Abstract
The agent of Lyme disease, Borrelia burgdorferi, has a number of outer membrane proteins that are differentially regulated during its life cycle. In addition to their physiological functions in the organism, these proteins also likely serve different functions in invasiveness and immune evasion. In borreliae, as well as in other bacteria, a number of membrane proteins have been implicated in binding plasminogen. The activation and transformation of plasminogen into its proteolytically active form, plasmin, enhances the ability of the bacteria to disseminate in the host. Outer membrane vesicles of B. burgdorferi contain enolase, a glycolytic-cycle enzyme that catalyzes 2-phosphoglycerate to form phosphoenolpyruvate, which is also a known plasminogen receptor in Gram-positive bacteria. The enolase was cloned, expressed, purified, and used to generate rabbit antienolase serum. The enolase binds plasminogen in a lysine-dependent manner but not through ionic interactions. Although it is present in the outer membrane, microscopy and proteinase K treatment showed that enolase does not appear to be exposed on the surface. However, enolase in the outer membrane vesicles is accessible to proteolytic degradation by proteinase K. Samples from experimentally and tick-infected mice and rabbits as well as from Lyme disease patients exhibit recognition of enolase in serologic assays. Thus, this immunogenic plasminogen receptor released in outer membrane vesicles could be responsible for external proteolysis in the pericellular environment and have roles in nutrition and in enhancing dissemination.
INTRODUCTION
Borrelia burgdorferi, the agent of Lyme disease, invades distant tissues from its site of entry in the skin (4, 14). Dissemination of the spirochetes is assisted by borrowed proteolytic activity that is incorporated into the outer membrane (OM) via specific receptors (17). Plasmin, a serine protease, is bound to the organism as plasminogen (PLG), a proenzyme present in body fluids that is activated by urokinase, a PLG activator. Once bound, plasmin can assist borreliae in degrading extracellular matrices and basement membranes, with the ultimate result of facilitating dissemination (21–23, 44, 59).
Borreliae have a close relationship with the host's PLG activation system. As mentioned above, these spirochetes can fix plasmin onto their OMs, but they can also stimulate cells of the innate immune response to produce PLG activators (19, 20, 32, 42, 46) and modulate PLG activation inhibitors (39). In a similar manner, borreliae are important stimulators for the production of matrix metalloproteinases as part of their proinflammatory repertoire (34, 35, 43). B. burgdorferi has an A-T rich genome with a corresponding abundance of lysines (31), which are the most common amino acids in PLG receptors. Thus, it is no surprise that B. burgdorferi has a number of molecules that can bind PLG, although not all molecules that bind PLG are biologically relevant. For example, lysine-rich OspA is a PLG receptor (33), but, other than in the initial stages of tick feeding, this interaction is not likely to be important in the dissemination within the mammalian host, as expression of OspA is downregulated (68). In contrast, OspC, a lipoprotein expressed after ticks begin to feed and in the early stages of infection of the mammal, is also a PLG receptor and one more likely to have biological relevance (28, 38, 48). Other known PLG receptors of B. burgdorferi include the Erp lipoproteins (12).
Relapsing fever borreliae (RFB) also bind PLG and use plasmin for dissemination as well (36, 59). Recent investigations have shown that both B. burgdorferi (10, 40) and RFB (41, 65, 67) have molecules that function as receptors for multiple ligands. Complement regulator-acquiring surface proteins of B. hermsii and B. burgdorferi (CspA) can bind extracellular matrices, factor H, and PLG (40), although multiple binding occurs in some instances via distinct nonoverlapping domains (65). Factor H binding protein A of an RFB also binds factor H and PLG (41). It is remarkable that these molecules have the dual purposes of protection against complement and fixing of an active plasmin onto the surface for dissemination in borreliae.
PLG is a single-chain glycoprotein that is inactive until cleaved by PLG activators to form plasmin. The active enzyme consists of five kringle domains, each with three disulfide bonds that contain the lysine binding sites and the catalytic domain. PLG binding is an important part of the pathogenesis of infections by Gram-positive bacteria, notably Staphylococcus and Streptococcus species. Of the several PLG receptors present in these bacteria, enzymes of the glycolytic system expressed on the surface have been studied in detail. Two glycolytic enzymes from Gram-positive bacteria have been implicated in the binding of PLG (5, 6). GAPDH (glyceraldehyde-3-phosphate dehydrogenase) and enolase (phosphopyruvate dehydratase) are expressed in the cytosol of bacterial cells, where they perform their traditional enzymatic functions in glycolysis (53, 54, 57, 63). In the case of enolase, this function is to catalyze 2-phosphoglycerate to phosphoenolpyruvate. However, there is evidence of the presence of enolase on the surface of Gram-positive (5, 62) and Gram-negative (69) bacteria, fungi, and protozoa (3, 58). The surface location of enolase in several types of prokaryotic and eukaryotic cells is intriguing, since this enzyme does not have known cell surface protein motifs such as a signal peptidase cleavage site, cell wall anchors or sequences, or membrane-spanning domains (61). Nonetheless, the α-enolase of Streptococcus pneumoniae, S. pyogenes, and group A and oral streptococci binds PLG through both terminal and internal lysine residues (7, 24, 63). The molecular recognition of PLG by the internal and terminal lysines is well conserved among the bacteria as well as protozoa (Trichomonas) and Candida. The internal and terminal lysine residues of the enolase of B. burgdorferi are also conserved, suggesting that this enzyme could be an important PLG receptor in this organism. Furthermore, the enolases of other bacteria are immunogenic, suggesting that this could also be true for B. burgdorferi.
Outer membrane vesicles (OMV) are released naturally by Gram-negative bacteria. Bulges of OM evaginate with periplasmic components (9, 26). OMV represent a considerable portion of the bacterial cell, and recent proteomic studies have now shown a significant representation of cytosolic and inner membrane molecules (51). OMV have been considered a part of the stress response (56), and their functions and roles in infection have been recently reviewed (30). Among the most important functions are the release of toxins and virulence factors, interaction with other bacteria and host cells, and modulation of the host response (30). The Borrelia OMV have been studied both in cultured organisms and in vivo. Borrelia OMV induce B cell responses under experimental conditions (74) and can bind to the endothelium (70). The OMs of B. burgdorferi have been characterized with respect to their lipoprotein and glycolipid contents (64) and the presence of both lipoproteins and nonlipidated molecules (71). Extensive shedding of OMV from green fluorescent protein (GFP)-labeled spirochetes during blood feeding in ticks was noted. An in vivo study showed that, depending of the conditions to which the organisms are exposed in feeding ticks, release of OMV can be induced and can also be decreased (27). Cryoelectron tomography studies of B. burgdorferi have shown that OMV are released near sites of cell division (47) and as a result of the action of bactericidal antibodies (49, 50). In this study, we document the presence of enolase in OMV and demonstrate that this glycolytic enzyme binds PLG in a lysine-dependent manner, is immunogenic, and does not appear to be exposed on the surface of the intact organism. The role of enolase in the OMV could be that of fixing plasmin in the peribacterial environment.
MATERIALS AND METHODS
Bacteria, cultures, and sera from laboratory animals and Lyme disease patients.
B. burgdorferi strain B31 was grown in complete BSK-II medium at 33°C. A New Zealand White rabbit (Charles River, Wilmington, MA) was intradermally inoculated with 50 μg of recombinant enolase in complete Freund's adjuvant and periodically given a booster inoculation of the recombinant protein in incomplete Freund's adjuvant to develop a measurable antibody response. Sera from C3H/HeN mice (Jackson Laboratories, Bar Harbor, ME) infected intradermally with 2 × 104 spirochetes were collected at various intervals after inoculation. Serum from a rabbit infected with B. burgdorferi were collected following the attachment of infected Ixodes scapularis females. Sera from patients with disseminated Lyme disease (with joint or nervous system involvement) and with previous reactive serology results were randomly selected from our serum bank for determination of their reactivity to recombinant enolase.
Vesicle isolation purification and mass spectrometry.
B. burgdorferi bacteria were harvested by centrifugation for 12 min at 4,000 × g and incubated at 37°C in fresh complete BSK-II media for 2 h followed by centrifugation. The supernatants were collected and filtered twice using 0.22-μm-pore-size Steriflip filters (Millipore, Billerica, MA). OMV in the supernatants were pelleted by ultracentrifugation for 1 h at 100,000 × g and resuspended in 40% OptiPrep (Axis Shield, Norton, MA). A discontinuous gradient was made following the manufacturer's instructions and centrifuged for 16 h at 100,000 × g. The OMV floated to the interface between the 20% layer and the 25% layer, where a white band was visualized. The OptiPrep gradient fractions were collected in 1-ml aliquots. The purified (based on similar protein contents) OMV were pooled from the 15%, 20%, and 25% fractions. These fractions were removed from the OptiPrep solution by resuspension in 50 ml of 20 mM HEPES (pH 7.5) and were centrifuged for 1 h at 100,000 × g. The pelleted, purified OMV were resuspended in 20 mM HEPES (pH 7.5). The OMV were analyzed for protein content using a combination of two-dimensional liquid chromatography and tandem mass spectrometry (MS/MS) or multidimensional protein identification technology (MudPIT).
Recombinant protein.
N-terminal polyhistidine-tagged enolase was generated in pET-28a(+) vector (EMD Chemicals Inc., Gibbstown, NJ) by amplification of the enolase gene (bb0337 [31]) by the use of primers EnolNterF (5′-CATATGGGTTTTCACATTTATGA-3′) and EnolNterR (5′-CTCGAGAATTTTTTGTTTAATAGAATA-3′) (The Midland Certified Reagent Company, Midland, TX) followed by digestion with restriction enzymes NcoI (New England BioLabs, Ipswich, MA) and XhoI (New England BioLabs). The resultant plasmid's insert was sequenced on both strands to ensure that no mutation had occurred during the PCR or cloning procedures.
The recombinant enolase was expressed in Escherichia coli Rossetta (DE3) (Novagen, Madison, WI) upon induction with IPTG (isopropyl-β-d-thiogalactopyranoside). The culture was subjected to sonication and the debris pelleted by centrifugation. The recombinant protein, in the soluble fraction, was purified following a His-Trap procedure. Protein purification was assessed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by Coomassie brilliant blue staining. Protein concentrations were measured by a bicinchoninic acid protein assay. (Thermo Fisher Scientific, Rockford, IL.)
Western blot assays.
A total of 3 μg of the recombinant enolase and 108 proteinase K (PK)-treated B. burgdorferi bacteria as well as B. burgdorferi grown at 34°C (pH 6.4 or pH 7.6) and at 23°C (pH 7.6) or the OMV fraction isolated using a discontinuous gradient was resuspended in 20 μl of phosphate-buffered saline (PBS) and received 10 μl of 3× SDS-PAGE sample buffer with 2-mercaptoethanol followed by boiling for 5 min. The components of each sample were separated by electrophoresis using 12.5% SDS-PAGE, and the proteins were transferred to nitrocellulose membranes (GE Healthcare, Piscataway, NJ). The antibodies used in the different experiments were rabbit antienolase serum, murine monoclonal anti-DnaK (mouse IgG1) antibody (16), murine monoclonal anti-OspA (mouse IgG1) antibody (16), murine monoclonal antiflagellin (mouse IgG1) antibody (18), and rabbit anti-OspC serum (28). The Western blots were resolved with infrared-labeled goat anti-rabbit IgG IRDye700CW and goat anti-mouse IgG IRDye800CW (Rockland Immunochemicals, Gilbertsville, PA) and visualized by scanning with an Odyssey infrared imaging system (LiCor Biosciences, Lincoln, NE).
ELISA.
Microtest 96-well enzyme-linked immunoassay (ELISA) plates (BD Biosciences, Bedford, MA) were coated overnight at 4°C with human PLG (Sigma-Aldrich, St. Louis, MO) (10 μg/ml), recombinant enolase (10 μg/ml), Borrelia whole lysate (10 μg/ml), or bovine serum albumin (BSA) (10 μg/ml) in coating buffer (50 mM NaCO3, 50 mM NaHCO3). After three PBS washes, the ELISA plates were blocked with 1% BSA for 1 h at 37°C and washed again. Recombinant enolase, PLG (Sigma), rabbit sera (1:100 in PBS), mouse sera (1:100 in PBS), or human sera (1:100 in PBS) were added and incubated for 1 h at 37°C and washed with PBS. Several ELISA experiments were carried out as follows. (i) For detection of enolase bound to PLG, rabbit antienolase serum was incubated for 1 h and washed three times with PBS followed by incubation with a goat anti-rabbit IgG-alkaline phosphatase conjugate (Sigma) at 37°C for 1 h. (ii) For detection of the PLG bound to enolase, rabbit anti-human PLG (Boehringer, Rheim, Germany) was incubated for 1 h and washed three times with PBS followed by incubation with a goat anti-rabbit IgG-alkaline phosphatase conjugate (Sigma) at 37°C for 1 h. (iii) For detection of rabbit, mouse, or human antibodies to the recombinant enolase or to the whole B. burgdorferi cell lysate, goat anti-rabbit IgG-alkaline phosphatase (Sigma), goat anti-mouse IgG-alkaline phosphatase (Sigma), and goat anti-human IgG-alkaline phosphatase (KPL, Gaithersburg, MA) were each incubated for 1 h at 37°C. Finally, the wells were washed three times with PBS before the alkaline phosphatase substrate (Sigma) was added. Plates were incubated at 37°C, and optical densities (OD) were read using a SpectraMax M2 ELISA plate reader (Molecular Devices, Sunnyvale, CA).
For experiments that evaluated the role of ionic interactions in enolase binding to PLG, increasing concentrations (0 to 400 mM) of NaCl (Sigma) were added to the plates together with the PLG. For assays analyzing the role of heparin binding domains in the enolase-PLG interaction, increasing concentrations (0 to 500 IU) of heparin (Sigma) were used. To determine the role of lysines in the enolase-PLG interaction, the lysine analog ε-aminocaproic acid (Sigma) (1 mM) was added with PLG to the enolase-coated plates.
Borrelia immunofluorescence assay.
B. burgdorferi bacteria were harvested from the mid-log-phase culture, washed with cold PBS (Invitrogen, Carlsbad, CA), and incubated in Hank's balanced salt solution (HBSS) (Invitrogen) with rabbit antienolase serum (1/100) for 1 h at 33°C to determine whether the enolase binds the surface of intact borreliae. After being washed in cold PBS (Invitrogen), the spirochetes were added to the wells of a Teflon-coated indirect immunofluorescence assay (IFA) slide (Erie Scientific, Portsmouth, NH), dried at 33°C, and fixed in 100% methanol. A 1:1,000 solution of goat anti-rabbit IgG conjugated to fluorescein isothiocyanate (FITC) (Abcam, Cambridge, MA) was added to each well followed by incubation for 1 h in a wet chamber. Slides were washed with PBS, dried before the slow-fade mounting medium (Invitrogen) was placed on the slides, and viewed using a Nikon Eclipse E400 microscope.
Immunogold detection of enolase by transmission electron microscopy (TEM).
B. burgdorferi was incubated with rabbit antienolase serum in HBSS for 1 h at 33°C. The spirochetes were fixed to polyvinyl formal-coated grids (Ted Pella, Redding, CA) in 1% glutaraldehyde (Sigma)–PBS and blocked for 30 min at room temperature in 1% BSA (Sigma). Grids were then placed in a 1% BSA suspension containing goat anti-rabbit IgG gold-labeled antibodies (Jackson Immunochemicals, West Grove, PA) at a dilution of 1:250 for 1 h at room temperature. Subsequently, grids were prepared for analysis by negative-stain TEM as previously described (50). For some experiments, spirochetes derived from dialysis chambers within the peritoneal cavity of rats (1) were used for detection of enolase. These spirochetes were the generous gift of Melissa Caimano and Dustin Radolf, University of Connecticut, and were fixed to the grids in the manner described above for the cultured organism.
PK treatments.
Proteinase K (PK) treatment of whole B. burgdorferi bacteria was done in a manner similar to methods published previously (13, 25, 29). B. burgdorferi cells were harvested from complete BSK-II medium and washed with HBSS at 4°C. After the last wash, the spirochetes were resuspended in HBSS, divided into equal aliquots of approximately 108 cells, centrifuged, and then resuspended in 1 ml (each) of Dulbecco's PBS (Invitrogen) containing 5 mM MgCl2 (PBS-Mg) alone or PBS-Mg with PK (Boehringer) at a concentration of 50 or 250 μg/ml. This was followed by incubation with gentle agitation for 1 h at room temperature (23°C). Proteolysis was stopped by centrifugation followed by two washes in HBSS containing a protease inhibitor cocktail (EDTA-free; Roche Diagnostics, Indianapolis, IN). Pellets were resuspended in 50 μl of PBS-Mg, and each received 12.5 μl of 5× SDS-PAGE sample buffer with 2-mercaptoethanol and was then boiled for 5 min. The entire content of each sample was subjected to 12.5% SDS-PAGE, and the protein was transferred to nitrocellulose. Enolase was detected using hyperimmune rabbit antiserum prepared against the recombinant protein. Outer surface protein A (OspA) and periplasmic flagellar protein FlaB were detected by use of monoclonal antibodies CB10 and CB1, respectively (18). Secondary antibodies were infrared-labeled goat anti-rabbit IgG IRDye700CW and goat anti-mouse IgG IRDye800CW (Rockland). Bands were visualized by scanning with an Odyssey infrared imaging system (LiCor Biosciences, Lincoln, NE).
For digestion of recombinant enolase, 4 μg of purified protein was incubated for 1 h at room temperature (23°C) with or without PK (250 μg/ml) in 40 μl of PBS-Mg. Proteolysis was stopped by addition of 10 μl of 5× SDS-PAGE sample buffer and boiling for 5 min. Following 12.5% SDS-PAGE of the entire volume for each sample, bands were visualized by Coomassie brilliant blue staining.
For proteolytic digestion of OMV with PK, 10 μg of purified OMVs was treated with a range of PK concentrations for 1 h at (23°C) and developed as described above for intact spirochetes and recombinant enolase. Controls included the use of an antibody to OspA and rabbit anti-HtrA serum.
RNA extraction and quantitative real-time PCR.
B. burgdorferi cultures were grown to a density of 108 spirochetes/ml under two different sets of conditions (33°C at pH 6.4 and 23°C at pH 7.6), and RNA was extracted using TRI reagent (MRC Inc., Cincinnati, OH) following the manufacturer's instructions. The RNA was treated once at 37°C for 60 min with DNase I (Roche) to remove any contaminating DNA. A final purification step using an RNeasy Midi kit (Qiagen, Valencia, CA) was carried out to eliminate DNase I as well as any trace of DNA contamination. Total RNA was quantified using an ND-1000 spectrophotometer (NanoDrop Products, Wilmington, DE). The RNA samples devoid of contaminating DNA were reverse transcribed to cDNA with TaqMan reverse transcription reagents (Applied Biosystems, Carlsbad, CA). Real-time PCRs were set up using a reaction mixture (Applied Biosystems) and primers specific to flaB (bb0147/342RT [5′-AGAGCAACTTACAGACGAAAT-3′] and bb0147/472RT [5′-AGTGATGCTGGTGTGTTAAT-3]) and enolase (bb0337; bb0337/680RT [5′-TAAAGAAGGCAGGATATGAAC-3′] and bb0337/814RT [5′-GCCCAATATTCAACCAT-3′]) at a final concentration of 2 μM and were used in an experimental procedure with a 7500 Fast real-time PCR system (Applied Biosystems). Preincubation cycle parameters were as follows: 1 cycle at 95°C for 5 min followed by 40 cycles of 95°C for 15 s, 55°C for 6 s, and 72°C for 6 s.
Statistics.
Data were analyzed using Welch's test with the GraphPad InStat 3.10 statistical program (GraphPad Software, Inc., San Diego, CA).
RESULTS
Enolase and other PLG receptors are present in OMV.
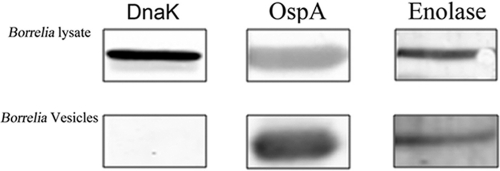
The protein content of the OMV of B. burgdorferi was analyzed by mass spectrometry in two separate experiments (Table 1). The two major proteins detected were OspB and OspA, which accounted for 53.6% and 12.5% of the total OMV spectra, respectively. Interestingly, the third most abundant protein detected was GroEL (bb0560), a cytosolic heat shock protein homolog that has also been associated with binding of membrane proteins in Borrelia (60) and other bacteria (11, 37). Two proteins from the glycolytic cycle, enolase (bb0337) and glyceraldehyde-3-phosphate dehydrogenase (bb0057), were also found (2). Both of these enzymes are known PLG receptors in several bacterial species (5, 6, 69). The OMV mass spectrometry results were confirmed by Western blot analysis (Fig. 1), where reactivity of an OM protein, OspA, was detected but no reactivity to DnaK, a known cytosolic molecule, was detected. CspA, also a PLG receptor (40), was also present in the OMV. It is of interest that the OMV have multiple molecules that can bind PLG in a known lysine-dependent manner.
Table 1.
Proteins identified in outer membrane vesicles in two independent experiments (A and B) using mass spectrometry in order of abundance
| Putative identification | Gene | MM (kDa)a | % in expt: |
|
|---|---|---|---|---|
| A | B | |||
| Outer surface protein B (OspB) | BBA16 | 32 | 53.611 | 43.710 |
| Outer surface protein A (OspA) | BBA15 | 29 | 12.558 | 10.801 |
| 60-kDa chaperonin (GroL) | BB0649 | 59 | 4.171 | 3.425 |
| Periplasmic serine protease DO (HtrA) | BB0104 | 53 | 2.064 | 2.042 |
| Enolase | BB0337 | 47 | 1.449 | 1.295 |
| Carboxyl-terminal protease (Ctp) | BB0359 | 54 | 1.361 | 1.339 |
| Surface lipoprotein P27 (p27) | BBA60 | 32 | 1.207 | 1.164 |
| BbCRASP-1 protein | BBA68 | 29 | 1.164 | 1.054 |
| Putative uncharacterized protein | BBB08 | 25 | 1.054 | 1.054 |
| Basic membrane protein A (BmpA) | BB0383 | 37 | 1.076 | 1.032 |
| Glyceraldehyde-3-phosphate dehydrogenase | BB0057 | 36 | 0.944 | 1.054 |
| Outer membrane protein P13 (p13) | BB0034 | 19 | 1.054 | 0.944 |
| Outer surface 22-kDa lipoprotein | BB0365 | 22 | 0.966 | 0.944 |
| Oligopeptide ABC transporter (OppA-3) | BB0330 | 62 | 0.703 | 0.922 |
| P66 protein (p66) | BB0603 | 68 | 0.681 | 0.900 |
MM, molecular mass.
Fig 1.
Detection of enolase in an outer membrane vesicle preparation by Western blot analysis. Rabbit antienolase serum (1/100) was used to detect the presence of the protein in a purified outer membrane vesicle preparation. Monoclonal antibodies to OspA and DnaK were used as controls.
The generation of His tag enolase was performed as described in Materials and Methods, and the different pools obtained were analyzed by SDS-PAGE to confirm the presence and purity of the protein before they were used to immunize a rabbit. Briefly, the enolase gene (bb0337) was amplified and the amplicon, as well as the plasmid, purified and treated with restriction enzymes. Both products were purified again to eliminate the small cut fragment and ligated together to generate the plasmid with the enolase gene insertion. Plasmid DNAs from different clones were tested by PCR with primers that target both the plasmid and the insertion to confirm the presence of the enolase gene. One was sequenced and used for the protein purification.
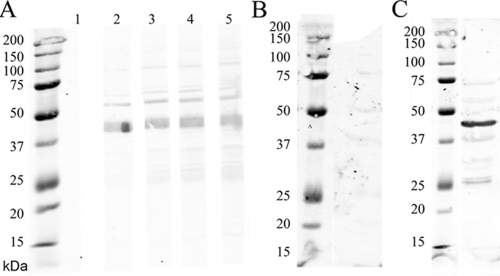
The preimmunization serum of a rabbit used to develop antibodies showed no reactivity to the recombinant enolase (Fig. 2A, lane 1) and developed antibodies after immunization (Fig. 2A, lanes 2 to 5). Likewise, preimmunization rabbit serum exhibited no reactivity to whole Borrelia cell lysate (Fig. 2B) but recognized native enolase after immunization (Fig. 2C).
Fig 2.
Rabbit antienolase serum recognizes both recombinant and native enolase. (A) Immunoblot showing recognition of the recombinant enolase (3 μg/lane). Lane 1, preimmunization rabbit serum does not recognize recombinant enolase; lanes 2 to 5, results for rabbit antienolase serum obtained at intervals in the immunization period. (B) Immunoblot. Preimmunization rabbit serum (1/100) does not react to whole Borrelia lysate. (C) Rabbit antienolase serum recognized native enolase in B. burgdorferi whole lysate. Molecular mass markers are shown at the left of each panel.
B. burgdorferi enolase binds PLG in a dose-dependent manner.
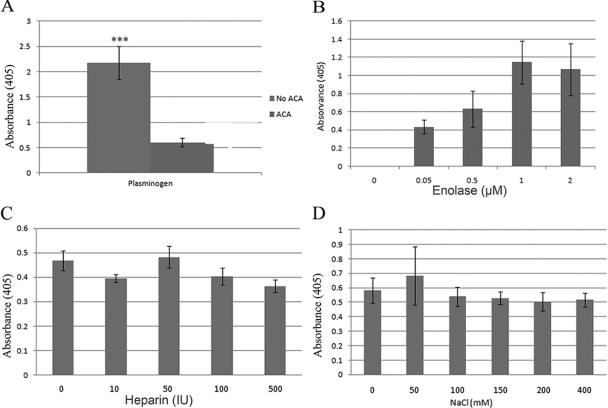
Previous studies have shown that a variety of Borrelia proteins are capable of binding PLG, enhancing the ability of the spirochete to disseminate (21, 23). The presence of enolase in the membrane (60) and in OMV suggests that it is localized to the OM of the spirochete and that it could therefore play a role in PLG binding. PLG (and BSA as a negative control) was immobilized onto ELISA plates and incubated with recombinant enolase. The binding was measured by ELISA. Addition of ε-aminocaproic acid, a known lysine analog, significantly (P < 0.001) inhibited the binding of enolase to PLG, indicating the dependency of this receptor-ligand interaction on the presence of lysine residues (Fig. 3A). In addition, lysines play an important role in PLG binding, interacting with the kringle domains present in the proenzyme. Enolase is a lysine-rich protein (40 residues, or 9.2% of 433 amino acids [31]), a characteristic shared by many other Borrelia proteins. Indeed, protein alignment of the enolase of B. burgdorferi (AAC66719) with that of Streptococcus pneumoniae (CAC83091), a known PLG receptor, showed homology with both internal and terminal PLG binding domains, suggesting a conserved function for interacting with host proteases (alignments not shown). Assays using different concentrations of PLG showed that the enolase bound PLG in a dose-dependent manner (Fig. 3B). In contrast, neither heparin nor NaCl inhibited the binding of enolase to PLG, indicating that the interaction is not dependent on ionic forces (Fig. 3C and D).
Fig 3.
(A) Enolase binds plasminogen in a lysine-dependent manner. Binding of human plasminogen to immobilized enolase in the presence and absence of ε-aminocaproic acid (ACA). ***, P < 0.001. (B) Enolase binds plasminogen in a dose-dependent manner. Binding was analyzed by ELISA after coating the plates with human plasminogen (10 μg/ml) and adding different concentrations (0 to 2 μM) of enolase. Bound enolase was detected using rabbit antienolase serum. BSA was used as the negative control, and its optical density (absorbance) was used as the blank. Values are represented as binding values minus background. (C) Binding of enolase (10 μg/ml) to immobilized plasminogen (10 μg/ml) in the presence of increasing concentrations (0 to 50 IU) of heparin was not inhibited. (D) Binding of enolase (10 μg/ml) to immobilized plasminogen (10 μg/ml) performed with increasing concentrations (0 to 400 mM) of NaCl was not inhibited. Data represent the means of the results of three separate experiments.
Localization of the enolase in the membrane.
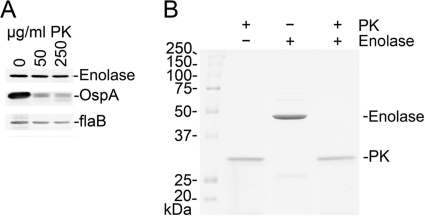
An indirect immunofluorescence assay (IFA), transmission electronic microscopy (TEM), and proteinase K treatment followed by Western blot analysis were performed to assess the location of the enolase in the Borrelia membrane. The TEM images collected from cultured and mouse-derived spirochetes after using the rabbit antienolase serum and labeling with 6-nm-diameter gold particles showed minor binding, but the preimmunization rabbit serum showed the same level of binding, indicating a nonspecific result (data not shown). IFA confirmed the TEM data. Identical results were obtained with spirochetes grown in dialysis chambers, indicating that host adaptation did not result in changes in enolase expression or localization within the organism (data not shown). PK treatment using whole bacteria (Fig. 4A) did not degrade the enolase. In both cases, a rapid degradation of OspA was observed; however, the enolase band remained constant and no changes were observed even at high concentrations (250 μg/ml) of protease. FlaB was used as a negative control, since it is in the periplasmic space and therefore should not be affected by PK treatment. Also, proteinase K treatment of the recombinant enolase was performed to show that enolase is actually degraded by the protease (Fig. 4B). Taken together, these results indicate that the enolase of B. burgdorferi is not expressed on the surface of the OM.
Fig 4.
Digestion of whole B. burgdorferi and recombinant enolase by proteinase K. (A) B. burgdorferi (108) were treated for 1 h at room temperature (23°C) with proteinase K (PK) at the concentrations indicated and analyzed by SDS-PAGE and Western blot to determine whether portions of enolase were surface exposed. Individual gel lanes received approximately 3 × 107 control or PK-treated spirochetes. (B) Recombinant enolase (4 μg) was incubated for 1 h at room temperature in PBS–5 mM MgCl2 with or without PK (250 μg/ml), separated by SDS-PAGE, and stained with Coomassie brilliant blue. The results shown are from a representative experiment.
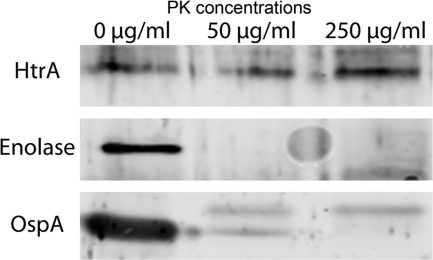
PK treatment of OMV followed by Western blot analysis to determine whether enolase in the OMV is accessible to degradation by proteolysis was performed. PK degraded the enolase as well as OspA in the OMV (Fig. 5). HtrA (bb0104), the periplasmic serine protease, which was also detected in the OMV (Table 1), was not degraded by PK proteolysis.
Fig 5.
Digestion of purified Borrelia vesicles by proteinase K. Vesicle samples (10 μg per lane) were treated with proteinase K (PK) at the concentrations indicated and analyzed by Western blotting.
The enolase of B. burgdorferi is immunogenic.
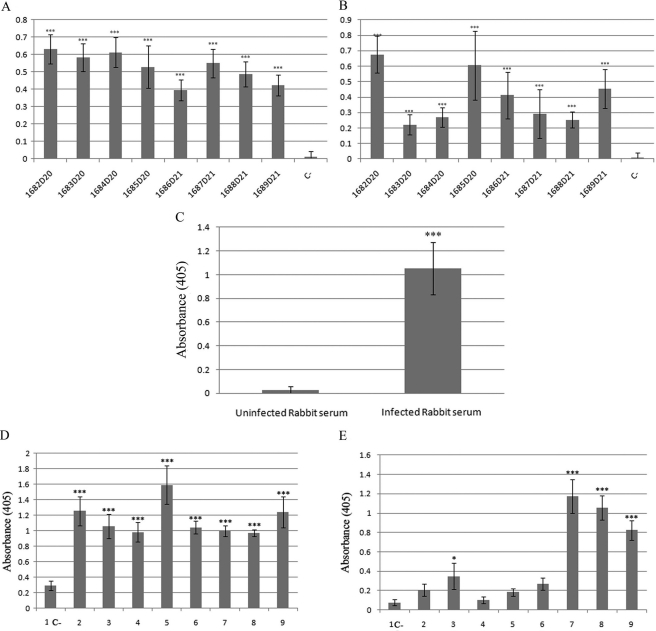
In several organisms, such as Trichomonas vaginalis, Candida albicans, and Streptococcus suis, enolase has been shown to be both surface exposed and immunogenic (58, 66, 75). To determine whether the Borrelia enolase could be immunogenic, sera from infected mice and from patients with Lyme disease were tested for reactivity to whole-cell Borrelia lysate and to recombinant enolase (Fig. 6A and B, respectively). In all cases, mouse sera that reacted against the whole Borrelia lysate also reacted with the recombinant enolase. This reactivity confirmed the immunogenicity of the enolase in experimental needle inoculations. Reactivity to enolase in rabbits also developed as the result of infections via tick bite (Fig. 6C), showing that the enolase triggered an antibody response during the course of a tick-borne infection.
Fig 6.
Enolase is an immunogenic enzyme. (A and B) Serologic reactivity of experimentally infected mice (1:100) to whole Borrelia lysate (A) and recombinant enolase (B). (C) Serologic reactivity of the serum of a rabbit infected with B. burgdorferi via tick bite to recombinant enolase. (D) Serologic reactivity of randomly selected sera from Lyme disease patients to Borrelia whole-cell lysate. (E) Serologic reactivity of sera from the same Lyme disease patients to recombinant enolase. Bars C, negative control representing pooled sera from mice, rabbits, and patients. ***, P < 0.001; *, P < 0.05.
Half of the group of serum samples from Lyme disease patients that exhibited reactivity to the whole-cell lysate (Fig. 6D) had high levels of reactivity to the recombinant enolase (OD at 405 nm [OD405] > 0.4), and the others had OD levels that were twice as high as that of the pooled-serum negative control (OD405 < 0.14) (Fig. 6E). These results indicate that enolase is recognized in the antibody repertoire in Lyme disease but at different levels of reactivity.
Enolase expression.
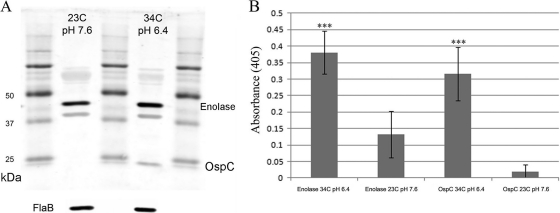
B. burgdorferi is maintained in an enzootic cycle alternating between tick vectors and vertebrate hosts. In the transition from the tick to the vertebrate milieu, the spirochete undergoes a dramatic switch in gene expression. The spirochete upregulates a large number of genes, most notably ospC, in response to the host milieu (73). The switch in gene expression can be mimicked in vitro by changing the temperature and pH (15, 68, 72). RNAs isolated from cultures grown under conditions of 23°C and pH 7.6 for tick experiments and 34°C and pH 6.4 for mammal experiments were used to carry out real-time PCR, which demonstrated that, after normalization to flaB, the genes were expressed at equal levels (PCR data not shown). To evaluate the possibility of posttranscriptional changes that could affect the overall quantity of enolase, Western blot analysis and ELISA were performed using OspC as the positive control and FlaB as the loading control. Using the temperature and pH combinations listed in Materials and Methods, differences in the enolase protein contents were detected by both methods (Fig. 7), suggesting that enolase is posttranscriptionally regulated under the different sets of conditions.
Fig 7.
The overall quantity of enolase protein is higher under conditions that mimic the conditions in the mammalian host. Panels A and B represent the results of Western blot analysis and ELISA, respectively, showing enolase and control levels. FlaB was used as a loading control. OspC was used as a positive control, since it is not expressed at 23°C and pH 7.6 but is expressed at 34°C and pH 6.4.
The levels of enolase seen at 34°C and pH 7.6 were similar to those seen at 34°C and pH 6.4, indicated that posttranscriptional regulation is dependent on temperature (data not shown).
DISCUSSION
Both pathogenic and nonpathogenic species of Gram-negative bacteria shed OMV (9, 45, 52, 55). Studies of OMV from diverse bacterial organisms support the idea of common functions: OMV allow bacteria to interact with each other within species, with other bacteria, and with eukaryotic cells. Both characterization of OMV functions and biochemical analysis have demonstrated a role in the transport of active virulence factors to host cells. OMV generated by pathogenic bacteria contain adhesins and toxins that mediate bacterial binding and invasion, cause cytotoxicity, and modulate the host immune response (30, 51). Therefore, OMV participate in host-pathogen interactions and are potent bacterial virulence factors. In Borrelia, the mechanisms of release of OMV have not been well characterized, but their existence and regulated release has been documented in several in vivo and in vitro studies (27, 64, 70, 74). Here we list the most abundant protein components of the OMV of B. burgdorferi as identified by mass spectrometry in two independent experiments. The results demonstrated that OspA and OspB are the two major components of the OMV and that OspB is at least four times as abundant as OspA. Other surface proteins detected among the 15 most abundant proteins were P27 (bba60), CspA (bba68), BmpA (bb383), P13 (bb0034), outer surface 22-kDa protein (bb0365), and P66 (bb0603). Interestingly, other putative cytoplasmic proteins such as GroEL (bb0649), GAPDH (glyceraldehyde-3-phosphate-dehydrogenase) (bb0057), and enolase (bb0337) were also detected.
Of particular note in the protein composition of the OMV is the presence of several known PLG receptors (OspA, OspB, and CspA) for Borrelia and several known PLG receptors (including GAPDH and enolase, both enzymes of the glycolysis pathway) for other bacteria (5, 6, 53, 54, 56, 57, 61, 63). Borreliae have an extensive association with the mammalian PLG activation system. B. burgdorferi and relapsing fever Borrelia fix plasmin onto their surfaces in the appropriate orientation so that it can function as a bound protease that assists the spirochetes in dissemination (23, 36, 59) and in degradation of matrix protein (22). Fixing plasmin to the Borrelia surface is only one aspect of the association with the PLG activation system. B. burgdorferi can induce the production of both urokinase PLG activator (uPA) and its receptor, uPAR (19, 20, 42). In this phase of the association, uPA fixed to the surface of inflammatory cells can activate the PLG bound to the Borrelia, with the possible result of enhancing dissemination of the spirochetes. Additional interactions include those involving PLG activation inhibitors (39) and metalloproteases (34, 35, 43). Few bacteria have such a complete association with a mammalian system. Therefore, it is of interest that the OMV of B. burgdorferi contain multiple PLG receptors that, given the proper orientation of the proteolytic activity, can further enhance these interactions.
The issue concerning how GAPDH and enolase are exported to the surface of the cell wall of Gram-positive organisms is an area of much research interest. These glycolytic enzymes do not have cell wall anchor sequences or obvious signal peptidase cleavage sites for export to the outer cell wall or for specific release into the environment. Like its counterparts in Gram-positive bacteria, the enolase of B. burgdorferi lacks the motifs that would be consistent with their possible transfer to a surface location in the outer membrane. Furthermore, the sequence of enolase of B. burgdorferi has been found to have homology to the internal and terminal PLG binding sequences of other bacterial enolases (8, 24).
Nonetheless, the outer-surface location of the enolase of Gram-positive organisms is well established, and evidence for its presence in the OMV of B. burgdorferi is presented here. However, there is one major difference between the location of the enolase in B. burgdorferi and its location in Gram-positive bacteria. We were not able to demonstrate a surface location for enolase in cultured or in vivo B. burgdorferi by fluorescence or electron microscopy and by treatment with proteinase K. Instead, we have documented the presence of enolase in OMV by mass spectroscopy and by Western blot analysis. In addition, we have shown that enolase is accessible to degradation by PK proteolysis in the OMV. As mentioned before, the enolase lacks signal peptides or cell wall anchors, so it is unclear how it is transported to and associated with the membrane. Our results suggest that enolase is not anchored in the membrane of the live spirochete but that its position changes when it is released in OMV, becoming accessible for the PK and therefore for PLG as well.
Although the specific functions of the Borrelia OMV are not known, the presence of several PLG receptors, including enolase (whose binding to PLG has been characterized in this study), point to a role in fixing plasmin to the peribacterial environment. Plasmin-coated B. burgdorferi as well as relapsing fever Borrelia can disseminate better and faster than non-plasmin-coated organisms. Borrelia with plasmin on the surface can also degrade extracellular matrices to promote dissemination (23, 36, 59). Active proteolysis in the peribacterial environment induced by released OMV could promote further degradation of matrix proteins and possibly the production of more easily acquired peptides, leading to a function in bacterial nutrition. Such external proteolytic activity would require the presence of some or all of the PLG receptors on the surface of the OMV to provide the proper location and orientation for the plasmin. Note that there is no evidence for protease release by B. burgdorferi, thus enhancing the importance of the idea of borrowed proteolytic activity.
It is well established that, upon tick feeding, Borrelia undergoes shifts in gene expression that change the protein profiles of the OM and, therefore, the protein composition of the OMV. Expression of enolase (bb0337) in Borrelia does not change as a result of blood induction or temperature shift (73), as real-time PCR did not reveal any differences in enolase expression when we combined high temperature (34°C) and low pH (6.4). However, the overall quantity of the enzyme that we detected was significantly higher at 34°C than at 23°C, showing that there is posttranscriptional regulation of the enolase. The mechanism that governs this regulation is not known.
Regardless of its function(s) in the OMV, enolase is a B cell immunogen recognized by sera from experimentally infected mice (inoculated with cultured organisms via needle injection), tick-infected rabbits, and Lyme disease patients. In this regard, how this glycolytic enzyme is transported to the OMV and how it becomes an immunogen are important issues that link bacterial physiology and pathogenesis.
ACKNOWLEDGMENTS
This study was supported by grants RO1-AI-027044 and RO1-AR-040445 from the National Institutes of Health to J.L.B.
Richard T. Marconi generously provided the OspC type A gene in a pET46 LIC that was used to generate the recombinant OspC that was subsequently purified and used to develop antibodies for this study. Melissa Caimano and Justin Radolf generously provided the host-derived spirochetes. The Northeast Biodefense Center Protein Expression Core at Wadsworth Center provided the purified recombinant enolase and is supported by U54-AI-0715558 (Lipkin).
Footnotes
Published ahead of print 14 November 2011
REFERENCES
- 1. Akins DR, Bourell KW, Caimano MJ, Norgard MV, Radolf JD. 1998. A new animal model for studying Lyme disease spirochetes in a mammalian host-adapted state. J. Clin. Invest. 101: 2240–2250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Anda P, Gebbia JA, Backenson PB, Coleman JL, Benach JL. 1996. A glyceraldehyde-3-phosphate dehydrogenase homolog in Borrelia burgdorferi and Borrelia hermsii. Infect. Immun. 64: 262–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Angiolella L, et al. 1996. Identification of a glucan-associated enolase as a main cell wall protein of Candida albicans and an indirect target of lipopeptide antimycotics. J. Infect. Dis. 173: 684–690 [DOI] [PubMed] [Google Scholar]
- 4. Benach JL, et al. 1983. Spirochetes isolated from the blood of two patients with Lyme disease. N. Engl. J. Med. 308: 740–742 [DOI] [PubMed] [Google Scholar]
- 5. Bergmann S, Rohde M, Chhatwal GS, Hammerschmidt S. 2001. alpha-Enolase of Streptococcus pneumoniae is a plasmin(ogen)-binding protein displayed on the bacterial cell surface. Mol. Microbiol. 40: 1273–1287 [DOI] [PubMed] [Google Scholar]
- 6. Bergmann S, Rohde M, Hammerschmidt S. 2004. Glyceraldehyde-3-phosphate dehydrogenase of Streptococcus pneumoniae is a surface-displayed plasminogen-binding protein. Infect. Immun. 72: 2416–2419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bergmann S, Rohde M, Preissner KT, Hammerschmidt S. 2005. The nine residue plasminogen-binding motif of the pneumococcal enolase is the major cofactor of plasmin-mediated degradation of extracellular matrix, dissolution of fibrin and transmigration. Thromb Haemost. 94: 304–311 [DOI] [PubMed] [Google Scholar]
- 8. Bergmann S, et al. 2003. Identification of a novel plasmin(ogen)-binding motif in surface displayed alpha-enolase of Streptococcus pneumoniae. Mol. Microbiol. 49: 411–423 [DOI] [PubMed] [Google Scholar]
- 9. Beveridge TJ. 1999. Structures of gram-negative cell walls and their derived membrane vesicles. J. Bacteriol. 181: 4725–4733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bhide MR, et al. 2009. Complement factor H binding by different Lyme disease and relapsing fever Borrelia in animals and human. BMC Res. Notes 2: 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bochkareva ES, Solovieva ME, Girshovich AS. 1998. Targeting of GroEL to SecA on the cytoplasmic membrane of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 95: 478–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Brissette CA, et al. 2009. Borrelia burgdorferi infection-associated surface proteins ErpP, ErpA, and ErpC bind human plasminogen. Infect. Immun. 77: 300–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bunikis J, Barbour AG. 1999. Access of antibody or trypsin to an integral outer membrane protein (P66) of Borrelia burgdorferi is hindered by Osp lipoproteins. Infect. Immun. 67: 2874–2883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Burgdorfer W, et al. 1982. Lyme disease-a tick-borne spirochetosis? Science 216: 1317–1319 [DOI] [PubMed] [Google Scholar]
- 15. Carroll JA, Garon CF, Schwan TG. 1999. Effects of environmental pH on membrane proteins in Borrelia burgdorferi. Infect. Immun. 67: 3181–3187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Coleman JL, Benach JL. 1992. Characterization of antigenic determinants of Borrelia burgdorferi shared by other bacteria. J. Infect. Dis. 165: 658–666 [DOI] [PubMed] [Google Scholar]
- 17. Coleman JL, Benach JL. 2000. The generation of enzymatically active plasmin on the surface of spirochetes. Methods 21: 133–141 [DOI] [PubMed] [Google Scholar]
- 18. Coleman JL, Benach JL. 1989. Identification and characterization of an endoflagellar antigen of Borrelia burgdorferi. J. Clin. Invest. 84: 322–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Coleman JL, Benach JL. 2003. The urokinase receptor can be induced by Borrelia burgdorferi through receptors of the innate immune system. Infect. Immun. 71: 5556–5564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Coleman JL, Gebbia JA, Benach JL. 2001. Borrelia burgdorferi and other bacterial products induce expression and release of the urokinase receptor (CD87). J. Immunol. 166: 473–480 [DOI] [PubMed] [Google Scholar]
- 21. Coleman JL, et al. 1997. Plasminogen is required for efficient dissemination of B. burgdorferi in ticks and for enhancement of spirochetemia in mice. Cell 89: 1111–1119 [DOI] [PubMed] [Google Scholar]
- 22. Coleman JL, Roemer EJ, Benach JL. 1999. Plasmin-coated borrelia Burgdorferi degrades soluble and insoluble components of the mammalian extracellular matrix. Infect. Immun. 67: 3929–3936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Coleman JL, et al. 1995. Borrelia burgdorferi binds plasminogen, resulting in enhanced penetration of endothelial monolayers. Infect. Immun. 63: 2478–2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cork AJ, et al. 2009. Defining the structural basis of human plasminogen binding by streptococcal surface enolase. J. Biol. Chem. 284: 17129–17137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cox DL, et al. 1996. Limited surface exposure of Borrelia burgdorferi outer surface lipoproteins. Proc. Natl. Acad. Sci. U. S. A. 93: 7973–7978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Deatherage BL, et al. 2009. Biogenesis of bacterial membrane vesicles. Mol. Microbiol. 72: 1395–1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dunham-Ems SM, et al. 2009. Live imaging reveals a biphasic mode of dissemination of Borrelia burgdorferi within ticks. J. Clin. Invest. 119: 3652–3665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Earnhart CG, et al. 2010. Identification of residues within ligand-binding domain 1 (LBD1) of the Borrelia burgdorferi OspC protein required for function in the mammalian environment. Mol. Microbiol. 76: 393–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. El-Hage N, et al. 2001. Surface exposure and protease insensitivity of Borrelia burgdorferi Erp (OspEF-related) lipoproteins. Microbiology 147: 821–830 [DOI] [PubMed] [Google Scholar]
- 30. Ellis TN, Kuehn MJ. 2010. Virulence and immunomodulatory roles of bacterial outer membrane vesicles. Microbiol. Mol. Biol. Rev. 74: 81–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fraser CM, et al. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390: 580–586 [DOI] [PubMed] [Google Scholar]
- 32. Fuchs H, Simon MM, Wallich R, Bechtel M, Kramer MD. 1996. Borrelia burgdorferi induces secretion of pro-urokinase-type plasminogen activator by human monocytes. Infect. Immun. 64: 4307–4312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fuchs H, Wallich R, Simon MM, Kramer MD. 1994. The outer surface protein A of the spirochete Borrelia burgdorferi is a plasmin(ogen) receptor. Proc. Natl. Acad. Sci. U. S. A. 91: 12594–12598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gebbia JA, Coleman JL, Benach JL. 2001. Borrelia spirochetes upregulate release and activation of matrix metalloproteinase gelatinase B (MMP-9) and collagenase 1 (MMP-1) in human cells. Infect. Immun. 69: 456–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gebbia JA, Coleman JL, Benach JL. 2004. Selective induction of matrix metalloproteinases by Borrelia burgdorferi via toll-like receptor 2 in monocytes. J. Infect. Dis. 189: 113–119 [DOI] [PubMed] [Google Scholar]
- 36. Gebbia JA, Monco JC, Degen JL, Bugge TH, Benach JL. 1999. The plasminogen activation system enhances brain and heart invasion in murine relapsing fever borreliosis. J. Clin. Invest. 103: 81–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Goulhen F, et al. 1998. Subcellular localization and cytotoxic activity of the GroEL-like protein isolated from Actinobacillus actinomycetemcomitans. Infect. Immun. 66: 5307–5313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Grimm D, et al. 2004. Outer-surface protein C of the Lyme disease spirochete: a protein induced in ticks for infection of mammals. Proc. Natl. Acad. Sci. U. S. A. 101: 3142–3147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Haile WB, Coleman JL, Benach JL. 2006. Reciprocal upregulation of urokinase plasminogen activator and its inhibitor, PAI-2, by Borrelia burgdorferi affects bacterial penetration and host-inflammatory response. Cell Microbiol. 8: 1349–1360 [DOI] [PubMed] [Google Scholar]
- 40. Hallstrom T, et al. 2010. Complement Regulator-Acquiring Surface Protein 1 of Borrelia burgdorferi Binds to Human Bone Morphogenic Protein 2, Several Extracellular Matrix Proteins, and Plasminogen. J. Infect. Dis. 202: 490–498 [DOI] [PubMed] [Google Scholar]
- 41. Hovis KM, Freedman JC, Zhang H, Forbes JL, Marconi RT. 2008. Identification of an antiparallel coiled-coil/loop domain required for ligand binding by the Borrelia hermsii FhbA protein: additional evidence for the role of FhbA in the host-pathogen interaction. Infect. Immun. 76: 2113–2122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hovius JW, et al. 2009. The urokinase receptor (uPAR) facilitates clearance of Borrelia burgdorferi. PLoS Pathog. 5: e1000447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hu LT, et al. 2001. Host metalloproteinases in Lyme arthritis. Arthritis Rheum. 44: 1401–1410 [DOI] [PubMed] [Google Scholar]
- 44. Hu LT, Perides G, Noring R, Klempner MS. 1995. Binding of human plasminogen to Borrelia burgdorferi. Infect. Immun. 63: 3491–3496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kadurugamuwa JL, Beveridge TJ. 1997. Natural release of virulence factors in membrane vesicles by Pseudomonas aeruginosa and the effect of aminoglycoside antibiotics on their release. J. Antimicrob. Chemother. 40: 615–621 [DOI] [PubMed] [Google Scholar]
- 46. Klempner MS, Noring R, Epstein MP, McCloud B, Rogers RA. 1996. Binding of human urokinase type plasminogen activator and plasminogen to Borrelia species. J. Infect. Dis. 174: 97–104 [DOI] [PubMed] [Google Scholar]
- 47. Kudryashev M, et al. 2009. Comparative cryo-electron tomography of pathogenic Lyme disease spirochetes. Mol. Microbiol. 71: 1415–1434 [DOI] [PubMed] [Google Scholar]
- 48. Lagal V, Portnoi D, Faure G, Postic D, Baranton G. 2006. Borrelia burgdorferi sensu stricto invasiveness is correlated with OspC-plasminogen affinity. Microbes Infect. 8: 645–652 [DOI] [PubMed] [Google Scholar]
- 49. LaRocca TJ, et al. 2010. Cholesterol lipids of Borrelia burgdorferi form lipid rafts and are required for the bactericidal activity of a complement-independent antibody. Cell Host Microbe 8: 331–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. LaRocca TJ, et al. 2009. The bactericidal effect of a complement-independent antibody is osmolytic and specific to Borrelia. Proc. Natl. Acad. Sci. U. S. A. 106: 10752–10757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lee EY, Choi DS, Kim KP, Gho YS. 2008. Proteomics in gram-negative bacterial outer membrane vesicles. Mass Spectrom Rev. 27: 535–555 [DOI] [PubMed] [Google Scholar]
- 52. Li Z, Clarke AJ, Beveridge TJ. 1998. Gram-negative bacteria produce membrane vesicles which are capable of killing other bacteria. J. Bacteriol. 180: 5478–5483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lottenberg R, Broder CC, Boyle MD. 1987. Identification of a specific receptor for plasmin on a group A streptococcus. Infect. Immun. 55: 1914–1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lottenberg R, et al. 1992. Cloning, sequence analysis, and expression in Escherichia coli of a streptococcal plasmin receptor. J. Bacteriol. 174: 5204–5210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Mayrand D, Grenier D. 1989. Biological activities of outer membrane vesicles. Can. J. Microbiol. 35: 607–613 [DOI] [PubMed] [Google Scholar]
- 56. McBroom AJ, Kuehn MJ. 2007. Release of outer membrane vesicles by Gram-negative bacteria is a novel envelope stress response. Mol. Microbiol. 63: 545–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Miles LA, et al. 1991. Role of cell-surface lysines in plasminogen binding to cells: identification of alpha-enolase as a candidate plasminogen receptor. Biochemistry 30: 1682–1691 [DOI] [PubMed] [Google Scholar]
- 58. Mundodi V, Kucknoor AS, Alderete JF. 2008. Immunogenic and plasminogen-binding surface-associated alpha-enolase of Trichomonas vaginalis. Infect. Immun. 76: 523–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Nordstrand A, Shamaei-Tousi A, Ny A, Bergstrom S. 2001. Delayed invasion of the kidney and brain by Borrelia crocidurae in plasminogen-deficient mice. Infect. Immun. 69: 5832–5839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Nowalk AJ, Nolder C, Clifton DR, Carroll JA. 2006. Comparative proteome analysis of subcellular fractions from Borrelia burgdorferi by NEPHGE and IPG. Proteomics 6: 2121–2134 [DOI] [PubMed] [Google Scholar]
- 61. Pancholi V. 2001. Multifunctional alpha-enolase: its role in diseases. Cell. Mol. Life Sci. 58: 902–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Pancholi V, Fischetti VA. 1998. alpha-enolase, a novel strong plasmin(ogen) binding protein on the surface of pathogenic streptococci. J. Biol. Chem. 273: 14503–14515 [DOI] [PubMed] [Google Scholar]
- 63. Pancholi V, Fischetti VA. 1992. A major surface protein on group A streptococci is a glyceraldehyde-3-phosphate-dehydrogenase with multiple binding activity. J. Exp. Med. 176: 415–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Radolf JD, et al. 1995. Characterization of outer membranes isolated from Borrelia burgdorferi, the Lyme disease spirochete. Infect. Immun. 63: 2154–2163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Rossmann E, et al. 2007. Dual binding specificity of a Borrelia hermsii-associated complement regulator-acquiring surface protein for factor H and plasminogen discloses a putative virulence factor of relapsing fever spirochetes. J. Immunol. 178: 7292–7301 [DOI] [PubMed] [Google Scholar]
- 66. Sandini S, Melchionna R, Arancia S, Gomez MJ, La Valle R. 1999. Generation of a highly immunogenic recombinant enolase of the human opportunistic pathogen Candida albicans. Biotechnol. Appl. Biochem. 29(Pt 3): 223–227 [PubMed] [Google Scholar]
- 67. Schott M, Grosskinsky S, Brenner C, Kraiczy P, Wallich R. 2010. Molecular characterization of the interaction of Borrelia parkeri and Borrelia turicatae with human complement regulators. Infect. Immun. 78: 2199–2208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Schwan TG, Piesman J, Golde WT, Dolan MC, Rosa PA. 1995. Induction of an outer surface protein on Borrelia burgdorferi during tick feeding. Proc. Natl. Acad. Sci. U. S. A. 92: 2909–2913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Sha J, et al. 2009. Surface-expressed enolase contributes to the pathogenesis of clinical isolate SSU of Aeromonas hydrophila. J. Bacteriol. 191: 3095–3107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Shoberg RJ, Thomas DD. 1993. Specific adherence of Borrelia burgdorferi extracellular vesicles to human endothelial cells in culture. Infect. Immun. 61: 3892–3900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Skare JT, et al. 1995. Virulent strain associated outer membrane proteins of Borrelia burgdorferi. J. Clin. Invest. 96: 2380–2392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Stevenson B, Schwan TG, Rosa PA. 1995. Temperature-related differential expression of antigens in the Lyme disease spirochete, Borrelia burgdorferi. Infect. Immun. 63: 4535–4539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Tokarz R, Anderton JM, Katona LI, Benach JL. 2004. Combined effects of blood and temperature shift on Borrelia burgdorferi gene expression as determined by whole genome DNA array. Infect. Immun. 72: 5419–5432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Whitmire WM, Garon CF. 1993. Specific and nonspecific responses of murine B cells to membrane blebs of Borrelia burgdorferi. Infect. Immun. 61: 1460–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Zhang A, Xie C, Chen H, Jin M. 2008. Identification of immunogenic cell wall-associated proteins of Streptococcus suis serotype 2. Proteomics 8: 3506–3515 [DOI] [PubMed] [Google Scholar]