Abstract
Maintaining an appropriate cellular concentration of p53 is critical for cell survival and normal development in various organisms. In this study, we provide evidence that the human E2 ubiquitin-conjugating enzyme RAD6 plays a critical role in regulating p53 protein levels under both normal and stress conditions. Knockdown and overexpression of RAD6 affected p53 turnover and transcription. We showed that RAD6 can form a ternary complex with MDM2 and p53 that contributes to the degradation of p53. Chromatin immunoprecipitation (ChIP) analysis showed that RAD6 also binds to the promoter and coding regions of the p53 gene and modulates the levels of H3K4 and K79 methylation on local chromatin. When the cells were exposed to stress stimuli, the RAD6-MDM2-p53 ternary complex was disrupted; RAD6 was then recruited to the chromatin of the p53 gene, resulting in an increase in histone methylation and p53 transcription. Further studies showed that stress-induced p53 transcriptional activation, cell apoptosis, and disrupted cell cycle progression are all RAD6 dependent. Overall, this work demonstrates that RAD6 regulates p53 levels in a “yin-yang” manner through a combination of two distinct mechanisms in mammalian cells.
INTRODUCTION
The ubiquitin system plays a critical role in numerous cellular events, such as cell cycle regulation, DNA repair, stress responses, metabolic homeostasis, organelle biosynthesis, apoptosis, and gene expression (12, 17). The protein ubiquitin pathway involves a multistep ubiquitin thioester cascade, which requires the ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzymes (UBC or E2), and the assistance of a ubiquitin-protein ligase (E3). Polyubiquitination is thought to mark proteins for degradation, whereas monoubiquitination may have other functions (10).
Rad6 belongs to a group of E2 enzymes (20) that are involved in DNA damage repair by catalyzing the ubiquitination of different target proteins (18, 23, 27, 28, 34, 35, 48). It has been shown that Rad6 interacts with Rad18 to catalyze the monoubiquitination of PCNA (proliferating cell nuclear antigen) on lysine 164 (K164), thereby promoting the error-prone DNA damage repair pathway (4, 5, 6). However, another mechanism has been shown to respond to DNA damage; through this mechanism, a complex containing Ubc13-MMS2-Rad5/Rad18-Rad6 promotes the polyubiquitination of PCNA and activates the error-free repair pathway (18, 48). Mutations in the catalytic site of Rad6 have been shown to confer hypersensitivity to a variety of DNA damage agents (40, 57). The Rad6 mutant has been shown to cause slow growth, severe defects in induced mutagenesis, and hypersensitivity to UV, X-ray, and chemical mutagens (33, 58).
The human homologs of yeast Rad6, HHR6A/RAD6A and HHR6B/RAD6B (human homologs of Rad6), have nearly 70% sequence identity with yeast Rad6, and more than 90% sequence identity is shared between these two human homologs (27, 28). The products of both genes are able to complement the DNA repair and UV mutagenesis defects of the Saccharomyces cerevisiae Rad6 (27, 28). Both mammalian genes are expressed in all organs and tissues and are not subject to mitotic cell cycle regulation (50). The mouse and human HHR6B/RAD6B genes are autosomal, whereas HHR6A/RAD6A is located on the X-chromosome (27, 28). RAD6A-null female mice fail to produce offspring, whereas male mice lacking RAD6A are fertile (49). In contrast, the loss of RAD6B function leads to male sterility (50). When mice lack both homologs, they appear to be nonviable (49), supporting the existence of an essential role of RAD6 in normal development. However, the exact role of RAD6 in embryonic lethality is unclear.
Bre1 is a RING finger-containing E3 ligase, which was first reported by Wood et al. as a factor interacting with Rad6 and functioning as the E3 ligase for Rad6 in transcription (62). Recent studies have shown that Rad6 promotes the monoubiquitination of H2B at K123 (in yeast) or K120 (in mammals) and that, as a prerequisite, it regulates the methylation of histone H3 at lysine 4 (H3K4) and lysine 79 (H3K79) by interacting with the E3 ligase Bre1 (25, 35, 44, 54, 56, 62, 65). This function of Rad6 seems to be highly conserved, because depletion of the Drosophila melanogaster dRad6, a homolog of yeast Rad6, also resulted in the reduction of the trimethylation of H3K4 and H3K79 as well as the altered transcription of more than 800 genes (11).
Rad6 also participates in the protein degradation process by cooperating with a different E3 ligase (13, 57, 61). Drosophila DMP53 degradation clearly involves dRad6 acting through a ubiquitin-proteasome pathway (11). However, this function has not been demonstrated in mammals, although RAD6 has been shown to physically interact with p53 (39).
In this paper, we show, for the first time, that human RAD6 plays a dual role in controlling the protein degradation and transcription of p53. Under normal conditions, RAD6 forms a degradation complex with MDM2 and regulates the turnover of p53. Simultaneously, RAD6 is recruited to the promoter and coding regions of the p53 gene and regulates the H3K4 and H3K79 methylation levels at these regions. When cells are exposed to stress stimuli, the RAD6-MDM2-p53 degradation complex is disrupted, allowing more RAD6 to be recruited to the chromatin of the p53 gene, which further activates p53 transcription. These processes were shown to be RAD6 dependent. We propose that both mechanisms contribute to the increase in p53 protein levels under stress conditions. This work therefore provides novel insight into the understanding of the role of RAD6 in controlling p53 protein levels in mammalian cells.
MATERIALS AND METHODS
Cell culture and transfection.
HeLa cells were cultured at 37°C in Dulbecco modified Eagle medium (DMEM) (Gibco) supplemented with 10% fetal calf serum and antibiotics (penicillin and streptomycin) in a 5% CO2 incubator. Transfection of constructs into HeLa cells was performed with Lipofectamine 2000 (Invitrogen), according to the manufacturer's standard protocol.
Plasmid constructs.
pCMV-Myc and pEGFP-N1 (Clontech) plasmids expressing RAD6A and RAD6B were constructed by cloning RAD6A and RAD6B PCR products, which were amplified from HeLa cell cDNA, into the pCMV-Myc and pEGFP-N1 vectors. The HA-MDM2 plasmid was a kind gift from Zhenkun Lou. Plasmids expressing RAD6A-C88A and RAD6B-C88A mutants (in which cysteine-88 was mutated to alanine) were generated by creating a point mutation in the RAD6A and RAD6B plasmids according to the manufacturer's instructions (Takara). The p53 and MDM2 truncates were a kind gift from Fuchu He's laboratory.
RNAi knockdown of RAD6A, RAD6B, and MDM2 in HeLa cells.
Small interfering RNAs (siRNAs) against RAD6A, RAD6B, and MDM2 were designed and synthesized by the GenePharm company. The RNA interference (RNAi) efficiency was analyzed with a reverse transcription-PCR (RT-PCR) assay.
Transfection of siRNA into HeLa cells was performed according to the manufacturer's protocol (Invitrogen). Briefly, 3 μg of each siRNA was transfected with 8 μl Lipofectamine 2000 (Invitrogen) per well of a 6-well plate.
Coimmunoprecipitation (co-IP) analysis.
HeLa cells were transfected with Myc-tagged RAD6A and RAD6B and hemagglutinin (HA)-tagged MDM2 using Lipofectamine 2000 (Invitrogen). After 48 h, cells were harvested, washed with ice-cold phosphate-buffered saline (PBS), resuspended in ATM lysis buffer (containing 100 mM Tris-Cl [pH 7.5], 150 mM NaCl, 0.2 mM EDTA, 20% glycerol, 0.4% NP-40, 2% Tween 20, and 0.2 mM phenylmethylsulfonyl fluoride [PMSF]), and sonicated on ice 10 times for 3 s each time, with 30% efficiency. The cell lysates were incubated with normal mouse IgG (Santa Cruz) (as a negative control), anti-p53 antibodies (DO-1; Santa Cruz), or anti-Myc antibodies (Zhongshan Golden Bridge) at 4°C overnight. Protein A/G-agarose beads were then added, and the solution was incubated for another 3 h, followed by centrifugation to harvest the agarose beads after they were washed 5 times with lysis buffer. The precipitated proteins were released by boiling in loading buffer and resolved by SDS-PAGE (15%). Immunoblot analyses were performed with antibodies against the Myc tag, ubiquitin, p53, or MDM2.
Cellular fractionation.
Cytoplasmic and nuclear extracts were differentially prepared as described by Jiao et al. (21). Briefly, HeLa cells were transfected with Myc-RAD6 for 48 h, harvested in PBS, and washed twice with buffer A (10 mM HEPES [pH 7.9], 1.5 mM MgCl2, 10 mM KCl, 1 mM PMSF, and 5 mM dithiothreitol [DTT]) before the pellet was resuspended in 3 volumes of buffer A+ (buffer A with an additional 0.1% NP-40). After incubation in buffer A+ for 5 min, the cells were centrifuged and the supernatant, representing the cytoplasmic extract, was removed. The pellet was resuspended in 3 volumes of buffer C (20 mM HEPES [pH 7.9], 420 mM NaCl, 1.5 mM MgCl2, 25% glycerol, 1 mM PMSF, and 5 mM DTT) and incubated on ice for 15 min. Finally, after the mixture was pelleted, the supernatant, representing the soluble fraction of the nuclear proteins, was removed (the concentration of NaCl was adjusted to 120 mM with buffer D, which contained 20 mM HEPES [pH 7.9], 1 mM PMSF, and 5 mM DTT).
Two-step coimmunoprecipitation.
Two-step coimmunoprecipitation was performed essentially according to the procedures described by Rui et al. (52). Briefly, HeLa cells were transfected with Myc-RAD6A and Myc-RAD6B. Nontransfected HeLa cells were used as a negative control for the first immunoprecipitation. At 48 h after transfection, the cells were lysed with ATM lysis buffer, sonicated briefly, and centrifuged. The supernatant was incubated with an anti-Myc antibody bound to protein A/G-agarose beads for 4 h at 4°C. The beads were washed with lysis buffer three times, and the Myc-RAD6 protein complex was eluted with 300 μl of lysis buffer containing 250 mM NaCl and Myc peptide for 3 h at 4°C. The second immunoprecipitation was performed using 150 μl of eluate from the first immunoprecipitation with 350 μl of lysis buffer containing 464 mM NaCl and 2 μg of an anti-p53 (DO-1) antibody or the control IgG, followed by the addition of protein A/G-agarose beads.
Western blot analysis.
HeLa cells were lysed in ATM lysis buffer (containing 100 mM Tris-Cl [pH 7.5], 150 mM NaCl, 0.2 mM EDTA, 20% glycerol, 0.4% NP-40, 2% Tween 20, and 0.2 mM PMSF). The protein concentration in the supernatant was measured with a bicinchoninic acid (BCA) assay kit (Calbiochem). SDS-PAGE was then performed using a 15% gel to resolve the proteins. Different amounts of total protein were loaded in each experiment to facilitate the detection of different target proteins: 50 μg/lane for p53, 20 μg/lane for Myc-RAD6, and 20 μg/lane for actin. After electrophoresis, proteins were transferred onto polyvinylidene difluoride (PVDF) membranes (Amersham) and hybridized with primary antibodies at the following dilutions: p53 (DO-1; Santa Cruz), 1:2,000; Myc tag (Zhongshan Golden Bridge, 1:2,000); HA tag (Zhongshan Golden Bridge), 1:2,000; and actin (Zhongshan Golden Bridge), 1:2,000. The horseradish peroxidase (HRP)-labeled secondary antibodies (Zhongshan Golden Bridge) were all used at a dilution of 1:2,000. An ECL detection system (Amersham) was used to detect the signals on the membranes.
Immunofluorescence staining.
Immunofluorescence staining was performed as described by Ni et al. (45). The primary antibody used in this study was anti-p53 (DO-1; Santa Cruz) at 1:50. DAPI (4′,6′-diamidino-2-phenylindole) (Sigma) was used at a concentration of 1 × 10−4 μg/μl. The secondary antibody coupled to Texas Red (1:100) was purchased from the Zhongshan Golden Bridge Company, China. Images were photographed with a laser scanning confocal microscope (Leica) with a 100× oil immersion objective.
ChIP.
Chromatin immunoprecipitation (ChIP) was performed according to the published protocols of Upstate and Ni et al. (45).
RT-PCR assay.
A total of 4 × 106 HeLa cells were lysed to isolate total RNA using the TRIzol reagent (Invitrogen), according to the manufacturer's instructions. Reverse transcription was performed as described by Ni et al. (45). Total RNA (5 μg) was reverse transcribed to synthesize cDNA in a volume of 20 μl (Moloney murine leukemia virus [M-MLV] reverse transcriptase; Takara). For each 25-μl PCR mixture, 1 μl of cDNA was used for 20 to 25 cycles. PCR products (10 μl) were loaded onto a 2% agarose gel, stained with ethidium bromide, and photographed.
Apoptosis assay.
HeLa cell apoptosis assays were performed according to the manufacturer's standard protocol (KeyGEN). Briefly, 5 × 105 HeLa cells were harvested and resuspended in 500 μl binding buffer. Resuspended cells were treated with 5 μl each of annexin V-fluorescein isothiocyanate (FITC) and propidium iodide (PI) solutions. Cells were then incubated for 5 min in the dark and subjected to flow cytometric analysis. The annexin V-positive cells were defined as the apoptotic cells.
Cell cycle analysis.
The flow cytometric analysis of HeLa cells was conducted according to the manual provided with the PI flow kit (KeyGEN). Briefly, the cells were harvested and stained with the PI solution for 15 min. Fluorescence was measured using a FACSCalibur apparatus (Becton Dickinson). Data collection and analysis were performed using CellQuest software.
RESULTS
Human RAD6 regulates the degradation of the p53 protein through the ubiquitin-proteasome pathway.
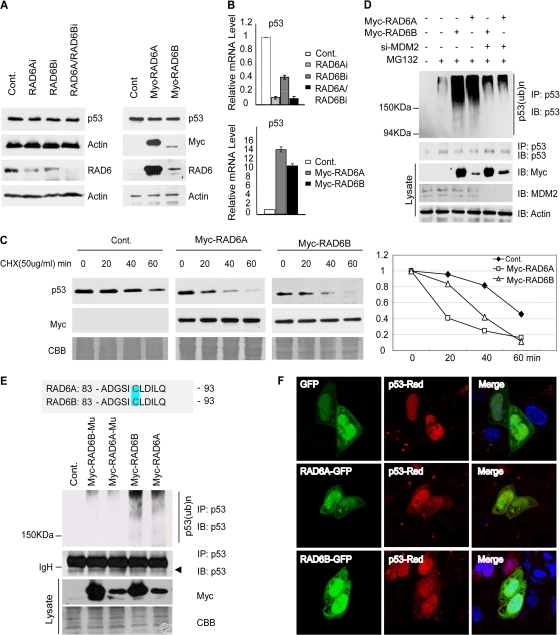
A previous study showed an interaction between p53 and RAD6 in mammalian cells (39). Our most recent work further showed that dRad6 regulates DMP53 turnover in Drosophila melanogaster (11). These results required further testing to determine whether RAD6 plays a conserved role in the regulation of p53 turnover in mammalian cells. We therefore tested the effect of RAD6 on p53 protein levels through the depletion and overexpression of RAD6 in human cells (the RNAi efficiencies of RAD6A and RAD6B are shown in Fig. 1A). Surprisingly, altering RAD6 expression did not have any obvious effect on p53 protein levels (Fig. 1A). We next investigated the mRNA levels of p53 under these conditions and found that the level of p53 RNA was decreased when RAD6 was depleted and was increased when RAD6 was overexpressed (Fig. 1B). These results support a possible role for RAD6 in the transcriptional control of p53.
Fig 1.
RAD6 regulates p53 degradation through the ubiquitin-proteasome pathway. (A) Neither the depletion nor the overexpression of RAD6 has an obvious effect on p53 protein levels. HL-7702 cells (left) or HeLa cells (right) were transfected with RAD6 siRNAs or Myc-RAD6 constructs as indicated. The immunoblot was stained with anti-p53 or anti-Myc antibodies plus antiactin antibodies as a loading control. (B) Depletion or overexpression of RAD6 affects the p53 mRNA level. HL-7702 cells were treated with nonspecific control (Cont.) or RAD6 siRNAs (RAD6Ai or RAD6Bi) (top). HL-7702 cells transfected with Myc control, Myc-RAD6A, or Myc-RAD6B plasmids are indicated on the bottom. Total RNA was prepared and subjected to real-time RT-PCR analysis using specific primers for p53 and GAPDH (as a control). (C) Overexpression of RAD6 decreases the half-life of p53 in HL-7702 cells. HL-7702 cells were transfected with or without Myc-RAD6 and subsequently treated with cycloheximide (CHX) for the indicated periods of time. Immunoblots were stained with antibodies against p53 or the Myc tag (to visualize Myc-RAD6). Coomassie brilliant blue (CBB) staining is shown as an internal control. (D) RAD6 overexpression promotes p53 ubiquitination in an MDM2-dependent manner. HL-7702 cells were transfected with (+) or without (−) Myc-RAD6 in the presence (+) or absence (−) of MDM2 siRNA and treated with dimethyl sulfoxide (DMSO) (−) or MG132. An anti-p53 antibody was used to visualize the amount of precipitated p53 and ubiquitinated p53. The expression of Myc-RAD6 (Myc) and MDM2 is shown in the lower panels. (E) RAD6 ubiquitin-conjugating enzymatic activity is required for its ubiquitination of p53. HL-7702 cells were transfected with or without (Cont.) Myc-RAD6 constructs (Myc-RAD6A or Myc-RAD6B) or their Myc-RAD6-C88A mutants (Myc-6A-C88A or Myc-6B-C88A) and treated with DMSO (Cont.) or MG132. Anti-p53 and antiubiquitin antibodies were used to visualize the amounts of precipitated p53 and ubiquitinated p53. The expression of Myc-RAD6 mutants (Myc) is shown in the lower panels. (F) RAD6 overexpression promotes the cytoplasmic localization of p53. H1299 cells transfected with green fluorescent protein (GFP) or RAD6-GFP together with the p53-DsRed2 construct were subjected to an immunofluorescence assay.
To further investigate whether RAD6 has any effect on p53 degradation, a chase analysis experiment was performed. Both control cells and transfected cells expressing high levels of RAD6 (RAD6A and RAD6B) were treated with 50 μg/ml cycloheximide (CHX) for the indicated times. Cells were then lysed and subjected to Western blot analysis. The results showed that overexpression of RAD6 caused a decrease in the half-life of p53 (Fig. 1C), confirming that RAD6 affects p53 degradation. We also determined the mRNA levels of p53 following this treatment, and we discovered that overexpression of either RAD6A or RAD6B increases the RNA levels of p53, which is consistent with the results shown in Fig. 1B. CHX treatment did not have any obvious influence on the transcription of p53 (data not shown).
Our recent work (11) showed that dRad6 regulates the ubiquitination of DMP53 in Drosophila; we therefore investigated whether RAD6 plays a similar role in mammalian cells. We tested this possibility using an in vivo ubiquitination assay. HL-7702 cells were transfected with or without Myc-RAD6 plasmids in the presence or absence of MDM2 siRNA and 25 μM MG132 for 8 h. These cells were harvested, lysed, and further subjected to immunoprecipitation (IP) with an anti-p53 antibody under denaturing conditions. IP lysates were then immunoblotted with anti-p53 antibodies. The results showed that the overexpression of RAD6A/RAD6B promotes p53 ubiquitination in an MDM2-dependent manner (Fig. 1D).
Cysteine-88 of the RAD6 in Saccharomyces cerevisiae is required for its enzymatic activity (58); we therefore examined whether mutating cysteine-88 in Homo sapiens has any effect on the ubiquitination of p53 (as shown in Fig. 1D). RAD6-C88A mutant plasmids (cysteine-88 was replaced with alanine) were constructed, and HL-7702 cells were transfected with either the Myc control plasmid or the Myc-RAD6 or Myc-RAD6-C88A mutant plasmid, with or without 25 μM MG132, for 8 h. The cells were lysed and subjected to IP with an anti-p53 antibody under denaturing conditions. IP lysates were then immunoblotted with anti-p53 antibodies. The results showed that the overexpression of the RAD6 mutant failed to promote p53 ubiquitination (Fig. 1E), demonstrating that cysteine-88 of RAD6 is critical for p53 ubiquitination in human cells.
It has been shown that the ubiquitination of p53 acts as a signal for its cytoplasmic translocation (37, 66). As a result, we further investigated whether RAD6 has any effect on p53 subcellular localization. Cells were transfected with pEGFP-N1 or pEGFP-RAD6 plasmid together with pDsRed2-p53 plasmid for 48 h. Cells were then harvested and stained with DAPI. As shown in Fig. 1F, overexpression of RAD6 promoted the cytoplasmic translocation of p53 in H1299 cells. This result is also consistent with the observed increase in the ubiquitination of p53 following RAD6 overexpression (Fig. 1D).
RAD6 forms a ternary complex with MDM2 and p53 that is independent of its enzymatic activity.
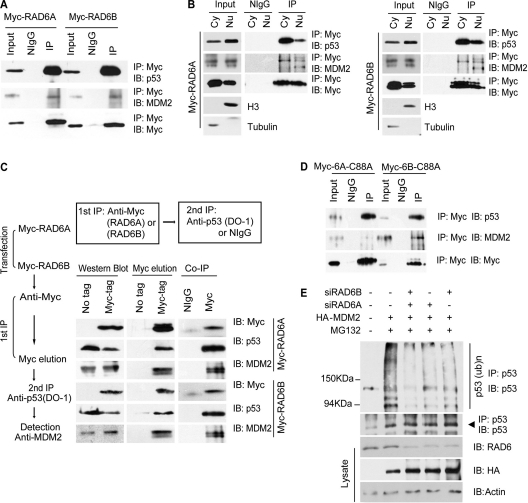
MDM2 is believed to be the primary factor that regulates p53 turnover (16, 19, 31). We therefore investigated whether the ubiquitination of p53 by RAD6 requires the incorporation of MDM2. Constructs expressing Myc-RAD6A/B were transfected into HeLa cells. The interaction between MDM2, p53, and Myc-RAD6A/B was confirmed by coimmunoprecipitation experiments with an anti-Myc antibody (Fig. 2A). The results showed that MDM2 and p53 can be immunoprecipitated by Myc-RAD6 proteins, suggesting that RAD6 interacts with MDM2 and p53 in vivo. The result was also confirmed using antibodies against endogenous proteins (data not shown). Next, we determined whether these interactions occur in the cytoplasm or the nucleus. Coimmunoprecipitation experiments, using cytoplasmic and nuclear fractions, showed that MDM2 and p53 are immunoprecipitated by Myc-RAD6 proteins (Fig. 2B). These results showed that RAD6 is able to interact with MDM2 and p53 in both the cytoplasm and the nucleus in mammalian cells.
Fig 2.
RAD6 forms a ternary complex with MDM2 and p53. (A) RAD6 interacts with p53 and MDM2 in vivo. HeLa cells were transfected with Myc-RAD6 plasmids. The immunoblots were stained with anti-p53, anti-MDM2, or anti-Myc antibodies to visualize these proteins. The result showed that p53 (upper panel) and MDM2 (middle panel) could be precipitated by an anti-Myc antibody. The antibody against the Myc tag is indicated in the lower panel. (B) The interaction between RAD6, MDM2, and p53 occurs in both the cytoplasm and the nucleus. HeLa cells were transfected with Myc-RAD6A and Myc-RAD6B plasmids. Cytoplasmic (Cy) and nuclear (Nu) extracts were differentially prepared, and the coimmunoprecipitation experiment was performed with an anti-Myc antibody. The immunoblots were stained with anti-p53, anti-MDM2, or anti-Myc antibodies to visualize these proteins. The results showed that p53 and MDM2 could be precipitated by an anti-Myc antibody in both the cytoplasm and the nucleus. The antibody against the Myc tag is indicated. (C) RAD6 forms a ternary complex with MDM2 and p53. A two-step coimmunoprecipitation experiment was employed to test for ternary complex formation. The procedures of the two-step coimmunoprecipitation are outlined on the left. HeLa cells were transfected with or without (no tag) Myc-RAD6. The first immunoprecipitation was performed with an anti-Myc antibody. The complex was eluted using the Myc peptide. The second step of immunoprecipitation used an anti-p53 antibody or the control mouse IgG (NIgG) to precipitate the complex. Protein samples from each step were then separately subjected to Western blot analysis using anti-Myc, anti-p53, and anti-MDM2 antibodies. The experiment was repeated with essentially the same results. (D) RAD6 ubiquitin-conjugating enzymatic activity is not required for the interaction with MDM2 and p53. HeLa cells were transfected with Myc-RAD6 mutants, Myc-RAD6-C88A (Myc-6A-C88A or Myc-6B-C88A), and coimmunoprecipitation experiments were employed using an anti-Myc antibody. The anti-p53 (upper panel) and anti-MDM2 (middle panel) antibodies were used to visualize these proteins in the Western blot analysis. The antibody against the Myc tag is indicated in the lower panel. (E) MDM2-induced p53 ubiquitination is dependent on RAD6. HL-7702 cells were transfected with (+) or without (−) HA-MDM2 in the presence (+) or absence (−) of RAD6 siRNAs and treated with DMSO (−) or MG132. The anti-p53 antibody was used to visualize the amounts of precipitated p53 and ubiquitinated p53. The expressions of p53 and HA-MDM2 (HA) are shown in the lower panels.
To determine whether RAD6, MDM2, and p53 are present in the same complex, two-step coimmunoprecipitation experiments were performed (Fig. 2C). HeLa cells were transfected with Myc-RAD6 plasmids. Nontransfected HeLa cells were used as a negative control. In the first immunoprecipitation step, anti-Myc was used to pull down RAD6, and the Myc peptide (GenScript) was used to elute the complex. The eluate was then immunoprecipitated with an anti-p53 antibody or a control IgG, followed by Western blotting to detect MDM2. As shown in Fig. 2C, MDM2 was present in the final immunoprecipitate but not in the control sample, confirming that RAD6, p53, and MDM2 exist in a ternary complex.
Next, we investigated whether the enzymatic activity of RAD6 is required for its interaction with MDM2 and p53. Our results in Fig. 1E show that cysteine-88 is required for RAD6 enzymatic activity, and the mutation of cysteine to alanine abolished the ubiquitination of p53. We therefore used the same mutant construct to test whether cysteine-88 of RAD6 is required for its interaction with MDM2 and p53. HeLa cells were transfected with Myc-RAD6-C88A mutants and cultured for 48 h. The cells were then lysed and subjected to IP with an anti-Myc antibody. IP lysates were further immunoblotted with anti-MDM2 or anti-p53 antibodies. The results showed that the mutation of cysteine-88 of RAD6 did not affect the interaction of RAD6 with MDM2 and p53 (Fig. 2D), implying that the enzymatic activity of RAD6 is not required for their interaction.
RAD6 plays an essential role in the function of the ternary complex in p53 ubiquitination.
We next examined whether the presence of RAD6 is essential for MDM2-induced p53 ubiquitination. HL-7702 cells were transfected with or without the HA-MDM2 plasmid in the presence or absence of RAD6 (RAD6A and RAD6B) siRNAs and 25 μM MG132 for 8 h. The harvested cells were lysed and subjected to IP with an anti-p53 antibody under denaturing conditions. IP lysates were then immunoblotted with an anti-p53 antibody. The results showed that the overexpression of MDM2 promotes p53 ubiquitination and that this occurs in a RAD6-dependent manner (Fig. 2E, compare lanes 2 to 5). It was reported that UbcH5c is an E2 ubiquitin-conjugating enzyme for MDM2-catalyzed p53 ubiquitination (19). We therefore next compared the effects of RAD6 and UbcH5c on p53 ubiquitination. The results indicated that RAD6 and UbcH5c function to similar extents in p53 ubiquitination (data not shown).
Taken together, our results show that RAD6 forms a functional ternary complex with MDM2 and p53 and that the ubiquitination of p53 requires the presence of all three members of this complex (Fig. 1D and 2).
The TAD of p53 is required for the RAD6-p53 interaction.
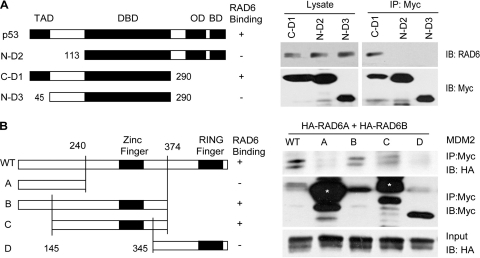
To identify the domains of p53 that interact with RAD6, we prepared constructs expressing fragments of p53, including a fragment without the N-terminal domain of Dmp53 (N-D2, lacking residues 1 to 132), a fragment without the C-terminal domain of p53 (C-D1, lacking residues 291 to 393), and a fragment without the transcriptional activation domain (TAD) or the C-terminal domain (residues 291 to 393) of p53 (N-D3). These constructs were transfected into the human lung carcinoma cell line H1299, a cell line that lacks endogenous p53. Coimmunoprecipitation experiments were performed using a mouse anti-Myc antibody (indicating p53 fragments). Immunoblotting was performed with antibodies against Myc or RAD6, as indicated. The results show that the TAD domain is required for the interaction between RAD6 and p53 (Fig. 3A), which is consistent with our previous results (11).
Fig 3.
Domains of p53 and MDM2 required for its interaction with RAD6. (A) Myc-tagged p53 truncates were transfected into H1299 cells for 48 h, and cells were subsequently harvested and lysed for a co-IP assay using an antibody against Myc. Mouse anti-Myc and rabbit anti-RAD6 antibodies used for Western blotting are indicated. (B) Myc-tagged MDM2 and its truncates were transfected into the H1299 cell line together with HA-RAD6 constructs (HA-RAD6A or HA-RAD6B) for 48 h, and cells were subsequently harvested and lysed for a co-IP assay with an antibody against Myc. Mouse anti-Myc and anti-HA antibodies used for Western blotting are indicated. The asterisks indicate nonspecific bands.
Previous studies demonstrated that MDM2 interacts with p53 via the TAD domain of p53 (9, 41), and the results raised a possibility that the interaction between RAD6 and p53 was mediated by MDM2. We therefore examined this hypothesis under MDM2 knockdown condition. Our results show that the interaction between RAD6 and p53 was indeed inhibited when MDM2 was depleted in HL-7702 cells (data not shown).
Determination of the regions in MDM2 required for the RAD6-MDM2 interaction.
To determine the regions of interaction between MDM2 and RAD6, a series of Myc-tagged MDM2 deletion mutants were constructed, as indicated in Fig. 3B. These constructs were transfected into H1299 cells together with HA-RAD6A and HA-RAD6B constructs. Coimmunoprecipitation experiments were performed using a mouse anti-Myc antibody (indicating MDM2 fragments). Immunoblotting was performed with antibodies against Myc or HA tag, as indicated. The result showed that MDM2 mutants B and C retained their ability to form a complex with RAD6 (Fig. 3B). However, MDM2 mutants A and D lost their ability to interact with RAD6 (Fig. 3B). This finding indicates that the region around amino acids (aa) 240 to 345 in MDM2 (including the zinc finger domain) is critical for its interaction with RAD6 (Fig. 3B).
RAD6 regulates the mRNA level of p53 by affecting histone H3 methylation.
We observed that RAD6 regulates the ubiquitination and degradation of p53 in mammalian cells (Fig. 1C and D and 2E); however, RAD6 ubiquitination of p53 did not affect the total protein levels of p53 (Fig. 1B), suggesting that RAD6 may also be involved in the transcription of the p53 gene. We therefore examined the changes in the mRNA level of p53 under altered RAD6 expression levels. Cells were transfected with siRNAs against RAD6 or Myc-RAD6 constructs for 48 h. The total RNA was extracted, and quantitative RT-PCR (qRT-PCR) analysis was employed using specific primers for p53 or GAPDH (glyceraldehyde-3-phosphate dehydrogenase) (as a control). The results showed that knockdown of RAD6 expression by siRNA significantly decreased p53 transcription (Fig. 1B, upper panel), while overexpression of RAD6 increased p53 transcription (Fig. 1B, lower panel).
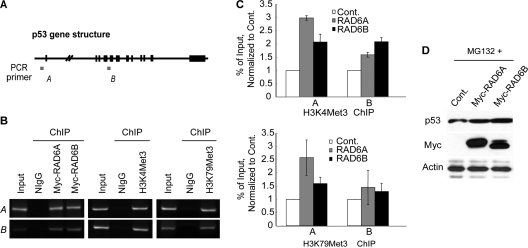
We next investigated how RAD6 regulates the mRNA level of p53. RAD6 has been shown to affect H3K4 and H3K79 trimethylation (23). H3K4 methylation is often associated with transcriptionally active genes (8, 53). We first determined whether there was any direct binding of RAD6 or association of H3K4me3 and K79me3 at the p53 promoter (Fig. 4A, region labeled A) and 5′ coding regions of p53 (Fig. 4A, region labeled B). HeLa cells were transfected with Myc-RAD6s for 48 h. Cells were collected and used to perform chromatin immunoprecipitation (ChIP) analyses with specific antibodies. Our results showed that RAD6, H3K4me3 and K79me3 bind at both the promoter and 5′ coding regions of the p53 gene (Fig. 4B). Similar results were obtained when ChIP assay was performed using antibodies against endogenous RAD6 proteins (data not shown).
Fig 4.
RAD6 regulates the transcription of p53. (A) Gene structure of p53. This diagram was drawn according to the NCBI gene searching results. The sites used for the design of the primers for chromatin immunoprecipitation (ChIP) are indicated (regions labeled A and B). (B) RAD6, H3K4me3, and H3K79me3 are enriched at the promoter (panels A) and 5′ coding (panels B) regions of the p53 gene. HeLa cells were transfected with (left panels) or without (middle and right panels) RAD6. The ChIP analysis showed that RAD6, H3K4me3, and K79me3 were enriched at these p53 gene regions. (C) Overexpression of RAD6 increases the levels of H3K4me3 and K79me3 at the promoter (bars A) and 5′ coding (bars B) regions of the p53 gene in HeLa cells. HeLa cells were transfected with or without (Cont.) Myc-RAD6. The ChIP-qPCR analysis showed that the overexpression of RAD6 resulted in increased levels of H3K4me3 and H3K79me3 at these regions of the p53 gene. (D) RAD6 overexpression increases p53 protein levels following proteasome inhibition. HeLa cells were transfected with Myc control (Cont.) or Myc-RAD6 plasmids and treated with MG132 (25 μM) for 12 h. The anti-p53 antibody was used to visualize the amount of p53. The expression of Myc-RAD6 (Myc) is shown in the lower panel.
To test whether changes in H3K4 and H3K79 methylation of the p53 gene correlate with the concentration of RAD6, another ChIP-qPCR analysis was performed using cells overexpressing RAD6. Our results confirmed an increase in H3K4 and H3K79 trimethylation at the promoter and 5′ coding regions of the p53 gene (Fig. 4C), supporting the finding that RAD6 increases the mRNA level of p53, possibly by modulating the level of H3 methylation of the chromatin of the p53 gene. We further investigated the effect of altered RAD6 expression on p53 protein levels under conditions of inhibition of its degradative function with the proteasome-specific inhibitor MG132. An increase of p53 protein levels was observed under this treatment (Fig. 4D), which was consistent with our hypothesis.
The RAD6-MDM2-p53 ternary complex is disrupted under stress conditions.
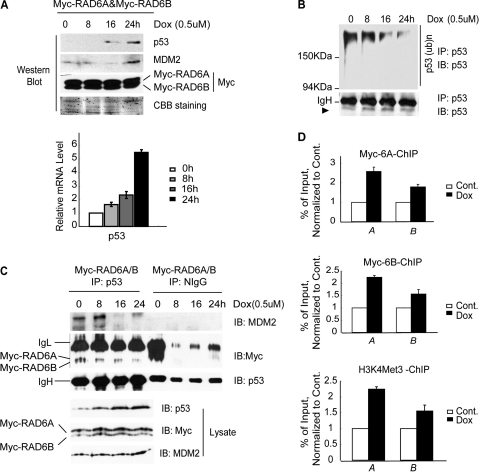
It has been reported that p53 is upregulated in the presence of the stress stimulus doxorubicin (23). We investigated whether the upregulation of p53 was caused by an alteration of the function of the ternary complex. HeLa cells were therefore transfected with Myc-RAD6A and -B for 48 h and treated with doxorubicin for the indicated times. Cell lysates were prepared for Western blot assays to verify the increase in p53 protein levels after doxorubicin treatment (Fig. 5A, upper panels). The ubiquitination of p53 was significantly inhibited in a time-dependent manner after doxorubicin treatment (Fig. 5B), as expected. The MDM2 protein levels were also increased, while the expression of RAD6 was not significantly affected (Fig. 5A, upper panels).
Fig 5.
RAD6 functions in the process of stress stimulus-induced p53 upregulation. (A) Doxorubicin (Dox) treatment causes the upregulation of p53 in HeLa cells. HeLa cells were transfected with the Myc-RAD6 plasmid and treated with doxorubicin (0.5 μM) for the indicated times. The immunoblot was stained with anti-p53, anti-MDM2, or anti-Myc antibodies. The histogram at the bottom shows the real-time RT-PCR analysis of the transcriptional levels of p53. (B) Doxorubicin (Dox) treatment inhibits the ubiquitination of p53 in HeLa cells. HeLa cells were treated with doxorubicin (0.5 μM) for different lengths of time. Coimmunoprecipitation analysis was employed using an anti-p53 antibody. The anti-p53 antibody was also used to visualize the amounts of precipitated p53 (lower panel) and ubiquitinated p53 (upper panel). (C) Doxorubicin (Dox) treatment inhibits the formation of the RAD6-MDM2-p53 degradation complex. HeLa cells were transfected with the Myc-RAD6 plasmid and treated with doxorubicin (0.5 μM) for the indicated periods of time. A coimmunoprecipitation analysis was employed using an anti-p53 antibody. The anti-p53, anti-MDM2, and anti-Myc antibodies were used to visualize the amounts of precipitated p53, MDM2, and RAD6, respectively (upper panels). The gel on the right shows an identical control IP using normal mouse IgG (NIgG). The expression levels of p53, MDM2, and RAD6 are shown in the lower panels. (D) Doxorubicin (Dox) treatment promotes the recruitment of RAD6 and H3K4me3 to the promoter (bars A) and 5′ coding (bars B) regions of the p53 gene. HeLa cells were transfected with the Myc-RAD6 plasmid and treated with or without (Cont.) doxorubicin (0.5 μM) for 24 h. The ChIP-qPCR analysis showed that doxorubicin treatment resulted in increased levels of RAD6 and H3K4me3 at these regions of the p53 gene.
We next investigated whether there were changes in the RAD6-MDM2-p53 degradation complex following doxorubicin treatment. HeLa cells were transfected with Myc-RAD6A and -B for 48 h and treated with doxorubicin for the indicated times. Cell lysates were prepared and subjected to co-IP assay with an anti-p53 antibody. The IP lysates were immunoblotted with anti-MDM2, anti-Myc, or anti-p53 antibodies to visualize MDM2, RAD6 (Myc-RAD6A and Myc-RAD6B), or p53, respectively. The results showed that considerably less MDM2 and RAD6 was precipitated by p53 after doxorubicin treatment (Fig. 5C, upper panels). These results support the finding that the functional RAD6-MDM2-p53 ternary complex is disrupted under doxorubicin treatment in a time-dependent manner.
Stress promotes the recruitment of RAD6 to the chromatin of the p53 gene and upregulates local histone methylation.
We noticed that doxorubicin treatment also promoted an increase in the p53 mRNA level in a time-dependent manner (Fig. 5A, lower panels). This observation raised the question of whether the increase in p53 mRNA level was correlated to the recruitment of RAD6 to the p53 promoter and subsequent altered histone methylation of the p53 gene. Therefore, we examined the RAD6 and H3K4me3 levels at the promoter and 5′ coding regions of the p53 gene. HeLa cells transfected with Myc-RAD6A and -B for 48 h were treated with or without doxorubicin for 24 h. Subsequently, a ChIP-qPCR analysis was performed using specific antibodies. Specific primers for the p53 promoter (bars A) and 5′ coding regions (bars B) were used for this assay. The results showed that doxorubicin treatment promotes both the recruitment of RAD6 to the chromatin of the p53 gene and the increases in the H3K4me3 concentrations at these regions (Fig. 5D).
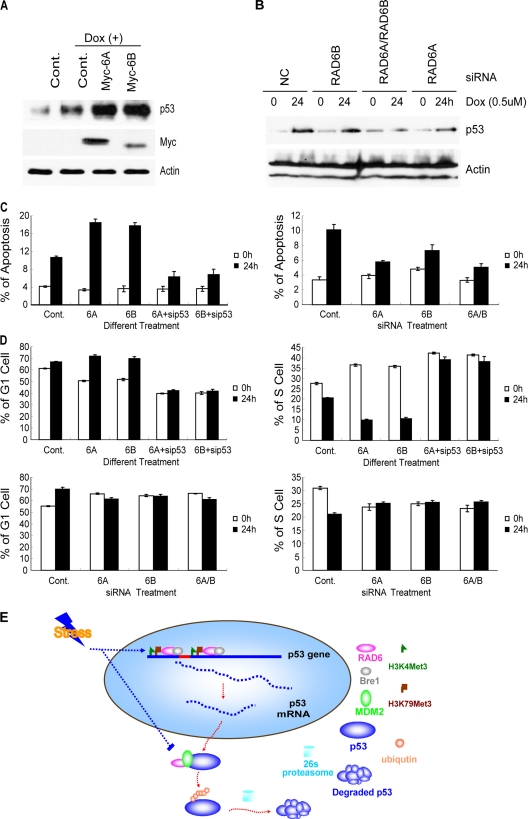
To further confirm the role of RAD6 in p53 transcriptional activation under stress conditions, we tested the expression of p53 under the condition of RAD6 overexpression and RAD6 depletion. Cells transfected with Myc-RAD6 constructs (Fig. 6A) or with a nonspecific control or RAD6-specific siRNAs for 48 h (Fig. 6B) were treated with or without doxorubicin for 24 h. Cells were lysed and subjected to a Western blot assay. The results showed that overexpression of RAD6 promotes the doxorubicin-induced increase of p53 (Fig. 6A), while depletion of RAD6 inhibits the doxorubicin-induced increase in p53 protein levels (Fig. 6B), which is consistent with our prediction. We also checked the mRNA level of p53 following these treatments and found that RAD6 overexpression promotes the doxorubicin-induced increase in p53 RNA level (data not shown), while depletion reduced the RNA levels of p53 under both normal and doxorubicin treatment conditions (data not shown).
Fig 6.
The loss of RAD6 affects apoptosis and cell cycle progression following stress stimuli. (A) RAD6 overexpression promotes the upregulation of the p53 protein level under stress stimuli. HL-7702 cells were transfected with Myc control (Cont.) or Myc-RAD6 constructs (Myc-RAD6A or Myc-RAD6B) and treated with or without doxorubicin (Dox) for 24 h. The cell extracts were subjected to Western blot analysis using antibodies against p53 and actin. (B) Knockdown of RAD6A and RAD6B expression inhibits the upregulation of p53 induced by doxorubicin treatment. HeLa cells were transfected with nonspecific control (Cont.) or RAD6 (RAD6Ai or RAD6Bi) siRNAs and treated with or without (0) doxorubicin (Dox) for 24 h. The cell extracts were subjected to Western blot analysis using antibodies against p53 and actin. (C) Alteration of RAD6 expression affects the stress stimulus-induced cell apoptosis in a p53-dependent manner. HL-7702 cells were transfected with Myc control (Cont.) or Myc-RAD6 constructs with or without p53 siRNA in the presence (24 h) or absence (0 h) of doxorubicin as indicated (left). HL-7702 cells were treated with nonspecific control (Cont.) or RAD6 siRNAs in the presence (24 h) or absence (0 h) of doxorubicin (right). Cells were harvested and doubly labeled with annexin V-FITC and PI before analysis by flow cytometry. The experiments were repeated 3 independent times. (D) The alteration of RAD6 expression affects the p53 upregulation-induced cell cycle changes (G1 phase arrest and reduction of the number of cells in S phase) triggered by stress stimuli. HL-7702 cells were transfected with Myc control (Cont.) or Myc-RAD6 constructs with or without p53 siRNA in the presence (24 h) or absence (0 h) of doxorubicin as indicated (upper panels). HL-7702 cells were treated with nonspecific control (Cont.) or RAD6 siRNAs in the presence (24 h) or absence (0 h) of doxorubicin (lower panels). Cells were harvested and stained with PI solution before analysis by flow cytometry. The experiments were repeated 3 independent times. (E) The pathways by which RAD6 regulates p53 to maintain its expression at an appropriate level. Under normal conditions, RAD6 maintains a low level of p53 protein expression through two pathways. One is the ubiquitin-proteasome pathway, and the other is the transcriptional regulation pathway. The ubiquitin-proteasome pathway is executed by a collaboration between RAD6 and MDM2. The transcriptional regulation pathway is activated through the direct recruitment of RAD6 to regions of the p53 gene (e.g., the promoter and 5′ coding regions), and it alters the H3K4me3 and K79me3 levels at these regions. However, when cells encounter stress stimuli (e.g., doxorubicin treatment), both of these pathways are affected. The RAD6-MDM2-p53 degradation complex is disrupted, inhibiting the degradation of p53. Simultaneously, more RAD6 is recruited to the promoter and 5′ coding regions of the p53 gene, and the levels of H3K4me3 and K79me3 are increased at these regions. As a result, the transcription of p53 is enhanced. Taken together, these results demonstrate that RAD6 plays a dual role in the regulation of p53 protein levels, and the two pathways function in opposing manners.
RAD6 is required for stress-induced apoptosis and cell cycle alteration.
Because RAD6 is involved in the regulation of p53 expression and previous studies have shown that p53 is involved in apoptosis and cell cycle regulation, we investigated whether RAD6 has any impact on doxorubicin-induced apoptosis and cell cycle alteration. HL-7702 cells transfected with Myc control or Myc-RAD6 constructs together with or without p53 siRNA for 48 h were treated with or without doxorubicin for 24 h. Cells were harvested and subjected to apoptosis assay using fluorescence-activated cell sorting (FACS). The results showed that the overexpression of RAD6 promoted the doxorubicin-induced apoptosis in a p53-dependent manner (Fig. 6C, left panel). We next examined the effect of RAD6 depletion on the doxorubicin-induced cell apoptosis. HL-7702 cells were transfected with nonspecific control or RAD6-specific siRNAs for 48 h and then treated with or without doxorubicin for 24 h. Cells were harvested and subjected to apoptosis assay using FACS. The results showed that the knockdown of RAD6 expression inhibited doxorubicin-induced apoptosis (Fig. 6C, right panel).
It has been shown that p53 upregulation induces G1 phase arrest and reduces the number of cells in S phase (59, 67). Thus, we subsequently performed a FACS assay to test the impact of RAD6 on cell cycle progression. Our results showed that doxorubicin-induced G1 phase arrest and the reduction of the number of cells in S phase were promoted by RAD6 overexpression and in a p53-dependent manner, while they were inhibited by the knockdown of RAD6 (Fig. 6D). Consistently, both of these effects on apoptosis and cell cycle progression by RAD6 overexpression and knockdown were also correlated with changes in p53 protein levels following these treatments (Fig. 6A and B).
Overall, these results confirm an essential role of RAD6 in stress-induced apoptosis and cell cycle progression.
DISCUSSION
RAD6 functions as an important regulator of p53 turnover in mammals.
The crucial tumor suppressor p53 (7, 15) plays a critical role in suppressing genome instability, which is a driving force of cancer progression (32, 36). Mutation or altered function of p53 is found in more than half of all cancer cases and is highly related to different types of tumorigenesis (1, 22, 60). p53 also plays a critical role in other cellular events, including cell cycle regulation, senescence, DNA repair, cell apoptosis, and the pluripotency of stem cells (30, 68). Posttranscriptional modifications, including phosphorylation and acetylation, are known to be critical for p53 stabilization and activation (30, 38). The ubiquitin-proteasome degradation pathway appears to be important for maintaining a low cellular level of p53 in normal cells (9, 41). The interaction between E3 ligase MDM2 (mouse double minute protein 2) and p53 is believed to be responsible for the rapid turnover of p53 (16, 19, 31), which regulates p53 through mono- or polyubiquitination in mammalian cells.
In this work, we provide direct evidence that RAD6, an E2 ligase, promotes the ubiquitination and degradation of p53 in human cells (Fig. 1). This finding is consistent with a previous study performed in a cell-free system, which showed that Rad6 could mediate the ubiquitination of p53; however, a direct effect of RAD6 on p53 degradation was not investigated (39). The impact of RAD6 on the ubiquitination of p53 is also supported by our analysis using the cysteine-88-to-alanine mutant (C88A, a loss-of-function mutant). The C88A mutation clearly failed to ubiquitinate p53, in contrast to the wild-type RAD6 protein (Fig. 1E). The interaction of RAD6 with MDM2 and p53 in mammalian cells and the subsequent formation of a ternary complex (Fig. 2), as well as the requirement of RAD6 for MDM2-mediated ubiquitination of p53, provide further support for a role of RAD6 in regulating p53 ubiquitination. Knockdown of RAD6 expression significantly decreased p53 ubiquitination levels (Fig. 2E). Together with our most recent study with Drosophila (11), in which we demonstrated that dRad6 regulates the ubiquitination and degradation of DMP53, this work suggests that the role of RAD6 in p53 turnover is conserved between flies and humans.
RAD6 has two transcriptional variants, RAD6A and RAD6B, in mammalian cells. The high homology (more than 95% similarity) between these two molecules suggests that RAD6A and RAD6B may play redundant roles in mammalian cells. Previous studies have shown that RAD6A and RAD6B have redundant functions in H2B ubiquitination and transcriptional activation, and RAD6A protein levels are higher than RAD6B levels in human cells (26, 27, 57). In this work, we found that these two molecules had no obvious differences in their effects on p53 activity, which may explain the observation that single RAD6A or RAD6B knockout mutations in mice are not lethal; lethality was observed only when both genes were deleted simultaneously (2, 3, 49).
Mechanism of RAD6 in the control of p53 mRNA level.
In contrast to a negative role for RAD6 in the control of p53 protein concentrations, we found that RAD6 directly associates with the chromatin of p53 and positively correlates with its mRNA level. Loss of human RAD6 resulted in a significant decrease in the mRNA level of p53 (Fig. 1B). Using a ChIP assay, we investigated the binding of RAD6 and H3 methylation on specific sequences of the p53 gene. We found that RAD6 binds the promoter and 5′ coding regions of the p53 gene, increasing the H3 methylation levels in these regions (Fig. 4). In Drosophila, depletion of the dRad6 gene caused a dramatic increase in p53 protein levels, whereas depletion of dRad6 had no obvious effect on the transcription of the DMP53 gene; these data suggest that there is a functional difference between the roles of dRad6 and RAD6 in transcription.
How does RAD6 regulate the p53 transcriptional process? Previous studies have shown that RAD6/HHR6 preferentially localizes to the euchromatin of chromosomes, suggesting that the protein is associated with transcriptionally active regions (26). Furthermore, expression of HHR6A/HHR6B has been shown to be elevated in mouse spermatids, coinciding with the developmental steps during which a large degree of chromatin modification occurs (26). More direct evidence of the role of RAD6 in transcriptional activation was shown by Roeder's group; they showed that human RAD6, through the interaction with PAF-bound hBRE1 and the recruitment of RAD6-hBRE1 to the Pol II transcription machinery, was recruited to genes undergoing transcription (23, 25). The subsequent H2B ubiquitination by the Rad6/Bre1 complex stimulated further changes in H3K4 and K79 methylation; this methylation, promoted by the Rad6/Bre1 complex, has been demonstrated in many different species (11, 24, 47, 62, 64, 69) and likely contributes to the activation of the p53 gene.
RAD6-induced H3K4 methylation is believed to be regulated by COMPASS (complex of protein associated with Set1), and H3K79 methylation is thought to be regulated by Dot1 (14, 44, 55, 56, 63). The Cps35 subunit of COMPASS mediates the cross talk between H2B monoubiquitination and H3 methylation (35). COMPASS can mono-, di-, and trimethylate lysine 4 of histone H3 (29, 42, 43, 46, 51), a marker known to be associated with actively transcribed genes.
Function of RAD6 under stress conditions.
Stress stimuli are known to cause changes in p53 protein levels in many different organisms (59, 67). We therefore investigated whether RAD6 plays a role in regulating p53 in the presence of stress stimuli. We tested this in cells treated with doxorubicin, because this treatment has been shown to stimulate an increase in p53 protein levels (Fig. 5) (23). Our results suggest that both the transcriptional and the posttranscriptional mechanisms of RAD6 participate in the response to doxorubicin treatment. Interestingly, more RAD6 was recruited to the chromatin of the p53 gene following doxorubicin treatment, which increased the local H3K4 and H3K79 methylation (Fig. 5). It is unclear how the stress signals modulate the binding of RAD6 to chromatin. The chromatin-bound RAD6 may originate from the disrupted RAD6-MDM2-p53 complex following cellular stress, causing its relocation to the p53 gene. Alternatively, it is possible that stress stimuli modify RAD6 to enhance its chromatin binding activity. The exact mechanism remains to be elucidated.
In summary, we have demonstrated a novel function for the E2 ubiquitin-conjugating enzyme RAD6 in the control of both the p53 protein levels and the transcriptional regulation of p53 in mammalian cells. We propose that RAD6 acts in a transcriptional and posttranscriptional manner to maintain p53 at an appropriate level under both normal and stress conditions.
ACKNOWLEDGMENTS
This work was supported by National 973 grants from the Ministry of Science and Technology (2007CB948101, 2009CB825603, and 2011CB965300).
Footnotes
Published ahead of print 14 November 2011
REFERENCES
- 1. Allton K, et al. 2009. Trim24 targets endogenous p53 for degradation. Proc. Natl. Acad. Sci. U. S. A. 106: 11612–11616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Baarends WM, et al. 2007. Increased phosphorylation and dimethylation of XY body histones in the Hr6b-knockout mouse is associated with derepression of the X chromosome. J. Cell Sci. 120: 1841–1851 [DOI] [PubMed] [Google Scholar]
- 3. Baarends WM, et al. 2003. Loss of HR6B ubiquitin-conjugating activity results in damaged synaptonemal complex structure and increased crossing-over frequency during the male meiotic prophase. Mol. Cell. Biol. 23: 1151–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bailly V, Lamb J, Sung P, Prakash S, Prakash L. 1994. Specific complex formation between yeast RAD6 and RAD18 proteins: a potential mechanism for targeting RAD6 ubiquitin-conjugating activity to DNA damage sites. Genes Dev. 8: 811–820 [DOI] [PubMed] [Google Scholar]
- 5. Bailly V, Lauder S, Prakash S, Prakash L. 1997. Yeast DNA repair proteins Rad6 and Rad18 form a heterodimer that has ubiquitin conjugating, DNA binding, and ATP hydrolytic activities. J. Biol. Chem. 272: 23360–23365 [DOI] [PubMed] [Google Scholar]
- 6. Bailly V, Prakash S, Prakash L. 1997. Domains required for dimerization of yeast Rad6 ubiquitin-conjugating enzyme and Rad18 DNA binding protein. Mol. Cell. Biol. 17: 4536–4543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Baker SJ, et al. 1989. Chromosome 17 deletions and p53 gene mutations in colorectal carcinomas. Science 244: 217–221 [DOI] [PubMed] [Google Scholar]
- 8. Berger SL. 2007. The complex language of chromatin regulation during transcription. Nature 447: 407–412 [DOI] [PubMed] [Google Scholar]
- 9. Brooks CL, Gu W. 2006. p53 ubiquitination: Mdm2 and beyond. Mol. Cell 21: 307–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chau V, et al. 1989. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science 243: 1576–1583 [DOI] [PubMed] [Google Scholar]
- 11. Chen S, Wei HM, Lv WW, Wang DL, Sun FL. 2011. The E2-ligase dRad6 regulates Dmp53 turnover in Drosophila. J. Biol. Chem. 286: 9020–9030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ciechanover A. 1994. The ubiquitin-proteasome proteolytic pathway. Cell 79: 13–21 [DOI] [PubMed] [Google Scholar]
- 13. Dohmen RJ, Madura K, Bartel B, Varshavsky A. 1991. The N-end rule is mediated by the UBC2(RAD6) ubiquitin-conjugating enzyme. Proc. Natl. Acad. Sci. U. S. A. 88: 7351–7355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dover J, et al. 2002. Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J. Biol. Chem. 277: 28368–28371 [DOI] [PubMed] [Google Scholar]
- 15. Finlay CA, Hinds PW, Levine AJ. 1989. The p53 proto-oncogene can act as a suppressor of transformation. Cell 57: 1083–1093 [DOI] [PubMed] [Google Scholar]
- 16. Haupt Y, Maya R, Kazaz A, Oren M. 1997. Mdm2 promotes the rapid degradation of p53. Nature 387: 296–299 [DOI] [PubMed] [Google Scholar]
- 17. Hochstrasser M. 1995. Ubiquitin, proteasomes, and the regulation of intracellular protein degradation. Curr. Opin. Cell Biol. 7: 215–223 [DOI] [PubMed] [Google Scholar]
- 18. Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. 2002. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419: 135–141 [DOI] [PubMed] [Google Scholar]
- 19. Honda R, Tanaka H, Yasuda H. 1997. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 420: 25–27 [DOI] [PubMed] [Google Scholar]
- 20. Jentsch S, McGrath JP, Varshavsky A. 1987. The yeast DNA repair gene RAD6 encodes a ubiquitin-conjugating enzyme. Nature 329: 131–134 [DOI] [PubMed] [Google Scholar]
- 21. Jiao R, et al. 2004. Physical and functional interaction between the Bloom's syndrome gene product and the largest subunit of chromatin assembly factor 1. Mol. Cell. Biol. 24: 4710–4719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kastan MB, et al. 1991. Levels of p53 protein increase with maturation in human hematopoietic cells. Cancer Res. 51: 4279–4286 [PubMed] [Google Scholar]
- 23. Kim J, et al. 2009. RAD6-mediated transcription-coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell 137: 459–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kim J, Hake SB, Roeder RG. 2005. The human homolog of yeast BRE1 functions as a transcriptional coactivator through direct activator interactions. Mol. Cell 20: 759–770 [DOI] [PubMed] [Google Scholar]
- 25. Kim J, Roeder RG. 2009. Direct Bre1-Paf1 complex interactions and RING finger-independent Bre1-Rad6 interactions mediate histone H2B ubiquitylation in yeast. J. Biol. Chem. 284: 20582–20592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Koken MH, et al. 1996. Expression of the ubiquitin-conjugating DNA repair enzymes HHR6A and B suggests a role in spermatogenesis and chromatin modification. Dev. Biol. 173: 119–132 [DOI] [PubMed] [Google Scholar]
- 27. Koken MH, et al. 1991. Structural and functional conservation of two human homologs of the yeast DNA repair gene RAD6. Proc. Natl. Acad. Sci. U. S. A. 88: 8865–8869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Koken M, et al. 1991. Dhr6, a Drosophila homolog of the yeast DNA-repair gene RAD6. Proc. Natl. Acad. Sci. U. S. A. 88: 3832–3836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Krogan NJ, et al. 2002. COMPASS, a histone H3 (lysine 4) methyltransferase required for telomeric silencing of gene expression. J. Biol. Chem. 277: 10753–10755 [DOI] [PubMed] [Google Scholar]
- 30. Kruse JP, Gu W. 2009. Modes of p53 regulation. Cell 137: 609–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kubbutat MH, Jones SN, Vousden KH. 1997. Regulation of p53 stability by Mdm2. Nature 387: 299–303 [DOI] [PubMed] [Google Scholar]
- 32. Lane DP. 1992. Worrying about p53. Curr. Biol. 2: 581–583 [DOI] [PubMed] [Google Scholar]
- 33. Lawrence CW. 1982. Mutagenesis in Saccharomyces cerevisiae. Adv. Genet. 21: 173–254 [DOI] [PubMed] [Google Scholar]
- 34. Lee KY, Myung K. 2008. PCNA modifications for regulation of post-replication repair pathways. Mol. Cells 26: 5–11 [PMC free article] [PubMed] [Google Scholar]
- 35. Lee JS, et al. 2007. Histone crosstalk between H2B monoubiquitination and H3 methylation mediated by COMPASS. Cell 131: 1084–1096 [DOI] [PubMed] [Google Scholar]
- 36. Levine AJ. 1997. p53, the cellular gatekeeper for growth and division. Cell 88: 323–331 [DOI] [PubMed] [Google Scholar]
- 37. Li M, et al. 2003. Mono- versus polyubiquitination: differential control of p53 fate by Mdm2. Science 302: 1972–1975 [DOI] [PubMed] [Google Scholar]
- 38. Lu X. 2005. p53: a heavily dictated dictator of life and death. Curr. Opin. Genet. Dev. 15: 27–33 [DOI] [PubMed] [Google Scholar]
- 39. Lyakhovich A, Shekhar MP. 2003. Supramolecular complex formation between Rad6 and proteins of the p53 pathway during DNA damage-induced response. Mol. Cell. Biol. 23: 2463–2475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lyakhovich A, Shekhar MP. 2004. RAD6B overexpression confers chemoresistance: RAD6 expression during cell cycle and its redistribution to chromatin during DNA damage-induced response. Oncogene 23: 3097–3106 [DOI] [PubMed] [Google Scholar]
- 41. Michael D, Oren M. 2003. The p53-Mdm2 module and the ubiquitin system. Semin. Cancer Biol. 13: 49–58 [DOI] [PubMed] [Google Scholar]
- 42. Miller T, et al. 2001. COMPASS: a complex of proteins associated with a trithorax-related SET domain protein. Proc. Natl. Acad. Sci. U. S. A. 98: 12902–12907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nagy PL, Griesenbeck J, Kornberg RD, Cleary ML. 2002. A trithorax-group complex purified from Saccharomyces cerevisiae is required for methylation of histone H3. Proc. Natl. Acad. Sci. U. S. A. 99: 90–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ng HH, Xu RM, Zhang Y, Struhl K. 2002. Ubiquitination of histone H2B by Rad6 is required for efficient Dot1-mediated methylation of histone H3 lysine 79. J. Biol. Chem. 277: 34655–34657 [DOI] [PubMed] [Google Scholar]
- 45. Ni JQ, Liu LP, Hess D, Rietdorf J, Sun FL. 2006. Drosophila ribosomal proteins are associated with linker histone H1 and suppress gene transcription. Genes Dev. 20: 1959–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nislow C, Ray E, Pillus L. 1997. SET1, a yeast member of the trithorax family, functions in transcriptional silencing and diverse cellular processes. Mol. Biol. Cell 8: 2421–2436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pavri R, et al. 2006. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell 125: 703–717 [DOI] [PubMed] [Google Scholar]
- 48. Pfander B, Moldovan GL, Sacher M, Hoege C, Jentsch S. 2005. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature 436: 428–433 [DOI] [PubMed] [Google Scholar]
- 49. Roest HP, et al. 2004. The ubiquitin-conjugating DNA repair enzyme HR6A is a maternal factor essential for early embryonic development in mice. Mol. Cell. Biol. 24: 5485–5495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Roest HP, et al. 1996. Inactivation of the HR6B ubiquitin-conjugating DNA repair enzyme in mice causes male sterility associated with chromatin modification. Cell 86: 799–810 [DOI] [PubMed] [Google Scholar]
- 51. Roguev A, et al. 2001. The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4. EMBO J. 20: 7137–7148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rui Y, et al. 2004. Axin stimulates p53 functions by activation of HIPK2 kinase through multimeric complex formation. EMBO J. 23: 4583–4594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ruthenburg AJ, Allis CD, Wysocka J. 2007. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol. Cell 25: 15–30 [DOI] [PubMed] [Google Scholar]
- 54. Shema E, et al. 2008. The histone H2B-specific ubiquitin ligase RNF20/hBRE1 acts as a putative tumor suppressor through selective regulation of gene expression. Genes Dev. 22: 2664–2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Shilatifard A. 2006. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu. Rev. Biochem. 75: 243–269 [DOI] [PubMed] [Google Scholar]
- 56. Sun ZW, Allis CD. 2002. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 418: 104–108 [DOI] [PubMed] [Google Scholar]
- 57. Sung P, Berleth E, Pickart C, Prakash S, Prakash L. 1991. Yeast RAD6 encoded ubiquitin conjugating enzyme mediates protein degradation dependent on the N-end-recognizing E3 enzyme. EMBO J. 10: 2187–2193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sung P, Prakash S, Prakash L. 1990. Mutation of cysteine-88 in the Saccharomyces cerevisiae RAD6 protein abolishes its ubiquitin-conjugating activity and its various biological functions. Proc. Natl. Acad. Sci. U. S. A. 87: 2695–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Tian C, et al. 2009. KRAB-type zinc-finger protein Apak specifically regulates p53-dependent apoptosis. Nat. Cell Biol. 11: 580–591 [DOI] [PubMed] [Google Scholar]
- 60. Vousden KH, Lu X. 2002. Live or let die: the cell's response to p53. Nat. Rev. Cancer 2: 594–604 [DOI] [PubMed] [Google Scholar]
- 61. Watkins JF, Sung P, Prakash S, Prakash L. 1993. The extremely conserved amino terminus of RAD6 ubiquitin-conjugating enzyme is essential for amino-end rule-dependent protein degradation. Genes Dev. 7: 250–261 [DOI] [PubMed] [Google Scholar]
- 62. Wood A, et al. 2003. Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Mol. Cell 11: 267–274 [DOI] [PubMed] [Google Scholar]
- 63. Wood A, Schneider J, Dover J, Johnston M, Shilatifard A. 2003. The Paf1 complex is essential for histone monoubiquitination by the Rad6-Bre1 complex, which signals for histone methylation by COMPASS and Dot1p. J. Biol. Chem. 278: 34739–34742 [DOI] [PubMed] [Google Scholar]
- 64. Wood A, Schneider J, Dover J, Johnston M, Shilatifard A. 2005. The Bur1/Bur2 complex is required for histone H2B monoubiquitination by Rad6/Bre1 and histone methylation by COMPASS. Mol. Cell 20: 589–599 [DOI] [PubMed] [Google Scholar]
- 65. Yamashita K, Shinohara M, Shinohara A. 2004. Rad6-Bre1-mediated histone H2B ubiquitylation modulates the formation of double-strand breaks during meiosis. Proc. Natl. Acad. Sci. U. S. A. 101: 11380–11385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Yuan J, Luo K, Zhang L, Cheville JC, Lou Z. 2010. USP10 regulates p53 localization and stability by deubiquitinating p53. Cell 140: 384–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Zhang Y, et al. 2010. Negative regulation of HDM2 to attenuate p53 degradation by ribosomal protein L26. Nucleic Acids Res. 38: 6544–6554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zhao T, Xu Y. 2010. p53 and stem cells: new developments and new concerns. Trends Cell Biol. 20: 170–175 [DOI] [PubMed] [Google Scholar]
- 69. Zhu B, et al. 2005. Monoubiquitination of human histone H2B: the factors involved and their roles in HOX gene regulation. Mol. Cell 20: 601–611 [DOI] [PubMed] [Google Scholar]