Abstract
All positive-strand RNA viruses replicate their genomes in association with rearranged intracellular membranes such as single- or double-membrane vesicles. Brome mosaic virus (BMV) RNA synthesis occurs in vesicular endoplasmic reticulum (ER) membrane invaginations, each induced by many copies of viral replication protein 1a, which has N-terminal RNA capping and C-terminal helicase domains. Although the capping domain is responsible for 1a membrane association and ER targeting, neither this domain nor the helicase domain was sufficient to induce replication vesicle formation. Moreover, despite their potential for mutual interaction, the capping and helicase domains showed no complementation when coexpressed in trans. Cross-linking showed that the capping and helicase domains each form trimers and larger multimers in vivo, and the capping domain formed extended, stacked, hexagonal lattices in vivo. Furthermore, coexpressing the capping domain blocked the ability of full-length 1a to form replication vesicles and replicate RNA and recruited full-length 1a into mixed hexagonal lattices with the capping domain. Thus, BMV replication vesicle formation and RNA replication depend on the direct linkage and concerted action of 1a's self-interacting capping and helicase domains. In particular, the capping domain's strong dominant-negative effects showed that the ability of full-length 1a to form replication vesicles was highly sensitive to disruption by non-productively titrating lattice-forming self-interactions of the capping domain. These and other findings shed light on the roles and interactions of 1a domains in replication compartment formation and support prior results suggesting that 1a induces replication vesicles by forming a capsid-like interior shell.
INTRODUCTION
Positive-strand RNA viruses are the largest genetic class of viruses and include many clinically important human pathogens as well as animal and plant pathogens. Postive-strand RNA virus genome replication and transcription occur in organelle-like structures that organize replication factors and templates and protect them from host defenses (11, 35, 38). These novel RNA replication compartments are induced by virus-specific membrane rearrangements such single- and double-membrane vesicles or appressed membranes or both (20, 29, 31, 44, 52, 53).
Brome mosaic virus (BMV), a member of the alphavirus-like superfamily of human, animal, and plant viruses, has been extensively studied as a model for positive-strand RNA virus RNA replication. BMV's genome is divided among three capped RNAs. RNA3 is dispensable for RNA replication but encodes cell-to-cell movement of protein 3a and the coat protein, both required for systemic spread of virus infection (6, 36). RNA1 and RNA2 encode RNA replication factors 1a and 2apol, respectively. 1a is a multifunctional protein with key roles in the assembly and function of the viral RNA replication complexes. 1a contains an N-proximal RNA capping domain (2, 3, 30) and a C-terminal NTPase/RNA helicase-like domain (referred to here as the helicase domain) (58) separated by a short proline-rich sequence with little predicted secondary structure, which may be a flexible spacer (10) (Fig. 1A). 2apol has a central RNA-dependent RNA polymerase-like domain and an N-terminal domain that interacts with the 1a helicase domain (8, 26).
Fig 1.
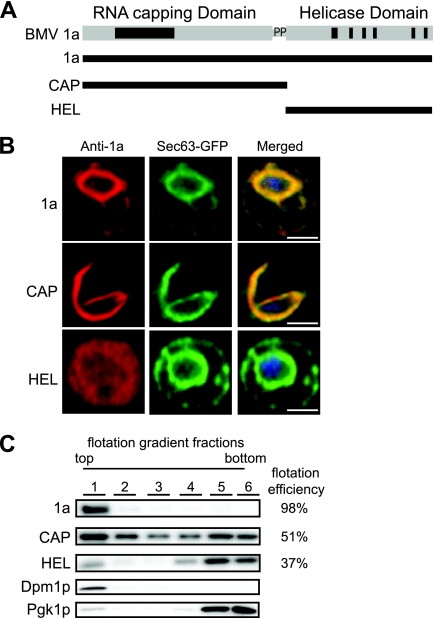
CAP is mainly responsible for 1a membrane association. (A) The CAP fragment contains the capping domain and the proline-rich linker region, and the HEL fragment contains the NTPase/helicase-like domain. (B) Fluorescence microscopy images of cells expressing wt 1a, CAP, or HEL and Sec63-GFP, an ER marker. TO-PRO-3 was used to stain DNA (blue). Bars, 2 μm. (C) Distribution of 1a, CAP, HEL, PGK (cytosolic protein control), and Dpm1p (ER luminal protein control) in membrane flotation gradients. Representative Western blots using anti-1a, anti-PGK, and anti-Dpm1p antisera are shown.
With or without other viral factors, 1a localizes to endoplasmic reticulum (ER) membranes (47, 48) and induces 60- to 80-nm vesicular ER invaginations or spherules (50). 1a also recruits 2apol and viral RNA templates to these spherules (8, 9, 39, 47, 48), which then serve as compartments or miniorganelles for RNA replication (50). The roles of 1a and 2apol in the assembly and function of these spherule replication compartments have parallels to those of Gag and Pol in the membrane-enveloped capsids of retrovirus virions (1, 50). Such retrovirus capsids are composed of hexameric arrays of Gag that curve to closure by incorporating pentameric (16) or gap (7) discontinuities. The high multiplicity of 1a in spherules (50) and its strong membrane association (10) and self-interaction (40) suggest that 1a might induce membrane invagination by forming a capsid-like shell similar to that of Gag. Consistent with this, confocal fluorescence shows that 1a accumulates in discrete, expanding ER patches during infection, implying that extended 1a-1a interactions occur in vivo on ER membranes (48). Similarly, replicase proteins from many other positive-strand RNA viruses can also multimerize (13, 19, 34, 41, 57). For example, the tobacco mosaic virus (TMV) 126-kDa protein, a 1a homolog, contains a helicase domain that forms hexamers (19).
Intriguingly, distinct membrane rearrangements can be induced by altering the balance of 1a-1a, 1a-2apol, and 1a-membrane interactions. In particular, increasing the levels of 2apol shifts 1a-induced membrane rearrangements from spherular invaginations to karmella-like, multilayer stacks of appressed double-membrane layers that support RNA replication as efficiently as spherules (51).
Linked RNA capping and helicase domains homologous to those of 1a are conserved across the alphavirus-like superfamily, but in some superfamily members, such as Sindbis and Semliki Forest viruses, the nonstructural proteins are translated as a polyprotein and are ultimately proteolytically processed into two separate proteins (56). For Sindbis virus and Semliki Forest virus, formation of the membrane spherules is induced by the partially processed nonstructural protein P123 and nsp4; however, synthesis of double-stranded RNA (dsRNA) is an essential prerequisite of their formation (14, 52). To shed further light on the roles and interactions of these domains, we expressed BMV 1a capping and helicase domains alone, together, or in combination with full-length 1a and used confocal and electron microscopy (EM) and complementary biochemical methods to assess the effects on membrane targeting, membrane rearrangement, and RNA replication. We show that spherular replication compartment formation and RNA replication require both the 1a capping and helicase domains and that these domains must be directly linked. Nevertheless, in vivo, the 1a capping domain alone targeted ER membranes, formed membrane-linked hexagonal lattices of tubules or stacked rings, and induced layering of ER double membranes. Additionally, the capping domain had strong dominant-negative effects on full-length 1a, blocking 1a's induction of spherular replication compartments and RNA replication and instead recruiting 1a into a hexagonal lattice. The 1a helicase domain failed to associate with membranes by itself but was recruited by full-length 1a to the ER, without interfering with spherule formation or RNA replication. These results provide new mechanistic insights into 1a domains and interactions involved in BMV RNA replication complexes and in RNA replication.
MATERIALS AND METHODS
Yeast and plasmids.
Saccharomyces cerevisiae strain YPH500 and culture conditions were as described previously (25). BMV 1a and the 1a fragment derivatives were expressed under the control of the GAL1 promoter by the use of pB1YT3 (2) or the class I (396/400/407A) or class II (K403/K406A) derivatives (33). The coding sequences of BMV 1a fragments of the capping domain (CAP) (amino acids [aa] 1 to 557) and the helicase-like domain (HEL) (aa 558 to 961) were PCR amplified with pB1YT3 as the template. FLAG or hemagglutinin (HA) tag sequences were cloned into the C terminus of full-length 1a or the CAP or HEL fragments by the use of standard molecular biology procedures. PCR fragments with each of the tags were digested with PacI and BamHI and cloned into pB1YT3 or pB1YT3L to replace the corresponding wild-type (wt) fragments. 2apol was expressed from pB2CT15 (ADH1 promoter) (25). ADH1-driven CAP fused to green fluorescent protein (GFP) was previously described (10). BMV RNA3 was expressed under the control of a CUP1 promoter from pB3VG128H or under the control of a GAL1 promoter from pB3MS82; both RNA3 derivatives have a four-nucleotide insertion in the coat protein gene that abolishes expression of the coat protein (54). The Sec63-GFP fusion protein was expressed from plasmid pWSECG, a derivative of pJK59 (gift from P. Silver, Department of Biological Chemistry and Molecular Pharmacology, Harvard University).
Membrane flotation assay.
Ten optical density units of yeast cells grown to mid-logarithmic phase to an optical density at 600 nm (OD600) of 10 were subjected to spheroplasting and resuspended in 350 μl of buffer TNE (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 5 mM EDTA, 5 mM benzamidine, 1 mM phenylmethylsulfonyl fluoride [PMSF], and aprotinin, leupeptin, and pepstatin A [10 μg/ml each]). Spheroplasts were lysed via 25 passes through a 22-gauge, 4-cm-long needle. Total lysates were centrifuged for 5 min at 4°C at 500 × g to remove cell debris, and 250 μl of the supernatants was mixed with 500 μl of 60% OptiPrep (Axis-Shield, Oslo, Norway). Density gradient centrifugation was performed for 2 h at 55,000 rpm using a Beckman TLS55 rotor and 600 μl of each sample overlaid by 1.4 ml of 30% OptiPrep and 100 μl of lysis buffer (58). After centrifugation, 6 fractions were collected from the top to the bottom of the gradient. For protein detection, samples were boiled in sodium dodecyl sulfate (SDS) loading buffer prior to SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting.
RNA and protein analysis.
Total yeast RNA isolation by the hot phenol method, Northern blot analysis, total protein extractions and Western blot analysis, and anti-1a, anti-2apol, anti-Dpm1, and anti-Pgk1 antibodies were as described previously (8, 10, 25). Mouse and rabbit anti-FLAG antibodies were purchased from Sigma, anti-GFP mouse polyclonal antibody was purchased from Molecular Probes, mouse anti-PDI was acquired from Abcam, and rabbit and mouse anti-HA antibodies were purchased from Santa Cruz and Roche, respectively. Northern blots were imaged on a Typhoon 9200 Imager (Amersham Biosciences, Piscataway, NJ). Band intensities were analyzed by using ImageQuant software (Molecular Dynamics, Piscataway, NJ).
Chemical cross-linking experiments.
Two optical density units of cells were harvested, pellets were resuspended in 5 ml of growth media containing 1% formaldehyde, samples were incubated at 30°C for 10 min, the cross-linking reaction was quenched by adding 2.5 M glycine for a final concentration of 0.125 M, and samples were incubated at 30°C for 10 min. Total proteins were extracted and analyzed in 3% to 8% Tris-acetate gels.
Immunofluorescence and confocal microscopy.
Confocal microscopy was performed as described previously (58). Briefly, yeast cells expressing wt 1a or the 1a fragments or both and/or Sec63-GFP were fixed with 4% formaldehyde, subjected to spheroplasting with lyticase, and permeabilized with 0.1% Triton X-100. Spheroplasts were then stained by using rabbit anti-1a serum, mouse anti-PDI, rabbit anti-FLAG, or mouse anti-HA antibodies or combinations thereof followed by anti-rabbit or anti-mouse secondary antibodies conjugated to Texas Red, Alexa Fluor 488, or Alexa Fluor 647. For nuclear staining, a 10-min incubation with 300 nM DAPI (4′,6-diamidino-2-phenylindole) (Invitrogen) was added after secondary antibody incubation. Fluorescent images were acquired with a Nikon A1R Bio-Rad inverted confocal microscope system. Projection images were created using ImageJ (http://rsb.info.nih.gov/ij/).
Electron microscopy.
Samples were prepared for electron microscopy as described previously (50). In brief, yeast cells were fixed for 1 h with 2% glutaraldehyde and 4% paraformaldehyde, washed, and postfixed for 1 h with 1% OsO4 and 1% uranyl acetate. Cells then were dehydrated via a series of stepwise increasing ethanol concentrations ranging from 50% to 100% and infiltrated and embedded with Spurrs resin. Samples were sectioned and placed on nickel grids, washed, incubated for 15 min in 2% glutaraldehyde, poststained with 8% uranyl acetate and Reynold's lead citrate, and viewed with a Philips CM120 microscope. For immunogold EM experiments, yeast cells were fixed for 1 h with 0.5% glutaraldehyde and 4% paraformaldehyde, postfixed in 0.1% OsO4 for 15 min, dehydrated in a series of stepwise increasing ethanol concentrations, and embedded in LR White resin (Polysciences, Inc., Warrington, PA). Grids were blocked with 0.5% gelatin, immunostained with rabbit anti-1a or mouse anti-GFP serum and 10-nm gold-labeled secondary antibodies, poststained, and analyzed by transmission EM (TEM) as described above.
Fatty acid analysis.
Total fatty acid (FA) from 10 optical density units of yeast was extracted and converted to methyl esters as described earlier (37). FA species were separated by gas-liquid chromatography according to chain length and degree of saturation and were identified by retention time. The molar amount of each species was measured by using a flame ionization detector (37).
RESULTS
The 1a capping domain is responsible for perinuclear ER membrane targeting.
To track the contributions of the capping (CAP) and helicase-like (HEL) domains of 1a to ER membrane association and reorganization, we divided 1a into two fragments. This division is similar to that of the final processed forms of alphavirus nonstructural proteins nsP1 and nsP2, which have RNA capping and NTPase/helicase activities, respectively (4, 18). nsP1, which is a peripheral membrane protein, acts as the sole membrane anchor of the replication complex (5). The CAP fragment contains the capping domain and a short proline-rich linker region, while the HEL fragment contains the NTPase/helicase domain (Fig. 1A). The CAP and HEL fragments each contain all sequences previously shown by yeast two-hybrid analysis to be required for CAP-CAP and intramolecular CAP-HEL interaction (40). Additionally, Escherichia coli-expressed and purified derivatives of CAP (aa 1 to 516) and HEL (aa 424 to 961) retain the ability to form an adduct with a guanine nucleotide (2, 30) and to hydrolyze ATP (58), respectively. Thus, both fragments maintain a structural conformation consistent with their function within full-length 1a. We first used confocal immunofluorescence microscopy to compare the subcellular localization of CAP and HEL with that of full-length 1a. 1a localized predominantly to the perinuclear ER, colocalizing almost completely with the Sec63-GFP ER marker (Fig. 1B). Likewise, CAP colocalized with Sec63-GFP, although only a portion of CAP was in the perinuclear ER whereas the rest formed wisp-like structures that extended into the cytoplasm (Fig. 1B). In contrast, HEL did not colocalize with Sec63-GFP and displayed a mostly diffused cytoplasmic localization (Fig. 1B).
To further assess the contribution of the CAP and HEL fragments to membrane association, lysates of yeast cells expressing 1a or the 1a fragments were loaded under flotation gradients, which upon centrifugation were fractionated and analyzed by SDS-PAGE and Western blotting using anti-1a antibodies. As a measure of membrane association, flotation efficiency was determined as previously described (33) as the percentage of protein present in the top two fractions. The flotation efficiency of 1a was ∼98% (Fig. 1C). Expressing CAP on its own reduced membrane association by about 2-fold, while the flotation efficiency of the HEL fragment was ∼35% (Fig. 1C). Combined, the confocal and flotation results reinforce the idea that CAP is responsible for ER targeting of 1a (10, 33).
The CAP domain induces formation of lattice-arrayed, hexagonal tubules.
As stated earlier, 1a is the only BMV protein necessary to induce formation of the 60- to 80-nm perinuclear ER invaginations in which BMV RNA replication occurs (Fig. 2A) (50). To test whether either half of 1a retained the ability to induce spherule formation, we examined yeast cells expressing CAP or HEL by EM.
Fig 2.
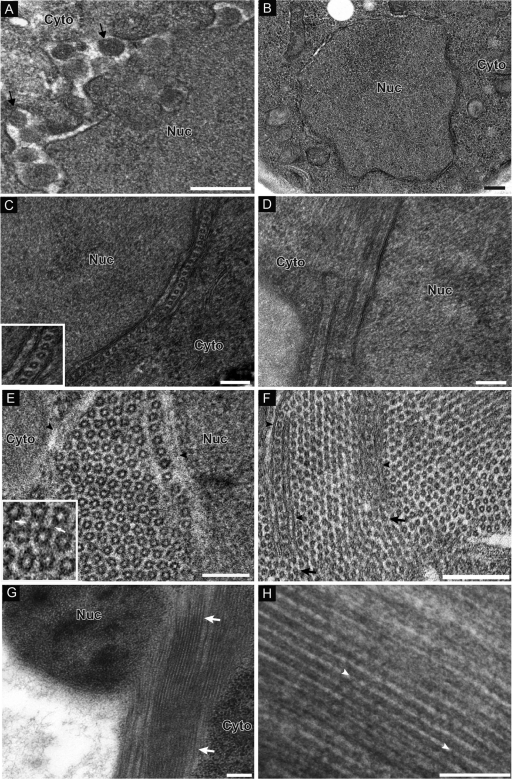
CAP fragment induces formation of ring/tubular structures. (A) Electron micrograph of yeast cells expressing 1a-induced spherules. Arrows point out individual spherular structures. (B) HEL does not induce any membrane rearrangements. (C) Micrograph of yeast cells expressing CAP. In one subset of cells, nuclei were surrounded by appressed layers of double-membrane ER. When cross-sections were viewed, hexameric ring-like structures were present in the intermembrane spaces. The inset shows a close-up view of rings. (D) In views of longitudinal section, there are lamina-like structures present between membrane bilayers. (E and F) Alternatively, hexagonal lattices that either were not immediately flanked by an ER membrane (E) or contained ER-like layers a few hundred nanometers in length isolating each side of a single row of rings (F) were present. In the inset in panel E, arrows point to filamentous material connecting adjacent rings. Arrowheads in panel F point out membrane protrusions within the hexagonal ring lattice. (G) Another subset of cells displayed long electron-dense tubular structures partially surrounding the nucleus. (H) Close-up view of tubules. Arrowheads point out electron-dense material connecting tubules running parallel to each other. Nuc, nucleus; Cyto, cytoplasm. Bars, 200 nm (A, B, C, and H) and 100 nm (D to G).
Yeast expressing the HEL domain did not display any detectable membrane rearrangements (Fig. 2B). In contrast, yeast expressing the CAP fragment displayed two distinct but related ultrastructural changes linked to membranes. In one subset of CAP-expressing cells, nuclei were surrounded by two to four appressed layers of double-membrane ER, whose cytoplasmically connected intermembrane spaces were 33 ± 2.1 nm wide (Fig. 2C and D). Within these cytoplasmic intermembrane spaces, approximately one-third of the sectioned views revealed a row of rings 16.8 ± 1.3 nm wide, regularly spaced 7.8 ± 1.2 nm from the membrane, with a center-to-center distance between neighboring rings of 24.8 ± 2.6 nm (Fig. 2C). In two-thirds of the sections, the intermembrane space lacked rings and presented a more laminar appearance, consisting of a denser, central region 17.6 ± 2.2 nm wide flanked on each side by more electron-lucent areas that were ∼7.9 ± 1.0 nm wide (Fig. 2D). In the next three paragraphs, we present evidence that these two views respectively represent cross-sections and side views of tubules.
The other novel ultrastructure in CAP-expressing cells involved extended arrays of rings 17.0 ± 1.4 nm wide arranged in a clearly hexagonal lattice (Fig. 2E). Close examination indicated that each ring consisted of six ∼4- to 5-nm-diameter dense objects clustered in a closely packed arrangement (Fig. 2E, inset). The individual appearances and center-to-center distance of two neighboring rings of 24.2 ± 1.7 nm were similar to those of the single rows of rings sandwiched between double-membrane ER layers (compare Fig. 2E with Fig. 2C). In the hexagonal arrays, though, the great majority of ∼14-nm rings were directly surrounded on six sides by other rings, without immediately flanking ER membranes; nevertheless, at least one edge of these hexagonal lattices was often flanked by membrane (Fig. 2E, arrowheads; the membrane appears grayish, as it was partially extracted). Moreover, occasionally ER-like, closely appressed double-membrane layers a few hundred nanometers in length invaded the lattice to isolate on each side a single row of rings (Fig. 2F, arrowheads) that appeared locally indistinguishable from the perinuclear, ER-sandwiched single rows of rings (Fig. 2C). However, at the point where these membrane protrusions ended, the row of rings so isolated melded apparently seamlessly into the surrounding hexagonal lattice of rings, which also flowed around and abutted the outer face of the isolating double membrane protrusion (Fig. 2F, arrows). This intersection of the two ultrastructural forms further underscored the similarities and potentially related nature of the ∼17-nm rings in each case. Higher-magnification micrographs showed that adjacent rings in the hexagonal lattice were frequently connected by filamentous material, perhaps explaining the regular ∼7-nm spacing separating each ring (Fig. 2E, inset).
In another subset of cells, nuclei were surrounded by electron-dense, tubule-like structures running parallel to each other that were 18.3 ± 1.4 nm wide and were separated by electron-lucent areas ∼7.9 ± 1.1 nm wide (Fig. 2G and H). These long tubular structures that partially surround the nucleus and extend into the cytoplasm are reminiscent of the wisp-like structures seen by confocal microscopy (Fig. 1B). Adjacent tubules appeared to be connected periodically by electron-dense, possibly proteinaceous bridges, perhaps explaining the regular spaces separating the tubules (Fig. 2H, arrowheads).
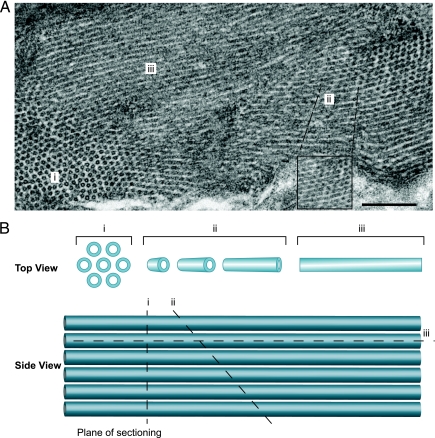
Additionally, in many sections the rings of the hexagonal lattice blended directly into elongated, parallel striations whose ∼18-nm-wide electron-dense regions, spaced by 7- to 8-nm electron-lucent regions (Fig. 3), resembled the tubules surrounding the nucleus and the laminar views of the perinuclear ER membrane layers (Fig. 2C and F). The nature and intermediates of this transition imply that the ordered rings represent perpendicular sections through an array of tubules. Though locally ordered in parallel arrays, a degree of flexibility in the array apparently allows the tubule angle to shift gradually across the hundreds of tubules in an EM field. Thus, as seen in Fig. 3, the view gradually shifts from perpendicular sections of tubules (region i; ordered, separated rings) to tubules sectioned at more oblique angles, revealing elongated, apparently stacked rings (region ii) and, eventually, tubules sectioned parallel to their main axis (region iii; striations).
Fig 3.
(A) Electron micrograph of yeast cell expressing CAP, displaying both ring and tubular structures. Depending on the plane of sectioning, the view gradually shifts from perpendicular sections of tubules (region I; ordered, separated rings) to tubules sectioned at more oblique angles, revealing elongated, apparently stacked rings (region ii) and, eventually, tubules sectioned parallel to their main axis (region iii; striations). The inset shows a higher-magnification image of region ii. (B) Schematic illustration of how the plane of sectioning through the tubules can determine the type of structure observed by EM. I, perpendicular section; ii, oblique section; iii, parallel section.
Taken together, these EM results show that CAP but not HEL induces membrane rearrangements that represent cross-sections and side views of tubules. In the next section, we show that these tubules or rings contain and are the primary localization site for CAP.
Immunogold labeling localizes CAP to tubules and rings.
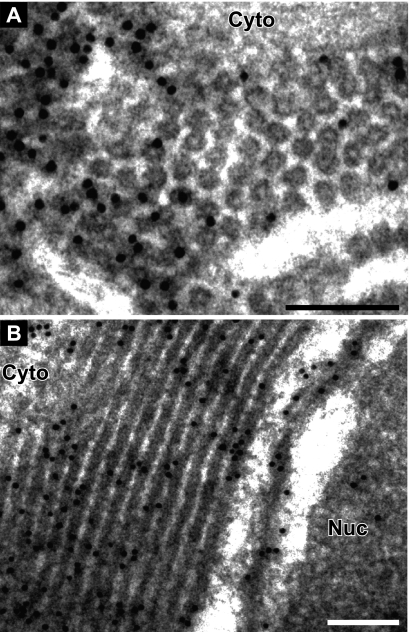
To more precisely localize CAP in relation to the rings and tubules, we performed immunogold EM with anti-1a antibodies. To preserve the antigenicity of 1a for the polyclonal antibody used, osmium fixation and staining were omitted, resulting in electron-lucent regions where membrane lipids were extracted during the dehydration steps (23). In cells expressing the CAP fragment only, 85% of all of the gold particles were in or on the rings surrounding the nucleus or in the hexagonal arrays, while only 8% were in the electron-lucent regions (Fig. 4A). Similarly, in sections displaying the laminar or “side” views of the tubules, 80% of all of the gold particles were in or on the electron-dense spaces between the extracted membranes (Fig. 4B). Another 10% of the gold particles were present within 10 to 20 nm of the electron-dense tubules, a distance that may be spanned by the primary and secondary antibodies linking the immunogold particles to their target epitopes (22). Notably, the ring/tubular structures remained intact despite lipid extraction due to the lack of osmium tetroxide fixation, further suggesting that they are mainly composed of proteins. Consistent with this, the homogeneity in the size of the rings suggests that they are made of components of a defined, consistent size. Taken together, these results suggest that CAP is the main component of the rings, but we cannot exclude the possibility that cellular components might be present as well.
Fig 4.
CAP localizes to ring/tubular structures. Immunogold EM localization of CAP fragment is shown. (A) Cross-view and (B) side view of rings/tubules induced by CAP. Nuc, nucleus; Cyto, cytoplasm. Bars, 100 nm.
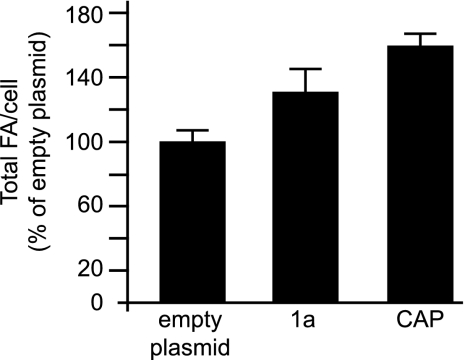
The presence of ring/tubular structures increases cellular fatty acid levels.
Expressing 1a alone increases total membrane lipid accumulation per cell by 25 to 33% (32). Since CAP was found not only in regular perinuclear ER but also in wisp-like extensions also labeled with the ER marker Sec63 (Fig. 1B), we wanted to determine whether formation of the CAP-induced tubules also increased total lipid levels. To test this, we used gas-liquid chromatography to analyze the fatty acid (FA) levels in yeast expressing empty plasmids, 1a, or CAP. As there were no significant changes in the relative compositions of individual lipid species, we compared total FA levels per cell by summing the molar amounts of various FA for each culture and expressing the resulting total FA value as a percentage of that of yeast transformed with empty plasmids. Agreeing with previous results, 1a expression increased total FA levels per cell by 33% (Fig. 5). Since 95% of all FA in yeast exist as fatty acyl chains of membrane lipids, total FA levels reflect yeast membrane content (46) and the 1a-induced increase in FA must represent an increase in membrane content. CAP expression increased total FA accumulation by 60% (Fig. 5). Thus, the increase in total FA accumulation must reflect the presence of double-membrane layers surrounding the ring/tubular structures, consistent with the confocal microscopy images that show that the 1a fragments colocalize with the ER marker Sec63 throughout (Fig. 1).
Fig 5.
Expression of CAP leads to an increase in total fatty acids per cell. Levels of total fatty acids per cell are shown as percentages of the total fatty acids per cell of yeast expressing empty plasmids. Each histogram shows averages and standard deviations of the results of three experiments.
CAP and HEL form multimers.
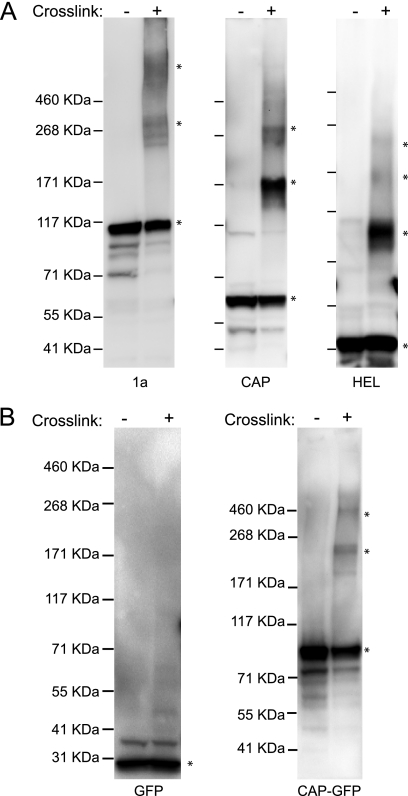
Prior yeast two-hybrid results showed that CAP and HEL can self-interact but did not address the level of this multimerization or reveal any HEL-HEL interaction (40). The formation of extended ring/tubule structures by CAP suggested that 1a fragments might multimerize to a high level. To address this, we used chemical cross-linking to examine the multimerization states of 1a, CAP, and HEL. Full-length 1a was used as a control, since yeast two-hybrid results show that 1a can self-interact (40). After cultures were treated with formaldehyde to cross-link any protein complexes, proteins were extracted and analyzed by SDS-PAGE and subjected to immunoblotting using anti-1a antibodies. In yeast expressing full-length 1a, we detected a major band migrating as expected for a 1a monomer (109 kDa), and this band was weaker in the formaldehyde-treated samples than in nontreated samples (Fig. 6A). Additionally, cross-linking caused the appearance of an SDS-resistant, 1a-containing band that electrophoresed to a position corresponding to a 1a trimer plus a higher-molecular-mass band that migrated more slowly than the 460-kDa marker, suggesting that the in vivo chemical cross-linking stabilized the higher-molecular-mass 1a-containing complexes during lysis and/or electrophoresis (Fig. 6A). Similarly, in yeast expressing CAP, a “ladder” of cross-linked bands whose molecular sizes were consistent with monomers and SDS-resistant trimers and hexamers became apparent (Fig. 6A). For HEL, we detected bands whose migration was consistent with monomers, trimers, and hexamers (Fig. 6A). It should be noted that cross-linking under the conditions used here is not saturating, as formaldehyde cross-links are reversible during standard sample preparation for SDS-PAGE analysis, which includes boiling in reducing Laemmli buffer; thus, the complexes observed represented only a portion of the interactions that exist in vivo (28). Omitting boiling or instead incubating the samples for 10 min at 50°C prior to gel loading resulted in the presence of a single band that did not migrate far from the gel well, presumably reflecting very-high-molecular-mass cross-linked complexes.
Fig 6.
1a, CAP, and HEL oligomerize. (A) Yeast expressing 1a, CAP, or HEL were treated with 1% paraformaldehyde to cross-link proteins, analyzed on 4% to 8% Tris-acetate gels, and immunoblotted with anti-1a antibody. Asterisks denote bands of interest. Numbers to the left of blots denote molecular-mass markers. (B) Yeast expressing free GFP or CAP fused to GFP were treated and analyzed as described for panel A.
To test whether proteins besides CAP might be involved in forming the observed higher-molecular-mass complexes (Fig. 6A, middle lanes), we performed control cross-linking experiments using monomeric GFP alone and fused to the CAP N terminus. Even after cross-linking with formaldehyde, only a monomeric band was observed for GFP alone (Fig. 6B). In contrast, bands corresponding to a monomer, trimer, and hexamer were present for CAP-GFP (Fig. 6B). The shifts in the migration of the CAP-GFP bands, relative to the bands produced by cross-linking CAP alone, corresponded to the expected multiples of the molecular mass of GFP (Fig. 6). These results show that CAP multimerizes into complexes containing three, six, and apparently more CAP monomers, consistent with the CAP-containing hexagonal ring lattices observed by EM.
Coexpressing CAP and HEL fragments in trans does not stabilize or replicate BMV RNA.
Most positive-strand RNA viruses encode multiple factors required for RNA replication, expressed from discrete open reading frames (ORFs) either by proteolytic cleavage of a single polyprotein or by readthrough or frameshift translation (56). For example, the nonstructural proteins of alphaviruses, including the nsP1 capping and nsP2 helicase proteins that are related to the 1a CAP and HEL fragments, are synthesized as polyprotein precursors that are then cleaved into individual components (56). Western blotting of yeast cells expressing 1a or CAP plus HEL showed that, when coexpressed in trans, the levels of CAP and HEL are very similar to those of full-length 1a (Fig. 7A). Accordingly, we tested whether the CAP and HEL fragments, coexpressed in trans, could complement each other to substitute for full-length 1a in replicating BMV RNA. In cells expressing BMV 1a and 2apol proteins, RNA3 is used as a template to make negative-strand RNA3, which is then used to make positive-strand RNA3 and subgenomic RNA4 (25). Yeast cells expressing 1a, 2apol, and RNA3 supported efficient viral RNA replication (Fig. 7B). In contrast, in cells expressing 2apol, RNA3, CAP, and HEL, RNA3 was not detectably amplified (Fig. 7B).
Fig 7.
Expressing CAP and HEL in trans failed to perform any full-length 1a functions. (A) Total protein was isolated from yeast expressing 1a or CAP plus HEL and analyzed by Western blotting using anti-1a antibodies. (B) Analysis of positive-strand RNA3 and RNA4 from yeast expressing 2apol, RNA3, and either wt 1a or a combination of the 1a fragments. Each set of conditions was tested in triplicate. (C) RNA3 levels in yeast expressing RNA3 alone or with wt 1a or with the 1a fragments. For panels B and C, total RNA was isolated from yeast cells and Northern blotting was used to examine the accumulation of RNA3 and RNA4 or RNA3 with a BMV RNA-specific probe. 18S rRNA was measured as a loading control. (D) EM of yeast cell coexpressing CAP and HEL. Bar, 200 nm. (E) Localization of GFP-tagged CAP and HA-tagged HEL expressed in trans. DNA was stained with DAPI (blue). Bar, 2 μm.
To address the inability to replicate the viral RNA, we examined whether CAP coexpressed with HEL could stabilize RNA3, a response closely linked to 1a-mediated recruitment of RNA3 to ER membrane sites of replication (12, 33, 50). In yeast cells, the half-life of RNA3 increases from 5 to 10 min in the absence of 1a to more than 3 h in the presence of 1a and increases RNA3 accumulation by 8- to 20-fold (24). As shown in Fig. 7C, GAL1 promoter-driven 1a increased RNA3 accumulation ∼10-fold. However, coexpressing the CAP and HEL fragments failed to stimulate RNA3 accumulation over the RNA3 levels in cells lacking 1a (Fig. 7C). Previous results suggest that 1a-mediated RNA recruitment and stabilization may include at least two distinguishable steps, a 1a-induced recruitment of RNA3 to membranes and a subsequent translocation of the RNA into a preformed replication compartment (58). Thus, even though CAP plus HEL might interact with RNA3, the tubules/rings may not physically protect RNA3 from degradation by cellular ribonucleases as well as spherules do. Thus, expressing the CAP and HEL fragments in trans abolished BMV replication, most likely due to the fact that expressing the fragments in trans failed to stabilize RNA3.
To determine whether coexpressing CAP and HEL restored spherule formation, we examined yeast cells by EM. In some instances, cells showed the presence of rings arranged in hexagonal lattices (data not shown) whereas other cells displayed appressed layers of double-membrane ER surrounding the nucleus that, when viewed in cross-section, contained rows of rings in the intermembrane space (Fig. 7D). In both cases, the rings were identical in size and morphology to those induced by CAP on its own (Fig. 2E). Similarly, as with CAP alone (Fig. 2F), approximately two-thirds of the cell sections expressing CAP plus HEL showed the perinuclear intermembrane spaces filled with a central, laminar electron-dense region flanked by some more electron-lucent regions (data not shown), corresponding to side views of CAP-containing tubules.
Immunofluorescence microscopy using GFP-tagged CAP and HA-tagged HEL revealed that HEL remained diffused throughout the cell whereas CAP formed a wisp-like structure that started around the perinuclear ER and extended into the cytoplasm (Fig. 7E). Thus, coexpression did not alter the localization of CAP and HEL (compare Fig. 1B and 7E), showing that CAP did not recruit HEL to the ER membrane.
CAP inhibits the ability of 1a to replicate BMV RNA in trans.
To investigate possible complementation or other types of genetic interactions between the 1a fragments and 1a, we tested whether coexpressing CAP or HEL with full-length 1a affected 1a-mediated RNA3 template recruitment and protection, which, as noted above, are reflected by 1a stimulation of viral RNA template accumulation. 1a caused RNA3 to accumulate to concentrations 10-fold higher than those observed in the absence of 1a expression (Fig. 8A, lanes 1 and 2). Coexpressing HEL with 1a yielded RNA3 accumulation identical to that of 1a alone (Fig. 8A, lane 4). Coexpressing CAP with 1a stimulated RNA3 accumulation ∼6-fold, which is a 40% decrease in RNA3 levels compared to those detected in the presence of 1a (Fig. 8A, lane 3).
Fig 8.
CAP expressed in trans inhibits the ability of wt 1a to stabilize and replicate BMV RNA. (A) Analysis of RNA3 levels in yeast expressing RNA3 alone or with wt 1a or with 1a and either CAP or HEL. (B) Positive-strand RNA3 and RNA4 from yeast expressing 2apol, RNA3, and 1a alone or 1a plus CAP or HEL. For panels A and B, total RNA was isolated from yeast cells and Northern blotting was used to examine the accumulation of RNA3 or RNA3 and RNA4 with a BMV RNA-specific probe. 18S rRNA was measured as a loading control. (C) Distribution of 1a, 1a fragments, and 2apol in membrane flotation gradient analyses of lysates from yeast cells coexpressing 2apol, 1a, and CAP or HEL. The flotation efficiency of each protein is shown on the right.
Since RNA3 accumulation in the absence of 2apol was affected only moderately, we tested whether coexpressing CAP or HEL with 1a had any effect on BMV RNA replication in the presence of 2apol. Strong signals were detected for positive-strand RNA3 and subgenomic RNA4 products in cells expressing 1a, 2apol, and RNA3 (Fig. 8B, lane 1). However, coexpressing CAP and 1a inhibited BMV replication by 96% (Fig. 8B, lane 2) whereas coexpressing 1a with HEL had no adverse effect on BMV replication (Fig. 8B, lane 3).
To further investigate the inhibition in BMV replication, we used flotation analyses to look at membrane association of 1a, the 1a fragments, and 2apol. When CAP and HEL were each coexpressed with 1a, their flotation efficiencies and that of 1a were similar to those seen when each was expressed on its own (Fig. 8C and 1B). In the presence of 1a plus CAP or HEL, virtually all of 2apol was membrane associated and accumulated to similar levels in all three cases (Fig. 8C). Thus, the CAP fragment partially interfered with 1a's ability to stabilize RNA3 and strongly interfered with BMV RNA replication even though 2apol was recruited to a membrane-associated state.
Coexpressing 1a and CAP leads to formation of tubule/ring structures.
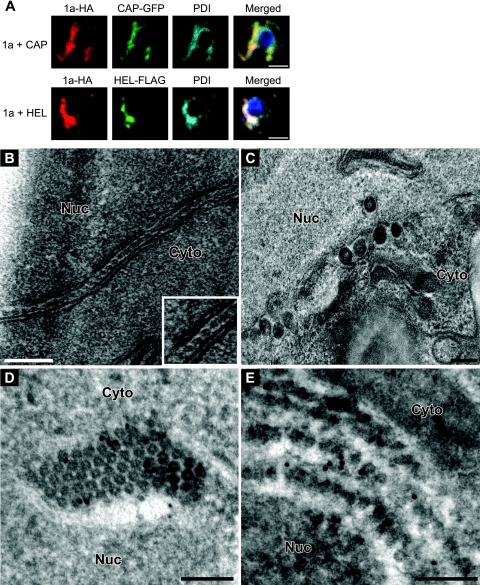
Immunofluorescence localization of 1a coexpressed with the 1a fragments is shown in Fig. 9A. In yeast coexpressing HA- and GFP-tagged versions of 1a and CAP, respectively, a portion of both signals was in the perinuclear ER while the rest was in a region of ER that stretched into the cytoplasm (Fig. 9A), a pattern quite distinct from that of 1a alone but identical to that of CAP expressed on its own (Fig. 1B). Moreover, the 1a and CAP signals completely colocalized with the ER marker PDI (Fig. 9A). In contrast, HEL-FLAG partially colocalized with 1a-HA at the perinuclear ER, but a portion of the signal was also diffused throughout the cytoplasm (Fig. 9A).
Fig 9.
CAP expressed in trans inhibits the ability of wt 1a to induce spherular structures. (A) Fluorescence microscopy images of cells coexpressing 1a-HA and CAP-GFP or 1a-HA and HEL-FLAG. PDI was used as an ER marker. Bars, 2 μm. (B) EM image of hexameric rings found between membrane layers in yeast coexpressing 1a and CAP. (C) Spherules 50 to 70 nm in diameter in yeast coexpressing 1a and HEL. (D and E) Anti-GFP (D) or anti-HA (E) immunogold EM labeling in yeast expressing CAP-GFP and 1a-HA. Nuc, nucleus; Cyto, cytoplasm. Bars, 200 nm (B and C) and 100 nm (D and E).
To determine the type of membrane rearrangement induced by coexpressing 1a with the 1a fragments, we examined yeast cells by EM. Expressing 1a and HEL together resulted in the formation of spherules that were similar in size and morphology to those induced by 1a (Fig. 9C). 1a plus CAP produced no spherules and instead induced the formation of double-membrane layers containing rings in the intermembrane space (Fig. 9B) as well as hexagonal arrays, while spherules were not detected (data not shown). To more precisely localize 1a and CAP in relation to the rings, we performed immunogold EM with antibodies recognizing HA-tagged 1a or GFP-tagged CAP and a secondary antibody conjugated to gold particles. In both cases, ∼85% of all gold particles were in the rings surrounding the nucleus, while only about 12% were in the electron-lucent regions separating the hexagonal arrays (Fig. 9D and E). Thus, CAP dramatically altered 1a's intrinsic functions for ER membrane rearrangement.
Class I mutations abolish spherule and tubule formation in the context of full-length 1a and CAP, respectively.
1a's membrane association with perinuclear ER membranes requires an amphipathic α-helix, helix A, that is located within the 1a CAP domain (33). In full-length 1a, one group of helix A mutants, termed class I, fail to induce ER spherules and have reduced membrane association, but such class I 1a mutants are over twice as effective as wt 1a at interacting with, stabilizing, and recruiting 2aPol to membranes (33). In contrast, class II mutants of helix A not only retain efficient ER membrane association but also increase RNA template recruitment and the frequency of 1a-induced membrane invaginations by 5-fold (33). To gain more insight into what interactions within CAP are necessary to form the tubules/rings, we made class I (L396/400/407A) or II (K403/406E) mutations within the context of the CAP fragment.
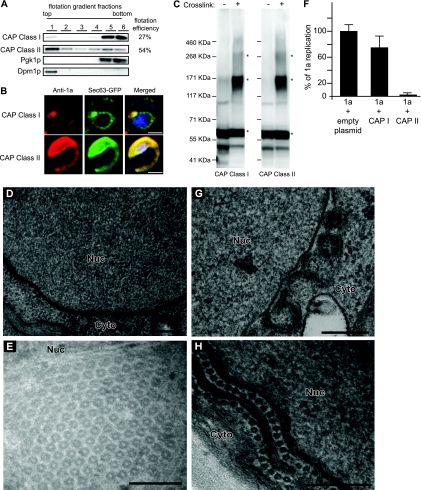
To assess the effect of class I or class II mutations on CAP membrane association, yeast lysates were loaded under flotation gradients, fractionated, and analyzed by SDS-PAGE and Western blotting using anti-1a antibodies. The flotation efficiency of CAP class I was 27% whereas that of CAP class II was 54%, similar to that of wt CAP (Fig. 10A). Confocal microscopy showed that only a portion of CAP class I localized to the perinuclear ER whereas the rest was dispersed throughout the cytoplasm (Fig. 10B). As was the case for wt CAP, CAP class II formed a wisp-like structure that colocalized with Sec63p-GFP both in the perinuclear ER and in a region of the ER that extended into the cytoplasm (Fig. 10B). Intriguingly, despite the fact that only a portion of CAP class I was membrane associated and localized properly to the perinuclear ER, its cross-linking pattern was identical to the CAP class II and wt CAP patterns in that it could assemble into SDS-resistant trimers and hexamers (Fig. 10C; compare to Fig. 6A).
Fig 10.
CAP with class I mutations multimerizes and partially localizes to ER but does not form tubules/hexamers. (A) Distribution of CAP with class I or II mutations. Flotation gradient and Western blotting analysis was performed as described for Fig. 1. (B) Fluorescence microscopy images of cells expressing CAP with class I or II mutations and Sec63-GFP, an ER marker. DAPI was used to stain DNA (blue). Bars, 2 μm. (C) Yeast expressing CAP class I or class II cross-linked as shown in Fig. 6. Asterisks denote bands of interest. Numbers to the left of blots denote molecular-mass markers. (D and E) Electron micrographs of yeast expressing CAP class I (D) or CAP class II (E). (F) Positive-strand RNA4 synthesis from yeast expressing 2apol, RNA3, and 1a alone or 1a plus CAP class I or CAP class II. Total RNA isolation and Northern blot analysis were done as described for Fig. 8. (G) Spherules 50 to 70 nm in diameter in yeast coexpressing 1a and CAP class I. (H) Image of hexameric rings found between membrane layers in yeast coexpressing 1a and CAP class II. Nuc, nucleus; Cyto, cytoplasm. Bars, 200 nm (D, G, and H) and 100 nm (E).
EM analysis revealed that CAP class I was not able to induce any visible membrane rearrangements (Fig. 10D) whereas CAP class II was able to form both hexagonal lattices (Fig. 10E) and tubules (data not shown). It should be noted that the rings and tubules formed by CAP class II were identical in size to and present at the same frequency as those made by wt CAP. Moreover, when coexpressed with 1a, 2apol, and RNA3, CAP class I was not able to significantly affect 1a's ability to replicate the viral RNA (Fig. 10F) or induce spherule formation (Fig. 10G). In contrast, coexpressing CAP class II with 1a resulted in a 97% decrease in BMV RNA replication (Fig. 10F), presumably by recruiting 1a into the hexagonal tubule lattices and inhibiting spherule formation (Fig. 10H).
Taken together, the results reported in this section show that mutations that affect spherule formation in the context of full-length 1a correlate with an inhibition of tubule formation in the context of the CAP fragment. Interestingly, mutations that result in an increase in spherule formation in the context of full-length 1a did not enhance the ability of CAP to make more hexagonal tubule lattices compared to wt CAP, implying that further interactions are involved in that phenotype.
DISCUSSION
Despite progress on a few fronts, the mechanisms by which positive-strand RNA viruses induce and maintain their membrane-bound RNA replication compartments remain largely unresolved (11). Here, we set out to identify further determinants of the invagination of vesicular RNA replication compartments or spherules by the 1a multifunctional BMV RNA replication protein. Our results show that 1a's induction of spherule RNA replication compartments requires the concerted action of both of its domains, CAP and HEL. Furthermore, they reveal that CAP and HEL individually multimerize to high levels, that CAP-CAP interactions drive the formation of a hexagonal lattice of interlinked tubules, and that CAP acts as a strong dominant-negative mutant to block spherule formation and RNA replication by wt 1a. Here we discuss these results and how they and other findings suggest specific mechanisms that may explain how CAP and HEL and their interactions contribute to forming membranous RNA replication compartments.
Replication compartment formation requires linked CAP and HEL.
1a consists of two domains, CAP and HEL (Fig. 1A), which are conserved throughout the alphavirus-like superfamily as parts of a single open reading frame. However, CAP and HEL have physically separable RNA capping (2, 30) and NTPase/helicase (27, 58) enzymatic functions and in the alphaviruses are separated by proteolytic processing into separate proteins, nsP1 and nsP2 (56). Moreover, CAP but not HEL directs localization to ER membranes (10).
Despite the separability of many functions of CAP and HEL, we found that neither CAP nor HEL alone duplicated 1a's ability to induce expression of the membrane spherules associated with bromovirus and alphavirus RNA replication. Moreover, although CAP and HEL each contained all sequences required for enzymatic activity (2, 30, 58), self-interaction (Fig. 2 and 6), and CAP-HEL interaction (40), coexpressing CAP and HEL also did not support spherule formation, stabilize RNA3 templates, or replicate the viral RNA (Fig. 7). Thus, CAP and HEL and their direct linkage are all essential for spherule formation and RNA replication.
This inability of separately expressed CAP and HEL to function synergistically in RNA replication is consistent with the failure of CAP to recruit HEL to ER membranes (Fig. 7D), and both findings are consistent with prior two-hybrid results implying that HEL and CAP can interact intramolecularly within full-length 1a but do not significantly interact intermolecularly between two 1a's (40). Among other functions, forming membrane-bounded replication compartments may require a direct linkage to allow CAP-HEL intramolecular interaction (40) and cooperation between CAP and HEL self-interactions (see also below) or for 1a-mediated recruitment of relevant host factors (12). Since CAP targets ER membranes whereas HEL interacts with BMV 2apol (26, 48) and BMV RNA (58), CAP-HEL linkage is also required to recruit BMV RNA polymerase and replication templates and likely also for later 1a-mediated steps in RNA replication such as RNA capping (2, 30).
CAP multimerization and hexagonal lattice formation.
Yeast two-hybrid assays revealed self-interaction of the 1a CAP domain and established the close connection of this interaction with RNA replication (40) but did not address the level of CAP multimerization. Our cross-linking results show that CAP self-interacts in vivo to form at least trimers and hexamers (Fig. 6A). Full-length 1a also formed trimers and higher-molecular-mass complexes (Fig. 6A). In vivo, CAP also induced two striking, related structures that appeared to differ only in the balance of CAP-CAP and CAP-membrane interactions. These were single rows of regularly spaced, parallel, ∼17-nm-diameter tubules flanked on either side by the ER membrane (Fig. 2F) and masses of similar parallel tubules, interlinked by ∼7-nm radial filaments into a hexagonal lattice and bounded at the edges only by flanking ER membranes (Fig. 2C and H). The relationship of these structures suggests that each face of the CAP-induced tubules is capable of a mutually exclusive interaction either with an ER membrane or with another tubule.
Strikingly, coexpressing CAP abolished wt 1a's ability to form spherules and to support RNA replication (Fig. 8B) while simultaneously recruiting 1a into hexagonal tubule lattices that also contained CAP (Fig. 9). Thus, the CAP fragment interacts with 1a as a strong dominant-negative mutant, and its titration of the lattice-forming interaction sites in 1a blocks spherule formation. Together, these results show that the CAP interactions that form the hexagonal lattice are highly relevant for spherule formation and RNA replication. Further strengthening this conclusion, a mutation that had been previously shown to affect spherule formation in the context of full-length 1a (33) correlated with an inhibition of tubule formation in the context of the CAP fragment (Fig. 10D). However, a different mutation that resulted in an increase in spherule formation in the context of full-length 1a (33) did not enhance the ability of CAP to make more hexagonal tubule lattices than wt CAP (Fig. 10E).
Resistance of the CAP-induced tubules to lipid depletion prior to EM (Fig. 4) implies that they are formed primarily from protein, while immunogold labeling showed that CAP localizes almost exclusively to the tubules and is a prominent tubule component (Fig. 4). Accordingly, the tubules' hexagonal arrangement (Fig. 2A, 2G, and 3) and potentially hexameric individual cross-sections (Fig. 2G, inset), plus CAP cross-linking to multimers that electrophorese as trimers and hexamers (Fig. 6A and B), suggest that the tubules might be built partly or largely from hexameric rings of CAP subunits. Since each CAP subunit would be ∼5 to 6 nm in diameter if globular, a hexamer of CAP subunits is consistent with the tubule ring cross-section (Fig. 2G inset), while, along the tubule length, periodic intertubule connections (Fig. 2D) and density variations (Fig. 3, top panel, inset) are consistent with the stacking of such rings to form the tubules. However, membranes appear to be an important component of the tubules/rings, as CAP expression led to a 66% increase in total lipid accumulation per cell (Fig. 5). The ability of CAP to modulate an increase in total lipid levels is understandable, as CAP is present on the membrane at high local concentrations, interacts with itself (Fig. 6), and accumulates on ER membranes (Fig. 1B). In this context, the greater increase in FA accumulation upon expression of CAP compared to full-length 1a might be due to the greater surface area the tubules occupy within the cell compared to spherules (compare the Sec63-GFP signals in Fig. 1B in cells expressing full-length 1a to those in cells expressing CAP).
Relation of CAP multimerization to spherule vesicle formation.
Localization and stoichiometry results imply that the membrane-bound, self-interacting 1a protein forms a shell lining BMV RNA replication vesicles (1, 50). Such a capsid-like shell would explain how 1a induces and maintains these replication vesicles. Related findings support the idea of a similar role for transmembrane, self-interacting FHV protein A in lining the mitochondrial FHV RNA replication vesicles (31). To form such a membrane-bending shell, 1a would need to use regular interactions to assemble a connected matrix such as the hexagonal arrays formed by retrovirus and other capsid proteins (7, 15). The CAP-induced hexagonal lattice (Fig. 2 and 3) shows that CAP is capable of such repeated interactions (Fig. 2G).
As a membrane binding, self-interacting protein that participates in invaginating membranes, CAP also has similarities to virion matrix proteins, including particular parallels with Marburg virus matrix protein VP40. In virions, such matrix proteins form a hexagonal lattice parallel to the membrane (17, 45, 49), but when VP40's N-terminal domain is expressed alone, its hexameric rings stack into tubules similar to those formed by the 1a CAP fragment (55). Thus, the interlinked CAP-induced tubules might similarly be built from CAP multimers and CAP interactions involved in forming normal replication compartments but whose final assembly would be perturbed for lack of guidance from other interactions mediated by the normally linked HEL domain (see also below).
In the hexagonal lattice revealed by transverse sections of the CAP-induced tubules, the electron-dense, CAP-containing rings are interlinked at a density of one ring per ∼480 nm2 (Fig. 2 to 4). Modeling the membrane-bound 1a proteins lining the ∼60- to 80-nm-diameter spherular BMV replication compartments (12, 50) as a hexagonal lattice of equivalent dimensions results in ∼33 hexamers roughly equivalent to 200 1a molecules per spherule, which compares well with previously reported estimates from EM and biochemical data (50). As in immature retrovirus virions, curvature of the membrane-associated lattice might be accommodated by occasional gap discontinuities (7).
HEL multimerization.
Prior two-hybrid assays revealed CAP-CAP and intramolecular CAP-HEL interactions but showed no indication of HEL-HEL interactions (40). We found that HEL was recruited to ER membranes by full-length 1a (Fig. 9A) but not by CAP alone (Fig. 7D), implying that the HEL domain of full-length 1a interacted with the independently expressed HEL fragment. Furthermore, our cross-linking results showed that HEL self-interacts to form trimers and larger complexes, including potential hexamers (Fig. 6). Intriguingly, like CAP, many helicases form hexamers (43), including the helicase domain of tobacco mosaic virus (TMV) 126-kDa protein (19), a 1a homolog (21). However, the fact that CAP alone did not recruit HEL does not itself rule out a possible CAP-HEL interaction in trans between full-length 1a molecules, as the presence or absence of the HEL domain could significantly affect the way CAP and HEL see one another in trans.
As noted above, 1a induction of RNA replication vesicles requires both CAP and HEL in a directly linked form. HEL-HEL as well as CAP-CAP interaction is likely required to form vesicles, since 1a-induced membrane rearrangement is shifted from vesicles to appressed membrane layers by overexpressing 2apol, a large (94-kDa) protein that interacts with the 1a HEL domain and likely interferes with HEL-HEL interaction (26, 39, 40, 51). Thus, in CAP and HEL, 1a contains two self-interacting, potentially hexamer-forming domains required for 1a-induced membrane invagination. This parallels the HIV capsid protein, which contains separately interacting regions that form two stacked hexamer rings whose size mismatch is thought to induce the Gag lattice and bound membrane to curve (7). Similar effects would explain why 1a requires directly linked CAP and HEL to direct membrane invagination.
In summary, our results show that both 1a domains are required and must act in concert to form spherule replication compartments, reveal that 1a can direct high-level multimerization, and implicate this multimerization in forming spherule replication compartments and likely other aspects of RNA replication. As replicase protein multimerization also appears important for RNA replication by at least some other positive-strand RNA viruses (13, 19, 34, 42, 57), the use of strong trans dominant-negative mutants and other techniques to interfere with such interactions, as seen here with the 1a CAP fragment, could be generally valuable approaches for blocking RNA replication complex formation and function.
ACKNOWLEDGMENTS
We thank Xiaofeng Wang and other members of our laboratory for helpful discussions. We also thank Benjamin August and Randall Massey of the University of Wisconsin Medical School Electron Microscopy Facility for assistance with EM and Lance Rodenkirch of the W. M. Keck Laboratory for Biological Imaging for assistance with confocal microscopy.
This project was supported by NIH grant GM35072. P.A. is an HHMI investigator. A.D. was partially supported by NIH training grants T32 GM07215 and T32 AI078985.
Footnotes
Published ahead of print 16 November 2011
REFERENCES
- 1. Ahlquist P. 2006. Parallels among positive-strand RNA viruses, reverse-transcribing viruses and double-stranded RNA viruses. Nat. Rev. Microbiol. 4:371–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ahola T, Ahlquist P. 1999. Putative RNA capping activities encoded by brome mosaic virus: methylation and covalent binding of guanylate by replicase protein 1a. J. Virol. 73:10061–10069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ahola T, den Boon JA, Ahlquist P. 2000. Helicase and capping enzyme active site mutations in brome mosaic virus protein 1a cause defects in template recruitment, negative-strand RNA synthesis, and viral RNA capping. J. Virol. 74:8803–8811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ahola T, Kaariainen L. 1995. Reaction in alphavirus mRNA capping: formation of a covalent complex of nonstructural protein nsP1 with 7-methyl-GMP. Proc. Natl. Acad. Sci. U. S. A. 92:507–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ahola T, Lampio A, Auvinen P, Kaariainen L. 1999. Semliki Forest virus mRNA capping enzyme requires association with anionic membrane phospholipids for activity. EMBO J. 18:3164–3172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Allison R, Thompson C, Ahlquist P. 1990. Regeneration of a functional RNA virus genome by recombination between deletion mutants and requirement for cowpea chlorotic mottle virus 3a and coat genes for systemic infection. Proc. Natl. Acad. Sci. U. S. A. 87:1820–1824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Briggs JA, et al. 2009. Structure and assembly of immature HIV. Proc. Natl. Acad. Sci. U. S. A. 106:11090–11095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen J, Ahlquist P. 2000. Brome mosaic virus polymerase-like protein 2a is directed to the endoplasmic reticulum by helicase-like viral protein 1a. J. Virol. 74:4310–4318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen J, Noueiry A, Ahlquist P. 2001. Brome mosaic virus protein 1a recruits viral RNA2 to RNA replication through a 5′ proximal RNA2 signal. J. Virol. 75:3207–3219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. den Boon JA, Chen J, Ahlquist P. 2001. Identification of sequences in brome mosaic virus replicase protein 1a that mediate association with endoplasmic reticulum membranes. J. Virol. 75:12370–12381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. den Boon JA, Diaz A, Ahlquist P. 2010. Cytoplasmic viral replication complexes. Cell Host Microbe 8:77–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Diaz A, Wang X, Ahlquist P. 2010. Membrane-shaping host reticulon proteins play crucial roles in viral RNA replication compartment formation and function. Proc. Natl. Acad. Sci. U. S. A. 107:16291–16296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dye BT, Miller DJ, Ahlquist P. 2005. In vivo self-interaction of nodavirus RNA replicase protein a revealed by fluorescence resonance energy transfer. J. Virol. 79:8909–8919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Frolova EI, Gorchakov R, Pereboeva L, Atasheva S, Frolov I. 2010. Functional Sindbis virus replicative complexes are formed at the plasma membrane. J. Virol. 84:11679–11695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ganser BK, Cheng A, Sundquist WI, Yeager M. 2003. Three-dimensional structure of the M-MuLV CA protein on a lipid monolayer: a general model for retroviral capsid assembly. EMBO J. 22:2886–2892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ganser-Pornillos BK, Yeager M, Sundquist WI. 2008. The structural biology of HIV assembly. Curr. Opin. Struct. Biol. 18:203–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ge P, et al. 2010. Cryo-EM model of the bullet-shaped vesicular stomatitis virus. Science 327:689–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gomez de Cedrón M, Ehsani N, Mikkola ML, Garcia JA, Kaariainen L. 1999. RNA helicase activity of Semliki Forest virus replicase protein NSP2. FEBS Lett. 448:19–22 [DOI] [PubMed] [Google Scholar]
- 19. Goregaoker SP, Culver JN. 2003. Oligomerization and activity of the helicase domain of the tobacco mosaic virus 126- and 183-kilodalton replicase proteins. J. Virol. 77:3549–3556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gosert R, Kanjanahaluethai A, Egger D, Bienz K, Baker SC. 2002. RNA replication of mouse hepatitis virus takes place at double-membrane vesicles. J. Virol. 76:3697–3708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Haseloff J, et al. 1984. Striking similarities in amino acid sequence among nonstructural proteins encoded by RNA viruses that have dissimilar genomic organization. Proc. Natl. Acad. Sci. U. S. A. 81:4358–4362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hayat MA. 1991. Colloidal gold. Academic Press, Inc., San Diego, CA [Google Scholar]
- 23. Hayat MA. 2000. Principles and techniques of electron microscopy: biological applications, 4th ed Cambridge University Press, Cambridge, United Kingdom [Google Scholar]
- 24. Janda M, Ahlquist P. 1998. Brome mosaic virus RNA replication protein 1a dramatically increases in vivo stability but not translation of viral genomic RNA3. Proc. Natl. Acad. Sci. U. S. A. 95:2227–2232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Janda M, Ahlquist P. 1993. RNA-dependent replication, transcription, and persistence of brome mosaic virus RNA replicons in S. cerevisiae. Cell 72:961–970 [DOI] [PubMed] [Google Scholar]
- 26. Kao CC, Ahlquist P. 1992. Identification of the domains required for direct interaction of the helicase-like and polymerase-like RNA replication proteins of brome mosaic virus. J. Virol. 66:7293–7302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Karpe YA, Lole KS. 2010. NTPase and 5′ to 3′ RNA duplex-unwinding activities of the hepatitis E virus helicase domain. J. Virol. 84:3595–3602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Klockenbusch C, Kast J. 2010. Optimization of formaldehyde cross-linking for protein interaction analysis of non-tagged integrin beta1. J. Biomed. Biotechnol. 2010:927585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Knoops K, et al. 2008. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 6:e226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kong F, Sivakumaran K, Kao C. 1999. The N-terminal half of the brome mosaic virus 1a protein has RNA capping-associated activities: specificity for GTP and S-adenosylmethionine. Virology 259:200–210 [DOI] [PubMed] [Google Scholar]
- 31. Kopek BG, Perkins G, Miller DJ, Ellisman MH, Ahlquist P. 2007. Three-dimensional analysis of a viral RNA replication complex reveals a virus-induced mini-organelle. PLoS Biol. 5:e220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lee WM, Ahlquist P. 2003. Membrane synthesis, specific lipid requirements, and localized lipid composition changes associated with a positive-strand RNA virus RNA replication protein. J. Virol. 77:12819–12828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liu L, et al. 2009. An amphipathic alpha-helix controls multiple roles of brome mosaic virus protein 1a in RNA replication complex assembly and function. PLoS Pathog. 5:e1000351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lyle JM, Bullitt E, Bienz K, Kirkegaard K. 2002. Visualization and functional analysis of RNA-dependent RNA polymerase lattices. Science 296:2218–2222 [DOI] [PubMed] [Google Scholar]
- 35. Miller S, Krijnse-Locker J. 2008. Modification of intracellular membrane structures for virus replication. Nat. Rev. Microbiol. 6:363–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mise K, Ahlquist P. 1995. Host-specificity restriction by bromovirus cell-to-cell movement protein occurs after initial cell-to-cell spread of infection in nonhost plants. Virology 206:276–286 [DOI] [PubMed] [Google Scholar]
- 37. Miyazaki M, Kim HJ, Man WC, Ntambi JM. 2001. Oleoyl-CoA is the major de novo product of stearoyl-CoA desaturase 1 gene isoform and substrate for the biosynthesis of the Harderian gland 1-alkyl-2,3-diacylglycerol. J. Biol. Chem. 276:39455–39461 [DOI] [PubMed] [Google Scholar]
- 38. Novoa RR, et al. 2005. Virus factories: associations of cell organelles for viral replication and morphogenesis. Biol. Cell 97:147–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. O'Reilly EK, Paul JD, Kao CC. 1997. Analysis of the interaction of viral RNA replication proteins by using the yeast two-hybrid assay. J. Virol. 71:7526–7532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. O'Reilly EK, Wang Z, French R, Kao CC. 1998. Interactions between the structural domains of the RNA replication proteins of plant-infecting RNA viruses. J. Virol. 72:7160–7169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Panavas T, Hawkins CM, Panaviene Z, Nagy PD. 2005. The role of the p33:p33/p92 interaction domain in RNA replication and intracellular localization of p33 and p92 proteins of Cucumber necrosis tombusvirus. Virology 338:81–95 [DOI] [PubMed] [Google Scholar]
- 42. Pata JD, Schultz SC, Kirkegaard K. 1995. Functional oligomerization of poliovirus RNA-dependent RNA polymerase. RNA 1:466–477 [PMC free article] [PubMed] [Google Scholar]
- 43. Patel SS, Picha KM. 2000. Structure and function of hexameric helicases. Annu. Rev. Biochem. 69:651–697 [DOI] [PubMed] [Google Scholar]
- 44. Pedersen KW, van der Meer Y, Roos N, Snijder EJ. 1999. Open reading frame 1a-encoded subunits of the arterivirus replicase induce endoplasmic reticulum-derived double-membrane vesicles which carry the viral replication complex. J. Virol. 73:2016–2026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rao Z, et al. 1995. Crystal structure of SIV matrix antigen and implications for virus assembly. Nature 378:743–747 [DOI] [PubMed] [Google Scholar]
- 46. Rattray JB, Schibeci A, Kidby DK. 1975. Lipids of yeasts. Bacteriol. Rev. 39:197–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Restrepo-Hartwig M, Ahlquist P. 1999. Brome mosaic virus RNA replication proteins 1a and 2a colocalize and 1a independently localizes on the yeast endoplasmic reticulum. J. Virol. 73:10303–10309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Restrepo-Hartwig MA, Ahlquist P. 1996. Brome mosaic virus helicase- and polymerase-like proteins colocalize on the endoplasmic reticulum at sites of viral RNA synthesis. J. Virol. 70:8908–8916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ruigrok RW, et al. 2000. Structural characterization and membrane binding properties of the matrix protein VP40 of Ebola virus. J. Mol. Biol. 300:103–112 [DOI] [PubMed] [Google Scholar]
- 50. Schwartz M, et al. 2002. A positive-strand RNA virus replication complex parallels form and function of retrovirus capsids. Mol. Cell 9:505–514 [DOI] [PubMed] [Google Scholar]
- 51. Schwartz M, Chen J, Lee WM, Janda M, Ahlquist P. 2004. Alternate, virus-induced membrane rearrangements support positive-strand RNA virus genome replication. Proc. Natl. Acad. Sci. U. S. A. 101:11263–11268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Spuul P, et al. 2011. Assembly of alphavirus replication complexes from RNA and protein components in a novel trans-replication system in mammalian cells. J. Virol. 85:4739–4751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Suhy DA, Giddings TH, Jr., Kirkegaard K. 2000. Remodeling the endoplasmic reticulum by poliovirus infection and by individual viral proteins: an autophagy-like origin for virus-induced vesicles. J. Virol. 74:8953–8965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sullivan ML, Ahlquist P. 1999. A brome mosaic virus intergenic RNA3 replication signal functions with viral replication protein 1a to dramatically stabilize RNA in vivo. J. Virol. 73:2622–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Timmins J, et al. 2003. Oligomerization and polymerization of the filovirus matrix protein VP40. Virology 312:359–368 [DOI] [PubMed] [Google Scholar]
- 56. van der Heijden MW, Bol JF. 2002. Composition of alphavirus-like replication complexes: involvement of virus and host encoded proteins. Arch. Virol. 147:875–898 [DOI] [PubMed] [Google Scholar]
- 57. Wang QM, et al. 2002. Oligomerization and cooperative RNA synthesis activity of hepatitis C virus RNA-dependent RNA polymerase. J. Virol. 76:3865–3872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wang X, et al. 2005. Brome mosaic virus 1a nucleoside triphosphatase/helicase domain plays crucial roles in recruiting RNA replication templates. J. Virol. 79:13747–13758 [DOI] [PMC free article] [PubMed] [Google Scholar]