Abstract
Since posttranslational modification (PTM) by the small ubiquitin-related modifiers (SUMOs) was discovered over a decade ago, a huge number of cellular proteins have been found to be reversibly modified, resulting in alteration of differential cellular pathways. Although the molecular consequences of SUMO attachment are difficult to predict, the underlying principle of SUMOylation is altering inter- and/or intramolecular interactions of the modified substrate, changing localization, stability, and/or activity. Unsurprisingly, many different pathogens have evolved to exploit the cellular SUMO modification system due to its functional flexibility and far-reaching functional downstream consequences. Although the extensive knowledge gained so far is impressive, a definitive conclusion about the role of SUMO modification during virus infection in general remains elusive and is still restricted to a few, yet promising concepts. Based on the available data, this review aims, first, to provide a detailed overview of the current state of knowledge and, second, to evaluate the currently known common principles/molecular mechanisms of how human pathogenic microbes, especially viruses and their regulatory proteins, exploit the host cell SUMO modification system.
INTRODUCTION
Small ubiquitin-related modifier (SUMO) was initially identified as reversible, proteinogenic posttranslational modification (PTM) by different laboratories in the mid-1990s (13, 116, 122, 125, 135). Today, it is classified as a member of the ubiquitin-like proteins (Ubls) due to its structural and sequence similarities to ubiquitin (89, 169); however, the surface properties of SUMO are quite distinct. Interestingly, it appears that the characteristic determinants of PTM by Ubls are phylogenetically ancient and may have evolved from biosynthetic pathways via repeated rounds of gene duplication and diversification (75). Consequently, SUMO is expressed by all eukaryotes but is absent from prokaryotes/archaea. Lower eukaryotes have a single SUMO gene, whereas plants and vertebrates express several SUMO paralogues.
In vertebrates, two subfamilies, namely, SUMO-1 and SUMO-2/3 proteins, are known. SUMO-2 and SUMO-3 are commonly referred to as SUMO-2/3 due to 98% sequence similarity and the lack, to date, of clearly distinguishable functional differences. Although members of each subfamily are highly similar, SUMO-1 and SUMO-2/3 share only about 50% amino acid sequence identity, although all are ∼100-residue proteins containing significant primary sequence homology to ubiquitin in the C terminus (∼20%) and a short unstructured N-terminal stretch (11, 128).
Recent research has shown important differences in the molecular functionalities of mammalian SUMO-1 and SUMO-2/3 proteins. The latter is present in higher levels than SUMO-1, whereas the unconjugated pool of SUMO-1 is lower than that of SUMO-2/3. Intriguingly, SUMO-2 and SUMO-3 can be conjugated to target proteins in a chain-wise fashion due to internal SUMO conjugation motifs (SCMs), whereas SUMO-1 lacks this ability. Moreover, some results suggest a certain degree of paralogue specificity for SUMO conjugation to distinct substrates in vivo (163), indicating differential roles in cell metabolism that are yet to be clearly defined. In humans, a fourth gene codes for SUMO-4; however, it is unclear whether its product can be conjugated to other proteins in vivo (140).
In principle, SUMO conjugation to diverse SCMs occurs by an enzymatic mechanism similar to ubiquitination (Fig. 1). However, the single E2 enzyme Ubc9 is a key component of the SUMO conjugation system and is essential for viability in most eukaryotes (5, 72, 132, 134). Although Ubc9 represents the only known E2 enzyme so far, and is therefore of unique importance for the SUMOylation pathway, it appears to additionally mediate regulatory functions in cellular metabolism independently of its E2 enzymatic activity (28, 85, 97, 144, 164, 179).
Fig 1.
Scheme of SUMO maturation, activation, conjugation, and ligation. All different SUMO isoforms are expressed as immature precursors with a variable C-terminal stretch (2 to 11 amino acids) after an essential GG motif. After maturation via the sentrin-specific proteases (SENPs) (129), the SUMO protein is activated in an ATP-dependent step by conjugation to the E1 heterodimer (Aos1/Uba2). SUMO is subsequently transferred to the unique E2 enzyme Ubc9, which covalently attaches the modifier to the ε-amino group of a target lysine residue in the presence of an E3 SUMO ligase. However, Ubc9 itself can bind the SCM signature sequence of target proteins and induce SUMO modification without an E3 ligase, indicating that the E3 enzyme may play an integral role in ensuring proper substrate specificity rather than exclusive stimulation of enzymatic conjugation itself (81). Currently, four different extensions of the classic consensus SUMO conjugation motif (SCM; ψ-K-x-E/D) have been identified: the phosphorylation-dependent sumoylation motif (PDSM; ψ-K-x-E/D-xx-pSP) (74), the negatively charged amino-acid-dependent sumoylation motif [NDSM; ψ-K-x-E-x(2-5)-E/D-x (2)-E/D] (198), an inverted SUMO conjugation motif (iSCM; E/D-x-K-ψ) (121), and the hydrophobic cluster SUMOylation motif (HCSM) (121).
Some evidence implicates misregulated SUMOylation in tumorigenesis, with detectable overexpression of the E2 conjugating enzyme Ubc9 in some human malignancies or a specific SUMO isopeptidase in others (10, 40, 43, 68, 82, 126, 127, 203). In addition, SUMOylation might be linked to neurodegenerative diseases, such as Huntington's, Alzheimer's, and Parkinson's diseases (166), and to type 1 diabetes (6, 18, 172).
Despite these proposed functions, the molecular consequences of SUMOylation for a target are difficult to predict. In general, it can be said that the underlying principle of SUMOylation is to alter a modified substrate's inter- and/or intramolecular interactions and hence its stability, localization, or activity. Some of the downstream consequences may be mediated by effectors via noncovalent SUMO interaction motifs (SIMs) (88, 173). Thus, SUMO modification of a target protein provides an additional interaction platform for recruiting SIM-containing effector proteins.
The observation that multiple cellular pathways are extensively regulated by SUMO modification, while only a low percentage of effector proteins are modified, currently represents a most puzzling question, aptly termed the SUMO enigma (71). One model suggests that SUMO is rapidly deconjugated after triggering the formation of stable protein complexes, thereby allowing global, long-lasting control of proteins via a labile, short-lived modification (71).
The biological functions of the SUMO system have been covered in many excellent reviews describing its involvement in transcriptional regulation, maintenance of genome integrity, promyelocytic leukemia protein-nuclear body (PML-NB) formation, DNA repair, subcellular localization, ubiquitin-mediated proteolysis, nuclear transport, signal transduction, and tumorigenesis (59, 69, 80, 180, 184). Intriguingly, the particular subnuclear structures called PML-NBs or nuclear domain 10 (ND10) have been implicated in comparable cellular mechanisms; the structural integrity/regulation of these accumulations and their associated proteins depend on PTM with SUMO. Besides being SUMOylated at three specific residues, the main protein component, PML, also contains a SIM (171), contributing to establishing a complex three-dimensional (3D) structure (101, 171, 187).
It has been proposed that over 165 known cellular proteins can be dynamically targeted to PML-NBs, in part depending on their SUMO modification and/or whether they contain a SIM (182). Moreover, the importance of PML-NBs as well as that of associated factors is highlighted by the fact that they are targeted by multiple viral proteins during infection as part of an intrinsic viral defense mechanism (183; reviewed in references 46, 47, and 178).
In summary, it is not surprising that various intracellular pathogens have evolved to take advantage of the conserved host cell SUMOylation machinery, by either modulating essential components or being targets of this PTM themselves (15, 190). This review focuses on elucidating the different principles of how human pathogenic microbes, especially viruses and their regulatory proteins, exploit the host cell SUMO modification system.
DNA VIRUSES
Currently, most research into how the SUMO modification system is involved in virus infection has been performed using DNA viruses. Many observations are inevitably linked to PML-NBs, since these structures have been described as an intrinsic antiviral defense mechanism, especially for nuclear replicating DNA viruses, and are considered to represent the nuclear SUMOylation hot spot (46, 47, 178, 183).
ADENOVIRIDAE
The avian adenovirus chicken embryo lethal orphan (CELO) represents one of the best analyzed model systems for modulating the cellular SUMO modification system so far (14, 31, 152), although it does not represent a human pathogen. After the initial discovery of the anti-apoptotic Gam-1 protein (32), which is functionally similar to the human adenovirus early region 1B 19-kilodalton (E1B-19K) protein, it soon became clear that Gam-1 induces the loss of PML-NBs, and in general leads to the deregulation of the SUMO pathway (33, 36). Intriguingly, the viral protein blocks cellular SUMOylation by a mechanism involving inhibition of E1-SUMO thioester-intermediate formation in vitro as well as proteasomal reduction of both E1 and E2 proteins (16). The mechanism of ubiquitin-dependent reduction of the heterodimer E1 via the CELO Gam-1 protein represents an as yet unique viral strategy to counteract the SUMOylation pathway (17).
In human adenoviruses, three proteins of a commonly established laboratory strain, human adenovirus serotype 5 (HAdV5), have been linked to the SUMO pathway. Based on conservation of the catalytic triad, structural data, and enzymatic processing of viral protein precursors (106, 113), the proteases of adenovirus, poxvirus, and African swine fever virus (ASFV) have been described as members of a new class of related cysteine proteases, which include the SUMO proteases (Ulps/sentrin-specific proteases [SENPs]) (110, 129).
HAdV5 encodes a set of early regulatory proteins, and the proteins have been identified to interact with enzymes or resemble substrates of the SUMOylation pathway. Initial experiments showed that E1A binds to murine Ubc9 (70), and these results were subsequently confirmed for the human ortholog. Interestingly, Yousef et al. further identified a specific amino acid sequence within the conserved region 2 of E1A which is necessary and sufficient to interact with the N-terminal region of the SUMO E2 enzyme (200). However, the interaction of these two proteins does not alter global SUMOylation within A549 or U2OS cells, nor is it essential for oncogenic transformation of p53-negative mouse embryonic fibroblasts (MEFs) by E1A. Although some evidence indicated that E1A interferes with polySUMOylation (200), whether and to what extent E1A mediates some of its potent transcriptional regulatory functions by modulating host cell SUMOylation are subjects for further studies (58).
In contrast, E1B-55K is itself a substrate for the host cell SUMO modification system, harboring a classical SCM around lysine 104 (44, 45). Indeed, PTM of E1B-55K is known to be involved in several aspects of viral protein function, such as functional inactivation of the tumor suppressor protein p53, and proteasomal degradation of the chromatin remodeling factor Daxx. In isolation, the modulation of these two cellular proteins subsequently determines the oncogenic potential of the E1B-55K protein in primary mammalian cells (44, 45, 167, 168). Moreover, during virus infection, E1B-55K subnuclear localization and interaction with certain isoforms of PML seemingly depend on SUMO modification of the viral protein (92, 191), although another early viral protein counteracts E1B-55K PTM via an as yet unknown mechanism (107). Recently published results show that HAdV5 E1B-55K itself acts as a SUMO E3 ligase, inducing the SUMOylation of p53 in vitro and in vivo and thereby promoting inactivation of the tumor suppressor protein via spatial restriction to PML-NBs (147).
In summary, these results indicate that HAdVs are intimately connected to the SUMOylation system, although the functional consequences for individual regulatory proteins as well as the viruses themselves are diverse and far from being understood.
HERPESVIRIDAE
The link between herpesviruses and SUMO modification is related to the herpesviruses' mode of counteracting the antiviral properties of PML-NBs. For example, during the initial stages of HSV-1 infection, the incoming viral genomes rapidly associate with newly established ND10-like nuclear accumulations. This occurs independently of de novo viral protein expression and actively restricts viral infection (50, 51, 54, 114). Based on available data with additional prototype members of different herpesvirus subgroups, it is reasonable to conclude that diverse molecular mechanisms have evolved to converge for the efficient counteracting of PML-NB-mediated antiviral activities.
HSV-1 (Alphaherpesvirinae).
Inactivation of PML-NBs during early stages of herpes simplex virus 1 (HSV-1) infection depends on expression of the immediate-early (IE) protein ICP0, which rapidly localizes to PML-NB subnuclear structures and disrupts them (22, 23, 29, 49, 123, 124). However, in the absence of ICP0, incoming viral DNA rapidly associates with de novo-formed ND10-like structures containing antiviral proteins such as PML itself, Sp100, and Daxx, a process that efficiently represses ongoing viral gene expression (39). Intriguingly, ICP0 is capable of inducing proteasomal degradation of PML, SUMO-modified PML, and the PML-associated protein Sp100 (8, 48, 130, 145), which is assumed to be a principal mechanism for HSV-1 to counteract the ND10-mediated antiviral defense barrier (52, 53).
Most recent findings provide substantial evidence that ICP0 functions by exploiting a more common cellular pathway that links SUMO modification with ubiquitin-dependent protein turnover rather than by specific targeting of individual proteins (19). Mammalian SUMO-targeted ubiquitin ligases (STUbLs), such as RNF4, resemble a recently described class of RING finger ubiquitin ligases that preferentially target SUMOylated proteins via their SIMs, thus providing a functional link between SUMOylation and ubiquitin-dependent degradation (60, 99, 148, 153, 176, 177, 181, 188, 196). Intriguingly, ICP0 possesses several STUbL-like properties, which enables it to induce global proteasomal reduction of SUMOylated proteins, particularly SUMO-modified PML (19). This observation provides a fascinating explanation of how the viral ICP0 protein may inhibit ND10 antiviral functions by depleting ND10 of its SUMOylated components and, consequently, eliminating recruitment of several repressive factors, such as Sp100 and Daxx.
VZV (Alphaherpesvirinae).
Similar to the closely related alphaherpesvirus HSV-1, efficient varicella-zoster virus (VZV) replication depends on the inactivation of PML-NBs (98, 157), although the molecular mechanism involved remains elusive. The VZV ORF61 protein resembles the functional orthologue of ICP0; however, it does not induce proteasomal degradation of PML by its RING domain but rather triggers an approximately 5-fold reduction in absolute PML-NB numbers in vitro (157).
Interestingly, colocalization and subsequent disruption of ND10 depends on a functional SIM motif within the viral ORF61 protein, facilitating efficient viral replication in human skin xenografts (186). Taken together, these results indicate that the interaction between SIM-containing viral regulatory proteins and SUMO-modified PML is conserved within the Alphaherpesvirinae, allowing them to target PML-NBs for dispersal, whereas the RING domain has variable functions, which nevertheless might include degradation of certain PML-associated antiviral proteins.
HCMV (Betaherpesvirinae).
Human cytomegalovirus (HCMV) encodes two major IE gene products that are thought to play key roles in initiating lytic replication by inhibiting PML-NB's repressive activity toward foreign DNA. However, HCMV IE1 induces nuclear dispersal of these structures in a functionally different way than that of the HSV-1 ICP0/VZV ORF61 protein. IE1 localizes to a mixture of nuclear diffuse and punctate PML-positive accumulations at initial stages of infection, shifting to an exclusively diffuse distribution at later times. In contrast, IE2 remains juxtaposed to nuclear accumulations that resemble preexisting PML-NBs (3, 87). Obviously, the two viral proteins share an intrinsic capability to associate with PML-NBs, but the functional consequences remain clearly distinct.
HCMV IE2 is a well-characterized transcriptional regulator mediating either repression of its own promoter (73, 149) or activation of other viral early promoters (66, 117), and its transactivating function is believed to allow progression of the viral replicative cycle. Interestingly, IE2 has been identified to interact with the E2 enzyme Ubc9, becoming modified at two specific lysine residues (K175, K180) by different SUMO isoforms (4, 76, 162).
In contrast to the notion that SUMO attachment may facilitate targeting of IE2 to PML-NBs during transient transfection, subnuclear targeting of the viral protein occurs independently of this PTM. However, reporter gene experiments imply that the transactivation capabilities of IE2 depend on its SUMOylation, possibly providing an additional interaction platform for recruiting the necessary cellular cofactors, such as TAF12 (4, 9, 76, 91).
Recent data further indicate that IE2 colocalizes and interacts with the SUMO E3 ligase protein inhibitor of activated STAT-1 (PIAS1), which significantly augments SUMO modification and transcriptional activity of the viral protein during exogenous overexpression (105). Biochemical evidence additionally confirmed that the IE2 protein contains a SIM motif, which is apparently necessary for efficient SUMO modification of the viral protein (12, 91). Consistently, mutations in the SIM and/or the SUMOylation sites impaired IE2-mediated transactivation of the viral early promoters.
Notably, replication of IE2 mutant viruses was severely repressed in normal human fibroblasts. Closer analysis of viral growth indicated that the replication defect correlates with a low-level accumulation of SUMO-modified IE2 (12, 91). Although these results are interesting, further work is required to evaluate the importance of IE2 PTM by SUMO in vivo, since studies with alternative laboratory HCMV strains provide somewhat conflicting results (12, 103).
Similarly, the IE1 protein is apparently modified by SUMO-1 at lysine 450, although phosphorylation of the viral protein may negatively regulate SUMO attachment (162, 174). However, both wild-type (wt) IE1 and the SUMOylation-deficient mutant IE1K450R not only are capable of localizing to and disrupting PML-NBs but also show no significant differences in protein stability, transactivation, or virus growth (174).
Importantly, in contrast to the HSV-1 ICP0-mediated mechanism of PML modulation, IE1-induced loss of high-molecular-weight PML is resistant to proteasome inhibitors, and no reduction of PML itself is observed (104, 130). Hence, it is not yet clear how IE1 limits SUMO modification of PML, although several potential direct or indirect mechanisms have been discussed (83, 104).
At present, two main downstream consequences of IE1 modification have been described. First, an IE1-SUMOylation-deficient virus mutant grew more slowly and produced a lower yield in human fibroblasts than that of wild-type virus, suggesting that SUMO modification is required for IE1 activity. However, intranuclear localization, PML-NB disruption, and transactivation of viral promoters were not influenced, although substantially reduced IE2 transcript and protein levels were observed (133). Second, SUMOylation of IE1 was found to inhibit binding to STAT2 and therefore repression of interferon (IFN)-responsive genes, resulting in about three orders of magnitude less viral growth after beta interferon stimulation. Hence, it appears that IE1 SUMOylation impairs the ability of the viral protein to interfere with cellular interferon signaling (77).
Many of the basic aspects described for HCMV IE proteins apply to the corresponding proteins of the closely related betaherpesvirus human herpesvirus 6 (HHV-6); however, more detailed studies are necessary to draw any final conclusions (63, 64).
EBV (Gammaherpesvirinae).
Similar to the alpha- and betaherpesviruses discussed above, the gammaherpesvirus Epstein-Barr virus (EBV) encodes an immediate-early protein called Zta (BZLF1), which is capable of disrupting PML-NBs. Besides manipulating the host cell, this viral protein is required for efficient expression of the EBV early genes and plays an essential role in lytic replication. Zta itself is a substrate for the host cell SUMOylation system involving SUMO-1, SUMO-2, and SUMO-3, although the modified residue K12 is not part of the classical consensus motif ψ-K-x-E/D (2, 67, 131).
Interestingly, it has been suggested that Zta outcompetes PML for the limiting pool of endogenous SUMO molecules, thereby inducing the loss of high-molecular-weight PML, i.e., a functionally different path from that of IE1 or ICP0. However, since SUMOylation-deficient mutants of Zta remain partially capable of dispersing PML-NBs, other mechanisms must be taken in account (2, 67). Moreover, modification with SUMO itself does not influence the stability or localization of the viral protein, as shown with ZtaK12A or ZtaK12R, but seemingly decreases the viral protein's transactivation abilities on certain viral promoters. Since Zta serves as one molecular switch in EBV's alternative life cycles, it has been suggested that SUMO modification of the viral protein decreases this protein's ability to transactivate viral gene expression and, consequently, favors triggering the establishment of latency rather than lytic replication (1, 67).
A recent model proposed by Murata et al. convincingly links the attenuation of Zta's transcriptional activity by SUMO with the recruitment of cellular HDAC3/HDAC7 proteins, since treatment with the HDAC inhibitor trichostatin A completely reverses the inhibitory effect of Zta SUMOylation (131). In contrast, the EBV-encoded protein kinase, EBV-PK, somehow counteracts SUMO modification of Zta, indicating that the interplay of these viral proteins and the host cell SUMOylation machinery is an important factor in the equilibrium of the lytic versus latent status of EBV (67).
The second immediate-early protein Rta of EBV is also SUMOylated at three specific lysine residues (K19, K213, K517). These results, together with the observation that Rta binds to the E2 enzyme Ubc9 and several PIAS proteins in vitro and in vivo, renders Rta a bona fide substrate of the SUMOylation system, although this protein does not contain any classical SCM (25, 112). Interestingly, Rta modification augments the capacity of the viral protein to transactivate certain promoters, although the mechanism remains undefined (25).
Last, EBNA3C is known to be modified by SUMO-1, SUMO-2, and SUMO-3. However, PTM is necessary not for the transactivation potential of the viral protein but rather to direct its subnuclear localization to PML-NBs (161).
KSHV (Gammaherpesvirinae).
The K-bZIP protein of Kaposi's sarcoma-associated herpesvirus (KSHV), among the earliest protein to be expressed after acute infection or reactivation of the latent genome, resembles the structural analogue of the EBV transcription factor Zta. K-bZIP appears to be modified by all three SUMO isoforms at residue K158 in vitro and in vivo, evidently modified in a chain-wise fashion by SUMO-2/3. However, like EBV Zta and HCMV IE1/IE2, PTM does not influence protein stability or localization of K-bZIP to PML-NBs (78). On the other hand, experiments including a SUMOylation-deficient mutant of K-bZIP confirmed that SUMOylation is largely but not exclusively responsible for transcriptional transrepression by K-bZIP and that this activity depends on a physical interaction with enzymatically active Ubc9 (78, 93). Although the detailed molecular mechanism is open for debate, it has been proposed that K-bZIP functions as a viral SUMO ligase or SUMO adaptor, allowing recruitment of Ubc9 to KSHV promoter sites, followed by transcriptional repression.
Consistent with this theory, Chang et al. recently showed that K-bZIP harbors a SIM that binds exclusively to SUMO-2/3 and not to SUMO-1 (26). Through this motif, K-bZIP induces SUMO modification of itself as well as certain interaction partners, such as the tumor suppressor proteins p53 and pRB, thus representing the first-described viral SUMO isoform-specific E3 ligase. Hence, K-bZIP is recruited to p53 target sites via its SIM and affects multiple p53 downstream genes in a SUMO-dependent manner (26). These results provide a new aspect for interpreting the molecular mode of K-bZIP induction of growth arrest during viral infection by alteration of the SUMOylation level of certain host cell factors.
In parallel, the viral LANA2 protein was shown to exploit the SUMO modification system in a functionally different way to ensure cell viability in KSHV-infected primary effusion lymphoma (PEL) cells (118, 119). In the presence of LANA2, PML is decreased in vivo via a proteasome-dependent mechanism involving enhanced modification of PML by SUMO-2 and ubiquitin, ultimately leading to significant alterations in PML-NB structure. The evidence provided by Marcos-Villar et al. argues that PML reduction is achieved by SUMO-dependent ubiquitinylation, although final proof for the involvement of STUbLs such as RNF4 remains to be shown (118, 119). Moreover, this activity requires the functional integrity of both the LANA2 SIM and several lysine residues that resemble the amino acids responsible for covalent attachment of SUMO-1 and SUMO-2 (118). These results suggest that KSHV has developed a fascinating mechanism to counteract PML-NB functions, which may in fact play a significant role in KSHV-mediated tumorigenesis.
Papillomaviridae.
To date, several studies with human papillomavirus (HPV) and bovine papillomavirus (BPV) clearly show an association of these DNA viruses with the host cell SUMO modification system. Indeed, both viral E1 (vE1) and vE2 proteins seem to interact with Ubc9 and to be targets for covalent SUMO attachment. Initial studies of HPV type 16 (HPV16) (199) and BPV (155) clearly identified an association of vE1 with the Ubc9 enzyme, although the functional consequences remain poorly defined. Analysis of different mutants as well as the conservation of critical residues identified the binding domain for Ubc9 on vE1 of BPV, while comprehensive mutational studies support the notion that SUMO attachment is important for nuclear accumulation of vE1 and, thus, efficient replication of certain papillomaviruses (155, 156). Moreover, initial evidence indicates that the PIAS family of E3 ligases may be involved in regulating vE1 SUMOylation (160). Recent data somewhat questions the exclusive dependency of vE1's molecular function on the SUMOylation pathway or interaction with Ubc9 (56), making this a subject of further studies to determine whether the proposed mechanisms are globally applicable to the different virus types and how experimental systems influence the respective results.
Intriguingly, HPV vE2 is readily SUMOylated in vitro and in vivo, with a preference for SUMO-2/3 conjugation in vivo. Apparently, exogenous overexpression of SUMO-2/3 with Ubc9 significantly stabilizes vE2 protein levels, whereas SUMO-1 fails to do so. This increase in stability is specific for vE2 but does not depend on covalent attachment of SUMO-2/3 to the viral protein, since inactivation of the predominantly modified lysine residue does not abolish the observed effect (193–195). This mechanism argues against simple competition between ubiquitin and SUMO for covalent attachment sites, rather suggesting that it is mediated through SUMOylation of a secondary cellular target(s).
A comparable stabilization effect has been described for the minor capsid protein L2 (120), which plays a critical role not only in generating infectious viral particles but also during early stages of HPV infection (165). L2 is covalently modified by SUMO-1 and, preferentially, SUMO-2/3 at lysine 35, thereby drastically enhancing the half-life of the capsid protein (120). However, only un-SUMOylated L2 was capable of interacting with the second capsid protein L1, demonstrating that SUMO modification has a detrimental effect on L2's capacity to bind L1.
Apparently, L2 is also capable of inducing a short-lived change in the SUMOylation status of host cell proteins by specifically upregulating endogenous SUMO-2/3 modification (120), although the mechanistic background remains unknown. Intriguingly, during keratinocyte differentiation, both SUMO-2/3 and Ubc9 are gradually upregulated, whereas SUMO-1 levels are unaltered (41). It is fascinating that the vE2/L2 protein levels, and presumably viral protein functions, may be directly linked to the levels of the host cell SUMOylation enzymes that in turn are coupled with the differentiation process in infected skin keratinocytes.
Poxviridae.
Currently, two proteins of vaccinia virus (VV) have been reported to associate with the host cell SUMOylation machinery. Initially, the E3L protein was identified as interacting with SUMO-1 in a yeast two-hybrid screen (159), and recently published results confirm the presence of a SIM within the viral protein. The SIM is required for efficient SUMO modification by SUMO-1 and SUMO-2 at lysine 40/99, protein stability, and nuclear localization of the viral E3L protein (62). However, mutational inactivation of E3L SUMOylation does not decrease efficient viral replication, although PTM of the viral protein by SUMO appears to impair transcriptional activation of p53 via a so far unknown mechanism.
The VV A40R protein is a nonstructural, type II membrane glycoprotein possibly involved in forming so-called cytoplasmic mini-nuclei that resemble endoplasmic reticulum (ER)-enwrapped virus factories for cytoplasmic replicating poxviruses. Interestingly, A40R is the only viral protein described so far that appears to be quantitatively modified by SUMO-1 at lysine 95. In this context, SUMO modification prevents self-association and insolubility of the viral protein, allowing proper cytoplasmic targeting of A40R to the viral replication sites. Although it has been suggested that SUMOylation of A40R may play a role in forming VV's cytoplasmic replication sites, as well as in viral replication itself and/or late viral gene transcription (143), further studies are needed to draw a final conclusion.
In addition, the poxvirus I7 protease shares functional and structural similarities with the SENP proteins, suggesting that I7 and the SENP proteins can be grouped into a specific kind of cysteine protease family (110, 129).
RNA VIRUSES
With a few exceptions, the role of SUMOylation for RNA viruses is poorly defined, although significant findings do suggest that a functional linkage is almost undeniable. Therefore, some of the following sections are limited to illustrating specifically described examples of the prototype members of certain virus families rather than describing the detailed functional consequences of SUMO modification.
Bunyaviridae.
In 2003, three studies provided the first evidence of an association between proteins from bunyaviruses and the host cell SUMOylation machinery (84, 102, 115). Although different species of the genus Hantavirus were used (Hantaan virus [HTNV], Tula virus [TULV], Seoul virus [SEOV]), the collective data identified the nucleocapsid protein (NP) as interacting with SUMO-1, Ubc9, and certain cellular E3 ligases of the PIAS family (PIAS1, PIASxβ), suggesting an evolutionarily conserved mechanism across the different species. While NP contains several conserved SCMs, no covalent SUMO attachment could be detected (84, 102, 115) even though both proteins were found to colocalize in perinuclear aggregates. Pertinently, separate overexpression of Ubc9 and NP indicated prominent localization of both proteins to the perinuclear region, where viral replication and assembly is thought to occur during productive infection. Although certain links between the host cell SUMOylation machinery and hantavirus in general are evident, the functional consequences for the virus life cycle remain to be clarified.
Coronaviridae.
Similar to Hantavirus NP, the severe acute respiratory syndrome coronavirus (SARS-CoV) nucleocapsid (N) protein is known to form the ribonucleocapsid together with the genomic viral RNA. However, multiple functions have been postulated for this multifunctional protein throughout the virus life cycle, including inhibition of host cell proliferation and/or apoptosis. Biochemical and mutagenesis studies by Li et al. showed that the SARS-CoV N protein is posttranslationally modified by SUMO-1 at lysine 62 (108, 109). Functionally, SUMOylation seems to promote homo-oligomerization of N, which might have a profound impact on the formation of viral ribonucleoprotein (RNP) complexes and nucleocapsid assembly. In line with this observation, two recently published studies identified the N protein as interacting with the E2 enzyme Ubc9 (55, 108). Unusually, this interaction is independent of the N protein SCM but depends on an evolutionarily conserved serine/arginine-rich motif, suggesting that different species of coronavirus may interact with Ubc9. Although this interaction may regulate the activity of Ubc9, affecting downstream signaling via modulation of cellular SUMOylation, more comprehensive studies are needed for resilient results.
Filoviridae.
Ebola Zaire virus (EBOV) is one of the most pathogenic human viruses known, with fatality rates as high as 90%. Dendritic cells (DCs) and macrophages are the main initial targets of infection, and counteracting the type I IFN response represents one of the main pathogenicity mechanisms of EBOV (7). Mechanistically, the viral protein VP35 induces covalent modification of IFN regulatory factor 7 (IRF7) and IRF3 by SUMO-1 and SUMO-2/3 via the cellular E3 ligase PIAS1 (7, 27, 96). This process involves binding of VP35 to PIAS1 and the subsequent SUMOylation of IRF3/IRF7, which significantly decreases IFN promoter activity. The present proposed model of exploiting a cellular E3 SUMO ligase and subsequently inactivating the host cell interferon system represents an ingenious mechanism used by these viruses to subvert basic host regulation pathways and promote efficient replication.
Flaviviridae.
Dengue viruses (DENV) are important mosquito-borne human pathogens, representing one of the leading causes of morbidity and mortality in subtropical and tropical areas. A recent study identified the cellular Ubc9 protein as an interaction partner of the envelope (env) protein of DENV serotype 2 both in vitro and in vivo (34). Specifically, this interaction depends on residues K51 and K241 of env; however, preliminary experiments showed neither association with SUMO-1 nor SUMO modification of env itself. Moreover, some evidence points to overexpression of Ubc9 inducing slight changes in the localization of the viral protein during cotransfection. However, it remains to be determined to what extent this affects the observed reduction in plaque formation of DENV in the presence of exogenous Ubc9 (34).
Orthomyxoviridae.
To date, very little is known about the importance of SUMO modification for the replication of orthomyxoviruses, in particular their most prominent member, influenza virus. Initial evidence for an association came from two large-scale screenings for cellular proteins that were either required for efficient influenza multiplication (94) or able to interact with various influenza proteins (170). Recently published results indicate that at least five viral proteins (NS1, PB1, NP, M1, NS2) are capable of being SUMO modified in vitro, and four of these (NS1, PB1, NS1, M1) seem to be modified in vivo during productive infection (141, 142, 192, 197).
Comprehensive in vitro analysis and in silico comparisons indicate that the SCMs involved are conserved among human and avian strains, pointing to an evolutionarily conserved mechanism (142, 197). Intriguingly, influenza virus is capable of triggering a global increase in cellular SUMOylation, whereas in turn, global knockdown of Ubc9 by short hairpin RNA (shRNA) drastically impairs influenza virus production (192), clearly indicating a functional role of the SUMO modification system in efficient infection.
It appears that PTM of the M1 protein at lysine 242 is essential for assembly of the vRNP-M1 complex, efficient nuclear export, and consequently, virus morphogenesis/release (192). NS1 itself interacts with the E2 enzyme Ubc9 to possibly promote its own SUMOylation at lysine 219/221. Although the molecular consequences of PTM for most of the viral proteins remain enigmatic, SUMO modification of NS1 has been implicated in protein stability and enhanced virus growth (197). Together, the data strongly suggest a more complex interaction between this cellular PTM system and influenza virus, although it remains possible that SUMOylation exerts an indirect effect on influenza infection simply by regulating type I IFN production.
Paramyxoviridae.
Parainfluenza virus 5 (PIV5) is a prototype member of paramyxoviruses, which include important animal and human pathogens, such as measles and mumps virus. The viral P protein is an essential cofactor of the viral RNA-dependent RNA polymerase, which has been shown to be modified within a classic SCM by SUMO-1, but not SUMO-2 or SUMO-3. However, since PIV5 PK254R still shows residual SUMOylation, additional conjugation sites other than K254 are likely (175).
When monitoring of the activity of a viral minigenome, elimination of P protein PTM significantly decreased viral gene expression, an effect that could not be directly attributed to structural changes in the PIV5 RNA-dependent RNA polymerase. Similar observations about the role of K254 in SUMO conjugation and its importance for genome activity, mRNA synthesis, and viral protein expression have been made during virus infection (175). It therefore appears that modification of P is necessary for efficient viral transcription by the PIV5 RNA-dependent RNA polymerase, probably by recruiting as yet unknown host cell factors to the viral transcription sites.
Picornaviridae.
Enterovirus 71 (EV71), which is closely related to poliovirus, is a member of the family Picornaviridae that mainly causes infections in early childhood and is seen only in rare outbreaks in adults. Typically for these viruses, a large polyprotein synthesized from the viral RNA is processed via viral proteases such as EV71 protease 3C. Recent work by Chen et al. identified EV71 3C as a substrate of the SUMO modification system, and modification at lysine 52 can be seen both in vitro and in vivo during virus infection (30). Besides the observation that SUMO-1 as well as SUMO-2 are attached to EV71 3C, the data also support for the first time the assumption that endogenous SUMO-1 itself is capable of forming poly-SUMO chains, despite its lack of an internal SCM, unlike SUMO-2 or SUMO-3. Furthermore, Ubc9 was identified as an interaction partner of EV71 3C, seemingly contributing to efficient 3C SUMOylation itself.
Intriguingly, PTM seems to augment ubiquitination and significantly reduce the half-life of the viral protein, whereas mutational inactivation of the SCM completely reversed these phenotypes (30). More significantly, modification at K52 attenuated 3C-mediated substrate cleavage of endogenous cellular proteins, apoptosis induction, and viral neurovirulence in mice. Evidence from clinically isolated strains support the hypothesis that SUMO modification and the Ubc9/3C interaction have important medical implications, while posttranslational modification of 3C serves as a host cell defense mechanism counteracted by evolving site-specific EV71 escape mutants (30). Although coxsackievirus B5 influences the host cell SUMOylation system to some extent (61), far more research is necessary to define the role of this PTM for picornaviruses in general.
Retroviridae.
Since HIV-1 represents one of the best-studied retroviruses due to its pathogenicity in humans, it has been characterized for PTM by SUMO. The C-terminal part of the HIV-1 gag polyprotein contains a small proline-rich domain that matures via the viral protease into p6. In the virion of HIV-1, p6 is the major phosphoprotein, mediating late steps in virion budding (57). In this context, it has been suggested that p6 serves as a docking site, and its mono-ubiquitination is necessary for recruiting the respective host cell components during the process of virion release (139, 150, 151, 185).
However, recently p6 was found to interact with SUMO-1 and the conjugation enzyme Ubc9, and further biochemical evidence showed that covalent modification occurs at lysine 27 at an SCM conserved among most HIV strains (65). Upon its overexpression, free SUMO-1 was incorporated into nascent virions, causing a 5-fold decrease in the infectivity of these particles. The inhibitory effect depends on covalent SUMO modification of p6, whereas incorporation of SUMO-1 occurs via a mechanism independent of p6 lysine, since virus infection with a p6K27 mutant is resistant to the detrimental effects of SUMO-1 despite comparable levels of incorporated SUMO-1. Although the molecular mechanism remains unclear, it has been suggested that SUMOylation may compete with monoubiquitination of p6, thereby inhibiting proper recruitment/incorporation of host cell factors during the budding process of infectious virions. Consequently, these viruses display defects in early stages of infection (65).
More recent studies elaborated on previous findings initially from Mason-Pfizer monkey virus (MPMV) (189) and Moloney murine leukemia virus (MoMuLV) (201), similarly showing that the gag protein of HIV-1 interacts with Ubc9 (79). Jaber et al. demonstrated that RNA interference (RNAi)-mediated knockdown of Ubc9 resulted in normal numbers of virions; however, these particles were up to 10-fold less infectious than those produced in the presence of Ubc9 (79). Comprehensive biochemical studies indicate that the amount of gp120 packaged into particles is significantly reduced in Ubc9 RNAi-treated cells compared to the amount in control cells. These results indicate that Ubc9 functions during the virus assembly process, particularly in the processing/packaging of mature envelope glycoproteins. However, experiments with a trans-dominant-negative mutant of Ubc9 suggest that the protein may function independently of its E2 SUMO-conjugating activity and, consequently, in a SUMO-independent manner (79).
In addition, it has been reported that the HIV-1 integrase (IN) is modified by all three isoforms of SUMO, and mutational inactivation of the phylogenetically conserved SCMs leads to reduced infectivity and slower replication kinetics in both epithelial and T cell lines (202). Cells infected with viruses harboring mutations in the SCMs of IN showed significantly fewer integration events than wt-infected control cells. Since the enzymatic activity of the HIV-1 integrase was not affected by the respective mutations, it has been suggested that SUMO attachment might serve as an additional interface on IN, again altering its affinity for certain cofactors that are required for efficient integration of the HIV genome into the host cell (202). Further studies will be required to clarify the molecular mechanisms by which the cellular SUMOylation system participates in the control of HIV-1, although sophisticated techniques provide strong evidence for an important role of the host cell SUMO modification system in HIV-1 replication in general (21, 95).
Besides HIV-1-mediated immunodeficiency, two other human diseases, namely, adult T-cell leukemia and human T-cell leukemia virus type 1 (HTLV-1)-associated myelopathy/spastic paraparesis, are caused by the human pathogenic retrovirus HTLV-1. Both diseases have been linked to expression of the viral regulatory protein Tax, a potent transcriptional activator of viral genes and host cell genes. Interestingly, Tax has been described as a viral oncogene with profound tumorigenic potential in mice and in primary lymphocytes or fibroblasts. To a large extent, Tax's oncogenic properties are associated with its ability to constitutively activate NF-κB signaling (20, 86, 111, 146).
Two separate studies have impressively shown that Tax is a substrate for the SUMO modification machinery, regulating its subcellular partitioning between the nucleus and cytoplasm depending on different PTMs (90, 100). Although SUMOylation of Tax is not sufficient for activating the NF-κB pathway, PTM modification is absolutely necessary to enable Tax to relocalize some components of the NF-κB pathway to specific subnuclear structures (90, 100). Furthermore, Tax induces the SUMOylation of NF-κB essential modulator (NEMO) in these characteristic Ubc9-positive subnuclear domains (90). These data indicate that PTM by SUMO dictates Tax's intracellular fate at the level of trafficking and localization, although the precise functional significance of this in relation to the NF-κB pathway remains to be identified.
EXTRA- AND INTRACELLULAR BACTERIA
In the last few years, several impressive studies have convincingly linked the host cell SUMO modification system with various extra- and intracellular bacteria. Although this field is still in its infancy, SUMOylation seemingly plays an important role in the infectivity and pathogenicity of bacteria. Yersinia species encode a type III secretion system and several effector proteins known as Yersinia outer proteins (Yops) that alter crucial signaling pathways inhibiting phagocytosis and the immune response in animal and plant hosts (37, 136).
Bioinformatic predictions suggest that YopJ has a secondary structure very similar to that of the adenovirus protease (AVP) or the yeast ubiquitin-like protein protease 1 (Ulp-1), both prototypes of cysteine proteases such as the mammalian SENPs. Interestingly, Orth et al. showed that this enzymatic activity of YopJ family proteins induces a reduction in conjugated and unconjugated SUMO-1 in host cells. Hence, it has been suggested that YopJ's interference with SUMO modification in general might represent a conserved mechanism interfering with diverse cellular signaling pathways (38, 110, 138). Since many different bacterial pathogens encode YopJ homologues (e.g., Salmonella, Rhizobium), further studies must determine the interplay with the host cell SUMO machinery.
Notably, the human pathogen Listeria monocytogenes uses a mechanistically different approach to decrease the levels of cellular SUMO-conjugated proteins. The bacterial virulence factor listeriolysin O (LLO) induces proteasome-independent degradation of the E2 enzyme Ubc9 and proteasome-dependent degradation of some SUMOylated proteins. In line with the observation that membrane association and the pore-forming activity of LLO are essential for Ubc9 degradation, the mechanistically related bacterial toxins perfringolysin (PFO; Clostridium perfringens) and pneumolysin (PLO; Streptococcus pneumoniae) show a comparable decrease in cellular Ubc9 levels (35, 158). Thus, other extracellular pathogens are seemingly able to target the SUMO modification system with mechanisms similar to that of L. monocytogenes, arguing for an evolutionarily conserved strategy in some pathogenic bacteria to increase infectivity by restricting SUMO-dependent host cell responses.
CONCLUDING REMARKS
SUMO modification is an important and widely used reversible modification system within eukaryotic cells. Every possible consequence has been described for target SUMOylation; however, a simplified view is that it influences mainly protein stability, subcellular localization, or transcriptional activity. Future discoveries about the molecular mechanisms of mono-/polySUMOylation by different SUMO paralogues, the components/motifs involved, and the respective downstream consequences are needed in order to define the role of this PTM in cell metabolism in sufficient detail. As with most successful regulatory pathways, viruses have undeniably evolved multiple strategies to exploit the host cell SUMOylation system.
Currently, most data related to SUMO modification and DNA viruses are inevitably linked to PML-NBs, since these structures provide a unifying framework as a nuclear SUMOylation hot spot and an intrinsic, antiviral defense barrier (46, 47, 178). Consequently, over three dozen viruses that show a certain degree of association with these subnuclear structures have now been identified (183). However, the mode of modulating and/or counteracting such interactions is best analyzed for adenoviruses and herpesviruses (Fig. 2).
Fig 2.
Modulation of PML-NBs by DNA viruses (Adenoviridae, Herpesviridae). The different adeno- and herpesvirus proteins known to be associated with PML-NBs are shown at the top, functional alterations of the cellular SUMOylation pathway are highlighted (gray), and the resulting alterations in PML-NB morphology are illustrated at the bottom. Detailed information and references are given in the text.
Unlike many other viral proteins, the CELO Gam-1 protein is capable of global deregulation of SUMOylation by decreasing the amount of SUMO E1 and E2 enzymes. Naturally, it is possible that this mechanism also decreases endogenous PML levels and disrupts PML-NBs; however, the precise molecular mechanism remains elusive (36). For HAdV5, several early regulatory proteins have been described to be associated with ND10, PML-associated proteins, and SUMOylation. Nonetheless, the substantial reorganization of PML-NBs by E4orf3 in so-called track-like structures currently remains a unique mechanism of ND10 inactivation (24, 42, 154) and a mechanistic mystery.
In contrast, several mechanisms of ND10 inactivation and/or deregulation of SUMO modification have been described for prototype members of herpesviruses (Fig. 2). Both the HSV-1 ICP0 and VZV ORF61 proteins contain SIMs, which enable them to efficiently localize to PML-NBs during the early phase of infection and induce their disruption. However, mechanistically, ICP0 acts as a STUbL-like protein and preferentially induces the proteasome-dependent loss of SUMO-modified PML, PML itself, and certain PML-associated proteins, whereas ORF61 triggers a proteasome-independent decrease in the total number of PML-NBs.
Intriguingly, the KSHV protein LANA2 phenotypically uses the same approach to impair PML-NB function as ORF61; however, the mechanism depends on an intact LANA2 SIM proteasomal degradation and most likely occurs via a STUbL-like mode, as for ICP0. Besides, it is possible that the identified SIMs within ICP0 and ORF61 additionally enable the viral proteins to target and selectively inactivate SUMO-modified PML components (e.g., Daxx, ATRX, Sp100) known to participate in the innate cellular defense barrier toward viral infection.
Both HCMV and EBV do not induce reduction of endogenous PML but rather trigger the loss of PML SUMOylation and, consequently, the disintegration of PML-NB structures, leading to a diffuse distribution within the nucleus of infected host cells. Yet again, the mechanistic aspects of PML-NB inactivation by betaherpesviruses remain for future studies.
Although the precise mechanisms involved remain unknown, SUMO modification of viral proteins themselves seems to play a major role in the transcriptional capabilities of IE1, IE2, Zta, and Rta. Unfortunately, it is still difficult to predict the functional outcome of SUMOylation modulation by viral proteins on transcription regulation in general. In part, this is due to the promiscuity of PTM by SUMO for transcription factor regulation, either inducing activation or repression, depending on the corresponding transcription factor. Moreover, PTM by SUMO significantly influences the localization and/or interaction capabilities of transcription factors, providing an additional interaction platform for recruiting cofactors, such as TAF12 in the case of IE2. Therefore, evaluations of additional common principles of how viruses exploit the cellular SUMOylation systems, particularly in the field of RNA viruses, are subjects of ongoing studies.
Although a remarkable amount of data have been generated using different model systems (Fig. 3), we are just beginning to understand the basic principles of the intimate relationship between pathogens and the cellular SUMOylation system. Nevertheless, this relationship will definitely provide further significant insights for molecular biology in general and may represent a promising target for drug-based interference in particular.
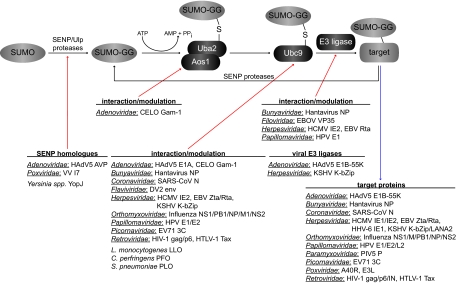
Fig 3.
Links between human pathogens and the host cell SUMOylation system. The scheme illustrates the links between proteins of human pathogenic microbes and the corresponding components of the SUMO modification system. The respective proteins are grouped according to the corresponding virus families/bacteria and listed alphabetically. Detailed information and references are given in the text.
ACKNOWLEDGMENTS
We apologize for having to omit citations or detailed discussions of many important observations and publications, in particular those that used plant and animal pathogens, due to space limitations.
The Heinrich Pette Institute is supported by the Freie und Hansestadt Hamburg and the Bundesministerium für Gesundheit.
Footnotes
Published ahead of print 9 November 2011
REFERENCES
- 1. Adamson AL. 2005. Effects of SUMO-1 upon Epstein-Barr virus BZLF1 function and BMRF1 expression. Biochem. Biophys. Res. Commun. 336:22–28 [DOI] [PubMed] [Google Scholar]
- 2. Adamson AL, Kenney S. 2001. Epstein-Barr virus immediate-early protein BZLF1 is SUMO-1 modified and disrupts promyelocytic leukemia bodies. J. Virol. 75:2388–2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ahn JH, Hayward GS. 1997. The major immediate-early proteins IE1 and IE2 of human cytomegalovirus colocalize with and disrupt PML-associated nuclear bodies at very early times in infected permissive cells. J. Virol. 71:4599–4613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ahn JH, Xu Y, Jang WJ, Matunis MJ, Hayward GS. 2001. Evaluation of interactions of human cytomegalovirus immediate-early IE2 regulatory protein with small ubiquitin-like modifiers and their conjugation enzyme Ubc9. J. Virol. 75:3859–3872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Alkuraya FS, et al. 2006. SUMO1 haploinsufficiency leads to cleft lip and palate. Science 313:1751. [DOI] [PubMed] [Google Scholar]
- 6. Aribi M. 2008. Candidate genes implicated in type 1 diabetes susceptibility. Curr. Diabetes Rev. 4:110–121 [DOI] [PubMed] [Google Scholar]
- 7. Babich A, Feldman LT, Nevins JR, Darnell JE, Weinberger C. 1983. Effect of adenovirus on metabolism of specific host mRNAs: transport control and specific translation discrimination. Mol. Cell. Biol. 3:1212–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bailey D, O'Hare P. 2002. Herpes simplex virus 1 ICP0 co-localizes with a SUMO-specific protease. J. Gen. Virol. 83:2951–2964 [DOI] [PubMed] [Google Scholar]
- 9. Barrasa MI, Harel N, Yu Y, Alwine JC. 2003. Strain variations in single amino acids of the 86-kilodalton human cytomegalovirus major immediate-early protein (IE2) affect its functional and biochemical properties: implications of dynamic protein conformation. J. Virol. 77:4760–4772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bawa-Khalfe T, Cheng J, Lin SH, Ittmann MM, Yeh ET. 2010. SENP1 induces prostatic intraepithelial neoplasia through multiple mechanisms. J. Biol. Chem. 285:25859–25866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bayer P, et al. 1998. Structure determination of the small ubiquitin-related modifier SUMO-1. J. Mol. Biol. 280:275–286 [DOI] [PubMed] [Google Scholar]
- 12. Berndt A, Hofmann-Winkler H, Tavalai N, Hahn G, Stamminger T. 2009. Importance of covalent and noncovalent SUMO interactions with the major human cytomegalovirus transactivator IE2p86 for viral infection. J. Virol. 83:12881–12894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Boddy MN, Howe K, Etkin LD, Solomon E, Freemont PS. 1996. PIC 1, a novel ubiquitin-like protein which interacts with the PML component of a multiprotein complex that is disrupted in acute promyelocytic leukaemia. Oncogene 13:971–982 [PubMed] [Google Scholar]
- 14. Boggio R, Chiocca S. 2005. Gam1 and the SUMO pathway. Cell Cycle 4:533–535 [DOI] [PubMed] [Google Scholar]
- 15. Boggio R, Chiocca S. 2006. Viruses and sumoylation: recent highlights. Curr. Opin. Microbiol. 9:430–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Boggio R, Colombo R, Hay RT, Draetta GF, Chiocca S. 2004. A mechanism for inhibiting the SUMO pathway. Mol. Cell 16:549–561 [DOI] [PubMed] [Google Scholar]
- 17. Boggio R, Passafaro A, Chiocca S. 2007. Targeting SUMO E1 to ubiquitin ligases: a viral strategy to counteract sumoylation. J. Biol. Chem. 282:15376–15382 [DOI] [PubMed] [Google Scholar]
- 18. Bohren KM, Nadkarni V, Song JH, Gabbay KH, Owerbach D. 2004. A M55V polymorphism in a novel SUMO gene (SUMO-4) differentially activates heat shock transcription factors and is associated with susceptibility to type I diabetes mellitus. J. Biol. Chem. 279:27233–27238 [DOI] [PubMed] [Google Scholar]
- 19. Boutell C, et al. 2011. A viral ubiquitin ligase has substrate preferential SUMO targeted ubiquitin ligase activity that counteracts intrinsic antiviral defence. PLoS Pathog. 7:e1002245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Boxus M, Willems L. 2009. Mechanisms of HTLV-1 persistence and transformation. Br. J. Cancer 101:1497–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Brass AL, et al. 2008. Identification of host proteins required for HIV infection through a functional genomic screen. Science 319:921–926 [DOI] [PubMed] [Google Scholar]
- 22. Burkham J, Coen DM, Hwang CB, Weller SK. 2001. Interactions of herpes simplex virus type 1 with ND10 and recruitment of PML to replication compartments. J. Virol. 75:2353–2367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Burkham J, Coen DM, Weller SK. 1998. ND10 protein PML is recruited to herpes simplex virus type 1 prereplicative sites and replication compartments in the presence of viral DNA polymerase. J. Virol. 72:10100–10107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Carvalho T, et al. 1995. Targeting of adenovirus E1A and E4-ORF3 proteins to nuclear matrix-associated PML bodies. J. Cell Biol. 131:45–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chang LK, et al. 2004. Post-translational modification of Rta of Epstein-Barr virus by SUMO-1. J. Biol. Chem. 279:38803–38812 [DOI] [PubMed] [Google Scholar]
- 26. Chang PC, et al. 2010. Kaposi's sarcoma-associated herpesvirus (KSHV) encodes a SUMO E3 ligase that is SIM-dependent and SUMO-2/3-specific. J. Biol. Chem. 285:5266–5273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chang TH, et al. 2009. Ebola Zaire virus blocks type I interferon production by exploiting the host SUMO modification machinery. PLoS Pathog. 5:e1000493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chang YL, et al. 2007. Regulation of nuclear receptor and coactivator functions by the carboxyl terminus of ubiquitin-conjugating enzyme 9. Int. J. Biochem. Cell Biol. 39:1035–1046 [DOI] [PubMed] [Google Scholar]
- 29. Chelbi-Alix MK, de Thé H. 1999. Herpes virus induced proteasome-dependent degradation of the nuclear bodies-associated PML and Sp100 proteins. Oncogene 18:935–941 [DOI] [PubMed] [Google Scholar]
- 30. Chen SC, et al. 2011. Sumoylation-promoted enterovirus 71 3C degradation correlates with a reduction in viral replication and cell apoptosis. J. Biol. Chem. 286:31373–31384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chiocca S. 2007. Viral control of the SUMO pathway: Gam1, a model system. Biochem. Soc. Trans. 35:1419–1421 [DOI] [PubMed] [Google Scholar]
- 32. Chiocca S, Baker A, Cotten M. 1997. Identification of a novel antiapoptotic protein, GAM-1, encoded by the CELO adenovirus. J. Virol. 71:3168–3177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chiocca S, et al. 2002. Histone deacetylase 1 inactivation by an adenovirus early gene product. Current Biol. 12:594–598 [DOI] [PubMed] [Google Scholar]
- 34. Chiu MW, Shih HM, Yang TH, Yang YL. 2007. The type 2 dengue virus envelope protein interacts with small ubiquitin-like modifier-1 (SUMO-1) conjugating enzyme 9 (Ubc9). J. Biomed. Sci. 14:429–444 [DOI] [PubMed] [Google Scholar]
- 35. Citro S, Chiocca S. 2010. Listeria monocytogenes: a bacterial pathogen to hit on the SUMO pathway. Cell Res. 20:738–740 [DOI] [PubMed] [Google Scholar]
- 36. Colombo R, Boggio R, Seiser C, Draetta GF, Chiocca S. 2002. The adenovirus protein Gam1 interferes with sumoylation of histone deacetylase 1. EMBO Rep. 3:1062–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cornelis GR. 2002. The Yersinia Ysc-Yop virulence apparatus. Int. J. Med. Microbiol. 291:455–462 [DOI] [PubMed] [Google Scholar]
- 38. Cornelis GR, Denecker G. 2001. Yersinia lead SUMO attack. Nature Med. 7:21–23 [DOI] [PubMed] [Google Scholar]
- 39. Cuchet-Lourenço D, et al. 2011. SUMO pathway dependent recruitment of cellular repressors to herpes simplex virus type 1 genomes. PLoS Pathog. 7:e1002123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Deng H, et al. Over-accumulation of nuclear IGF-1 receptor in tumor cells requires elevated expression of the receptor and the SUMO-conjugating enzyme Ubc9. 2011. Biochem. Biophys. Res. Commun. 404:667–671 [DOI] [PubMed] [Google Scholar]
- 41. Deyrieux AF, Rosas-Acosta G, Ozbun MA, Wilson VG. 2007. Sumoylation dynamics during keratinocyte differentiation. J. Cell Sci. 120:125–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Doucas V, et al. 1996. Adenovirus replication is coupled with the dynamic properties of the PML nuclear structure. Genes Dev. 10:196–207 [DOI] [PubMed] [Google Scholar]
- 43. Dunnebier T, et al. 2010. Polymorphisms in the UBC9 and PIAS3 genes of the SUMO-conjugating system and breast cancer risk. Breast Cancer Res. Treat 121:185–194 [DOI] [PubMed] [Google Scholar]
- 44. Endter C, Hartl B, Spruss T, Hauber J, Dobner T. 2005. Blockage of CRM1-dependent nuclear export of the adenovirus type 5 early region 1B 55-kDa protein augments oncogenic transformation of primary rat cells. Oncogene 24:55–64 [DOI] [PubMed] [Google Scholar]
- 45. Endter C, Kzhyshkowska J, Stauber R, Dobner T. 2001. SUMO-1 modification required for transformation by adenovirus type 5 early region 1B 55-kDa oncoprotein. Proc. Natl. Acad. Sci. U. S. A. 98:11312–11317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Everett RD. 2001. DNA viruses and viral proteins that interact with PML nuclear bodies. Oncogene 20:7266–7273 [DOI] [PubMed] [Google Scholar]
- 47. Everett RD, Chelbi-Alix MK. 2007. PML and PML nuclear bodies: implications in antiviral defence. Biochimie 89:819–830 [DOI] [PubMed] [Google Scholar]
- 48. Everett RD, et al. 1998. The disruption of ND10 during herpes simplex virus infection correlates with the Vmw110- and proteasome-dependent loss of several PML isoforms. J. Virol. 72:6581–6591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Everett RD, et al. 1997. A novel ubiquitin-specific protease is dynamically associated with the PML nuclear domain and binds to a herpesvirus regulatory protein. EMBO J. 16:1519–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Everett RD, Murray J. 2005. ND10 components relocate to sites associated with herpes simplex virus type 1 nucleoprotein complexes during virus infection. J. Virol. 79:5078–5089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Everett RD, Murray J, Orr A, Preston CM. 2007. Herpes simplex virus type 1 genomes are associated with ND10 nuclear substructures in quiescently infected human fibroblasts. J. Virol. 81:10991–11004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Everett RD, Parada C, Gripon P, Sirma H, Orr A. 2008. Replication of ICP0-null mutant herpes simplex virus type 1 is restricted by both PML and Sp100. J. Virol. 82:2661–2672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Everett RD, et al. 2006. PML contributes to a cellular mechanism of repression of herpes simplex virus type 1 infection that is inactivated by ICP0. J. Virol. 80:7995–8005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Everett RD, Sourvinos G, Leiper C, Clements JB, Orr A. 2004. Formation of nuclear foci of the herpes simplex virus type 1 regulatory protein ICP4 at early times of infection: localization, dynamics, recruitment of ICP27, and evidence for the de novo induction of ND10-like complexes. J. Virol. 78:1903–1917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Fan Z, et al. 2006. SARS-CoV nucleocapsid protein binds to hUbc9, a ubiquitin conjugating enzyme of the sumoylation system. J. Med. Virol. 78:1365–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Fradet-Turcotte A, Brault K, Titolo S, Howley PM, Archambault J. 2009. Characterization of papillomavirus E1 helicase mutants defective for interaction with the SUMO-conjugating enzyme Ubc9. Virology 395:190–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Freed EO. 2002. Viral late domains. J. Virol. 76:4679–4687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Frisch SM, Mymryk JS. 2002. Adenovirus-5 E1A: paradox and paradigm. Nat. Rev. Mol. Cell Biol. 3:441–452 [DOI] [PubMed] [Google Scholar]
- 59. Geiss-Friedlander R, Melchior F. 2007. Concepts in sumoylation: a decade on. Nat. Rev. Mol. Cell Biol. 8:947–956 [DOI] [PubMed] [Google Scholar]
- 60. Geoffroy MC, Jaffray EG, Walker KJ, Hay RT. 2010. Arsenic-induced SUMO-dependent recruitment of RNF4 into PML nuclear bodies. Mol. Biol. Cell 21:4227–4239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Gomes R, Guerra-Sa R, Arruda E. 2010. Coxsackievirus B5 induced apoptosis of HeLa cells: effects on p53 and SUMO. Virology 396:256–263 [DOI] [PubMed] [Google Scholar]
- 62. González-Santamaría J, et al. 2011. Regulation of vaccinia virus E3 protein by small ubiquitin-like modifier proteins. J. Virol. 85:12890–12900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Gravel A, Dion V, Cloutier N, Gosselin J, Flamand L. 2004. Characterization of human herpesvirus 6 variant B immediate-early 1 protein modifications by small ubiquitin-related modifiers. J. Gen. Virol. 85:1319–1328 [DOI] [PubMed] [Google Scholar]
- 64. Gravel A, Gosselin J, Flamand L. 2002. Human herpesvirus 6 immediate-early 1 protein is a sumoylated nuclear phosphoprotein colocalizing with promyelocytic leukemia protein-associated nuclear bodies. J. Biol. Chem. 277:19679–19687 [DOI] [PubMed] [Google Scholar]
- 65. Gurer C, Berthoux L, Luban J. 2005. Covalent modification of human immunodeficiency virus type 1 p6 by SUMO-1. J. Virol. 79:910–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Hagemeier C, Walker SM, Sissons PJ, Sinclair JH. 1992. The 72K IE1 and 80K IE2 proteins of human cytomegalovirus independently trans-activate the c-fos, c-myc and hsp70 promoters via basal promoter elements. J. Gen. Virol. 73:2385–2393 [DOI] [PubMed] [Google Scholar]
- 67. Hagemeier SR, et al. 2010. Sumoylation of the Epstein-Barr virus BZLF1 protein inhibits its transcriptional activity and is regulated by the virus-encoded protein kinase. J. Virol. 84:4383–4394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Han Y, et al. 2010. SENP3-mediated de-conjugation of SUMO2/3 from promyelocytic leukemia is correlated with accelerated cell proliferation under mild oxidative stress. J. Biol. Chem. 285:12906–12915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Hannoun Z, Greenhough S, Jaffray E, Hay RT, Hay DC. 2010. Post-translational modification by SUMO. Toxicology 278:288–293 [DOI] [PubMed] [Google Scholar]
- 70. Hateboer G, et al. 1996. mUBC9, a novel adenovirus E1A-interacting protein that complements a yeast cell cycle defect. J. Biol. Chem. 271:25906–25911 [DOI] [PubMed] [Google Scholar]
- 71. Hay RT. 2005. SUMO: a history of modification. Mol. Cell 18:1–12 [DOI] [PubMed] [Google Scholar]
- 72. Hayashi T, et al. 2002. Ubc9 is essential for viability of higher eukaryotic cells. Exp. Cell Res. 280:212–221 [DOI] [PubMed] [Google Scholar]
- 73. Hermiston TW, Malone CL, Stinski MF. 1990. Human cytomegalovirus immediate-early two protein region involved in negative regulation of the major immediate-early promoter. J. Virol. 64:3532–3536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Hietakangas V, et al. 2006. PDSM, a motif for phosphorylation-dependent SUMO modification. Proc. Natl. Acad. Sci. U. S. A. 103:45–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Hochstrasser M. 2009. Origin and function of ubiquitin-like proteins. Nature 458:422–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Hofmann H, Floss S, Stamminger T. 2000. Covalent modification of the transactivator protein IE2-p86 of human cytomegalovirus by conjugation to the ubiquitin-homologous proteins SUMO-1 and hSMT3b. J. Virol. 74:2510–2524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Huh YH, et al. 2008. Binding STAT2 by the acidic domain of human cytomegalovirus IE1 promotes viral growth and is negatively regulated by SUMO. J. Virol. 82:10444–10454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Izumiya Y, et al. 2005. Kaposi's sarcoma-associated herpesvirus K-bZIP represses gene transcription via SUMO modification. J. Virol. 79:9912–9925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Jaber T, et al. 2009. Human Ubc9 contributes to production of fully infectious human immunodeficiency virus type 1 virions. J. Virol. 83:10448–10459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Johnson ES. 2004. Protein modification by SUMO. Annu. Rev. Biochem. 73:355–382 [DOI] [PubMed] [Google Scholar]
- 81. Johnson ES, Gupta AA. 2001. An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell 106:735–744 [DOI] [PubMed] [Google Scholar]
- 82. Kaikkonen S, et al. 2009. SUMO-specific protease 1 (SENP1) reverses the hormone-augmented SUMOylation of androgen receptor and modulates gene responses in prostate cancer cells. Mol. Endocrinol. 23:292–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Kang H, et al. 2006. Inhibition of SUMO-independent PML oligomerization by the human cytomegalovirus IE1 protein. J. Gen. Virol. 87:2181–2190 [DOI] [PubMed] [Google Scholar]
- 84. Kaukinen P, Vaheri A, Plyusnin A. 2003. Non-covalent interaction between nucleocapsid protein of Tula hantavirus and small ubiquitin-related modifier-1, SUMO-1. Virus Res. 92:37–45 [DOI] [PubMed] [Google Scholar]
- 85. Kaul S, Blackford JA, Jr, Cho S, Simons SS., Jr 2002. Ubc9 is a novel modulator of the induction properties of glucocorticoid receptors. J. Biol. Chem. 277:12541–12549 [DOI] [PubMed] [Google Scholar]
- 86. Kehn K, et al. 2004. Mechanisms of HTLV-1 transformation. Front. Biosci. 9:2347–2372 [DOI] [PubMed] [Google Scholar]
- 87. Kelly C, Van Driel R, Wilkinson GW. 1995. Disruption of PML-associated nuclear bodies during human cytomegalovirus infection. J. Gen. Virol. 76:2887–2893 [DOI] [PubMed] [Google Scholar]
- 88. Kerscher O. 2007. SUMO junction—what's your function? New insights through SUMO-interacting motifs. EMBO Rep. 8:550–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Kerscher O, Felberbaum R, Hochstrasser M. 2006. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu. Rev. Cell Dev. Biol. 22:159–180 [DOI] [PubMed] [Google Scholar]
- 90. Kfoury Y, et al. 2011. Tax ubiquitylation and SUMOylation control the dynamic shuttling of Tax and NEMO between Ubc9 nuclear bodies and the centrosome. Blood 117:190–199 [DOI] [PubMed] [Google Scholar]
- 91. Kim ET, Kim YE, Huh YH, Ahn JH. 2010. Role of noncovalent SUMO binding by the human cytomegalovirus IE2 transactivator in lytic growth. J. Virol. 84:8111–8123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Kindsmüller K, et al. 2007. Intranuclear targeting and nuclear export of the adenovirus E1B-55K protein are regulated by SUMO1 conjugation. Proc. Natl. Acad. Sci. U. S. A. 104:6684–6689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Kobayashi S, et al. 2004. Ubc9 interacts with chicken ovalbumin upstream promoter-transcription factor I and represses receptor-dependent transcription. J. Mol. Endocrinol. 32:69–86 [DOI] [PubMed] [Google Scholar]
- 94. König R, et al. 2010. Human host factors required for influenza virus replication. Nature 463:813–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. König R, et al. 2008. Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell 135:49–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Kubota T, et al. 2008. Virus infection triggers SUMOylation of IRF3 and IRF7, leading to the negative regulation of type I interferon gene expression. J. Biol. Chem. 283:25660–25670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Kurtzman AL, Schechter N. 2001. Ubc9 interacts with a nuclear localization signal and mediates nuclear localization of the paired-like homeobox protein Vsx-1 independent of SUMO-1 modification. Proc. Natl. Acad. Sci. U. S. A. 98:5602–5607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Kyratsous CA, Silverstein SJ. 2009. Components of nuclear domain 10 bodies regulate varicella-zoster virus replication. J. Virol. 83:4262–4274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Lallemand-Breitenbach V, et al. 2008. Arsenic degrades PML or PML-RARalpha through a SUMO-triggered RNF4/ubiquitin-mediated pathway. Nat. Cell Biol. 10:547–555 [DOI] [PubMed] [Google Scholar]
- 100. Lamsoul I, et al. 2005. Exclusive ubiquitination and sumoylation on overlapping lysine residues mediate NF-kappaB activation by the human T-cell leukemia virus tax oncoprotein. Mol. Cell. Biol. 25:10391–10406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Lang M, et al. 2010. Three-dimensional organization of promyelocytic leukemia nuclear bodies. J. Cell Sci. 123:392–400 [DOI] [PubMed] [Google Scholar]
- 102. Lee BH, et al. 2003. Association of the nucleocapsid protein of the Seoul and Hantaan hantaviruses with small ubiquitin-like modifier-1-related molecules. Virus Res. 98:83–91 [DOI] [PubMed] [Google Scholar]
- 103. Lee HR, Ahn JH. 2004. Sumoylation of the major immediate-early IE2 protein of human cytomegalovirus Towne strain is not required for virus growth in cultured human fibroblasts. J. Gen. Virol. 85:2149–2154 [DOI] [PubMed] [Google Scholar]
- 104. Lee HR, et al. 2004. Ability of the human cytomegalovirus IE1 protein to modulate sumoylation of PML correlates with its functional activities in transcriptional regulation and infectivity in cultured fibroblast cells. J. Virol. 78:6527–6542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Lee JM, et al. 2003. PIAS1 enhances SUMO-1 modification and the transactivation activity of the major immediate-early IE2 protein of human cytomegalovirus. FEBS Lett. 555:322–328 [DOI] [PubMed] [Google Scholar]
- 106. Lee P, Hruby DE. 1994. Proteolytic cleavage of vaccinia virus virion proteins: mutational analysis of the specificity determinants. J. Biol. Chem. 269:8616–8622 [PubMed] [Google Scholar]
- 107. Lethbridge KJ, Scott GE, Leppard KN. 2003. Nuclear matrix localization and SUMO-1 modification of adenovirus type 5 E1b 55K protein are controlled by E4 Orf6 protein. J. Gen. Virol. 84:259–268 [DOI] [PubMed] [Google Scholar]
- 108. Li FQ, Xiao H, Tam JP, Liu DX. 2005. Sumoylation of the nucleocapsid protein of severe acute respiratory syndrome coronavirus. FEBS Lett. 579:2387–2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Li Q, Xiao H, Tam JP, Liu DX. 2006. Sumoylation of the nucleocapsid protein of severe acute respiratory syndrome coronavirus by interaction with Ubc9. Adv. Exp. Med. Biol. 581:121–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Li SJ, Hochstrasser M. 1999. A new protease required for cell-cycle progression in yeast. Nature 398:246–251 [DOI] [PubMed] [Google Scholar]
- 111. Li XH, Gaynor RB. 2000. Mechanisms of NF-kappaB activation by the HTLV type 1 tax protein. AIDS Res. Hum. Retroviruses 16:1583–1590 [DOI] [PubMed] [Google Scholar]
- 112. Liu ST, et al. 2006. Sumoylation of Rta of Epstein-Barr virus is preferentially enhanced by PIASxbeta. Virus Res. 119:163–170 [DOI] [PubMed] [Google Scholar]
- 113. López-Otín C, Simon-Mateo C, Martinez L, Vinuela E. 1989. Gly-Gly-X, a novel consensus sequence for the proteolytic processing of viral and cellular proteins. J. Biol. Chem. 264:9107–9110 [PubMed] [Google Scholar]
- 114. Lukashchuk V, Everett RD. 2010. Regulation of ICP0-null mutant herpes simplex virus type 1 infection by ND10 components ATRX and hDaxx. J. Virol. 84:4026–4040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Maeda A, et al. 2003. The intracellular association of the nucleocapsid protein (NP) of hantaan virus (HTNV) with small ubiquitin-like modifier-1 (SUMO-1) conjugating enzyme 9 (Ubc9). Virology 305:288–297 [DOI] [PubMed] [Google Scholar]
- 116. Mahajan R, Delphin C, Guan T, Gerace L, Melchior F. 1997. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell 88:97–107 [DOI] [PubMed] [Google Scholar]
- 117. Malone CL, Vesole DH, Stinski MF. 1990. Transactivation of a human cytomegalovirus early promoter by gene products from the immediate-early gene IE2 and augmentation by IE1: mutational analysis of the viral proteins. J. Virol. 64:1498–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Marcos-Villar L, et al. 2011. Covalent modification by SUMO is required for efficient disruption of PML oncogenic domains by Kaposi's sarcoma-associated herpesvirus latent protein LANA2. J. Gen. Virol. 92:188–194 [DOI] [PubMed] [Google Scholar]
- 119. Marcos-Villar L, et al. 2009. Kaposi's sarcoma-associated herpesvirus protein LANA2 disrupts PML oncogenic domains and inhibits PML-mediated transcriptional repression of the survivin gene. J. Virol. 83:8849–8858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Marusic MB, Mencin N, Licen M, Banks L, Grm HS. 2010. Modification of human papillomavirus minor capsid protein L2 by sumoylation. J. Virol. 84:11585–11589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Matic I, et al. 2010. Site-specific identification of SUMO-2 targets in cells reveals an inverted SUMOylation motif and a hydrophobic cluster SUMOylation motif. Mol. Cell 39:641–652 [DOI] [PubMed] [Google Scholar]
- 122. Matunis MJ, Coutavas E, Blobel G. 1996. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J. Cell Biol. 135:1457–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Maul GG, Everett RD. 1994. The nuclear location of PML, a cellular member of the C3HC4 zinc-binding domain protein family, is rearranged during herpes simplex virus infection by the C3HC4 viral protein ICP0. J. Gen. Virol. 75:1223–1233 [DOI] [PubMed] [Google Scholar]
- 124. Maul GG, Guldner HH, Spivack JG. 1993. Modification of discrete nuclear domains induced by herpes simplex virus type 1 immediate early gene 1 product (ICP0). J. Gen. Virol. 74:2679–2690 [DOI] [PubMed] [Google Scholar]
- 125. Meluh PB, Koshland D. 1995. Evidence that the MIF2 gene of Saccharomyces cerevisiae encodes a centromere protein with homology to the mammalian centromere protein CENP-C. Mol. Biol. Cell 6:793–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Mo YY, Moschos SJ. 2005. Targeting Ubc9 for cancer therapy. Expert Opin. Ther. Targets 9:1203–1216 [DOI] [PubMed] [Google Scholar]
- 127. Moschos S. J, et al. 2010. Expression analysis of Ubc9, the single small ubiquitin-like modifier (SUMO) E2 conjugating enzyme, in normal and malignant tissues. Hum. Pathol. 41:1286–1298 [DOI] [PubMed] [Google Scholar]
- 128. Mossessova E, Lima CD. 2000. Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol. Cell 5:865–876 [DOI] [PubMed] [Google Scholar]
- 129. Mukhopadhyay D, Dasso M. 2007. Modification in reverse: the SUMO proteases. Trends Biochem. Sci. 32:286–295 [DOI] [PubMed] [Google Scholar]
- 130. Muller S, Dejean A. 1999. Viral immediate-early proteins abrogate the modification by SUMO-1 of PML and Sp100 proteins, correlating with nuclear body disruption. J. Virol. 73:5137–5143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Murata T, et al. 2010. Transcriptional repression by sumoylation of Epstein-Barr virus BZLF1 protein correlates with association of histone deacetylase. J. Biol. Chem. 285:23925–23935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Nacerddine K, et al. 2005. The SUMO pathway is essential for nuclear integrity and chromosome segregation in mice. Dev. Cell 9:769–779 [DOI] [PubMed] [Google Scholar]
- 133. Nevels M, Brune W, Shenk T. 2004. SUMOylation of the human cytomegalovirus 72-kilodalton IE1 protein facilitates expression of the 86-kilodalton IE2 protein and promotes viral replication. J. Virol. 78:7803–7812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Nowak M, Hammerschmidt M. 2006. Ubc9 regulates mitosis and cell survival during zebrafish development. Mol. Biol. Cell 17:5324–5336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Okura T, et al. 1996. Protection against Fas/APO-1- and tumor necrosis factor-mediated cell death by a novel protein, sentrin. J. Immunol. 157:4277–4281 [PubMed] [Google Scholar]
- 136. Orth K. 2002. Function of the Yersinia effector YopJ. Curr. Opin. Microbiol. 5:38–43 [DOI] [PubMed] [Google Scholar]
- 137. Reference deleted.
- 138. Orth K, et al. 2000. Disruption of signaling by Yersinia effector YopJ, a ubiquitin-like protein protease. Science 290:1594–1597 [DOI] [PubMed] [Google Scholar]
- 139. Ott DE, Coren LV, Chertova EN, Gagliardi TD, Schubert U. 2000. Ubiquitination of HIV-1 and MuLV Gag. Virology 278:111–121 [DOI] [PubMed] [Google Scholar]
- 140. Owerbach D, McKay EM, Yeh ET, Gabbay KH, Bohren KM. 2005. A proline-90 residue unique to SUMO-4 prevents maturation and sumoylation. Biochem. Biophys. Res. Commun. 337:517–520 [DOI] [PubMed] [Google Scholar]
- 141. Pal S, Rosas JM, Rosas-Acosta G. 2010. Identification of the non-structural influenza A viral protein NS1A as a bona fide target of the Small Ubiquitin-like MOdifier by the use of dicistronic expression constructs. J. Virol. Methods 163:498–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Pal S, Santos A, Rosas JM, Ortiz-Guzman J, Rosas-Acosta G. 2011. Influenza A virus interacts extensively with the cellular SUMOylation system during infection. Virus Res. 158:12–27 [DOI] [PubMed] [Google Scholar]
- 143. Palacios S, et al. 2005. Quantitative SUMO-1 modification of a vaccinia virus protein is required for its specific localization and prevents its self-association. Mol. Biol. Cell 16:2822–2835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Pan X, et al. 2006. Ubc9 interacts with SOX4 and represses its transcriptional activity. Biochem. Biophys. Res. Commun. 344:727–734 [DOI] [PubMed] [Google Scholar]
- 145. Parkinson J, Everett RD. 2000. Alphaherpesvirus proteins related to herpes simplex virus type 1 ICP0 affect cellular structures and proteins. J. Virol. 74:10006–10017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Peloponese JM, Jr, Kinjo T, Jeang KT. 2007. Human T-cell leukemia virus type 1 Tax and cellular transformation. Int. J. Hematol. 86:101–106 [DOI] [PubMed] [Google Scholar]
- 147. Pennella MA, Liu Y, Woo JL, Kim CA, Berk AJ. 2010. Adenovirus E1B 55-kilodalton protein is a p53-SUMO1 E3 ligase that represses p53 and stimulates its nuclear export through interactions with promyelocytic leukemia nuclear bodies. J. Virol. 84:12210–12225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Petrie K, Zelent A. 2008. Marked for death. Nat. Cell Biol. 10:507–509 [DOI] [PubMed] [Google Scholar]
- 149. Pizzorno MC, O'Hare P, Sha L, LaFemina RL, Hayward GS. 1988. trans-Activation and autoregulation of gene expression by the immediate-early region 2 gene products of human cytomegalovirus. J. Virol. 62:1167–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Pornillos O, Alam SL, Davis DR, Sundquist WI. 2002. Structure of the Tsg101 UEV domain in complex with the PTAP motif of the HIV-1 p6 protein. Nat. Struct. Biol. 9:812–817 [DOI] [PubMed] [Google Scholar]
- 151. Pornillos O, et al. 2002. Structure and functional interactions of the Tsg101 UEV domain. EMBO J. 21:2397–2406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Pozzebon M, Segre CV, Chiocca S. 2009. Inhibition of the SUMO pathway by Gam1. Methods Mol. Biol. 497:285–301 [DOI] [PubMed] [Google Scholar]
- 153. Prudden J, et al. 2007. SUMO-targeted ubiquitin ligases in genome stability. EMBO J. 26:4089–4101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Puvion-Dutilleul F, et al. 1995. Adenovirus infection induces rearrangements in the intranuclear distribution of the nuclear body-associated PML protein. Exp. Cell Res. 218:9–16 [DOI] [PubMed] [Google Scholar]
- 155. Rangasamy D, Wilson VG. 2000. Bovine papillomavirus E1 protein is sumoylated by the host cell Ubc9 protein. J. Biol. Chem. 275:30487–30495 [DOI] [PubMed] [Google Scholar]
- 156. Rangasamy D, Woytek K, Khan SA, Wilson VG. 2000. SUMO-1 modification of bovine papillomavirus E1 protein is required for intranuclear accumulation. J. Biol. Chem. 275:37999–38004 [DOI] [PubMed] [Google Scholar]
- 157. Reichelt M, et al. 2011. Entrapment of viral capsids in nuclear PML cages is an intrinsic antiviral host defense against varicella-zoster virus. PLoS Pathog. 7:e1001266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Ribet D, et al. 2010. Listeria monocytogenes impairs SUMOylation for efficient infection. Nature 464:1192–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Rogan S, Heaphy S. 2000. The vaccinia virus E3L protein interacts with SUMO-1 and ribosomal protein L23a in a yeast two hybrid assay. Virus Genes 21:193–195 [DOI] [PubMed] [Google Scholar]
- 160. Rosas-Acosta G, Langereis MA, Deyrieux A, Wilson VG. 2005. Proteins of the PIAS family enhance the sumoylation of the papillomavirus E1 protein. Virology 331:190–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Rosendorff A, et al. 2004. EBNA3C coactivation with EBNA2 requires a SUMO homology domain. J. Virol. 78:367–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Sadanari H, Yamada R, Ohnishi K, Matsubara K, Tanaka J. 2005. SUMO-1 modification of the major immediate-early (IE) 1 and 2 proteins of human cytomegalovirus is regulated by different mechanisms and modulates the intracellular localization of the IE1, but not IE2, protein. Arch. Virol. 150:1763–1782 [DOI] [PubMed] [Google Scholar]
- 163. Saitoh H, Hinchey J. 2000. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J. Biol. Chem. 275:6252–6258 [DOI] [PubMed] [Google Scholar]
- 164. Saltzman A, et al. 1998. hUBC9 associates with MEKK1 and type I TNF-alpha receptor and stimulates NFkappaB activity. FEBS Lett. 425:431–435 [DOI] [PubMed] [Google Scholar]
- 165. Sapp M, Bienkowska-Haba M. 2009. Viral entry mechanisms: human papillomavirus and a long journey from extracellular matrix to the nucleus. FEBS J. 276:7206–7216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Sarge KD, Park-Sarge OK. 2009. Sumoylation and human disease pathogenesis. Trends Biochem. Sci. 34:200–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Schreiner S, et al. 2011. Adenovirus type 5 early region 1B 55K oncoprotein dependent degradation of cellular factor Daxx is required for efficient transformation of primary rodent cells. J. Virol. 85:8752–8765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Schreiner S, et al. 2010. Proteasome-dependent degradation of Daxx by the viral E1B-55K protein in human adenovirus-infected cells. J. Virol. 84:7029–7038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Schwartz DC, Hochstrasser M. 2003. A superfamily of protein tags: ubiquitin, SUMO and related modifiers. Trends Biochem. Sci. 28:321–328 [DOI] [PubMed] [Google Scholar]
- 170. Shapira SD, et al. 2009. A physical and regulatory map of host-influenza interactions reveals pathways in H1N1 infection. Cell 139:1255–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Shen TH, Lin HK, Scaglioni PP, Yung TM, Pandolfi PP. 2006. The mechanisms of PML-nuclear body formation. Mol. Cell 24:331–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Sia C. 2005. Is a new immune response mediator in the NF-kappaB pathway—SUMO-4—related to type 1 diabetes? Rev. Diabet. Stud. 2:58–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Song J, Durrin LK, Wilkinson TA, Krontiris TG, Chen Y. 2004. Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc. Natl. Acad. Sci. U. S. A. 101:14373–14378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Spengler ML, Kurapatwinski K, Black AR, Azizkhan-Clifford J. 2002. SUMO-1 modification of human cytomegalovirus IE1/IE72. J. Virol. 76:2990–2996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Sun D, Xu P, He B. 2011. Sumoylation of the P protein at K254 plays an important role in growth of parainfluenza virus 5. J. Virol. 85:10261–10268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Sun H, Leverson JD, Hunter T. 2007. Conserved function of RNF4 family proteins in eukaryotes: targeting a ubiquitin ligase to SUMOylated proteins. EMBO J. 26:4102–4112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Tatham MH, et al. 2008. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat. Cell Biol. 10:538–546 [DOI] [PubMed] [Google Scholar]
- 178. Tavalai N, Stamminger T. 2008. New insights into the role of the subnuclear structure ND10 for viral infection. Biochim. Biophys. Acta 1783:2207–2221 [DOI] [PubMed] [Google Scholar]
- 179. Tomoiu A, Gravel A, Tanguay RM, Flamand L. 2006. Functional interaction between human herpesvirus 6 immediate-early 2 protein and ubiquitin-conjugating enzyme 9 in the absence of sumoylation. J. Virol. 80:10218–10228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Ulrich HD. 2009. The SUMO system: an overview. Methods Mol. Biol. 497:3–16 [DOI] [PubMed] [Google Scholar]
- 181. Uzunova K, et al. 2007. Ubiquitin-dependent proteolytic control of SUMO conjugates. J. Biol. Chem. 282:34167–34175 [DOI] [PubMed] [Google Scholar]
- 182. Van Damme E, Laukens K, Dang TH, Van Ostade X. 2010. A manually curated network of the PML nuclear body interactome reveals an important role for PML-NBs in SUMOylation dynamics. Int. J. Biol. Sci. 6:51–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Van Damme E, Van Ostade X. 2011. Crosstalk between viruses and PML nuclear bodies: a network-based approach. Front. Biosci. 17:2910–2920 [DOI] [PubMed] [Google Scholar]
- 184. Verger A, Perdomo J, Crossley M. 2003. Modification with SUMO: a role in transcriptional regulation. EMBO Rep. 4:137–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. VerPlank L, et al. 2001. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55(Gag). Proc. Natl. Acad. Sci. U. S. A. 98:7724–7729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Wang L, et al. 2011. Disruption of PML nuclear bodies is mediated by ORF61 SUMO-interacting motifs and required for varicella-zoster virus pathogenesis in skin. PLoS Pathog. 7:e1002157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Weidtkamp-Peters S, et al. 2008. Dynamics of component exchange at PML nuclear bodies. J. Cell Sci. 121:2731–2743 [DOI] [PubMed] [Google Scholar]
- 188. Weisshaar SR, et al. 2008. Arsenic trioxide stimulates SUMO-2/3 modification leading to RNF4-dependent proteolytic targeting of PML. FEBS Lett. 582:3174–3178 [DOI] [PubMed] [Google Scholar]
- 189. Weldon RA, Jr, Sarkar P, Brown SM, Weldon SK. 2003. Mason-Pfizer monkey virus Gag proteins interact with the human sumo conjugating enzyme, hUbc9. Virology 314:62–73 [DOI] [PubMed] [Google Scholar]
- 190. Wilson VG, Rangasamy D. 2001. Viral interaction with the host cell sumoylation system. Virus Res. 81:17–27 [DOI] [PubMed] [Google Scholar]
- 191. Wimmer P, et al. 2010. SUMO modification of E1B-55K oncoprotein regulates isoform-specific binding to the tumour suppressor protein PML. Oncogene 29:5511–5522 [DOI] [PubMed] [Google Scholar]
- 192. Wu CY, Jeng KS, Lai MM. 2011. The SUMOylation of matrix protein M1 modulates the assembly and morphogenesis of influenza A virus. J. Virol. 85:6618–6628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Wu YC, Bian XL, Heaton PR, Deyrieux AF, Wilson VG. 2009. Host cell sumoylation level influences papillomavirus E2 protein stability. Virology 387:176–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Wu YC, Deyrieux AF, Wilson VG. 2007. Papillomaviruses and the host SUMOylation system. Biochem. Soc. Trans. 35:1433–1435 [DOI] [PubMed] [Google Scholar]
- 195. Wu YC, Roark AA, Bian XL, Wilson VG. 2008. Modification of papillomavirus E2 proteins by the small ubiquitin-like modifier family members (SUMOs). Virology 378:329–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Xie Y, et al. 2007. The yeast Hex3.Slx8 heterodimer is a ubiquitin ligase stimulated by substrate sumoylation. J. Biol. Chem. 282:34176–34184 [DOI] [PubMed] [Google Scholar]
- 197. Xu K, et al. 2011. Modification of nonstructural protein 1 of influenza A virus by SUMO1. J. Virol. 85:1086–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Yang SH, Galanis A, Witty J, Sharrocks AD. 2006. An extended consensus motif enhances the specificity of substrate modification by SUMO. EMBO J. 25:5083–5093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Yasugi T, Vidal M, Sakai H, Howley PM, Benson JD. 1997. Two classes of human papillomavirus type 16 E1 mutants suggest pleiotropic conformational constraints affecting E1 multimerization, E2 interaction, and interaction with cellular proteins. J. Virol. 71:5942–5951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Yousef AF, et al. 2010. Identification of a molecular recognition feature in the E1A oncoprotein that binds the SUMO conjugase UBC9 and likely interferes with polySUMOylation. Oncogene 29:4693–4704 [DOI] [PubMed] [Google Scholar]
- 201. Yueh A, et al. 2006. Interaction of moloney murine leukemia virus capsid with Ubc9 and PIASy mediates SUMO-1 addition required early in infection. J. Virol. 80:342–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Zamborlini A, et al. 2011. Impairment of human immunodeficiency virus type-1 integrase SUMOylation correlates with an early replication defect. J. Biol. Chem. 286:21013–21022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Zhu S, Sachdeva M, Wu F, Lu Z, Mo YY. 2010. Ubc9 promotes breast cell invasion and metastasis in a sumoylation-independent manner. Oncogene 29:1763–1772 [DOI] [PMC free article] [PubMed] [Google Scholar]