Abstract
Genes encoding vanillin dehydrogenase (vdh) and vanillate O-demethylase (vanAB) were identified in Rhodococcus jostii RHA1 using gene disruption and enzyme activities. During growth on vanillin or vanillate, vanA was highly upregulated while vdh was not. This study contributes to our understanding of lignin degradation by RHA1 and other actinomycetes.
TEXT
Rhodococcus is a genus of catabolically versatile soil bacteria that belong to a mycolic acid-containing suborder of actinobacteria (18). The ability of rhodococci to transform a wide range of organic compounds and pollutants, combined with their robust growth and exceptional stress tolerance, has led to their use in a wide range of biotechnological applications (17). Rhodococcus jostii RHA1 was isolated from lindane-contaminated soil and was initially characterized for its potent polychlorinated biphenyl (PCB)-degrading properties (15). Subsequent genomic studies of RHA1 have provided important insights into the physiology and catabolic versatility of rhodococci and related actinobacteria (7, 18).
RHA1 was shown to transform lignin, the second-most-abundant polymer in the biosphere, producing a number of monocyclic phenolic compounds (2). This is consistent with RHA1's ability to degrade a wide range of such aromatic compounds (7, 18). Molecular genetic and biochemical studies have demonstrated that this ability to degrade lignin depends on DypB, one of two heme-containing dye-decolorizing peroxidases (DyPs) that RHA1 harbors (1). The bacterial degradation of lignin has significant potential in the transformation of biomass for a range of product streams, including cellulose-based biofuels, high-valued aromatic compounds, resins, and carbon fibers (4). One degradation product of lignin is vanillin, widely used in the food and fragrance industries. As part of our efforts to better understand the lignin-transforming capabilities of RHA1, we investigated the bacterium's vanillin degradation pathway.
Growth experiments.
RHA1 grew at rates of 0.041 absorbance unit (AU) h−1 and 0.035 AU h−1 on 1 mM vanillate and 1 mM vanillin, respectively, as the sole organic substrate in liquid M9 mineral media supplemented with Goodies (3) at 30°C. The lag phase on each of these liquid media was about 2 and 4 days, respectively. The growth of RHA1 was inhibited at higher concentrations of vanillin, but not vanillate, and was not detected in 10 mM vanillin. The bacterium did not grow on either benzaldehyde (in vapor phase) or o-vanillin. The presence of benzaldehyde in vapor phase also inhibited the growth of RHA1 on 1 mM vanillin and 3 mM pyruvate, suggesting that aromatic aldehydes may be toxic to RHA1.
Identification of putative vanillin catabolic genes.
Two enzymes involved in the bacterial catabolism of vanillin and vanillate have been previously identified: vanillin dehydrogenase transforms vanillin to vanillate in an NAD+-dependent fashion (12), and vanillate O-demethylase is a two-component Rieske oxygenase that transforms vanillate to protocatechuate (12). The oxygenase and reductase components of the vanillate O-demethylase are encoded by vanA and vanB, respectively.
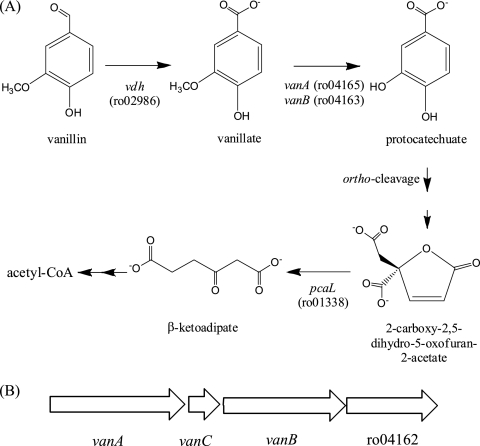
BLAST searches of the RHA1 genome revealed two vanillin dehydrogenase homologs, the predicted products of ro02986 (GI:111019975) and ro02797 (GI:111019788), which share 81% and 49% amino acid sequence identity, respectively, with vanillin dehydrogenase from Pseudomonas sp. strain HR199 (GI:1946288) (12). In contrast, only a single vanA homolog was identified, ro04165 (GI:111021144). The predicted product of this gene shares 35% amino acid sequence identity and 52% similarity with the vanillate O-demethylase oxygenase from Pseudomonas sp. strain HR199 (GI:3915234) (12). A vanB homolog, ro04163 (GI:111021142), occurs just downstream of vanA, whose predicted gene product shares 43% amino acid sequence identity with the vanillate O-demethylase reductase from Pseudomonas sp. strain HR199 (GI:3915246) (12). A second vanB homolog, ro02845 (GI:111019836), occurs elsewhere in the genome and shares 41% amino acid sequence identity with the reductase from Pseudomonas sp. strain HR199. In RHA1, vanA and vanB are separated by ro04164, which is predicted to encode a protein of 76 amino acid residues and annotated here as vanC. These three genes are arranged in a putative transcriptional unit under the control of a predicted IclR family transcriptional regulator encoded by ro04162 (Fig. 1B).
Fig 1.
(A) Vanillin catabolic pathway in RHA1. (B) Vanillate O-demethylase gene cluster. The vanA and vanB genes encode the oxygenase and reductase components, respectively, of vanillate O-demethylase. Gene ro04162 is predicted to encode an IclR family transcriptional regulator. Acetyl-CoA, acetyl coenzyme A.
Expression of vdh and van genes in RHA1.
We employed reverse transcription-quantitative PCR (RT-qPCR) to evaluate the expression of the putative vanillin catabolic genes during growth of RHA1 on each of 1 mM vanillin, 1 mM vanillate, and 3 mM pyruvate. Cells were grown in 125-ml liquid cultures to mid-log phase (optical density at 600 nm [OD600], ∼0.12), treated with “stop solution” and RNAprotect (Qiagen), and stored at −80°C, as described previously (16). RNA was extracted essentially as previously described (5). One microgram of purified RNA and 4 μl of qScript cDNA SuperMix (Quanta Bioscience, Gaithersburg, MD) in a total volume of 20 μl were used to synthesize cDNA according to the manufacturer's instructions. qPCR was performed using an Mx3000P real-time PCR system (Stratagene, La Jolla, CA). Twenty-microliter reaction mixtures contained 10 μl PerfeCTa qPCR FastMix, UNG, Low ROX (Quanta Bioscience, Gaithersburg, MD), 50 ng cDNA template, 10 nM probe, and 40 nM concentrations of each primer (forward and reverse). Thermocycling involved 2 min at 50°C, 3 min at 95°C, and 45 cycles in which each cycle consisted of 30 s at 95°C and 1 min 30 s at 65°C. The primers and TaqMan probes that were used are listed in Table S1 in the supplemental material. The relative levels of each gene transcript, R, were calculated from the crossing threshold of the qPCR, Cq, by using the equation R = 2−ΔΔCq. Briefly, the crossing threshold of the target gene, Cqtarget, was first normalized using the crossing threshold of the gene encoding DNA polymerase IV, CqpolIV, by using the equation ΔCq = Cqtarget − CqpolIV. The level of this gene had been previously identified as being relatively invariant across growth conditions (5). The normalized ΔCq of samples from cells grown on the carbon source of interest was then compared with that of samples from cells grown on pyruvate using the equation ΔΔCq = ΔCqsample − ΔCqpyr.
Among the tested genes, the vdh homolog ro02797 was most highly upregulated on vanillin and vanillate (Table 1). Somewhat unexpectedly, ro02986 was not detectably upregulated despite its higher sequence identity with a characterized vanillin dehydrogenase gene. The vanA gene was also upregulated on vanillin (210-fold) and vanillate (1,200-fold). In contrast, the second vanB homolog, ro02845, was not significantly upregulated during growth on either vanillin or vanillate. Finally, pcaL, which encodes a bifunctional β-ketoadipate enol-lactone hydrolase and γ-carboxymuconolactone decarboxylase in the β-ketoadipate pathway of RHA1 (10), was slightly upregulated during growth on vanillin or vanillate compared to its growth on pyruvate (Table 1). The lesser upregulation of pcaL likely reflects the constitutive expression of this gene in pyruvate-grown cells, as indicated by a previous proteomic study (10). Indeed, the degree of upregulation of pcaL agrees well with the finding that the protein was ∼5-fold more abundant in phthalate- and benzoate-grown cells than pyruvate-grown cells.
Table 1.
Transcriptional analysis of vanillin catabolic genes in strain RHA1a
| ORFb | Gene namec | Upregulation (fold) after growth on: |
|
|---|---|---|---|
| Vanillin | Vanillate | ||
| ro01338 | pcaL | 4.2 | 5.7 |
| ro02797 | 43,000 | 3,300 | |
| ro02986 | vdh | 0.4 | 1.3 |
| ro04163 | vanB | —d | — |
| ro04165 | vanA | 210 | 1,200 |
| ro02845 | 0.3 | 1.6 | |
Upregulation of the indicated genes was determined by using RT-qPCR in cells grown on 1 mM of the indicated growth substrate and comparing the growth to that of cells grown on pyruvate. Values within 10% of the presented ones were obtained under slightly different reaction conditions.
ORF, open reading frame.
pcaL, 3-oxoadipate enol-lactone hydrolase and 4-carboxymuconolactone. decarboxylase; vdh, vanillin dehydrogenase; vanB, vanillate O-demethylase oxygenase subunit; vanA, vanillate O-demethylase reductase subunit.
—, not significant.
Functional characterization.
To further characterize the vanillin catabolic genes, vanACB, vanB, vdh, ro02797, and ro02845 were each amplified by PCRand cloned into the pET-28a expression vector, and their sequences were confirmed. The oligonucleotides used in constructing these vectors are listed in Table S1 in the supplemental material, and the resulting plasmids are listed in Table S2 in the supplemental material. The encoded proteins were produced in Escherichia coli as described in the supplemental material, and their activities were investigated.
Crude extracts of E. coli expressing ro02986 or ro02797 (total protein, ∼15 mg) were incubated at room temperature in a total volume of 300 μl of 100 mM Tris-Cl, pH 8.5, containing 5 mM vanillin and 5 mM NAD+. Crude extracts of cells expressing ro02986 transformed all of the vanillin to vanillate within 30 min, as determined by thin-layer chromatography (TLC; solvent, chloroform-methanol [19:1]). In contrast, extracts of cells expressing ro02797 did not detectably transform vanillin. These cell extracts also did not transform benzaldehyde. The recombinant vanillin dehydrogenase encoded by ro02986 could be partially purified by (NH4)2SO4 precipitation, but it lost activity during attempted chromatographic steps. While these results do not exclude the possibility that the ro02797-encoded protein was produced in an inactive form, they nevertheless indicate that ro02986 encodes a vanillin dehydrogenase, which was therefore annotated as vdh.
Crude extracts of E. coli expressing vanACB only contained detectable levels of VanA. VanB was therefore produced using a separate construct. Crude extracts of these two cells were combined (∼1.5 mg total protein) and incubated at room temperature in a total volume of 300 μl of 100 mM Tris-Cl, pH 8.5, containing 5 mM vanillate and 5 mM NADH. After 16 h, all of the vanillate was transformed to protocatechuate, as determined by TLC (solvent, chloroform-methanol [4:1]). In contrast, a similar mixture of a crude extract of E. coli expressing vanACB and a crude extract of E. coli expressing ro02845 did not detectably transform vanillate. These results are in good agreement with the bioinformatic and transcriptomic analyses indicating that ro04165 and ro04163 encode the oxygenase and reductase components, respectively, of vanillate O-demethylase. Interestingly, a mixture of crude extracts of E. coli expressing vanA or vanB alone (∼1.5 mg protein each) also did not detectably transform vanillate. This result indicates that VanC is essential for vanillate O-demethylase activity. As VanC contains no sequence similarity to known electron transfer components or chaperones, its role remains unclear.
Targeted gene deletion.
To further validate the annotation of the vanillin degradation genes, vdh (ro02986) and vanA (ro04165) were disrupted by targeted mutagenesis using a sacB counterselection system as previously described (11, 14). The oligonucleotides used in constructing the mutants are listed in Table S1 in the supplemental material, and the plasmids generated in the process are listed in Table S2 in the supplemental material. The vdh mutant R. jostii RHA045 was unable to grow on 1 mM vanillin as the sole organic substrate. In contrast, the mutant grew on 1 mM vanillate with kinetics similar to that of RHA1. The mutant was complemented by cloning vdh into pTip type II (8) and using the resulting plasmid, pTipvdh, to transform RHA045. RHA045::vdh grew on 1 mM vanillin when incubated in the presence of 25 μg/ml chloramphenicol and 40 ng/ml thiostrepton. In contrast, RHA0045 was unable to grow on vanillin under similar conditions when transformed with the empty pTip plasmid. These results support the conclusion that ro02986 encodes the only physiologically relevant vanillin dehydrogenase in RHA1.
The vanA mutant R. jostii RHA046 did not grow on medium containing vanillin or vanillate as the sole organic substrate but was able to grow on pyruvate. The mutant was complemented using pTip containing vanA. RHA046::vanA grew on 1 mM vanillate, 25 μg/ml chloramphenicol, and 40 ng/ml thiostrepton. In contrast, RHA046 transformed with an empty pTip plasmid was unable to grow on vanillate. These results confirm the annotation of ro04165 as vanA.
Finally, we previously demonstrated that RHA1 can degrade protocatechuate by the β-ketoadipate pathway, encoded by the pca genes (10). The previously constructed pcaL mutant, RHA005, failed to grow on either vanillin or vanillate as the sole organic substrate but, compared to the wild type, exhibited normal growth on pyruvate. This result establishes that the protocatechuate generated from vanillate is degraded via the β-ketoadipate pathway in RHA1.
To our knowledge, this is the first study characterizing vanillin catabolic genes in a Rhodococcus species. Among the five rhodococcal genomes in GenBank, only Rhodococcus opacus B4 has the vanACB cluster (locus_tags ROP_40940, ROP_40930, and ROP_40920). The vanA gene does not occur in the genomes of the two Rhodococcus equi strains or the two Rhodococcus erythropolis strains. Among characterized VanA proteins, the rhodococcal enzymes share the highest amino acid sequence identity (68%) with VanA from Streptomyces sp. strain NL15-2K (GI:115391876) (9). Curiously, the streptomycete homolog catalyzes the 4-O demethylation of veratrate. In Comamonas testosteroni BR6020, separate isozymes catalyze the 3-O and 4-O demethylation of veratrate (13). Finally, vanC, a gene of unknown function, appears to be unique to the rhodococci: it does not occur in any of the characterized van gene clusters, including those from Streptomyces (9), Pseudomonas (12), and Comamonas (13).
The physiological role of ro02797 remains unclear. This gene, as well as ro02986, had originally been annotated as encoding benzaldehyde dehydrogenases. Although ro02797 was highly upregulated in RHA1 during growth on vanillin or vanillate, the encoded protein was unable to transform either vanillin or benzaldehyde. RHA1 can degrade many different aromatic compounds, including benzoate and vanillin. Potential substrates for the enzyme encoded by ro02797 include other aromatic aldehydes that are derived from lignin, such as veratraldehyde (3,4-dimethoxybenzaldehyde), phenylacetaldehyde, and cinnamaldehyde (6). Nevertheless, the apparent toxicity of aromatic aldehydes could potentially limit applications of the bacterial transformation of lignin.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a Discovery Grant from the Natural Sciences and Engineering Research Council (NSERC) of Canada to L.D.E.
We thank Nicolas Seghezzi for his assistance with growth studies.
Footnotes
Published ahead of print 4 November 2011
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Ahmad M, et al. 2011. Identification of DypB from Rhodococcus jostii RHA1 as a lignin peroxidase. Biochemistry 50:5096–5107 [DOI] [PubMed] [Google Scholar]
- 2. Ahmad M, et al. 2010. Development of novel assays for lignin degradation: comparative analysis of bacterial and fungal lignin degraders, Mol. Biosyst. 6:815–821 [DOI] [PubMed] [Google Scholar]
- 3. Bauchop T, Elsden SR. 1960. The growth of micro-organisms in relation to their energy supply. J. Gen. Microbiol. 23:457–469 [DOI] [PubMed] [Google Scholar]
- 4. Bugg TD, Ahmad M, Hardiman EM, Singh R. 2011. The emerging role for bacteria in lignin degradation and bio-product formation. Curr. Opin. Biotechnol. 22:394–400 [DOI] [PubMed] [Google Scholar]
- 5. Gonçalves ER, et al. 2006. Transcriptomic assessment of isozymes in the biphenyl pathway of Rhodococcus sp. strain RHA1. Appl. Environ. Microbiol. 72:6183–6193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Javor T, Buchberger W, Tanzcos I. 2000. Determination of low-molecular-mass phenolic and non-phenolic lignin degradation compounds in wood digestion solutions by capillary electrophoresis. Mikrochim. Acta 135:45–53 [Google Scholar]
- 7. McLeod MP, et al. 2006. The complete genome of Rhodococcus sp. RHA1 provides insights into a catabolic powerhouse. Proc. Natl. Acad. Sci. U. S. A. 103:15582–15587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nakashima N, Tamura T. 2004. Isolation and characterization of a rolling-circle-type plasmid from Rhodococcus erythropolis and application of the plasmid to multiple-recombinant-protein expression. J. Bacteriol. 70:5557–5568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nishimura M, Ishiyama D, Davies J. 2006. Molecular cloning of streptomyces genes encoding vanillate demethylase. Biosci. Biotechnol. Biochem. 70:2316–2319 [DOI] [PubMed] [Google Scholar]
- 10. Patrauchan MA, et al. 2005. Catabolism of benzoate and phthalate in Rhodococcus sp. strain RHA1: redundancies and convergence. J. Bacteriol. 187:4050–4063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Patrauchan MA, et al. 2008. Roles of ring-hydroxylating dioxygenases in styrene and benzene catabolism in Rhodococcus jostii RHA1. J. Bacteriol. 190:37–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Priefert H, Rabenhorst J, Steinbuchel A. 1997. Molecular characterization of genes of Pseudomonas sp. strain HR199 involved in bioconversion of vanillin to protocatechuate. J. Bacteriol. 179:2595–2607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Providenti MA, O'Brien JM, Ruff J, Cook AM, Lambert IB. 2006. Metabolism of isovanillate, vanillate, and veratrate by Comamonas testosteroni strain BR6020. J. Bacteriol. 188:3862–3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Roberts JN, et al. 2011. Characterization of DyP peroxidases from Rhodococcus jostii RHA1. Biochemistry 50:5108–5119 [DOI] [PubMed] [Google Scholar]
- 15. Seto M, et al. 1995. Multiple polychlorinated biphenyl transformation systems in the Gram-positive bacterium Rhodococcus sp. strain RHA1. Appl. Environ. Microbiol. 61:4510–4513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sharp JO, et al. 2007. An inducible propane monooxygenase is responsible for N-nitrosodimethylamine degradation by Rhodococcus sp. strain RHA1. Appl. Environ. Microbiol. 73:6930–6938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. van der Geize R, Dijkhuizen L. 2004. Harnessing the catabolic diversity of rhodococci for environmental and biotechnological applications. Curr. Opin. Microbiol. 7:255–261 [DOI] [PubMed] [Google Scholar]
- 18. Yam KC, Okamoto S, Roberts JN, Eltis LD. 2011. Adventures in Rhodococcus—from steroids to explosives. Can. J. Microbiol. 57:155–168 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.