitrogen and sulfur sources, respectively; these compounds are known to be assimilated by enzymes containing heme. The addition of proline and methionine as nitrogen and sulfur sources did not restore the growth of strain ALC on a medium containing acidified NO2− (see Fig. S4 in the supplemental material). The addition of ergosterol, an essential compound synthesized by the activity of hemoprotein, did not restore growth. These results indicated that the defective growth was not due to defective nitrate and sulfate assimilation or defective ergosterol synthesis and that hemC plays an important role in protecting A. nidulans against RNS stress.
We cultured the WT strain and strains PD1 and ALC in the presence of H2O2 (Fig. 1A, bottom) and found that H2O2 repressed the growth of ALC more than that of the WT. The WT strain and strain PD1 grew similarly on an H2O2-containing medium. These results indicated that normal levels of PBG-D expression as high as those in the WT are necessary and sufficient for fungal cells to resist oxidative stress caused by H2O2.
Human PBG-D is located both in the cytosol and in the nucleus, and its level is controlled by proteasome activity (11). We added the proteasome inhibitor MG-132 to A. nidulans cultures and found no difference in cellular PBG-D activity between MG-132-treated and control cells (data not shown). Most PBG-D activity was detected in the cytoplasmic fraction in A. nidulans cells; <1% of PBG-D activity was detected in the membrane, nuclear, and mitochondrial fractions (data not shown). This indicated that the mechanisms of PBG-D regulation differ between humans and the fungus.
Regulation of hemC.
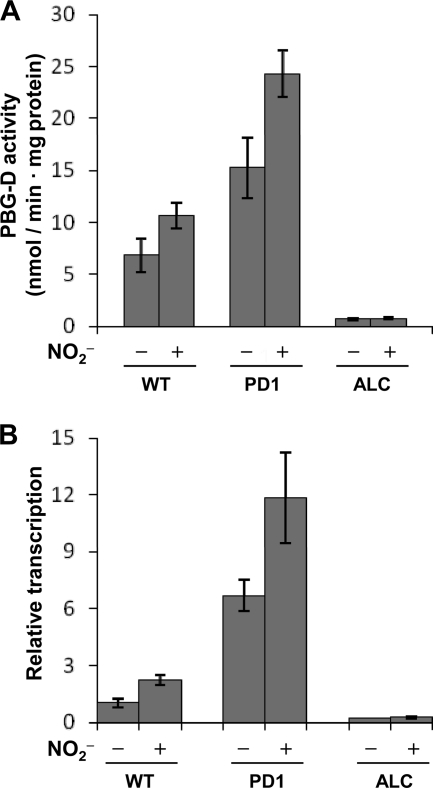
The WT strain produced 1.6-fold more PBG-D activity in cell extracts when cultured in the presence of 15 mM acidified NO2− than when cultured in its absence (Fig. 2A). Levels of the hemC transcripts measured by quantitative reverse transcription-PCR (RT-PCR) increased 2.3-fold upon the addition of acidified NO2− (Fig. 2B) and were comparable to the PBG-D activities, indicating that PBG-D was activated by RNS. We detected only a few hemC transcripts in strain ALC both in the presence and in the absence of acidified NO2− (Fig. 2B). Strain PD1 accumulated more hemC transcripts than the WT, and the addition of acidified NO2− increased hemC transcript levels (Fig. 2B). These transcript levels were comparable to the levels of PBG-D activity (Fig. 2A) and of growth (Fig. 1A) in the presence of acidified NO2−, confirming that PBG-D plays a role in RNS-resistant growth.
Fig 2.
Regulation of PBG-D in the WT strain and strains PD1 and ALC. Mycelia (1 g) were cultured in MM containing 10 mM proline, 10 mM methionine, and 10 μg ml−1 ergosterol with or without 15 mM NaNO2 (pH 5.5) for 12 h at 37°C. (A) PBG-D activities in cell extracts. (B) Relative ratios of hemC transcripts to actA transcripts (used for normalization). Data are means for three experiments, and error bars represent standard errors. P, <0.005.
PBG-D protects aconitase and respiration from RNS.
Aconitase is one of the most susceptible targets of RNS, because it has a particularly sensitive [4Fe-4S] cluster (8). When ALC was cultured with glucose, cellular aconitase activity was lowered by acidified NO2− to a greater extent than that in the WT strain or strain PD1 (Table 1). This indicated that PBG-D protected aconitase from RNS. The respiration mechanism is another RNS-susceptible target, and it contains a NO-sensitive terminal oxidase (3). When cells were cultured with glucose, respiration activity was more sensitive to acidified NO2− in the mitochondrial fractions of ALC than in those of the WT, whereas the inhibition was alleviated in PD1. The decreased respiratory activity of ALC in the absence of NO2− is most probably caused by the reduced cellular heme content (see below). These results indicated that PBG-D confers protection against damage to aconitase as well as against damage to cellular respiration caused by RNS.
Table 1.
Activities of aconitase and respiration in A. nidulans strains
| Strain | Activity (nmol min−1 mg−1)a |
|||
|---|---|---|---|---|
| Aconitase |
Respiration |
|||
| Without NO2− | With 15 mM NO2− | Without NO2− | With 15 mM NO2− | |
| WT | 170 ± 10 | 120 ± 10 | 80 ± 20 | 40 ± 10 |
| PD1 | 170 ± 10 | 170 ± 10 | 80 ± 20 | 60 ± 10 |
| ALC | 180 ± 20 | 90 ± 10 | 50 ± 10 | 20 ± 10 |
Mycelia were incubated for 12 h in 100 ml of MM (pH 5.5) with or without 15 mM NaNO2. Activities in cell extracts were measured. Values are means for three independent experiments ± standard errors.
Flavohemoglobins and nitrite reductase facilitate RNS damage tolerance.
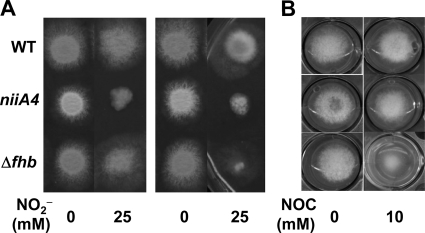
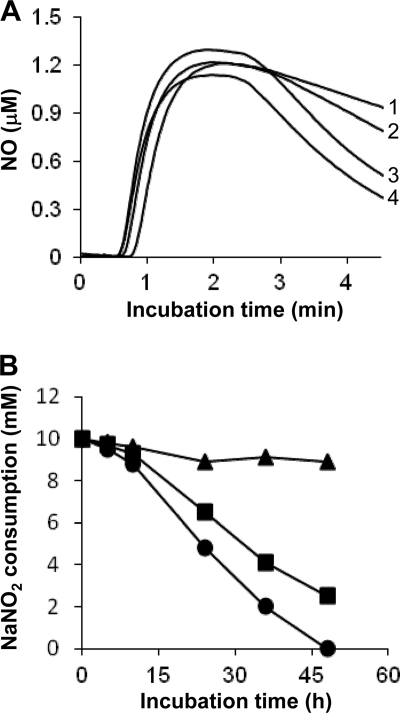
Aspergillus nidulans harbors two predicted paralogs that encode cytosolic FHbA and mitochondrial FHbB (32). We constructed an A. nidulans strain in which both paralogs were deleted (Δfhb) (see Fig. S3 in the supplemental material). The Δfhb and WT strains grew at similar rates on MM plates containing 25 mM acidified NO2− and 10 mM proline (nitrogen source) (Fig. 3A, left). The addition of 10 mM NOC-18 (an NO donor) to the MM plates retarded the growth of the Δfhb strain compared with that of the WT strain (Fig. 3B), indicating that fhbA and/or fhbB conferred protection against NO. A comparison of NO consumption by resting cells of these strains indicated that the Δfhb strain consumed NO at a lower rate than that of the WT (Fig. 4A). These results indicated that FHb detoxified NO and that the FHb-dependent system for NO tolerance is conserved among A. oryzae (40, 41) and other fungi (4, 21).
Fig 3.
Morphologies of colonies on MM plates. Conidia (1 × 105) of the WT strain, FGSC713 (niiA4), and the Δfhb mutant were incubated on MM plates containing 10 mM methionine, 10 μg ml−1 ergosterol, and the indicated nitrogen sources and NO donor (NOC-18) at 37°C for 48 h. (A) Effects of acidified NO2− (pH 5.5) on growth on MM plates containing 10 mM proline (left) or 5 mM tartrate ammonium (right) as the nitrogen source. (B) Effects of NOC-18 on growth on MM plates containing 10 mM proline as a nitrogen source.
Fig 4.
Consumption of NO and NO2− by A. nidulans strains. (A) Consumption of NO by resting cells. Aqueous NO was measured using an NO electrode in a control buffer without strains (line 1) or with WT (line 3), Δfhb (line 2), or FGSC713 (line 4) cells (5 mg each). The results are typical of those of triplicate experiments. (B) Nitrite consumption in MM (pH 5.5) containing 10 mM NaNO2, 10 mM proline, 10 mM methionine, and 10 μg ml−1 ergosterol at 37°C. Circles, WT; squares, Δfhb mutant; triangles, FGSC713. Data are means for three experiments, and standard errors are below 5%. P, <0.001.
The Δfhb strain grew more slowly than the WT on MM plates containing acidified NO2− when ammonium instead of proline served as the nitrogen source (Fig. 3A, right). This indicated that another gene(s) participates in acidified NO2− tolerance and that its expression is repressed by ammonium. One possible candidate is NiR, since it reduces NO2− (which we used as an NO donor) and is downregulated by ammonium (27). Strain FGSC713 (niiA4), lacking intact NiR, generated smaller colonies than the WT on MM plates containing acidified NO2− and proline, indicating that impairment of NiR caused hypersensitive growth in response to NO2− (Fig. 3A). Strain FGSC713 consumed NO2− far more slowly than the WT and Δfhb strains (Fig. 4B), whereas it consumed NO (Fig. 4A) and grew as fast as the WT in the presence of NOC-18 (Fig. 3B). These results indicated that NiR does not react with NO; rather, it consumes acidified NO2−.
PBG-D modulates the activities of FHb and NiR.
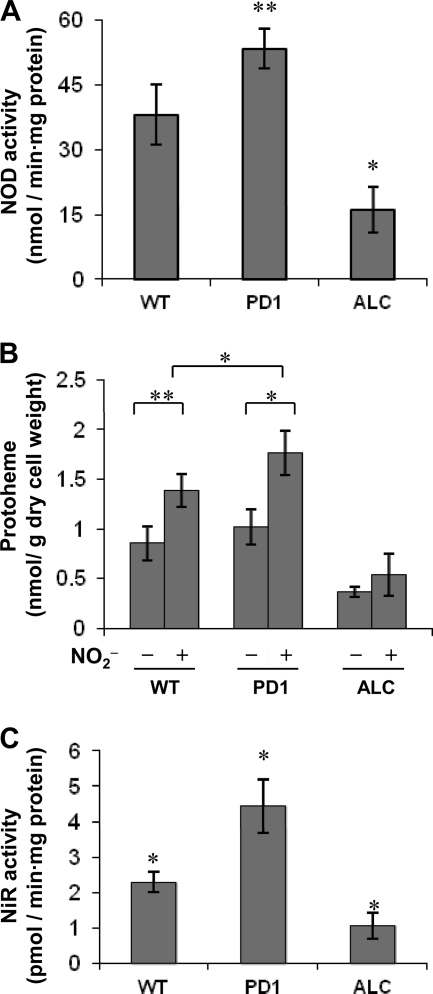
We measured the NOD activities of FHb in cell extracts of the WT strain and strains PD1 and ALC after culture with acidified NO2−. The results indicated that PD1 produced 1.4-fold more NOD activity than the WT (Fig. 5A). Strain ALC produced 0.5-fold less FHb activity than the WT (Fig. 5A), indicating that PBG-D positively regulates FHb activity. Quantitative RT-PCR analysis indicated that the amounts of fhbA and fhbB transcripts did not differ among the three strains (data not shown), suggesting that PBG-D upregulates FHb via a posttranscriptional mechanism. We measured levels of cellular protoheme, which is indispensable for the production of NOD activity by FHb, and found that PD1 and ALC cultured with acidified NO2− contained higher and lower levels of protoheme, respectively, than the WT cultured under the same conditions (Fig. 5B). This is comparable to the extent of growth tolerance of RNS (Fig. 1) and to the levels of PBG-D and NOD activities (Fig. 2A and 5A), suggesting that PBG-D supplies protoheme to FHb and protects cells from RNS. Notably, the WT accumulated 1.6-fold more protoheme after incubation with acidified NO2− than without it (Fig. 5B). This finding is consistent with the notion that acidified NO2− upregulates PBG-D (Fig. 2B), and it suggests that the protective mechanism is inducible. Notably, our findings suggest that RNS-dependent induction of the protective mechanism is independent of PBG-D activity: even though PBG-D activity is lower and is not inducible in strain ALC (Fig. 2A), protoheme synthesis still appears to be inducible by acidified NO2− (Fig. 5B).
Fig 5.
PBG-D modulates FHb and NiR activities. Mycelia of the WT strain and strains PD1 and ALC (4 g each) were transferred to 100 ml of MM containing 10 mM NaNO2, 10 mM proline, 10 mM methionine, and 10 μg ml−1 ergosterol (pH 5.5) and were incubated for 12 h at 37°C. (A) Activity of NOD in cell extracts; (B) protoheme levels in mycelia; (C) NiR activities in cell extracts. Data are means for three experiments, and error bars represent standard errors. ∗, P < 0.005; ∗∗, P < 0.01.
Siroheme is an essential cofactor of NiR that is synthesized via PBG-D (19). We could not quantify siroheme, probably because the amount was below the detection limit or because it is unstable in cell extracts. However, PD1 produced 2-fold more, and ALC produced 2-fold less, NiR activity than the WT (Fig. 5C), and the amounts of niiA transcripts did not differ among the three strains (data not shown), suggesting that PBG-D also upregulates NiR at the posttranscriptional level. This is consistent with the notion that PBG-D regulates NiR activity by regulating the siroheme supply.
DISCUSSION
This study demonstrates that A. nidulans hemC, encoding PBG-D, is a novel RNS defense gene that modulates the activities of FHb and NiR. To date, most studies on the RNS response, especially those on filamentous fungi, have focused on inducible adaptation mechanisms that are dispensable for normal growth. Examples are FHb and cytochrome P450nor, although the gene for the latter is absent in A. nidulans. Fungi synthesize and use heme for essential biological processes, such as respiration and sterol synthesis, like other organisms, and thus, heme synthesis has been regarded as a housekeeping mechanism. Our finding that hemC is essential for normal growth agrees with this notion. Our other finding, that upregulation of hemC confers RNS tolerance, implies that this housekeeping mechanism also responds to RNS and functions as a critical mechanism for fungal RNS tolerance.
The induction of hemC expression and PBG-D production (Fig. 2) accompanied the upregulation of protoheme synthesis in A. nidulans (Fig. 5B), indicating that the level of PBG-D limited heme synthesis under our experimental conditions. The synthesized protoheme serves as a cofactor for FHb and enables RNS tolerance, which implies that hemC responds to RNS and regulates protoheme synthesis for adaptation to nitrosative stress conditions. Reports have shown that RNS also induce the expression of genes for some fungal hemoproteins, including catalases and peroxidases, which facilitate tolerance of the oxidative stress caused by RNS (22, 25). The A. nidulans isozymes respond to and facilitate the tolerance of oxidative stress (13, 18), although their role in the RNS response remains obscure. However, they are likely to be produced in response to RNS, since RNS generate reactive oxygen species under physiological conditions (38). Thus, the upregulation of PBG-D in fungi exposed to RNS provides protoheme to induce the activity of these enzymes as well as FHb.
This study found that, as with other fungi, hemC is essential for the growth of A. nidulans cells under normal conditions. We also found that in contrast to nitrosative stress conditions, PBG-D overexpression had little effect on protoheme synthesis under normal conditions (Fig. 5B). This indicated that other enzymes in the heme biosynthetic pathway limit protoheme synthesis under normal conditions, in contrast to the rate-limiting role of PBG-D in A. nidulans exposed to RNS (this study) and in normally growing S. cerevisiae (12). Notably, we found more protoheme in A. nidulans exposed to RNS (Fig. 5B), suggesting that RNS upregulates not only PBG-D (Fig. 2) but also another enzyme(s) to facilitate sufficient protoheme production. One candidate for the latter is uroporphyrinogen III decarboxylase (Hem12p in S. cerevisiae), which might play a rate-limiting role second to that of PBG-D in S. cerevisiae, but this notion remains to be proven.
Although the mechanism of hemC induction by RNS remains obscure, we speculate that two transcription factors, NirA and AP-1, are involved in the mechanism. NirA is a specific factor inducing the transcription of the nitrate-utilizing genes niaD, niiA, and crnA. It binds to the 5′-CTCCGHGG-3′ consensus sequence in the promoters and activates the transcription of these genes in the presence of nitrate or NO2− (30). A sequence (5′-CTCCGCGT-3′) similar to the consensus sequence was identified in the hemC gene promoter, suggesting that NirA functions in RNS (acidified NO2−)-dependent induction. S. cerevisiae AP-1 (Yap1p) participates in the cellular response to oxidative stress and cytotoxic agents (23). No specific nucleotide sequence to which the A. nidulans AP-1 isozyme binds has been identified, although we found a sequence similar to the Yap1p recognition sequence (5′-TTACTAA-3′), which might regulate hemC expression by RNS, at the −367 position of the hemC gene promoter (5′-TTAATAA-3′). Direct evidence for the roles of these transcription factors in hemC regulation must await future investigation.
No functions for PBG-D in the RNS response have been reported on the basis of the transcriptomes of S. cerevisiae, C. albicans, and H. capsulatum (6, 14, 17, 25). This might be due to the relatively lower induction level of hemC transcription by RNS than of the genes characterized by the transcriptome analyses. This implies that phenotypic screening using a genomic library is advantageous for identifying novel genes involved in protection against RNS stress. Further screening should identify even more genes involved in fungal RNS tolerance, and an investigation into such genes should clarify the comprehensive mechanisms of such tolerance.
Supplementary Material
ACKNOWLEDGMENTS
We thank Norma Foster for critical reading of the manuscript.
This study was partly supported by the Bio-oriented Technology Research Advancement Institution and by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Footnotes
Published ahead of print 28 October 2011
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Bailey C, Arst HN., Jr 1975. Carbon catabolite repression in Aspergillus nidulans. Eur. J. Biochem. 51:573–577 [DOI] [PubMed] [Google Scholar]
- 2. Battersby AR, Fookes CJ, Matcham GW, McDonald E. 1980. Biosynthesis of the pigments of life: formation of the macrocycle. Nature 285:17–21 [DOI] [PubMed] [Google Scholar]
- 3. Brunori M, Giuffre A, Sarti P, Stubauer G, Wilson MT. 1999. Nitric oxide and cellular respiration. Cell. Mol. Life Sci. 56:549–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. de Jesus-Berrios M, et al. 2003. Enzymes that counteract nitrosative stress promote fungal virulence. Curr. Biol. 13:1963–1968 [DOI] [PubMed] [Google Scholar]
- 5. Fang FC. 1997. Perspectives series: host/pathogen interactions. Mechanisms of nitric oxide-related antimicrobial activity. J. Clin. Invest. 99:2818–2825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Flatley J, et al. 2005. Transcriptional responses of Escherichia coli to S-nitrosoglutathione under defined chemostat conditions reveal major changes in methionine biosynthesis. J. Biol. Chem. 280:10065–10072 [DOI] [PubMed] [Google Scholar]
- 7. Gardner AM, Helmick RA, Gardner PR. 2002. Flavorubredoxin, an inducible catalyst for nitric oxide reduction and detoxification in Escherichia coli. J. Biol. Chem. 277:8172–8177 [DOI] [PubMed] [Google Scholar]
- 8. Gardner PR, Costantino G, Szabo C, Salzman AL. 1997. Nitric oxide sensitivity of the aconitases. J. Biol. Chem. 272:25071–25076 [DOI] [PubMed] [Google Scholar]
- 9. Gardner PR, Gardner AM, Martin LA, Salzman AL. 1998. Nitric oxide dioxygenase: an enzymic function for flavohemoglobin. Proc. Natl. Acad. Sci. U. S. A. 95:10378–10383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Goretski J, Zafiriou OC, Hollocher TC. 1990. Steady-state nitric oxide concentrations during denitrification. J. Biol. Chem. 265:11535–11538 [PubMed] [Google Scholar]
- 11. Grunberg-Etkovitz N, Greenbaum L, Grinblat B, Malik Z. 2006. Proteasomal degradation regulates expression of porphobilinogen deaminase (PBGD) mutants of acute intermittent porphyria. Biochim. Biophys. Acta 1762:819–827 [DOI] [PubMed] [Google Scholar]
- 12. Hoffman M, Gora M, Rytka J. 2003. Identification of rate-limiting steps in yeast heme biosynthesis. Biochem. Biophys. Res. Commun. 310:1247–1253 [DOI] [PubMed] [Google Scholar]
- 13. Holdom MD, Hay RJ, Hamilton AJ. 1996. The Cu, Zn superoxide dismutases of Aspergillus flavus, Aspergillus niger, Aspergillus nidulans, and Aspergillus terreus: purification and biochemical comparison with the Aspergillus fumigatus Cu,Zn superoxide dismutase. Infect. Immun. 64:3326–3332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hromatka BS, Noble SM, Johnson AD. 2005. Transcriptional response of Candida albicans to nitric oxide and the role of the YHB1 gene in nitrosative stress and virulence. Mol. Biol. Cell 16:4814–4826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ickowicz Schwartz D, et al. 2004. Differentiation-dependent photodynamic therapy regulated by porphobilinogen deaminase in B16 melanoma. Br. J. Cancer 90:1833–1841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ilari A, Boffi A. 2008. Structural studies on flavohemoglobins. Methods Enzymol. 436:187–202 [DOI] [PubMed] [Google Scholar]
- 17. Justino MC, Vicente JB, Teixeira M, Saraiva LM. 2005. New genes implicated in the protection of anaerobically grown Escherichia coli against nitric oxide. J. Biol. Chem. 280:2636–2643 [DOI] [PubMed] [Google Scholar]
- 18. Kawasaki L, Wysong D, Diamond R, Aguirre J. 1997. Two divergent catalase genes are differentially regulated during Aspergillus nidulans development and oxidative stress. J. Bacteriol. 179:3284–3292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Leustek T, et al. 1997. Siroheme biosynthesis in higher plants. Analysis of an S-adenosyl-l-methionine-dependent uroporphyrinogen III methyltransferase from Arabidopsis thaliana. J. Biol. Chem. 272:2744–2752 [DOI] [PubMed] [Google Scholar]
- 20. Liu L, et al. 2001. A metabolic enzyme for S-nitrosothiol conserved from bacteria to humans. Nature 410:490–494 [DOI] [PubMed] [Google Scholar]
- 21. Liu L, Zeng M, Hausladen A, Heitman J, Stamler JS. 2000. Protection from nitrosative stress by yeast flavohemoglobin. Proc. Natl. Acad. Sci. U. S. A. 97:4672–4676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lushchak OV, Inoue Y, Lushchak VI. 2010. Regulatory protein Yap1 is involved in response of yeast Saccharomyces cerevisiae to nitrosative stress. Biochemistry (Mosc.) 75:629–664 [DOI] [PubMed] [Google Scholar]
- 23. Lushchak VI. 2011. Adaptive response to oxidative stress: bacteria, fungi, plants and animals. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 153:175–190 [DOI] [PubMed] [Google Scholar]
- 24. Marletta MA. 1993. Nitric oxide synthase: function and mechanism. Adv. Exp. Med. Biol. 338:281–284 [PubMed] [Google Scholar]
- 25. Nittler MP, Hocking-Murray D, Foo CK, Sil A. 2005. Identification of Histoplasma capsulatum transcripts induced in response to reactive nitrogen species. Mol. Biol. Cell 16:4792–4813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Osherov N, Mathew J, May GS. 2000. Polarity-defective mutants of Aspergillus nidulans. Fungal Genet. Biol. 31:181–188 [DOI] [PubMed] [Google Scholar]
- 27. Pateman JA, Rever BM, Cove DJ. 1967. Genetic and biochemical studies of nitrate reduction in Aspergillus nidulans. Biochem. J. 104:103–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Poock SR, Leach ER, Moir JW, Cole JA, Richardson DJ. 2002. Respiratory detoxification of nitric oxide by the cytochrome c nitrite reductase of Escherichia coli. J. Biol. Chem. 277:23664–23669 [DOI] [PubMed] [Google Scholar]
- 29. Poole RK, Hughes MN. 2000. New functions for the ancient globin family: bacterial responses to nitric oxide and nitrosative stress. Mol. Microbiol. 36:775–783 [DOI] [PubMed] [Google Scholar]
- 30. Punt PJ, et al. 1995. The intergenic region between the divergently transcribed niiA and niaD genes of Aspergillus nidulans contains multiple NirA binding sites which act bidirectionally. Mol. Cell. Biol. 15:5688–5699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Radi R, Beckman JS, Bush KM, Freeman BA. 1991. Peroxynitrite-induced membrane lipid peroxidation: the cytotoxic potential of superoxide and nitric oxide. Arch. Biochem. Biophys. 288:481–487 [DOI] [PubMed] [Google Scholar]
- 32. Schinko T, et al. 2010. Transcriptome analysis of nitrate assimilation in Aspergillus nidulans reveals connections to nitric oxide metabolism. Mol. Microbiol. 78:720–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shimizu M, Fujii T, Masuo S, Fujita K, Takaya N. 2009. Proteomic analysis of Aspergillus nidulans cultured under hypoxic conditions. Proteomics 9:7–19 [DOI] [PubMed] [Google Scholar]
- 34. Shoun H, Tanimoto T. 1991. Denitrification by the fungus Fusarium oxysporum and involvement of cytochrome P-450 in the respiratory nitrite reduction. J. Biol. Chem. 266:11078–11082 [PubMed] [Google Scholar]
- 35. Stach P, Einsle O, Schumacher W, Kurun E, Kroneck PM. 2000. Bacterial cytochrome c nitrite reductase: new structural and functional aspects. J. Inorg. Biochem. 79:381–385 [DOI] [PubMed] [Google Scholar]
- 36. Takasaki K, et al. 2004. Fungal ammonia fermentation, a novel metabolic mechanism that couples the dissimilatory and assimilatory pathways of both nitrate and ethanol. Role of acetyl CoA synthetase in anaerobic ATP synthesis. J. Biol. Chem. 279:12414–12420 [DOI] [PubMed] [Google Scholar]
- 37. Ullmann BD, et al. 2004. Inducible defense mechanism against nitric oxide in Candida albicans. Eukaryot. Cell 3:715–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wei T, Chen C, Hou J, Xin W, Mori A. 2000. Nitric oxide induces oxidative stress and apoptosis in neuronal cells. Biochim. Biophys. Acta 1498:72–79 [DOI] [PubMed] [Google Scholar]
- 39. Wink DA, et al. 1991. DNA deaminating ability and genotoxicity of nitric oxide and its progenitors. Science 254:1001–1003 [DOI] [PubMed] [Google Scholar]
- 40. Zhou S, et al. 2011. Functional analysis and subcellular location of two flavohemoglobins from Aspergillus oryzae. Fungal Genet. Biol. 48:200–207 [DOI] [PubMed] [Google Scholar]
- 41. Zhou S, et al. 2009. Cloning and characterization of two flavohemoglobins from Aspergillus oryzae. Biochem. Biophys. Res. Commun. 381:7–11 [DOI] [PubMed] [Google Scholar]
- 42. Zumft WG. 1997. Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 61:533–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zweier JL, Samouilov A, Kuppusamy P. 1999. Non-enzymatic nitric oxide synthesis in biological systems. Biochim. Biophys. Acta 1411:250–262 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.