Abstract
The budding yeast Cdc13-Stn1-Ten1 complex is crucial for telomere protection and has been proposed to resemble the RPA complex structurally and functionally. The Cdc13 homologues in Candida species are unusually small and lack two conserved domains previously implicated in telomere regulation, thus raising interesting questions concerning the mechanisms and evolution of these proteins. In this report, we show that the unusually small Cdc13 homologue in Candida albicans is indeed a regulator of telomere lengths and that it associates with telomere DNA in vivo. We demonstrated high-affinity telomere DNA binding by C. tropicalis Cdc13 (CtCdc13) and found that dimerization of this protein through its OB4 domain is important for high-affinity DNA binding. Interestingly, CtCdc13-DNA complex formation appears to involve primarily recognition of multiple copies of a six-nucleotide element (GGATGT) that is shared by many Candida telomere repeats. We also determined the crystal structure of the OB4 domain of C. glabrata Cdc13, which revealed a novel mechanism of OB fold dimerization. The structure also exhibits marked differences to the C-terminal OB fold of RPA70, thus arguing against a close evolutionary kinship between these two proteins. Our findings provide new insights on the mechanisms and evolution of a critical telomere end binding protein.
INTRODUCTION
The special structures located at the ends of linear eukaryotic chromosomes, known as telomeres, are critical for chromosome stability; they protect the terminal DNAs from degradation, end-to-end fusion and other abnormal transactions (6, 12, 31). Telomeric DNAs are bound by functionally important proteins through both DNA-protein and protein-protein interactions. In most organisms, telomeres comprise short repetitive G-rich sequences and terminate in 3′ overhangs referred to as G-tails. Even though the G-tails represent a shared feature of almost all telomeres, they appear to be bound by divergent protein complexes in different organisms. A widespread dimeric G-tail binding protein complex was first described in ciliated protozoa and named TEBPα/β in these organisms (36). Subsequent studies revealed orthologues of these proteins in both fission yeast and mammals (named Pot1-Tpz1 in fission yeast and POT1-TPP1 in mammals) (29, 43). By contrast, the G-tails of budding yeast are capped by a trimeric complex comprised of Cdc13, Stn1, and Ten1 (CST) (22). Genetic and structural analyses suggest that CST represents a telomere-specific RPA-like complex (9, 12, 40). Interestingly, even though CST proteins were initially thought to be confined to budding yeast, recent studies have uncovered Stn1 and Ten1 homologues in Schizosaccharomyces pombe as well as CST-like complexes in plants and mammals (24, 28, 41). Thus, in many organisms the CST complex may act as an alternative telomere end protection complex with functions overlapping or parallel to those of the POT1-TPP1 complex.
Among all the CST complexes, the structures and mechanisms of the Saccharomyces cerevisiae subunits are the most extensively characterized. S. cerevisiae Cdc13 (ScCdc13) is a multifunctional protein with a myriad of binding targets (Fig. 1A). It uses a C-terminal OB fold (DNA-binding domain [DBD]) to bind with high affinity and sequence specificity to the irregular, GT-rich repeats of S. cerevisiae telomeres (27). It also employs a recruitment domain (RD) to interact with the telomerase regulatory protein Est1, and this interaction promotes the recruitment of telomerase to chromosome ends and the activation of telomerase (5, 35). Moreover, we have recently shown that the N-terminal OB fold domain of ScCdc13 (OB1) mediates ScCdc13 dimerization and that this dimerization promotes Cdc13-Pol1 (the catalytic subunit of polymerase α [Pol α] interaction and regulates telomere length (39). Others have reported that dimerization may allow ScCdc13OB1 to bind DNA (26). In comparison to ScCdc13, fewer interaction partners have been identified for ScStn1 and ScTen1. Both ScStn1 and ScTen1 have been reported to bind telomere DNA with moderate to low affinity (9). ScStn1 is also known to interact with Pol12, another subunit of the Pol α complex (13). The multiplicity of interactions between CST and Pol α supports a role for CST in regulating telomere C-strand synthesis, which is thought to be mediated by Pol α (12).
Fig 1.
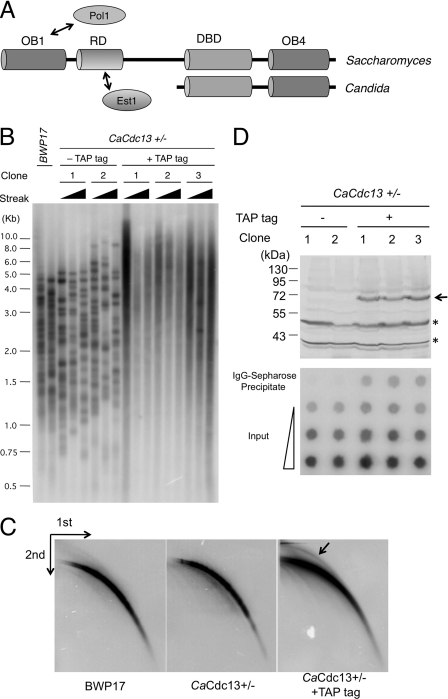
Domain organizations of Cdc13s and the role of Candida Cdc13 in telomere regulation. (A) The different domain organizations of Cdc13 homologues from Saccharomyces and Candida species are illustrated. The OB1 and RD domains of Saccharomyces Cdc13 have been shown to interact with Pol1 and Est1, respectively. (B) The wild type (BWP17) and strains heterozygous for CDC13 (with or without a C-terminal TAP tag) were passaged in YPD+uri and analyzed for telomere lengths. For each heterozygous CDC13+/− clone (with or without TAP tag), telomeres were examined after 3, 6, and 9 streaks on plates. (C) Chromosomal DNAs from the indicated strains were digested with AluI and NlaIII and then subjected to two-dimensional gel electrophoresis and Southern blotting to assess the levels of linear and circular telomeric DNAs (marked by an arrow). (D) (Top) The expression of TAP-tagged Cdc13 protein in extracts derived from the untagged and tagged strains were analyzed by Western blotting using antibodies directed against protein A. The positions of CaCdc13-TAP and two cross-reacting proteins are indicated by an arrow and two asterisks, respectively. (Bottom) Strains with or without TAP-tagged Cdc13 were subjected to ChIP analysis using IgG-Sepharose. The input (0.13, 0.64, and 3.2%) and precipitated DNAs (100%) were spotted on nylon filters and probed with labeled C. albicans telomere repeats.
As alluded to earlier, a provocative recent proposal concerning CST is that it represents a telomere-specific RPA complex (9). Indeed, we and others have shown a high degree of structural and functional resemblances between Stn1 and RPA32, as well as between Ten1 and RPA14 (9, 10, 34, 40). By contrast, existing data do not support a paralogous relationship between Cdc13 and RPA70, the largest subunits of the two complexes. Even though both Cdc13 and RPA70 consist of multiple OB fold domains, neither the first OB fold (OB1) nor the penultimate OB fold (DBD) of Cdc13 displays a strong similarity to the corresponding domain in RPA70 (39). However, because the structures of other domains of Cdc13 have not been resolved, the possibility remains that additional studies could provide supports for a paralogous relationship between Cdc13 and RPA70.
Our laboratories have employed Candida species as alternative model systems for understanding CST structure and mechanisms. The telomere repeat units of Candida species are unusual in being long, regular, and non-G-rich (25). Homologues of the CST proteins can nevertheless be readily identified in most Candida genomes (21, 42). In a previous report, we described the high-resolution structure of a complex of Candida tropicalis Stn1N and Ten1 and the functions of C. albicans Stn1 and Ten1 in telomere regulation (40). However, our analysis of the C. albicans Cdc13 (CaCdc13) homologue was hampered by the fact that the gene is essential for cell viability. Interestingly, many Cdc13 homologues in Candida species are noticeably smaller; they lack the N-terminal half of their S. cerevisiae counterpart and contain just two OB fold domains: DBD (responsible for DNA binding of ScCdc13) and OB4 (implicated in binding Stn1) (Fig. 1A) (21). Because the N-terminal half of ScCdc13 is responsible for dimerization and ScCdc13-Pol1 and ScCdc13-Est1 interaction, its absence in Candida Cdc13s raises fascinating questions concerning the mechanisms and evolution of these homologues. In this report, we provide evidence that the unusually small Cdc13 homologue in Candida albicans is indeed a regulator of telomere lengths and structure and that it associates with telomere DNA in vivo. We demonstrated high-affinity telomere DNA binding by C. tropicalis Cdc13 (CtCdc13) and found that this interaction requires a long DNA target site, as well as both the DBD and the OB4 domain. In addition, we showed that dimerization of CtCdc13 through its OB4 domain is important for high-affinity DNA binding. Moreover, we determined the crystal structure of the OB4 domain of C. glabrata Cdc13 (CgCdc13) and uncovered a novel mode of OB fold dimerization. Comparative structural analysis revealed marked differences between CgCdc13OB4 and the C-terminal OB fold of RPA70, arguing against a close evolutionary kinship between these two proteins. Our findings provide new insights on the mechanisms and evolution of Cdc13 and underscore the utility of investigating the CST complex in Candida species.
MATERIALS AND METHODS
Construction and growth of Candida strains.
The C. albicans strain BWP17 (ura3Δ::λimm434/ura3Δ::λimm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG) was used as the parental strain (45). The heterozygous CDC13+/− strain was generated by subjecting BWP17 to one round of transformation and 5-fluoroorotic acid (5-FOA) selection using a CDC13::hisG-URA3-hisG cassette (containing ∼630 bp of CDC13 upstream and ∼800 bp of downstream sequence) (7, 8). For making the strains with C-terminally tagged Cdc13, we first constructed a tagging plasmid by replacing the green fluorescent protein (GFP) fragment in pGFP-URA with a tandem affinity purification (TAP) fragment derived by PCR from pBS1479 (11, 37). This plasmid is named pTAP-URA3 and contains the TAP tag followed by the C. albicans ADH2 terminator and the URA3 selectable marker. A cassette consisting of the following elements was then obtained by PCR using appropriate primers and pTAP-URA3 as the template: 100 bp of the 3′ end of CaCDC13, the TAP tag, the ADH2 terminator, the URA3 marker, and 100 bp of the DNA downstream of the CaCDC13 termination codon. This tagging cassette was used to transform the CDC13+/− heterozygous strains. Correct transformants were selected on SD-ura plates and identified by PCR.
Sequence analysis.
Cdc13 homologues from Candida and Saccharomyces spp. were identified from NCBI (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and Broad Institute (http://www.broad.mit.edu/annotation/genome/candida_group/Blast.html) databases by BLAST or psi-BLAST searches. The multiple sequence alignment was generated using the PROMALS server (http://prodata.swmed.edu/promals/promals.php) and displayed using Boxshade (http://www.ch.embnet.org/software/BOX_form.html).
Telomere analyses.
The telomere length analysis and the two-dimensional gel analysis of circular and linear telomeric DNA were performed as previously described (46).
Gel electrophoretic mobility shift analysis.
Full-length CtCDC13 and individual domains (DBD, amino acids 1 to 195; OB4, amino acids 196 to 369) were cloned into the pSMT3 vector to enable the expression of His6-SUMO-Cdc13 fusion proteins. Because of the atypical translation of the CUG codon in Candida species, the CTG triplets encoding amino acids 33 and 132 of CtCdc13 were mutated to TCG to enable the expression of wild-type proteins in Escherichia coli (38). Following induction, extracts were prepared and the fusion proteins purified with Ni-nitrilotriacetic acid (NTA) chromatography as previously described (46). The fusion protein was cleaved by the ULP1 protease, and the Cdc13 fragment was purified away from the His6-SUMO tag by a second round of Ni-NTA affinity chromatography. Some of the DNA-binding reactions employed CtCdc13 that had been further purified over a glycerol gradient. Full-length CgCDC13 and its DBD (amino acids 404 to 594) were cloned into the pSMT3 vector and purified using the same method. Binding reactions contained 10 mM Tris-HCl (pH 8.0), 50 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol (DTT), and 5% glycerol. Following incubation at 25°C for 20 min, the reaction mixtures were electrophoresed through a nondenaturing polyacrylamide gel to resolve the free probe from the DNA-protein complex. Binding activity was analyzed using a Typhoon PhosphorImager and ImageQuant software (GE Healthcare). To examine the effect of dimerization on DNA binding, the following amino acids in three connecting loops in the CtCdc13 OB4 domain were mutated by QuikChange: SISE234-238 in LA1, TILDDR295-300 in L23, and KQKI358-361in L45. Each His6-SUMO-fused Cdc13 mutant protein was expressed in and purified from E. coli BL21(DE3). The binding activities of the mutant proteins were analyzed as described above.
Coexpression and GST pulldown assays.
The genes encoding full-length CtCdc13 and individual domains were transferred from the pSMT3 vector into the pGEX4T-2 vector (GE Healthcare). Each His6-SUMO-Cdc13 fusion protein was coexpressed with either the corresponding glutathione S-transferase (GST)–Cdc13 fusion protein or GST in E. coli BL21(DE3). To examine the roles of the connecting loops in CtCdc13OB4 dimerization, the following three sets of amino acids were mutated by QuikChange: SISE234-238 in LA1, TILDDR295-300 in L23, and KQKI358-361in L45. Each His6-SUMO-fused CtCdc13OB4 mutant protein was coexpressed with the corresponding GST-fused mutant protein in E. coli BL21(DE3). Following induction, extracts were prepared and subjected to GST pulldown assays. Briefly, ∼1 to 3 mg of each extract was incubated with 20 μl of glutathione-Sepharose beads in 300 μl of 1× phosphate-buffered saline (PBS) (10 mM Na2HPO4, pH 7.3, 1.8 mM KH2PO4, 140 mM NaCl, and 2.7 mM KCl) containing 10% glycerol and 0.1% Triton X-100. Following incubation at 25°C for 1 h, the beads were washed with 1 ml of the same buffer five times. Pulldown samples were analyzed by SDS-polyacrylamide gel electrophoresis, followed by staining with Coomassie brilliant blue R-250 or Western blotting.
ChIP.
Chromatin immunoprecipitation (ChIP) of TAP-tagged Cdc13 was carried out using the same procedure as described earlier for tagged Candida Rap1 (46).
Expression, purification, and crystallization of CgCdc13OB4.
CgCdc13OB4 was cloned into the pMST3 vector (a modified pET28b vector with the SUMO sequence cloned 3′ to the His6 tag [43]), and the resulting expression plasmid was transformed into E. coli BL21(DE3). After induction for 16 h with 0.1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) at 20°C, the cells were harvested by centrifugation and the pellets were resuspended in lysis buffer (50 mM Tris-HCl, pH 8.0, 50 mM NaH2PO4, 400 mM NaCl, 3 mM imidazole, 10% glycerol, 1 mM phenylmethylsulfonyl fluoride [PMSF], 0.1 mg/ml lysozyme, 2 mM 2-mercaptoethanol). The cells were then lysed by sonication and the cell debris was removed by ultracentrifugation. The supernatant was mixed with Ni-NTA agarose beads (Qiagen) and rocked for 2 h at 4°C before elution with 250 mM imidazole. Then, the Ulp1 protease was added, and the mixture was incubated for 12 h at 4°C to remove the His6-SUMO tag. CgCdc13OB4 was then further purified by passage through a Mono-Q ion-exchange column and by gel filtration chromatography on a Hiload Superdex75 (GE Healthcare) equilibrated with 25 mM Tris-HCl, pH 8.0, 150 mM NaCl, and 5 mM dithiothreitol (DTT). The purified CgCdc13OB4 was concentrated to 20 mg/ml and stored at −80°C.
Crystals of the wild-type protein were grown by the sitting-drop vapor diffusion method at 4°C. However, repeated attempts to obtain crystals of Se-Met-substituted wild-type CgCdc13OB4 were unsuccessful. Hence, several single Met-to-Leu point mutations of CgCdc13OB4 were evaluated for crystallization. Eventually, crystals of Se-Met-substituted M661L mutant protein were successfully grown at 4°C by the sitting-drop vapor diffusion method. The precipitant contained 32% PEG4000, 10 mM CaCl2, 0.1 M Tris-HCl, pH 7.4, and 0.2 M ammonium sulfate. Crystals were gradually transferred into a harvesting solution (0.2 M ammonium sulfate, 20% glycerol, 34% PEG 4000, 10 mM CaCl2, 0.1 M Tris-HCl, pH 7.4, and 10 mM DTT) before being flash-frozen in liquid nitrogen for storage and data collection under cryogenic conditions. A Se-Met single anomalous dispersion (SAD) (at Se peak wavelength) data set with a resolution of 2.0 Å was collected at beam line 21ID-D at APS and processed using HKL2000 (32). CgCdc13OB4 crystals belong to space group P21 and contain two CgCdc13OB4 molecules per asymmetric unit. Four selenium atoms were located and refined, and the SAD phases were calculated using SHARP (17). The initial SAD map was significantly improved by solvent flattening. A model was automatically built into the modified experimental electron density by using ARP/WARP (18). The model was then transferred into the native unit cell by rigid-body refinement and further refined using simulated-annealing and positional refinement in CNS (4), with manual rebuilding using program O (15). The final refined structure shows that Met661, located in the loop region between strands β2 and β3, is solvent exposed and makes no contributions to the dimer interface. Thus, the M661L mutation is unlikely to have any effect on protein folding, stability, or dimerization.
RESULTS
C. albicans Cdc13 localizes to telomeres and regulates telomere lengths in vivo.
In our previous report, we identified plausible homologues of each CST component in Candida and Saccharomyces genomes and investigated the functions and mechanisms of Candida albicans Stn1 and Ten1 proteins in telomere regulation. Unlike Stn1 and Ten1, C. albicans Cdc13 appears to be essential for cell viability, thus hampering analysis of its function (40). To ascertain a role for the putative CaCdc13 in telomere regulation, we first attempted to determine if the protein is associated with telomeres in vivo. To facilitate chromatin immunoprecipitation (ChIP), a TAP tag was fused to the C terminus of the single CaCDC13 allele in the heterozygote CaCDC13+/− strain background. Interestingly, several independently derived tagged strains were found to possess telomeres that were longer and more heterogeneously sized than those of the parental CaCDC13+/− strain (Fig. 1B). In addition, higher levels of extrachromosomal telomere circles (t-circles) were detected in several TAP-tagged strains (Fig. 1C and data not shown). Western analysis showed that the TAP-tagged CaCdc13 protein was well expressed (Fig. 1D). These observations are consistent with the idea that C-terminal tagging caused a partial loss of CaCDC13 function and that CaCdc13 suppresses abnormal telomere elongation and t-circle formation like CaStn1 and CaTen1. Nevertheless, the tagged strains exhibited normal growth, indicating that the CaCDC13-TAP allele can at least supply the essential function of the native gene.
We then analyzed the telomere association of CaCdc13-TAP by using ChIP with IgG-Sepharose, which interacts with the protein A epitope of the TAP tag. As shown in Fig. 1D, the CaCdc13-TAP protein in three independently generated, tagged strains exhibited significant cross-linking to telomeric DNA upon formaldehyde treatment, thus confirming the ability of Cdc13 to localize to telomeres in vivo. In contrast, no association of Cdc13 with the RPL11 promoter can be detected, suggesting that Cdc13 does not bind indiscriminately to all chromosomal locations (see Fig. S1 in the supplemental material). These results indicate that CaCdc13 is indeed a telomere-associated protein and argue that despite the absence of N-terminal domains, CaCdc13 acts directly at telomeres, possibly forming a CST complex with CaStn1 and CaTen1, which are known to be necessary for the maintenance of proper telomere lengths and structure (12, 19, 30, 40).
Telomere-specific DNA binding activity of C. tropicalis Cdc13.
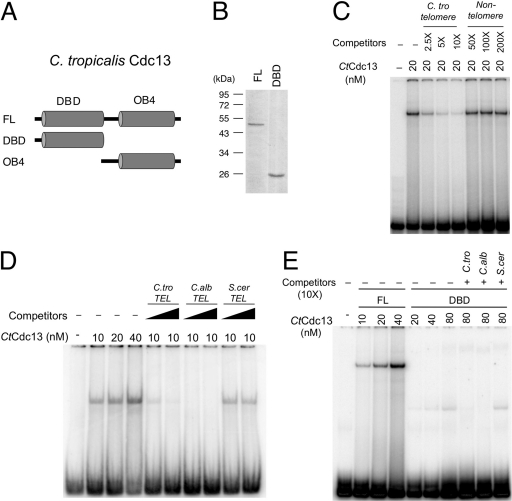
The remarkably high degree of telomere sequence divergence in the Candida clade raises an interesting question concerning the mechanisms of DNA recognition by Cdc13: how do highly homologous DNA-binding domains (i.e., the DBDs of Cdc13s) recognize such diverse sequence targets? To gain insights into the mechanisms of telomere DNA recognition, we attempted to characterize in detail the DNA-binding properties of small Cdc13s. Initial screening of protein expression and purification indicated that the Cdc13 protein from C. tropicalis, but not that from C. albicans, can be obtained in large quantities from E. coli in an active form. We therefore expressed and purified SUMO-fused CtCdc13 with a C-terminal FLAG tag in E. coli. After removal of the SUMO domain and further purification to near homogeneity, the full-length CtCdc13 protein was subjected to a series of electrophoretic mobility shift assays (EMSAs) to determine its DNA-binding affinity and sequence specificity (Fig. 2 and 3). For comparative purposes, the binding affinity and sequence specificity of the putative DBD of CtCdc13 were also determined. As expected, the full-length CtCdc13 protein binds to the C. tropicalis telomere repeats with high affinity (Kd [dissociation constant] of ∼40 nM) (Fig. 2D). The formation of the complex was concentration dependent, and all of the probes can be bound when sufficient amounts of proteins were added to the reaction (Fig. 2D; also see Fig. S2A in the supplemental material). DNA binding by CtCdc13 was also highly sequence specific, as revealed by a competition experiment; whereas an unlabeled telomeric competitor at a 2.5-fold molar excess substantially inhibited the formation of the labeled DNA-protein complex, a nontelomeric competitor had no effect even when present at a 200-fold molar excess (Fig. 2C). In addition, while the telomere repeats from both C. tropicalis and C. albicans (which differ from each other at 7/23 nucleotide positions) competed well in binding to CtCdc13, the purely GT repeat of the S. cerevisiae telomere sequence did not (Fig. 2D). These results indicate that CtCdc13 has a clear sequence preference for the Candida telomere repeats but that the recognition is not entirely species specific. Interestingly, the DBD of CtCdc13 exhibited the same sequence preference as that of the full-length protein but a significantly lower binding affinity (Fig. 2E; also see Fig. S2B) (Kd ≫ 320 nM), suggesting that the OB4 of CtCdc13 is important for DNA binding affinity but not for sequence specificity. We also analyzed the OB4 domain of CtCdc13 and found that this domain alone does not possess appreciable DNA-binding activity (data not shown). Thus, unlike the DBD of ScCdc13, which has an autonomous high-affinity telomere DNA-binding activity, the comparable domain of CtCdc13 does not, hinting at significant mechanistic differences (14, 20).
Fig 2.
Specific binding of Candida telomeric DNA by C. tropicalis [C.tro] Cdc13. (A) The C. tropicalis Cdc13 protein and the domains tested for DNA binding are illustrated. (B) Purified full-length (FL) CtCdc13 and the DBD were analyzed by SDS-PAGE and Coomassie staining. (C) CtCdc13 was incubated with 7.5 nM labeled C. tropicalis TEL-GX1.5B (see Fig. 3 for sequence), and the indicated competitor oligonucleotides (C. tro telomere, same as the probe; nontelomeric competitor,AATTGTCGACTTATGGAGCAATTCTTGTTAAACA). The resulting DNA-protein complexes were analyzed by native gel electrophoresis. The concentrations of CtCdc13 and the levels of the competitors relative to the probe for the reactions are listed at the top. (D) The indicated concentrations of full-length CtCdc13 were incubated with 7.5 nM probe consisting of two copies of the C. tropicalis telomere repeat (C.tro TEL-GX2). The resulting DNA-protein complexes were analyzed by native gel electrophoresis. The Kd for this DNA-protein interaction was estimated to be ∼40 nM based on the concentration of protein needed to reduce the free probe by 50% (four left lanes). Some assays also included excess unlabeled oligonucleotides consisting of various telomere repeat sequences. These competitor oligonucleotides were added at 2.5-fold or 10-fold molar excess. (E) The indicated concentrations of full-length CtCdc13 or DBD were incubated with 7.5 nM probe consisting of two copies of the C. tropicalis telomere repeat (C. tro TEL-GX2). The indicated competitor oligonucleotides were added at 10-fold molar excess. C.alb, C. albicans; S.cer, S. cerevisiae.
Fig 3.
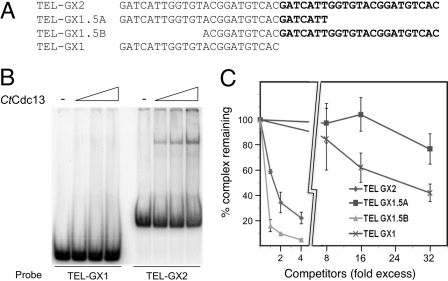
The effect of telomere DNA length on CtCdc13-DNA interaction. (A) The sequences of oligonucleotides used as probes or competitors in this series of assays are listed. (B) EMSAs were performed using increasing concentrations of CtCdc13 (0, 10, 20, and 40 nM) and either TEL-GX1 or TEL-GX2 as the probe. (C) Competition EMSAs were performed using 10 nM CtCdc13 and 7.5 nM labeled TEL-GX2. Increasing concentrations of four different competitor oligonucleotides were added, and the levels of DNA-protein complexes were quantified and plotted. Data are from three independent experiments.
Another recent survey revealed low-affinity DNA binding by the DBDs of C. albicans, C. parapsilosis, and C. glabrata Cdc13 homologues (23). To determine if other domains of these proteins might contribute to DNA binding (as was observed for CtCdc13), we attempted to examine the properties of full-length Cdc13s and the DBDs from these species. Thus far, we have been able to isolate only full-length CgCdc13 and its DBD (see Fig. S3A and B in the supplemental material). Interestingly, full-length CgCdc13 binds to the cognate telomere repeats with high affinity (Kd = ∼20 nM), whereas the DBD alone failed to form a stable complex with the same oligonucleotide (see Fig. S3C). In the DBD assays, broad smears were observed above the free probe, but few distinct bands could be detected, suggesting dissociation of the DBD-DNA complex during native gel electrophoresis. Hence, the CgCdc13 DBD alone appears to bind telomeric DNA but evidently requires other domains to form a stable complex. Like CtCdc13, DNA binding by the full-length CgCdc13 is highly sequence specific: in competition assays, >100-fold-higher concentrations of a nontelomeric oligonucleotide are needed to achieve the same degree of inhibition as with a telomeric oligonucleotide (unpublished data). We conclude that non-DNA-binding domains may modulate the DNA-binding properties of multiple Cdc13 homologues.
Characterization of the preferred DNA binding sites for CtCdc13.
In our initial assays, we used a probe (TEL-GX2) consisting of two C. tropicalis telomere repeat units. Remarkably, when a one-repeat probe (TEL-GX1) was tested in EMSA, no complex could be detected (Fig. 3A and B). Consistent with this finding, the one-repeat oligonucleotide was a much weaker competitor than the two-repeat oligonucleotide in competition assays (see Fig. S4 in the supplemental material). We tested multiple permutations of the one-repeat oligonucleotide as probes or competitors in EMSA and found that none of them exhibited strong binding to CtCdc13 (data not shown). Further EMSA analysis suggests that at least one extra copy of the right-half portion of the telomere repeat unit is required for high-affinity CtCdc13 binding (Fig. 3C). In particular, the TEL-GX1.5B oligonucleotide, which contains two iterations of the right half of the telomere repeat unit, binds CtCdc13 with an affinity similar to that of the TEL-GX2 oligonucleotide (Fig. 3C). By contrast, the TEL-GX1.5A oligonucleotide, which contains two iterations of the left half of the telomere repeat unit, exhibited a substantially reduced affinity.
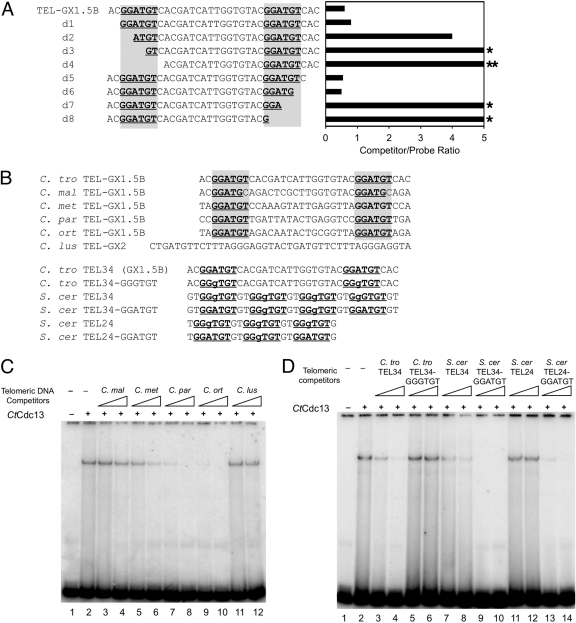
To investigate further the DNA length and sequence requirement for high-affinity binding by CtCdc13, we progressively removed 2 or 3 nucleotides (nt) from each end of TEL-GX1.5B, and tested the truncated oligonucleotides as competitors in the binding assays (Fig. 4A). As expected, large deletions from either end abrogated the ability of the oligonucleotide to serve as an efficient competitor (e.g., oligonucleotides d3, d4, d7, and d8). Interestingly, there are two iterations of a hexanucleotide element (GGATGT) in TEL-GX1.5B, and we frequently observed a significant reduction in binding affinity if this element was partially or completely deleted from either the 5′ or 3′ end (Fig. 4A). These results suggest that the GGATGT element may be a critical determinant of CtCdc13 binding and that two iterations of the element may be required for high-affinity binding. This notion is consistent with our finding that no permutation of a single telomere repeat unit can serve as an efficient probe or competitor (Fig. 3B and data not shown); all such permutations contain just one GGATGT element and hence would be expected to bind poorly to CtCdc13.
Fig 4.
The role of a 6-nucleotide element in CtCdc13-DNA interaction. (A) The listed oligonucleotides were used as competitors in EMSAs containing 10 nM CtCdc13 and 7.5 nM labeled TEL-GX2 DNA. The locations of the GGATGT sequence element required for high-affinity binding are highlighted. The amount of competitor oligonucleotide required to achieve a 50% reduction in complex formation was determined and plotted. Stars designate cases where a 4-fold excess competitor resulted in ∼30% and ∼0% reductions in complex formation, respectively. (B) The sequences of oligonucleotides used as competitors in EMSAs are displayed. The consensus GGATGT and the closely related GGgTGT elements are highlighted. (C) The indicated oligonucleotides (consisting of various Candida telomere repeats: C. maltosa [C. mal], C. metapsilosis [C. met], C. parapsilosis [C. par], C. orthopsilosis [C. ort], and C. lusitaniae [C. lus]) were used as competitors in EMSAs that contained 10 nM CtCdc13 and 7.5 nM labeled TEL-GX2 DNA. The competitors were added at levels 2.5- and 10-fold higher than that for the probe. (D) The indicated oligonucleotides (consisting of wild-type or mutated S. cerevisiae [S. cer] and C. tropicalis [C. tro] telomere repeats) were used as competitors in EMSAs that contained 10 nM CtCdc13 and 7.5 nM labeled TEL-GX2 DNA. The competitors were added at levels 2.5- and 10-fold higher than that for the probe.
To ascertain further the role of the GGATGT elements in promoting Cdc13 binding, we performed three sets of experiments. First, we tested the abilities of five heterologous Candida telomere oligonucleotides to act as competitors in EMSA (Fig. 4B and C). Three of the five oligonucleotides (C. metapsilosis, C. parapsilosis, and C. orthopsilosis) contain two iterations of the GGATGT elements. Consistent with the importance of this element, these three oligonucleotides acted as stronger competitors (Fig. 4C, compare lanes 3, 4, 11, and 12 with lanes 5 to 10). Second, we replaced two nucleotides in several S. cerevisiae oligonucleotides (which contain just G and T residues) to convert the closely related GGGTGT to GGATGT and tested the resulting oligonucleotides in competition assays (Fig. 4B and D). Remarkably, each of the converted oligonucleotides competed much more efficiently than the corresponding S. cerevisiae oligonucleotides, supporting a preference for the A nucleotide at the relevant position (Fig. 4D, lanes 7 to 14). However, we also noticed that the S. cerevisiae TEL34 oligonucleotide, which contains four GGGTGT elements, competed better than the S. cerevisiae TEL24 oligonucleotide, which contains three GGGTGT elements (Fig. 2C and 4D, compare lanes 7 and 8 with lanes 11 and 12). One possible explanation is that multiple copies of the GGGTGT sequence in an oligonucleotide can compensate for the lower affinity of the sequence element to allow significant binding. Finally, we converted the GGATGT elements in the C. tropicalis TEL34 oligonucleotide to GGGTGT and found the resulting oligonucleotide to have reduced affinity for CtCdc13 (Fig. 4D, lanes 3 to 6). Altogether, these observations argue strongly for the importance of the GGATGT element. However, it is also clear that other nucleotides outside of the consensus GGATGT elements modulate the affinity of CtCdc13-DNA interaction. For example, even though the C. orthopsilosis and C. metapsilosis oligonucleotides both have two GGATGT elements separated by 17 nucleotides, the former has a significantly higher affinity for CtCdc13 than the latter. Further studies will be necessary to define more completely the sequence preference of CtCdc13. Regardless of the outcomes of such studies, the requirement of an extended DNA target site (e.g., ∼30 nt of natural C. tropicalis telomere sequence [TEL-GX1.5B] and 24 nt of an artificial G-rich sequence S. cerevisiae TEL24-GGATGT]) for CtCdc13 clearly makes its DNA-binding properties distinct from those for ScCdc13, which recognizes a much shorter target (11 nt) (14).
The crystal structure of the Cdc13 OB4 dimer from C. glabrata.
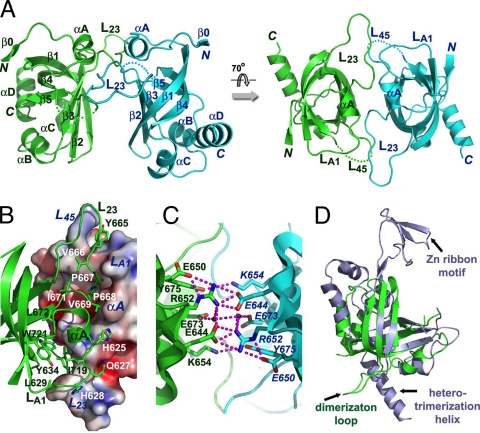
One way to account for the long DNA binding site (with duplicated consensus motif) and the involvement of the OB4 domain in CtCdc13-DNA interaction is to invoke OB4 dimerization. The binding of a dimeric Cdc13 complex to an extended and duplicated target site would be expected to enhance substantially the affinity of interaction. In support of this idea, the OB4 domains of CaCdc13 and CgCdc13 have been shown to self-associate in two-hybrid assays (39). However, the molecular basis of OB4 dimerization is unknown. In fact, even the notion that the C terminus of Cdc13 comprises an OB fold has not been experimentally verified. We therefore screened several Cdc13 OB4 domains for recombinant expression and crystallization. In the end, we were able to express and purify OB4 of CgCdc13 (residues 607 to 754) from E. coli and solved its crystal structure by single anomalous dispersion (SAD) method using Se-Met-substituted proteins at a resolution of 2.0 Å (Table 1). Indeed as predicted, the structure of CgCdc13OB4 is made of an OB fold with a slightly deformed central β-barrel sitting on a flat surface formed by three peripheral helices, αB, αC, and αD (Fig. 5A). Between strands β2 and β3, there is a long and extended loop, L23, which is essential for homodimerization of CgCdc13OB4 as described below (Fig. 5A). Given that the secondary structural elements of CgCdc13OB4 are among the most conserved regions revealed by sequence alignments (see Fig. S5 in the supplemental material), the crystal structure of CgCdc13OB4 supports the existence of a C-terminal OB fold in all Saccharomyces and Candida Cdc13 proteins.
Table 1.
Data collection, phasing, and refinement statistics
| Parametera | Valueb for CgCdc13OB4 |
|---|---|
| Data collection (Se peak) | |
| Space group | P21 |
| Cell dimensions | |
| a, b, c (Å) | 61.815, 38.745, 62.709 |
| a, b, g (°) | 90, 109.021, 90 |
| Wavelength (Å) | 0.97949 |
| Resolution (Å) | 100–2.0 |
| Rmerge | 0.064 (0.162) |
| I/σ | 53.7 (11.9) |
| Completeness (%) | 98.7 (92.3) |
| Redundancy | 7.2 (6.2) |
| Phasing | |
| Figure of merit (anomalous) | 0.26324 (acentric reflections); 0.10262 (centric reflections) |
| Phasing power (anomalous) | 1.781 |
| Refinement | |
| Resolution (Å) | 30–1.90 |
| No. of reflections | 22461 |
| Rwork/Rfree (%) | 0.2059/0.2382 |
| No. of atoms | |
| Protein | 2157 |
| Water | 132 |
| B-factors (Å2) | |
| Protein | 42.655 |
| Water | 46.244 |
| RMS deviations | |
| Bond lengths (Å) | 0.007 |
| Bond angles (°) | 1.075 |
Rmerge = Σ|I − <I>|/ΣI, where I is the observed intensity and <I> is the average intensity of multiple observations of symmetry-related reflections, where RMS is root mean square, phasing power = RMS (|FH|/E), |FH| is the heavy-atom structure factor amplitude, and E is the residual lack of closure error, figure of merit = <ΣP(α)eiα/ΣP(α)>, where α is the phase and P(α) is the phase probability distribution. R = Σ||Fo| − |Fc||/Σ|Fo|, where Fo and Fc are observed and calculated structure factor amplitudes, respectively. Rfree is calculated for a randomly chosen 5% of reflections; Rwork is calculated for the remaining 95% of reflections used for structure refinement.
Values in parentheses are for the highest-resolution shell.
Fig 5.
Structure of the C-terminal OB fold of C. glabrata Cdc13. (A) Ribbon diagram of two views of the CgCdc13OB4 dimer. The two subunits are colored in green and cyan, respectively. The secondary structural elements are labeled. The CgCdc13OB4 dimer at right is rotated by 70° about a horizontal axis relative to the dimer at left. (B) The hydrophobic dimer interface of CgCdc13OB4. One CgCdc13OB4 molecule is in surface representation and colored according to its electrostatic potential. The other molecule is in ribbon representation and colored in green. Side chains of residues in loops LA1, L23, and L45 important for dimerization are shown as stick models. (C) An extensive electrostatic interaction network is formed by a cluster of symmetry-related charged and polar residues on the β1-β2-β3 side of the barrel. The intermolecular hydrogen bonds are shown as dashed magenta lines. (D) Superposition of CgCdc13OB4 on the crystal structure of human RPA70C reveals that CgCdc13OB4 is not structurally similar to RPA70C (3). CgCdc13OB4 and RPA70C are colored in green and light blue, respectively. The superposition is based on the OB fold β-barrels of the proteins.
Consistent with previous two-hybrid and gel filtration chromatography results, there are two CgCdc13OB4 molecules in each asymmetric unit (39). The large solvent-accessible surface area buried by the dimer interface (∼2,420 Å) implies that CgCdc13OB4 exists as a dimer in solution prior to crystallization. The mode of dimerization is entirely distinct from that observed for ScCdc13OB1; whereas the symmetry dyad is perpendicular to the axis of the β-barrel and the two protomers are arranged end to end for ScCdc13OB1, the symmetry dyad is parallel to the axis of the β-barrel and the two protomers are arranged side to side for CgCdc13OB4 (Fig. 5A). The major driving force for dimer formation of CgCdc13OB4 is provided by hydrophobic contacts mediated by three connecting loops (Fig. 5A). Five resides in loop L23 (665YVPPV669) bind into a hydrophobic cleft formed by two loops, LA1 (between αA and β1) and L45 (between β4 and β5), from the other subunit in the dimer (Fig. 5B). In particular, Pro667 and Pro668 of one CgCdc13OB4 fit snugly into a complementary surface of the other molecule (Fig. 5B). In addition to these hydrophobic contacts, there is another interface involving a cluster of charged and polar residues (Glu644, Glu650, Arg652, Lys654, Glu673, and Tyr675) from strands β1, β2, and β3 of each subunit (Fig. 5C). Together with two ordered water molecules, these residues form an extensive and symmetric electrostatic interaction network with a total of 18 salt bridges and hydrogen bonds.
As described in the introduction, even though the Stn1-Ten1 subcomplex is clearly paralogous to RPA32-RPA14, the relationship between Cdc13 and RPA70 has remained unclear. Notably, RPA70 also contains a C-terminal OB fold (RPA70C) (39). Hence, we compared the structures of CgCdc13OB4 and RPA70C in order to glimpse their evolutionary relationship. Three-dimensional superposition analysis revealed several marked differences between the two domains outside the central β-barrel core, arguing against a close evolutionary kinship (Fig. 5D). RPA70C does not contain a long loop between strands β2 and β3 that is crucial for the dimerization of CgCdc13OB4 (Fig. 5D). On the other hand, CgCdc13OB4 lacks several features unique to RPA70C. First, RPA70C contains a zinc ribbon motif embedded in the OB fold between strands β1 and β2, which might play a role in single-stranded DNA binding (Fig. 5D). In contrast, strands β1 and β2 in CgCdc13OB4 are connected by a short two-residue loop. Second, the C-terminal helix in RPA70C protrudes away from the β-barrel core to interact with the other two components of the RPA complex, RPA32 and RPA14, through an intermolecular three-helix bundle (3). In contrast, the C-terminal helix of CgCdc13OB4, αD, is short and packs together with helices αB and αC (Fig. 5A). Hence, it is unlikely that CgCdc13OB4 interacts with Stn1 and Ten1 in the same manner as RPA70 does with its binding partners. Therefore, our comparative structural analysis does not support the idea of a common ancestry for RPA70 and Cdc13.
The dimerization of the OB4 domain in CtCdc13.
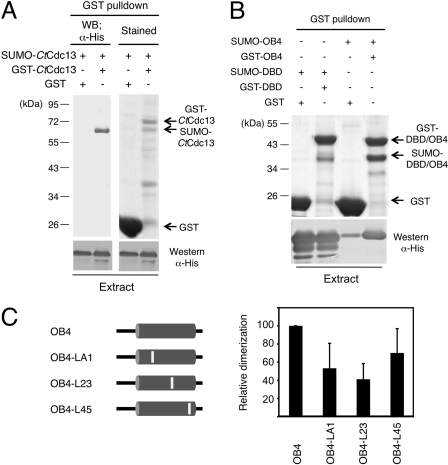
We next attempted to apply the insights derived from the CgCdc13OB4 dimer structure to the analysis of CtCdc13. First, we investigated the ability of CtCdc13 to form dimers. A SUMO-fused CtCdc13 with a His6 tag (SUMO-CtCdc13) and a GST-fused CtCdc13 (GST-CtCdc13) were coexpressed in E. coli. Cell extracts were prepared and subjected to pulldown assays using glutathione-Sepharose beads. As shown in Fig. 6A, GST-CtCdc13 but not GST alone can coprecipitate approximately equal amounts of SUMO-CtCdc13, supporting self-association. Additional pulldown assays using either the CtCdc13DBD or CtCdc13OB4 domain fusions revealed a much stronger self-association of the OB4 domain, suggesting that this domain is largely responsible for dimerization (Fig. 6B). Interestingly, the DBD also appears to be capable of self-association, at least when overproduced in E. coli. The physiologic relevance of this much weaker interaction remains to be determined.
Fig 6.
Self-association of CtCdc13. (A) (Top) The indicated proteins were coexpressed and subjected to GST pulldown analysis. The bound proteins were analyzed by SDS-PAGE and either Coomassie staining or Western blotting (WB) using anti-His tag antibodies. (Bottom) The levels of His-tagged SUMO-Cdc13 protein in the input extracts were analyzed by Western blotting. (B) (Top) The indicated proteins were coexpressed and subjected to GST pulldown analysis. The glutathione-Sepharose-bound proteins were analyzed by SDS-PAGE and Coomassie staining. (Bottom) The levels of His-tagged SUMO fusion proteins (SUMO-DBD or SUMO-OB4) in the input extracts were analyzed by Western blotting. (C) GST pulldown assays were performed using either wild-type or mutated OB4 domains fused to the GST and SUMO tags. The ratio of the SUMO fusion to GST fusion protein in each precipitated sample was quantified, normalized to the wild-type sample, and then plotted. The results are from three independent experiments.
We then attempted to identify dimerization-defective mutants of CtCdc13OB4 by using the structure of CgCdc13OB4 and a multiple sequence alignment of Cdc13 homologues as the guides. As described earlier, three connecting loops in CgCdc13OB4 (named LA1, L23, and L45) are largely responsible for forming the dimer interface. Notably, these loop residues are not well conserved in the Saccharomyces and Candida Cdc13 homologues (see Fig. S5 in the supplemental material). Nevertheless, we reasoned that divergent sequences may be compatible with dimerization and proceeded to replace multiple amino acid residues in each corresponding loop in CtCdc13OB4 to generate the LA1 (SISE234-238), L23 (TILDDR295-300), and L45 (KQKI358-361) mutants (see Fig. S5). The abilities of the mutated OB4 domains to self-associate were then tested in pulldown assays (Fig. 6C). As predicted, each mutant exhibited a significant reduction in self-association, with the LA1 and L23 mutant manifesting defects more severe (∼50–65% reduction) than those of the L45 mutant (∼30% reduction). Hence, despite the clear sequence differences between the loops of the CgCdc13 and CtCdc13 OB4 domains, these loops appear to mediate a conserved function in protein dimerization.
The role of dimerization on DNA binding by CtCdc13.
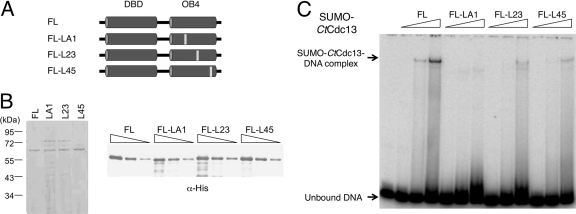
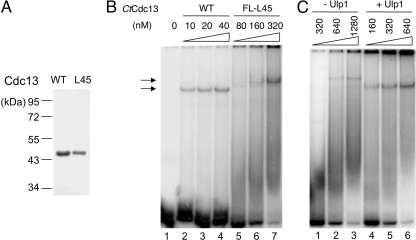
To investigate the role of dimerization on the DNA binding activity of Cdc13, we expressed full-length SUMO-tagged CtCdc13 proteins carrying the LA1, L23, and L45 mutations in E. coli (Fig. 7A). Because the LA1 and L23 mutants were expressed at relatively low levels and difficult to purify after ULP1 cleavage, we compared the DNA-binding properties of the SUMO-tagged proteins (Fig. 7B and C). Notably, all three mutant proteins exhibited reduced affinity for the C. tropicalis telomere repeats in comparison to the wild-type protein, suggesting that dimerization contributes to DNA (Fig. 7C). Because the L45 mutant is expressed at higher levels and can be purified in substantial quantities in the untagged form, we also performed a more detailed comparison between this mutant and wild-type protein following ULP1 cleavage and further purification (Fig. 8A and B). Interestingly, the L45 mutant evidently retained significant DNA-binding activity, as evidenced by decreasing signals for the free probe when substantial amounts of the protein were added to the binding reactions. However, a higher concentration of the mutant was needed to form the same level of complex as the wild-type protein. Moreover, a broad smear can be observed below the mutant protein-DNA complex, suggesting a significant dissociation of the complex during native gel electrophoresis (Fig. 8B). These observations support the notion that the L45 mutant binds telomeric DNA with reduced affinity and stability. Curiously, the presumptive L45 mutant-DNA complex has a reduced mobility in comparison to the wild-type complex, raising questions about its identity (Fig. 8B, compare lanes 2 to 4 and lanes 5 to 7). However, we observed a clear mobility difference between the protein-DNA complex formed by the SUMO-tagged mutant and that formed by the untagged mutant, indicating that the observed complexes were due to CtCdc13 rather than a contaminant (Fig. 8C). The altered mobility of the DNA-CtCdc13-L45 complex may be due to an altered conformation of the protein dimer.
Fig 7.
The effects of OB4 mutations on DNA binding by CtCdc13. (A) The locations of the OB4 mutations in the full-length CtCdc13 are illustrated. (B) (Left) Purified SUMO-CtCdc13 mutant proteins bearing substitutions in the OB4 domain were analyzed by SDS-PAGE and Coomassie staining. (Right) Serial dilutions of the purified protein preparations were analyzed by SDS-PAGE and Western blotting using anti-His antibodies. (C) EMSAs were performed using increasing concentrations (116, 232, and 464 nM) of SUMO-fused wild-type and mutated CtCdc13s bearing amino acid replacements in the OB4 domain and the TEL-GX1.5B probe (7.5 nM). FL, full length.
Fig 8.
DNA binding by the CtCdc13-L45 mutant. (A) The purified CtCdc13 and CtCdc13-L45 (without the SUMO tag) were analyzed by SDS-PAGE and Coomassie staining. (B) The indicated concentrations of wild-type and L45 mutant proteins were tested for binding to the TEL-GX2 probe. (C) The SUMO-CtCdc13-L45 fusion protein was incubated in the absence or presence of ULP1 protease, and then tested for binding to the TEL-GX2 probe. WT, wild type.
DISCUSSION
We have shown that the unusually small Cdc13 homologues in Candida species are indeed regulators of telomere lengths and thus orthologous to the prototypical Cdc13 first identified and characterized in S. cerevisiae. We further demonstrated that the small Cdc13s likely form dimers through a homotypic interaction between the OB4 domain and that this dimerization increases the affinity of Cdc13s for the Candida telomere repeats and enables the proteins to perform their telomere-dedicated functions. Our determination of the high-resolution structure of CgCdc13OB4 also underscored the remarkable versatility of OB fold domains in mediating protein-protein interactions. The evolutionary and mechanistic implications of these findings are discussed below.
Candida Cdc13s serve telomere-specific functions.
Our detailed analysis of the DNA-binding properties of CtCdc13 suggests that this protein has sufficient affinity and sequence specificity to interact with Candida telomeres in vivo and perform telomere-specific functions. This conclusion is supported by ChIP analysis of CaCdc13, which revealed telomere localization of this small Cdc13 in vivo. However, it is at odds with a recent report that posits a more general function for small Cdc13s in chromosome transactions (23). This alternative proposition was based on analyses of the DNA-binding properties of the DBDs from C. albicans, C. parapsilosis, and C. glabrata. All three DBDs exhibited low affinity (ranging from ∼100 to 600 nM) and sequence specificity for short telomeric oligonucleotides, leading the investigators to discount a telomere-specific function. Our results on CtCdc13 and CgCdc13 suggest that dimerization-assisted DNA binding may be quite prevalent among Cdc13 homologues and that the DNA-binding properties of the DBDs alone do not always reflect those of the full-length proteins.
The propensity of telomere proteins to dimerize.
A striking implication of the current report, when juxtaposed against previous findings, is that Cdc13 homologues have a propensity to dimerize and have evolved different modes of dimerization. As described earlier, whereas Saccharomyces and Kluyveromyces Cdc13s form dimers through their OB1 domains, Candida Cdc13s use the structurally quite distinct OB4 domains to mediate dimerization (26, 39). How can the distinct modes of dimerization evolve so readily for Cdc13 (and other telomere proteins such as TRF1, TRF2, and Taz1)? An attractive hypothesis invokes the colocalization of multiple molecules of a telomere-binding protein on the iterative telomere sequence (16, 21). The clustering of a protein greatly increases its local concentration and amplifies the effect of mutations on protein-protein interactions. In this setting, even a low free energy of interaction conferred by a few point mutations may lead to a substantial increase in the fraction of molecules that bind to each other, which may in turn enhance telomere protection sufficiently to allow for selection.
Another notable implication of the combined observations on Saccharomyces and Candida Cdc13 dimerization is that dimerization can serve different purposes in different organisms. In particular, dimerization of ScCdc13 is not required for high-affinity DNA binding; the ScCdc13OBDBD domain alone interacts with an 11-nt telomere oligonucleotide with a Kd in the picomolar range. Rather, dimerization of ScCdc13 has been shown to modulate its interactions with Pol1 and to regulate telomere lengths through additional mechanisms (39). Why then is dimerization of small Cdc13s necessary for high-affinity DNA binding? The answer to this puzzle may reside in the extraordinary telomere sequence divergence exhibited by Candida species (25). This sequence divergence presents a considerable challenge to Cdc13: to evolve suitable affinity and specificity for the different telomere repeats during a short evolutionary time span. However, the OBDBD domains of Candida Cdc13s align well with the corresponding domain in ScCdc13, and many of the residues implicated in ScCdc13-DNA interactions are conserved in the Candida proteins (data not shown) (1). Furthermore, phylogenetic analysis does not yield evidence of more-rapid evolution of OBDBD relative to OB4 of Candida Cdc13s (data not shown). Thus, instead of evolving unique recognition specificity for each telomere repeat, the Candida Cdc13s may have largely retained a universal preference for GT-rich sequence elements within the divergent repeats and use the duplicated binding domains in the dimeric protein complex to enhance binding affinity. Indeed, our preliminary analysis suggests that the GGATGT element, which is shared by many repeats and duplicated in the minimal high-affinity site, may be the key determinant of binding for CtCdc13 (Fig. 4). Investigation of the recognition specificities of other Cdc13s will be necessary to confirm or disprove the validity of our hypothesis. Regardless of the potential outcomes, comparative analysis of Candida Cdc13-DNA interactions promises to provide a useful paradigm for understanding the coevolution of DNA-binding proteins and their target sequences.
The versatility of OB fold domains in mediating protein-protein interactions.
Even though the OB fold domain was initially defined as an oligonucleotide/oligosaccharide-binding module, more-recent studies have highlighted the remarkable functional diversity of this protein fold and the myriad ways in which this fold can mediate protein-protein interactions (2, 39, 44). In keeping with this theme, our high-resolution structures of the ScCdc13OB1 dimer and the CgCdc13OB4 dimer revealed dramatically distinct modes of dimerization. In the case of OB1, the two protomers are arranged end to end, and the symmetry dyad is perpendicular to the axis of the β-barrel. By contrast, the CgCdc13OB4 dimer involves a 2-fold symmetry axis that runs parallel to the β-barrel axis and a side-to-side dimerization interface (Fig. 6A). It is also worth noting that despite our success in identifying dimerization mutants of CtCdc13OB4, the residues implicated in CgCdc13OB4 and CtCdc13OB4 dimerization are in fact not well conserved in other homologues (see Fig. S5 in the supplemental material). Hence, dramatically different sequences in the connecting loops of the Cdc13 OB4 domain are compatible with dimerization, making it extremely challenging to infer this biochemical property based on sequence analysis alone. It is tempting to speculate that the repeated utilization of OB fold domains in proteins associated with single-stranded telomeres may be due not only to its nucleic acid binding activity but also to its versatility in binding protein partners.
The evolutionary relationship between CST and RPA.
As described before, whereas there are compelling supports for structural and functional similarities between Stn1-Ten1 and RPA32-RPA14, the relationship between Cdc13 and RPA70 has remained unclear. Our results provide additional arguments against a close evolutionary kinship between Cdc13 and RPA70. Specifically, we showed that the last OB fold of Cdc13 does not resemble the corresponding domain in RPA70. Coupled with previous crystallographic and NMR analyses, we now have high-resolution structures of three domains in Cdc13, each of which proved to be quite different from its putative RPA70 counterpart. Thus, CDC13 may not have arisen through a duplication of the RPA70 gene and then undergone functional specialization. Rather, Cdc13 may have originated independently from a different OB fold-containing protein and been recruited later to the Stn1-Ten1 complex to enhance its function. This notion is supported by the apparent absence of a Cdc13 homologue in S. pombe, as well as the very disparate sizes and structures of mammalian and plant CTC1s, which are presumed functional equivalents of Cdc13 in these organisms (28, 41). Further analyses of Cdc13 and other large CST subunits should provide insights on the evolutionary origin and mechanistic diversity of these proteins.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by NIH grants (GM069507 to N.L. and GM083015-01 to M.L.), an American Cancer Society Research Scholar grant (to M.L.), and a STARR Cancer Consortium grant (to N.L.). M.L. is a Howard Hughes Medical Institute Early Career Scientist. The General Medicine and Cancer Institutes Collaborative Access Team has been funded in whole or in part with federal funds from the National Cancer Institute (grant Y1-CO-1020) and the National Institute of General Medical Science (grant Y1-GM-1104). Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under contract no. DE-AC02-06CH11357.
Footnotes
Published ahead of print 24 October 2011
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1. Anderson EM, Halsey WA, Wuttke DS. 2003. Site-directed mutagenesis reveals the thermodynamic requirements for single-stranded DNA recognition by the telomere-binding protein Cdc13. Biochemistry 42:3751–3758 [DOI] [PubMed] [Google Scholar]
- 2. Bochkarev A, Bochkareva E. 2004. From RPA to BRCA2: lessons from single-stranded DNA binding by the OB-fold. Curr. Opin. Struct. Biol. 14:36–42 [DOI] [PubMed] [Google Scholar]
- 3. Bochkareva E, Korolev S, Lees-Miller SP, Bochkarev A. 2002. Structure of the RPA trimerization core and its role in the multistep DNA-binding mechanism of RPA. EMBO J. 21:1855–1863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brunger AT, et al. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D. Biol. Crystallogr. 54:905–921 [DOI] [PubMed] [Google Scholar]
- 5. Chan A, Boule JB, Zakian VA. 2008. Two pathways recruit telomerase to Saccharomyces cerevisiae telomeres. PLoS Genet. 4:e1000236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. de Lange T. 2009. How telomeres solve the end-protection problem. Science 326:948–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Enloe B, Diamond A, Mitchell A. 2000. A single-transformation gene function test in diploid Candida albicans. J. Bacteriol. 182:5730–5736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fonzi W, Irwin M. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134:717–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gao H, Cervantes RB, Mandell EK, Otero JH, Lundblad V. 2007. RPA-like proteins mediate yeast telomere function. Nat. Struct. Mol. Biol. 14:208–214 [DOI] [PubMed] [Google Scholar]
- 10. Gelinas AD, et al. 2009. Telomere capping proteins are structurally related to RPA with an additional telomere-specific domain. Proc. Natl. Acad. Sci. U. S. A. 106:19298–19303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gerami-Nejad M, Berman J, Gale CA. 2001. Cassettes for PCR-mediated construction of green, yellow, and cyan fluorescent protein fusions in Candida albicans. Yeast 18:859–864 [DOI] [PubMed] [Google Scholar]
- 12. Giraud-Panis MJ, Teixeira MT, Geli V, Gilson E. 2010. CST meets shelterin to keep telomeres in check. Mol. Cell 39:665–676 [DOI] [PubMed] [Google Scholar]
- 13. Grossi S, Puglisi A, Dmitriev PV, Lopes M, Shore D. 2004. Pol12, the B subunit of DNA polymerase alpha, functions in both telomere capping and length regulation. Genes Dev. 18:992–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hughes TR, Weilbaecher RG, Walterscheid M, Lundblad V. 2000. Identification of the single-strand telomeric DNA binding domain of the Saccharomyces cerevisiae Cdc13 protein. Proc. Natl. Acad. Sci. U. S. A. 97:6457–6462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jones TA, Zou JY, Cowan SW, Kjeldgaard M. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47:110–119 [DOI] [PubMed] [Google Scholar]
- 16. Kuriyan J, Eisenberg D. 2007. The origin of protein interactions and allostery in colocalization. Nature 450:983–990 [DOI] [PubMed] [Google Scholar]
- 17. La Fortelle ED, Bricogne G. 1997. Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Methods Enzymol. 276:472–494 [DOI] [PubMed] [Google Scholar]
- 18. Lamzin VS, Perrakis A, Wilson KS. 2001. The ARP/WARP suite for automated construction and refinement of protein models, p 720–722 In Rossmann MG, Arnold E. (ed), International tables for crystallography, vol F. Crystallography of biological macromolecules. Kluwer Academic Publishers, Dordrecht, The Netherlands [Google Scholar]
- 19. Lin J-J, Zakian VA. 1996. The Saccharomyces CDC13 protein is a single-strand TG1-3 telomeric DNA binding protein in vitro that affects telomere behavior in vivo. Proc. Natl. Acad. Sci. U. S. A. 93:13760–13765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lin YC, Hsu CL, Shih JW, Lin JJ. 2001. Specific binding of single-stranded telomeric DNA by Cdc13p of Saccharomyces cerevisiae. J. Biol. Chem. 276:24588–24593 [DOI] [PubMed] [Google Scholar]
- 21. Lue NF. 2010. Plasticity of telomere maintenance mechanisms in yeast. Trends Biochem. Sci. 35:8–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lundblad V. 2006. Budding yeast telomeres, p 345–386 In de Lange T, Lundblad V, Blackburn E. (ed), Telomeres and telomerase, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 23. Mandell EK, Gelinas AD, Wuttke DS, Lundblad V. 2011. Sequence-specific binding to telomeric DNA is not a conserved property of the Cdc13 DNA binding domain. Biochemistry 50:6289–6291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Martin V, Du LL, Rozenzhak S, Russell P. 2007. Protection of telomeres by a conserved Stn1-Ten1 complex. Proc. Natl. Acad. Sci. U. S. A. 104:14038–14043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. McEachern MJ, Blackburn EH. 1994. A conserved sequence motif within the exceptionally diverse telomeric sequences of budding yeasts. Proc. Natl. Acad. Sci. U. S. A. 91:3453–3457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mitchell MT, et al. 2010. Cdc13 N-terminal dimerization, DNA binding, and telomere length regulation. Mol. Cell. Biol. 30:5325–5334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mitton-Fry RM, Anderson EM, Theobald DL, Glustrom LW, Wuttke DS. 2004. Structural basis for telomeric single-stranded DNA recognition by yeast Cdc13. J. Mol. Biol. 338:241–255 [DOI] [PubMed] [Google Scholar]
- 28. Miyake Y, et al. 2009. RPA-like mammalian Ctc1-Stn1-Ten1 complex binds to single-stranded DNA and protects telomeres independently of the Pot1 pathway. Mol. Cell 36:193–206 [DOI] [PubMed] [Google Scholar]
- 29. Miyoshi T, Kanoh J, Saito M, Ishikawa F. 2008. Fission yeast Pot1-Tpp1 protects telomeres and regulates telomere length. Science 320:1341–1344 [DOI] [PubMed] [Google Scholar]
- 30. Nugent CI, Hughes TR, Lue NF, Lundblad V. 1996. Cdc13p: a single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science 274:249–252 [DOI] [PubMed] [Google Scholar]
- 31. O'Sullivan RJ, Karlseder J. 2010. Telomeres: protecting chromosomes against genome instability. Nat. Rev. Mol. Cell Biol. 11:171–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Otwinowski Z, Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307–326 [DOI] [PubMed] [Google Scholar]
- 33. Palm W, de Lange T. 2008. How shelterin protects mammalian telomeres. Annu. Rev. Genet. 42:301–334 [DOI] [PubMed] [Google Scholar]
- 34. Paschini M, Mandell EK, Lundblad V. 2010. Structure prediction-driven genetics in Saccharomyces cerevisiae identifies an interface between the t-RPA proteins Stn1 and Ten1. Genetics 185:11–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pennock E, Buckley K, Lundblad V. 2001. Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell 104:387–396 [DOI] [PubMed] [Google Scholar]
- 36. Price CM, Cech TR. 1987. Telomeric DNA-protein interactions of Oxytricha macronuclear DNA. Genes Dev. 1:783–793 [DOI] [PubMed] [Google Scholar]
- 37. Rigaut G, et al. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030–1032 [DOI] [PubMed] [Google Scholar]
- 38. Santos M, Keith G, Tuite M. 1993. Non-standard translational events in Candida albicans mediated by an unusual seryl-tRNA with a 5′-CAG-3′ (leucine) anticodon. EMBO J. 12:607–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sun J, et al. 2011. Structural bases of dimerization of yeast telomere protein Cdc13 and its interaction with the catalytic subunit of DNA polymerase alpha. Cell Res. 21:258–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sun J, et al. 2009. Stn1-Ten1 is an Rpa2-Rpa3-like complex at telomeres. Genes Dev. 23:2900–2914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Surovtseva YV, et al. 2009. Conserved telomere maintenance component 1 interacts with STN1 and maintains chromosome ends in higher eukaryotes. Mol. Cell 36:207–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Teixeira MT, Gilson E. 2005. Telomere maintenance, function and evolution: the yeast paradigm. Chromosome Res. 13:535–548 [DOI] [PubMed] [Google Scholar]
- 43. Wang F, et al. 2007. The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature 445:506–510 [DOI] [PubMed] [Google Scholar]
- 44. Wang F, et al. 2010. Crystal structures of RMI1 and RMI2, two OB-fold regulatory subunits of the BLM complex. Structure 18:1159–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wilson RB, Davis D, Mitchell AP. 1999. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J. Bacteriol. 181:1868–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yu EY, Yen WF, Steinberg-Neifach O, Lue NF. 2010. Rap1 in Candida albicans: an unusual structural organization and a critical function in suppressing telomere recombination. Mol. Cell. Biol. 30:1254–1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.