Abstract
HIV CCR5 antagonists select for env gene mutations that enable virus entry via drug-bound coreceptor. To investigate the mechanisms responsible for viral adaptation to drug-bound coreceptor-mediated entry, we studied viral isolates from three participants who developed CCR5 antagonist resistance during treatment with vicriviroc (VCV), an investigational small-molecule CCR5 antagonist. VCV-sensitive and -resistant viruses were isolated from one HIV subtype C- and two subtype B-infected participants; VCV-resistant isolates had mutations in the V3 loop of gp120 and were cross-resistant to TAK-779, an investigational antagonist, and maraviroc (MVC). All three resistant isolates contained a 306P mutation but had variable mutations elsewhere in the V3 stem. We used a virus-cell β-lactamase (BlaM) fusion assay to determine the entry kinetics of recombinant viruses that incorporated full-length VCV-sensitive and -resistant envelopes. VCV-resistant isolates exhibited delayed entry rates in the absence of drug, relative to pretherapy VCV-sensitive isolates. The addition of drug corrected these delays. These findings were generalizable across target cell types with a range of CD4 and CCR5 surface densities and were observed when either population-derived or clonal envelopes were used to construct recombinant viruses. V3 loop mutations alone were sufficient to restore virus entry in the presence of drug, and the accumulation of V3 mutations during VCV therapy led to progressively higher rates of viral entry. We propose that the restoration of pre-CCR5 antagonist therapy HIV entry kinetics drives the selection of V3 loop mutations and may represent a common mechanism that underlies the emergence of CCR5 antagonist resistance.
INTRODUCTION
Human immunodeficiency virus entry is a competition at the target cell membrane between viral inactivation and successful binding to CD4 and a coreceptor, either CCR5 or CXCR4 (16, 30, 32, 33, 37, 38). Engagement of receptor and coreceptor by gp120 and the subsequent rearrangement of gp41, the HIV fusion glycopeptides, leads to fusion (18, 21, 29, 57, 60, 63). Small-molecule antagonists that bind CCR5 and inhibit gp120-CCR5 binding through an allosteric mechanism have been developed (9, 23, 51, 53, 56, 61). Maraviroc (MVC) is an FDA-approved CCR5 antagonist, and vicriviroc (VCV) is an investigational compound whose development has been halted (10, 47).
HIV can escape CCR5 antagonism through the outgrowth of preexisting CXCR4-using populations or by the selection of resistance mutations (2, 52, 58). Mutations in the third hypervariable loop (V3 loop) of HIV-1 gp120 most commonly cause CCR5 antagonist resistance, but no canonical sites or amino acid substitutions have been identified; resistance mutations from one isolate generally do not confer resistance when transferred into unrelated env backbones (12, 17, 24, 26, 27, 34–36, 49, 54). This varied nature of HIV CCR5 antagonist genotypic resistance contrasts with resistance patterns established in other antiretroviral drug classes, for which canonical mutations (M184V in reverse transcriptase for lamivudine, V38A in gp41 for enfuvirtide, etc.) always lead to resistance, regardless of viral genetic background. Resistance to small-molecule CCR5 antagonists typically manifests as a decrease in the maximal percent inhibition (MPI) achieved, not an increase in the half-maximal inhibitory concentration (IC50), and occurs by a noncompetitive mechanism as the virus adapts to use both native CCR5 and drug-bound coreceptor (40, 41, 48, 54, 59). There are notable exceptions to this paradigm: in vitro-derived mutations in the gp41 fusion peptide without V3 changes can cause CCR5 antagonist resistance, and a recent report described clinical MVC resistance that emerged by a competitive mechanism (1, 6).
The efficiency and kinetics of HIV entry have been associated with viral fitness (3, 19, 25, 42). Mutations in the coreceptor binding site modulate entry kinetics and some fusion inhibitor (enfuvirtide) resistance mutations have been shown to either reduce or correct the time to half-maximal viral fusion (t1/2max) (43, 45, 46). We hypothesized that V3 loop CCR5 antagonist resistance mutations would also affect viral entry rates. We used participants' plasma samples from ACTG 5211 (A5211), a phase IIb VCV study, to identify CCR5 antagonist-resistant viral isolates and applied a virus-cell β-lactamase (BlaM) entry assay to evaluate the impact of CCR5 antagonist resistance mutations on entry kinetics (4, 11, 54). This assay reliably measures the extent of virus-cell fusion despite the fact that complete fusion occurs in endosomes (5, 29). CCR5 antagonist-resistant isolates identified from three participants during VCV therapy developed different V3 mutations, yet all isolates displayed delayed entry kinetics that corrected to pretherapy levels in the presence of drug. Our findings suggest that CCR5 antagonist resistance mutations may be selected to maximize entry kinetics and viral fitness under drug selection pressure.
MATERIALS AND METHODS
Subject samples.
We identified vicriviroc (VCV)-resistant HIV-1 from three subjects treated with VCV through participation in AIDS Clinical Trials Group (ACTG) 5211 (A5211; ClinicalTrials.gov identifier NCT00082498), a phase IIb clinical trial of VCV, an investigational CCR5 antagonist (11, 54). Three to six time points were analyzed per subject, including study entry (week 0) and the time of maximal observed VCV resistance. All research involving human participants was approved by the Partners Human Research Committee and the relevant institutional review boards of all participating ACTG sites. Written informed consent was obtained from all study participants.
Cells and cell culture.
TZM-bl and U87-CD4-CCR5 cells were obtained from the NIH AIDS Research and Reference Reagent Program (ARRRP). 293T cells were obtained from the American Type Culture Collection (ATCC; Manassas, VA). TZM-bl and 293T cell lines were maintained in Dulbecco modified Eagle medium with l-glutamine (DMEM; Gibco, Invitrogen) supplemented with 10% fetal bovine serum (Invitrogen), 100 U/ml penicillin, and 100 μg/ml streptomycin (Cellgro; Mediatech) that was referred to as DMEM complete (DMEM-C). U87-CD4-CCR5 cells were grown in DMEM-C plus 300 μg/ml of Geneticin (G418, Sigma) and 1 μg/ml puromycin (MP Biomedicals) to maintain CD4 and CCR5 coreceptor expression. For all kinetics experiments, DMEM-C was replaced with phenol red-free DMEM (Gibco, Invitrogen) that was similarly supplemented.
Single-donor peripheral blood mononuclear cells (PBMC) from HIV-seronegative donors were obtained by Ficoll-Hypaque density gradient centrifugation. Prior to HIV infection, PBMC were stimulated with phytohemagglutinin (2 μg/ml; Gibco) for 3 days and maintained in R-20 medium (RPMI 1640 supplemented with 20% fetal bovine serum, 2 mM l-glutamine, 10 mM HEPES buffer, 100 U/ml recombinant human interleukin-2 [NIH ARRRP], 50 U/ml penicillin, and 50 μg/ml streptomycin). All cell cultures were maintained at 37°C and 5% CO2.
CCR5 antagonists and susceptibility testing.
Maraviroc and TAK-779 were obtained from the NIH ARRRP. VCV was a kind gift from B. M. Baroudy (formerly of Schering-Plough). Subjects enrolled in A5211 had VCV susceptibility testing performed by Monogram Biosciences using a PhenoSense entry inhibitor assay (59). Clonal susceptibility testing was performed as previously described (12, 54).
HIV-1 env cloning sequence analysis.
HIV-1 RNA was extracted from plasma samples, and the full-length env gene was amplified. To minimize any potential founder effect, four independent reverse transcription (RT) reactions and PCR amplifications were performed and combined for each time point. These purified amplicons were then ligated into a TOPO-TA vector (Invitrogen) and electroporated into TOP10 cells. Subclones were isolated and sequenced by conventional (Sanger) methods as described previously (54). Between 15 and 32 clones were sequenced per time point. All sequences were edited, aligned, and compiled with Geneious Pro (7).
Virus construction.
We used a yeast gap-repair homologous recombination system to generate recombinant HIV-1 that contained the dominant full-length env sequence of plasma virus obtained from subject 07 (Sub07) at weeks 0, 16, 19, and 28 as described previously (12, 54). Recombinant virus that incorporated HIV-1 envelopes from subjects 57 (Sub57) and 85 (Sub85) were constructed by a modification of a previously described method (15, 20). Briefly, the cytomegalovirus (CMV) promoter was amplified and attached to a 265-bp segment of the rev gene from pNL43 using overlap PCR. A second overlap PCR was then performed to link the CMV-rev segment to the cloned or uncloned env amplicon of interest. These CMV-rev-env amplicons were then cotransfected into 293T cells with an NL4-3 envelope-deleted vector (22).
To generate infectious virus that carried the BlaM-Vpr fusion protein, we transfected 293T cells with pCMV4-BlaM-Vpr kindly provided by W. C. Greene, pAdVantage (Promega), and either recombinant plasmid (Sub07 env genes) or CMV-rev-env amplicons (Sub57 and Sub85 env genes) together with the env gene-deleted NL4-3. Briefly, 8 × 106 293T cells were plated and cultured overnight at 37°C. On the day of transfection, cells were washed twice with phosphate-buffered saline (PBS; Cellgro; Mediatech) and the medium was replaced by fresh phenol red-negative DMEM-C and warmed to 37°C. We used the Fugene 6 protocol (Roche Molecular Biochemicals) to transfect cells as previously described (12, 54). Supernatants were collected 48 h later, passed through a 0.45-μm filter, centrifuged at 72,000 × g for 90 min at 4°C, aliquoted, and stored at −80°C until use. Viral titers were determined by p24 antigen enzyme-linked immunosorbent assay (ELISA; Perkin-Elmer, Boston, MA) and by endpoint dilution on TZM-bl cells as described previously (12, 54).
HIV-1 entry kinetics assays.
We used a well-validated BlaM activity assay to quantify virus-cell fusion in real time and investigate the effects of CCR5 antagonist resistance on entry kinetics (4, 29). A total of 5 × 104 cells/well (TZM-bl cells, U87-CD4-CCR5 cells, or single-donor PBMC) were grown overnight in 96-well black clear-bottom plates (Costar, Sigma-Aldrich) in DMEM-C without phenol red. On the day of the viral fusion experiment, target cells were washed with PBS and loaded with CCF2-AM dye (LiveBlazer; Invitrogen) for 1 h at room temperature. After two additional PBS washes, cells were chilled to 4°C and identical volumes of recombinant virus expressing the envelope of interest and carrying the BlaM-Vpr fusion, also at 4°C, were added to each well. Previous modeling demonstrated that normalized entry kinetics data do not depend on the amount of input virus, or multiplicity of infection (MOI) (30). Between 25 and 400 ng of p24 antigen was added to each well; for each experiment, an identical amount of virus was added. Cells and viruses were incubated at 4°C for 30 min and then transferred into a microplate reader (FLUOstar Optima; BMG Labtech); fusion was initiated by an immediate shift to 37°C. For experiments conducted in the presence of drug, cells were preincubated with 1 nM or 1 μM TAK-779, 500 nM VCV, or 1 μM MVC for 2 h at 37°C.
Fusion kinetics at 37°C were determined by real-time fluorimetry and monitored at 5-min intervals for 180 min. Entry was determined by measuring the shift from green to blue fluorescence, indicating β-lactamase cleavage of target cell CCF2. Fluorescence emissions were quantified using a 405-nm ± 20-nm excitation filter (Chroma Technology) and emission filters of 460 nm ± 50 nm (blue channel; Chroma) and 535 nm ± 25 nm (green channel; Chroma). Results were expressed as the ratio of blue to green emissions at each time point. These ratios were calculated after blue/green emissions from control CCF2-loaded cells without virus were subtracted at the corresponding time point.
The progression of entry was determined by plotting changes in the ratio of blue to green over time. To normalize the data, the maximum entry of each recombinant virus was determined by the time point at which maximum the blue-to-green ratio occurred and was maintained (plateau). The correlation between time and percentage of fusion was then calculated and curved fitted using GraphPad Prism 5 (GraphPad, La Jolla, CA).
Extent of fusion experiments.
For the enfuvirtide blockage experiments, targets cells were seeded in 96-well plates for 24 h. On the day of infection, HIV-1 viruses bearing BlaM-Vpr were used to bind on the surface of target cells. Plates were centrifuged at 3,000 rpm and 4°C for 30 min. For the temperature block procedure, fusion was initiated by transferring the plate into a 37°C incubator and stopped at specified time points by immediately placing the plate on ice. Enfuvirtide (1 μM) was added to each well at the specified time point. Quantification of fluorescence intensity was determined by using a microplate reader as described above.
Nucleotide sequence accession numbers.
The env nucleotide sequences used in this study have been deposited in GenBank under accession numbers EU663615 (Sub07sens), EU663618 (Sub07res), and JQ182772 to JQ182988 (Sub57sens/Sub57res and Sub85sens/Sub85res and all V3 loops submitted).
RESULTS
Identification of clinical CCR5 antagonist resistance.
We previously reported VCV resistance of an HIV-1 subtype C virus from a VCV-treated participant in ACTG A5211 (Sub07, previously referred to as subject 07J) (12, 54). Two additional VCV-resistant isolates have now been identified from HIV-1 subtype B-infected subjects. Vicriviroc resistance in plasma virus was identified after 144 weeks of VCV treatment in subject 57 (Sub57) and after 138 weeks in subject 85 (Sub85), with MPIs of 40% and 36%, respectively, in the Monogram entry susceptibility assay (Table 1). Sequence analysis of cloned env genes of the VCV-resistant viruses demonstrated mutations on both sides of the V3 stem, but the pattern of mutations differed between the isolates. The resistant virus from Sub57 (Sub57res) carried V3 mutations G306P, N308D, Y316W, G321A, and E322D, whereas the resistant virus from Sub85 (Sub85res) carried mutations S306P, N308D, R315K, A316V, G319A, and H330Y. Both resistant isolates carried a proline substitution at position 306 (X306P, where X is G or S) and the N308D mutation; the S306P mutation was noted previously for the VCV-resistant subtype C viruses from Sub07 (Sub07res) (54).
Table 1.
V3 loop mutations in HIV-1 subtype B-infected subjects 57 and 85a
| Week | Subject 57 |
Subject 85 |
||||
|---|---|---|---|---|---|---|
| MPI | No. of clones | V3 loop sequence | MPI | No. of clones | V3 loop sequence | |
| 0 | 94 | 20 | CTRPNNNTRRGINIGPGRAWYTTGEIIGDIRQAHC | 99 | 17 | CTRPNNNTRRSINIGPGRAWYGTDIIGDIRKAHC |
| 1 | ----------------E------------------ | 4 | --------------A-----------------Y- | |||
| 1 | -------A--------------------------- | 2 | --------------------------------Y- | |||
| 1 | ---------------S------------------- | |||||
| 1 | ------------------------K---------- | |||||
| 2 | 27 | ---------K------------------------- | ||||
| 4 | ----------------------------------- | |||||
| 48 | 89 | 10 | ------------------------D---------- | 94 | 19 | ----------N-D-------------------Y- |
| 8 | ----------D------------------------ | 12 | --------------------------------Y- | |||
| 4 | ----------P------------------------ | 1 | ----------N-D---------A---------Y- | |||
| 3 | ------------------------E---------- | |||||
| 1 | ------------S------------------K--- | |||||
| 106 | 13 | ----------P-D-------W--AD---------- | ||||
| 7 | ----------P-D----------AD---------- | |||||
| 1 | ----------P-D----------AD--------Y- | |||||
| 1 | ----------P-D-------W--AD-----G---- | |||||
| 1 | ----------P-D-------W--ADV--------- | |||||
| 1 | ----------P-D-------WP-AD---------- | |||||
| 1 | ----------P-D----------AD-T-------- | |||||
| 1 | ----------P-D----------AD--------Y- | |||||
| 1 | ----------P-D----------AD-------V-- | |||||
| 138 | 36 | 24 | ----------P-D----KV--A----------Y- | |||
| 7 | ----------P-D----KV--A------------ | |||||
| 144 | 40 | 14 | ----------P-D-------W--AD---------- | |||
| 1 | ----------P-D-------W--AD-------V-- | |||||
MPI, maximal percent inhibition; V3, third hypervariable loop of HIV-1 gp120.
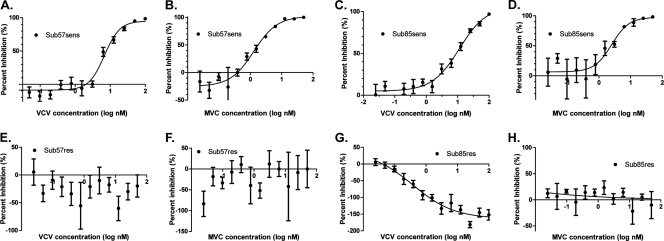
We next constructed recombinant viruses that incorporated the dominant full-length env gene obtained at week 0 (start of VCV therapy) and at later time points and performed susceptibility testing with a panel of CCR5 antagonists (54). Recombinant viruses expressing the week 0 env gene from Sub57 (Sub57sens) and Sub85 (Sub85sens) demonstrated MPIs of 100% to VCV, maraviroc (MVC), and TAK-779 (Fig. 1 and data not shown). Recombinant viruses expressing cloned env genes from VCV-resistant viruses (Sub57res and Sub85res, respectively) were cross-resistant to MVC and TAK-779. In TZM-bl cells, VCV and MVC MPIs for Sub85res were −163.0 and 1.0, respectively (Fig. 1G and H). An exact curve-fit for susceptibility testing of Sub57res with VCV and MVC was not possible due to a lack of convergence of the data; most data points were well below 0% inhibition. The Sub85res recombinant demonstrated markedly enhanced replication in the presence of VCV. Susceptibility testing with U87-CD4-CCR5 cells corroborated these findings, although the absolute MPI values differed. The VCV, MVC, and TAK-779 MPIs for Sub57res were 16.9, 43.0, and 27.5, respectively, and those for Sub85res were 39.2, 61.1, and 38.6, respectively. The MPIs of the VCV-resistant recombinant viruses were lower than the MPIs obtained by the PhenoSense entry inhibitor assay using the uncloned env gene from the corresponding time points (Table 1). Given the observed cross-resistance to all CCR5 antagonists tested, we initially used TAK-779 for subsequent kinetics experiments.
Fig 1.
CCR5 antagonist susceptibilities of Sub57 and Sub 85 virus. In each graph, the percentages of inhibition relative to the viral level in the no-drug control at various inhibitor concentrations are shown. (A) Sub57 baseline VCV susceptibility, (B) Sub57 baseline MVC susceptibility, (C) Sub85 baseline VCV susceptibility, (D) Sub85 baseline MVC susceptibility, (E) Sub57 week 144 VCV susceptibility, (F) Sub57 week 144 MVC susceptibility, (G) Sub85 week 138 VCV susceptibility, (H) Sub85 week 138 MVC susceptibility. Error bars represent the standard errors of the means of results from at least two experiments, each performed in triplicate. Nonlinear regression with a variable slope was used to estimate a fitted curve; the data for Sub57 week 144 susceptibilities did not converge.
Entry kinetics of CCR5 antagonist-resistant HIV-1.
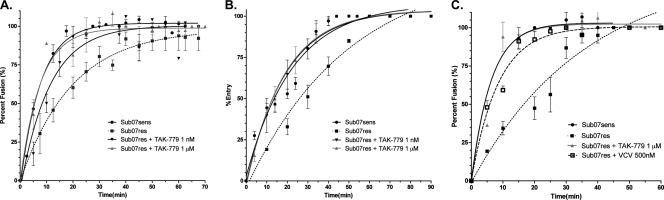
We first assessed the entry kinetics of VCV-susceptible (Sub07sens) and -resistant (Sub07res) HIV-1 from Sub07 (Fig. 2). When tested on U87-CD4-CCR5 cells, Sub07sens achieved maximal fusion in 40 min; the time to half-maximal fusion was 5.4 min (95% confidence interval [CI], 4.7 to 6.3 min). In contrast, Sub07res did not achieve maximal fusion even after 90 min, with a t1/2max of 13.6 min (95% CI, 14.8 to 29.3 min). The Sub07res virus also exhibited delayed entry into TZM-bl cells and PBMC relative to results for Sub07sens. The observed t1/2max for Sub07sens and Sub07res was 15.7 min (95% CI, 12.8 to 20.2 min) and 34.4 min (95% CI, 25.6 to 52.7), respectively, in TZM-bl cells and 4.3 min (95% CI, 3.1 to 7.4 min) and 7.7 min (95% CI, 5.9 to 11.3 min), respectively, in PBMC. The Sub07sens and Sub07res viruses reached maximal fusion with PBMC in 25 and 45 min, respectively, but required 90 min to do so in TZM-bl cells. The addition of TAK-779 at a nonsaturating concentration (1 nM) reduced the t1/2max for Sub07res on U87-CD4-CCR5 cells to 7.7 min (95% CI, 5.8 to 11.4 min), a rate that was intermediate between the values obtained for Sub07sens and Sub07res in the absence of drug. The addition of TAK-779 at concentrations expected to saturate CCR5 binding (1 μM) restored the fusion rates of Sub07res to a t1/2max that was indistinguishable from those of Sub07sens for all three cell types tested (Fig. 2). Faster fusion rates on PBMC were also seen with Sub07res in the presence of saturating VCV concentrations (500 nM) (Fig. 2C). Statistically significant differences in fusion rates between sensitive and resistant isolates, and between resistant isolates in the presence and absence of drug, were observed for all three subjects (nonoverlapping 95% confidence intervals) (Table 2).
Fig 2.
Clonal entry kinetics of Sub07 subtype C isolates. Entry kinetics in a virus-cell BlaM fusion assay was assessed over 60 to 90 min in (A) U87-CD4-CCR5 cells, (B) TZM-bl cells, and (C) PBMC isolated from a single donor. Data were fitted to a one-phase association binding equation, Y = Ymax(1−e−KX), and the time to half-maximal fusion (t1/2max) was calculated for each experimental condition. Error bars represent the standard errors of the means of results from at least two experiments, each performed in triplicate. The goodness-of-fit measure (r2) was between 0.90 and 0.99 for all curve fits with the exception of Sub07res + 1 μM TAK-779 in PBMCs; for this condition, r2 was 0.80.
Table 2.
Half-maximal entry times for CCR5 antagonist-sensitive and -resistant virusesa
| Virus | Clonal t1/2max (min) (95% CI) |
Population t1/2max (min) (95% CI) |
||
|---|---|---|---|---|
| U87-CD4-CCR5 | TZM-bl | Single-donor PBMC | TZM-bl | |
| Sub07sens | 5.4 (4.7–6.3) | 15.7 (12.8–20.2) | 4.3 (3.1–7.4) | ND |
| Sub07res | 13.6 (14.8–29.3) | 34.4 (25.6–52.7) | 7.7 (5.9–11.3) | ND |
| Sub07res + 1 nM TAK-779 | 7.7 (5.8–11.4) | 13.2 (10.8–17.0) | ND | |
| Sub07res + 1 μM TAK-779 | 5.2 (3.9–7.7) | 12.9 (10.7–16.1) | 4.2 (3.3–5.9) | ND |
| Sub57sens | ND | 3.9 (3.4–4.5) | ND | 12.0 (7.7–26.7) |
| Sub57res | ND | 11.4 (7.9–20.8) | ND | 17.1 (12.5–27.0) |
| Sub57res + 1 μM TAK-779 | ND | 5.5 (4.6–6.8) | ND | 7.8 (6.3–10.4) |
| Sub85sens | ND | 11.8 (10.4–13.2) | ND | 3.1 (2.5–4.1) |
| Sub85res | ND | 20.0 (17.8–22.2) | ND | 22.3 (20.9–23.6) |
| Sub85res + 1 μM TAK-779 | ND | 10.1 (9.5–10.6) | ND | 11.5 (10.2–12.7) |
t1/2max, time to half-maximal fusion; 95% CI; 95% confidence interval; ND, not done.
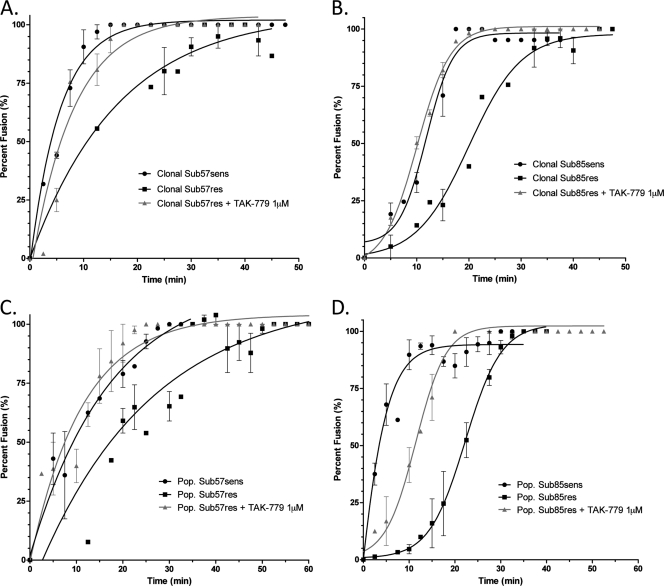
Similar results were obtained with recombinant viruses expressing CCR5 antagonist-resistant envelopes from HIV-1 subtype B (Fig. 3A and B). Viruses expressing the cloned env gene from VCV-susceptible HIV-1 from subjects 57 and 85 (Sub57sens and Sub85sens, respectively) reached maximal fusion in 20 min and had times to half-maximal fusion of 3.9 min (95% CI, 3.4 to 4.5 min) and 11.8 min (95% CI, 10.4 to 13.2), respectively. In contrast, recombinants expressing the VCV-resistant env genes from Sub57res and Sub85res required more than 50 min to achieve maximal fusion and demonstrated times to half-maximal fusion of 11.4 min (95% CI, 7.9 to 20.8) and 20.0 min (95% CI, 17.8 to 22.2 min), respectively. When tested in the presence of 1 μM TAK-779, the t1/2max for Sub57res was reduced to 5.5 min (95% CI, 4.6 to 6.8), and the t1/2max for Sub85res was reduced to 10.1 min (95% CI, 9.5 to 10.6 min). Of note, the entry kinetics of Sub85sens and Sub85res were best approximated by a biphasic sigmoid curve rather than the single-phase association curve used for fitting data from the Sub07 and Sub57 viruses.
Fig 3.
Entry kinetics of the subtype B Sub57 and Sub85 viral isolates. The entry of CCR5 antagonist-sensitive and -resistant isolates from Sub57 and Sub85 was assessed with TZM-bl cells in the presence and absence of 1 μM TAK-779. Recombinant viruses were generated with (A) Sub57 and (B) Sub85 dominant clonal envelopes or (C) Sub57 and (D) Sub85 population-derived env gene populations from baseline and resistant time points. Error bars represent the standard errors of the means of results from at least two experiments, each performed in triplicate. Nonlinear regression was used to estimate a fitted curve. Goodness-of-fit measures (r2) were greater than 0.95 for all curves except Sub57 population fits; r2 for those experiments ranged from 0.83 to 0.91.
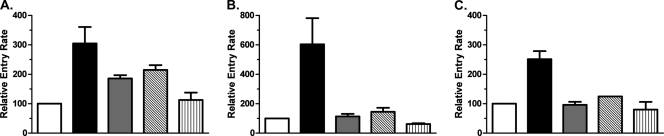
To verify that the fusion rates observed with recombinants expressing the dominant env clone were representative of the plasma HIV-1 population overall, recombinant viruses expressing uncloned HIV-1 env amplicons from Sub57 and Sub85 were also tested. Entry kinetics for recombinants carrying uncloned env amplicons obtained from Sub57 at weeks 0 and 144 showed lower fusion rates than recombinants expressing the cloned dominant env sequence from the corresponding time points, but in the absence of drug, the VCV-resistant viruses still showed a lower rate of fusion than the susceptible viruses (Fig. 3C and Table 2). As expected, the t1/2max for entry of the uncloned VCV-resistant virus was accelerated in the presence of 1 μM TAK-779 (Table 2). Similarly, recombinants expressing the uncloned VCV-sensitive env gene from Sub85 at week 0 had higher entry rates than the corresponding recombinants carrying the VCV-resistant env gene from week 138 (Fig. 3D and Table 2). The addition of 1 μM TAK-779, however, only partially accelerated the t1/2max of the resistant virus, to a value intermediate between the t1/2max values for the VCV-susceptible and resistant viruses in the absence of drug (Table 2). To increase the generalizability of these findings, fusion rates were tested with the cloned env gene in TZM-bl cells in the presence of MVC and VCV (Fig. 4). Data were normalized to the t1/2max observed for the drug-sensitive isolates from each subject, represented graphically as 100%. In these experiments, the addition of MVC, VCV, or TAK-779 had similar effects on entry kinetics rates. We saw no evidence of a correlation between relative entry rates and MPI in these experiments.
Fig 4.
Relative entry rates as a function of CCR5 antagonist. The entry rates of drug-resistant isolates in the presence and absence of saturating CCR5 antagonist concentrations, normalized to the half-maximal entry rates of subject-specific CCR5 antagonist-sensitive isolate, are shown. (A) Sub07, (B) Sub57, (C) Sub85. White bar, sensitive isolate; solid black bar, resistant isolate; gray bar, resistant isolate + 500 nM VCV; diagonally striped bar, resistant isolate + 1 μM MVC; vertically striped bar, resistant isolate + 1 μM TAK-779. Error bars represent the standard errors of the means of results from at least two experiments, each performed in triplicate.
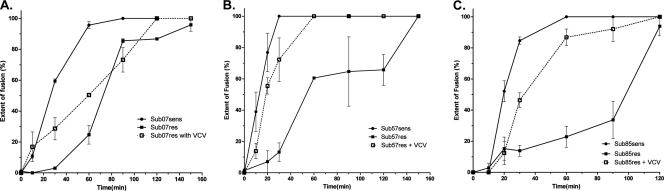
We next used a panel of small-molecule antagonists to investigate the effect of resistance on the total extent of virus fusion (Fig. 5). After incubation at 37°C, reactions were stopped at the indicated times by the addition of enfuvirtide; fluorescence intensity was measured. A control experiment with enfuvirtide added at time zero demonstrated no change in the blue-green fluorescence ratios over the duration of the experiment (data not shown). More fusion was observed for drug-sensitive viral isolates, relative to CCR5 antagonist-resistant clones. The addition of VCV to experiments with Sub07res, Sub57res, and Sub85res increased the extent of fusion in the first 60 min, a phenomenon also observed with TAK-779 (data not shown). Similar amounts of fusion were seen with sensitive and resistant isolates when incubation times were extended to 120 to 160 min.
Fig 5.
The extent of fusion deficits correct in the presence of a CCR5 antagonist. The relative extent of entry into U87-CD4-CCR5 cells in the presence and absence of VCV 500 nM is shown. Virus fusion with cells was stopped by adding enfuvirtide after the indicated incubation times. (A) Sub07 isolates, (B) Sub57 isolates, (C) Sub85 isolates. Error bars represent the standard errors of the means of results from at least two experiments, each performed in triplicate.
HIV-1 gp120 V3 loop mutations are sufficient for changes in fusion kinetics.
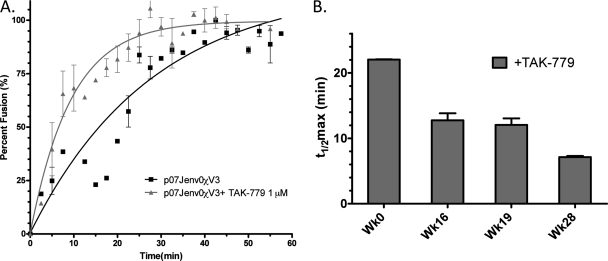
To demonstrate that the observed differences in t1/2max between VCV-susceptible and -resistant viruses from Sub07 were related to changes in the V3 loop, we constructed a recombinant HIV-1, denoted p07Jenv0χV3, that contained a chimeric env gene with the V3 loop from Sub07res substituted into the Sub07sens env gene. When tested with TZM-bl cells, the chimeric virus demonstrated a t1/2max for fusion of 19.5 min (95% CI, 13.6 to 34.4 min) in the absence of drug and 6.5 min (95% CI, 5.1 to 9.0 min) in the presence of 1 μM TAK-779, indicating that the mutant V3 loop conferred the delayed fusion kinetics observed for the Sub07res virus (Fig. 6A).
Fig 6.
The role of V3 loop mutations in CCR5 antagonist-resistant viral entry kinetics. Rates of Sub07-derived recombinant virus entry were determined in the presence and absence of TAK-779 for (A) the viral construct p07Jenv0χV3 that substituted the V3 loop of Sub07res into the env backbone of Sub07sens and (B) dominant env clones isolated at weeks 0 (Wk0; Sub07sens), 16, 19, and 28 (Sub07res) during VCV therapy. V3 mutations chronologically accumulated over 28 weeks and led to progressive reductions in the VCV MPI (54). The kinetics data were fitted to a one-phase association binding equation. Error bars in both panels represent the standard errors of the means of results from at least two experiments, each performed in triplicate.
Accumulation of V3 mutations increases fusion rates.
The accumulation of V3 loop mutations in HIV-1 from Sub07 over 28 weeks of VCV therapy conferred progressively higher levels of resistance and increased viral infectivity in the presence of drug (54). We hypothesized that this adaptation to VCV-bound CCR5 increased the rate of viral fusion. To determine if increasing VCV resistance corresponded to a trend toward faster entry in the presence of drug, we tested recombinant viruses that incorporated the dominant full-length env sequence from weeks 16 and 19, along with Sub07sens and Sub07res. Fusion kinetics experiments performed in the presence of 1 μM TAK-779 demonstrated a decreasing t1/2max of 22.0 ± 0.1 min (t1/2max ± standard error of the mean [SEM]), 12.8 ± 1.8 min, 12.0 ± 1.7 min, and 7.1 ± 0.03 min for Sub07sens, week 16 virus, week 19 virus, and Sub07res, respectively (Fig. 6B). Virus t1/2max declined as mutations accumulated in V3 and the MPI for VCV decreased.
DISCUSSION
We used VCV-resistant subtype B and C HIV-1 obtained from three subjects enrolled in a phase 2b study of VCV to explore the effects of CCR5 antagonist resistance on viral entry kinetics. All three VCV-resistant viruses were cross-resistant to MVC and TAK-779. In-house susceptibility testing was conducted with TZM-bl and U87 cell lines; MPIs for VCV observed with U87 cells more closely matched the MPIs obtained by a commercially available entry susceptibility assay, also performed with U87 cells. The time to high-level resistance ranged from 28 to 144 weeks of VCV therapy. Decreasing susceptibility to CCR5 antagonists correlated with an increasing number of mutations in the V3 loop of gp120. Different V3 mutations were selected in each case, with the exception of the proline substitution at position 306, which was common to all three resistant viruses.
Resistance to CCR5 antagonists has been described for HIV-1 from subjects with subtypes B, C, and D (6, 26, 27, 34, 35, 48, 54). Mutations in the V3 loop have been observed in most but not all cases. A proline substitution at position 306 is present only rarely in HIV-1 envelopes from patients who have not been treated with CCR5 antagonists. Incorporation of a proline in the V3 stem most likely distorts the conformation of the V3 loop in a way that facilitates interaction of the mutant gp120 with VCV-bound CCR5. Studies with site-directed mutations showed that presence of the G306P substitution together with other V3 mutations is necessary for complete resistance to VCV in the Sub07sens env gene, but when introduced as the sole substitution, the G306P mutation confers increased susceptibility to VCV (12). Of note, although the viruses we characterized were all cross-resistant to MVC, a proline substitution at position 306 has not been described for the limited number of isolates with primary MVC resistance reported to date (6, 55).
Recombinant viruses expressing envelopes of the CCR5 antagonist-resistant viruses exhibited delayed entry kinetics on CCR5-expressing CD4+ cell lines and on PBMC. Wild-type rates of entry were restored in the presence of the antagonist. The level of CD4 and CCR5 (or CXCR4) on the cell surface modulates the rate of virus entry. Hence, the choice of target cell type can influence the phenotypic manifestations of CCR5 antagonist resistance (14, 30, 40, 41, 44). The t1/2max for fusion of a given virus differed by cell type, but enhancement of entry kinetics of the resistant viruses in the presence of CCR5 antagonists was observed for all cell types tested. The fastest entry rates were noted for PBMC. Although surface expression of CCR5 on TZM-bl and U87-CD4-CCR5 cells is substantially greater than that that found on resting peripheral blood lymphocytes, interleukin-2 (IL-2)-stimulated PBMC, such as those used in the assays reported here, express levels of CCR5 comparable to or greater than the levels observed for CCR5-expressing U87 cells (13, 39, 44, 62).
Experiments with recombinant viruses expressing uncloned envelopes demonstrated slower entry kinetics than recombinants that incorporated a cloned VCV-sensitive or -resistant env gene. Recombinant viruses that express uncloned env genes represent the diversity of viruses found in the plasma and reflect the average entry rate of the circulating quasispecies. Experiments with V3 chimeras and with recombinants expressing envelopes with intermediate levels of VCV resistance demonstrated that alterations in entry kinetics correlated with changes in the V3 loop. Entry rates in the presence of VCV increased as V3 loop mutations were added into the evolving env backbone. The entry rates of the VCV-resistant viruses in the presence of drug were never higher than the rates of the pretreatment, VCV-susceptible viruses, suggesting that the virus had maximized fusion kinetics in the fitness landscape particular to each host.
We observed qualitative differences in the kinetics observed for isolates from different patients. In the presence of drug, the entry of viruses expressing uncloned Sub57res env genes was similar to the rate observed for viruses expressing uncloned Sub57sens env genes. In contrast, although entry of viruses expressing uncloned Sub85res env genes was increased in the presence of drug, entry remained slower than for viruses expressing uncloned Sub85sens env. These differences did not correlate with the degree of resistance as measured by MPIs; in a commercial entry susceptibility assay, the MPIs of uncloned Sub57res and Sub85 were similar. The MPI is a measure of resistance, but our data suggest that it is not a direct proxy for the mechanistic changes, in this case changes to viral entry rates, that underlie that resistance.
HIV-1 fusion is a temperature-sensitive process (8, 28, 31). In the virus entry assay we used, virus and target cells were combined at 4°C and the fusion process was initiated by rapid warming to 37°C. The entry rates we observed represent a total combined rate for all components of the fusion process: CD4 binding, coreceptor engagement, a competing viral inactivation process, and eventual fusion. Late fusion events occur at similar rates in different viral isolates, so it is likely that the differences we observed in entry kinetics reflect changes in the kinetics of binding to CD4, CCR5, or both (29). Increased coreceptor affinity accelerates viral fusion; because CCR5 antagonist-resistant HIV-1 uses the drug-bound CCR5, the V3 loop mutations that confer resistance must increase gp120 affinity for the antagonist-coreceptor complex (36, 40, 44, 54, 59). A clonal analysis of env sequences from Sub07 and Sub57, however, demonstrated that even after the virus acquired the capacity to use the drug-coreceptor complex, resistance mutations continued to accumulate.
Entry of HIV-1 is considered the rate-limiting step of viral replication. Lower rates of virus entry and decreased fitness have been described for HIV-1 resistant to the fusion inhibitor enfuvirtide (19, 22, 43, 45). We propose that the dominant selection pressure driving the evolution of CCR5 antagonist resistance is the restoration of wild-type rates of virus entry. To permit entry in the presence of drug, HIV-1 must adapt to a new CCR5 conformation equilibrium, acquire the ability to bind the drug-CCR5 complex, and then maximize entry rates and fitness. A longitudinal analysis of env sequences from Sub07 and Sub57 by deep sequencing and cloning, respectively, demonstrates the complexity of V3 forms that are generated as the virus explores sequence space in search of a new fitness peak (52). We speculate that the variety of mutations capable of conferring resistance to small-molecule CCR5 antagonists produces a common structural and functional effect: maximizing entry kinetics in the context of the available env background.
This study has several limitations. Our analysis is based on observations from three CCR5 antagonist-resistant isolates that developed during VCV therapy; different results might be observed with additional isolates or with clinical isolates that developed primary maraviroc resistance. All samples were cross-resistant to MVC, however, and we endeavored to increase the generalizabilty of our findings by determining entry kinetics of subtype B and C HIV-1 in multiple cell types. To date, only a limited number of clinical isolates with CCR5 antagonist resistance have been studied. The entry kinetics curve fitting for Sub85res in the presence and absence of drug differed from that of other isolates and confounds a direct comparison of half-maximal entry rates across isolates. It is possible that sampling additional time points before the t1/2max would have demonstrated sigmoidal rate curves for all resistant isolates. We emphasize that a reproducible pattern of delayed CCR5 antagonist-resistant virus entry that corrects with drug was seen for all isolates, regardless of kinetics. Different HIV-1 strains exhibit various rates of virus entry, and it is possible, given the plasticity of the envelope, that other mechanisms of CCR5 antagonist resistance will be identified (30, 50). Studies to determine the entry kinetics of a VCV-resistant virus that carries mutations in the fusion domain of gp41 would be of interest in this regard. Our data suggest that the restoration of entry kinetics, with its resultant fitness implications, may ultimately drive HIV adaptation to small-molecule CCR5 antagonists.
ACKNOWLEDGMENTS
We thank Warner C. Greene for his kind gift of the pCMV4-BlaM-Vpr fusion plasmid. We thank Noriaki Hosoya for technical assistance in the completion of these experiments and the entire A5211 protocol team for their efforts during the trial.
A.M.N.T. was supported by NIH grant K08 AI081547. T.J.H. was supported by the KL2 Medical Research Investigator Training (MeRIT) program of Harvard Catalyst, The Harvard Clinical and Translational Science Center (UL1 RR025758). This study was supported in part by the AIDS Clinical Trials Group funded by the National Institute of Allergy and Infectious Diseases, NIH (grant AI068636, AIDS Clinical Trials Group), and NIH grants AI069419, Cornell CTU; AI051966 to R.M.G.; and AI055357 to D.R.K., and the National Center for Research Resources (grant RR024996, Cornell CTSC; grant RR016482 to D.R.K.).
Footnotes
Published ahead of print 16 November 2011
REFERENCES
- 1. Anastassopoulou CG, Ketas TJ, Klasse PJ, Moore JP. 2009. Resistance to CCR5 inhibitors caused by sequence changes in the fusion peptide of HIV-1 gp41. Proc. Natl. Acad. Sci. U. S. A. 106:18–5323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Archer J, et al. 2009. Detection of low-frequency pretherapy chemokine (CXC motif) receptor 4 (CXCR4)-using HIV-1 with ultra-deep pyrosequencing. AIDS 23:09–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ball SC, et al. 2003. Comparing the ex vivo fitness of CCR5-tropic human immunodeficiency virus type 1 isolates of subtypes B and C. J. Virol. 77:21–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cavrois M, De Noronha C, Greene WC. 2002. A sensitive and specific enzyme-based assay detecting HIV-1 virion fusion in primary T lymphocytes. Nat. Biotechnol. 20:51–1154 [DOI] [PubMed] [Google Scholar]
- 5. Cavrois M, Neidleman J, Yonemoto W, Fenard D, Greene WC. 2004. HIV-1 virion fusion assay: uncoating not required and no effect of Nef on fusion. Virology 328:36–44 [DOI] [PubMed] [Google Scholar]
- 6. Delobel P, et al. 2010. Shift in phenotypic susceptibility suggests a competition mechanism in a case of acquired resistance to maraviroc. AIDS 24:82–1384 [DOI] [PubMed] [Google Scholar]
- 7. Drummond AJ, et al. 2011. Geneious, v5.4.6. Biomatters Ltd., Auckland, New Zealand [Google Scholar]
- 8. Frey S, et al. 1995. Temperature dependence of cell-cell fusion induced by the envelope glycoprotein of human immunodeficiency virus type 1. J. Virol. 69:1462–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Garcia-Perez J, et al. 2011. New insights into the mechanisms whereby low molecular weight CCR5 ligands inhibit HIV-1 infection. J. Biol. Chem. 286:4978–4990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gulick RM, et al. 2008. Maraviroc for previously treated patients with R5 HIV-1 infection. N. Engl. J. Med. 359:29–1441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gulick RM, et al. 2007. Phase 2 study of the safety and efficacy of vicriviroc, a CCR5 inhibitor, in HIV-1-infected, treatment-experienced patients: AIDS clinical trials group 5211. J. Infect. Dis. 196:304–312 [DOI] [PubMed] [Google Scholar]
- 12. Henrich TJ, et al. 2010. Evolution of CCR5 antagonist resistance in an HIV-1 subtype C clinical isolate. J. Acquir. Immune Defic. Syndr. 55:420–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Heredia A, et al. 2008. Reduction of CCR5 with low-dose rapamycin enhances the antiviral activity of vicriviroc against both sensitive and drug-resistant HIV-1. Proc. Natl. Acad. Sci. U. S. A. 105:20476–20481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Johnston SH, et al. 2009. A quantitative affinity-profiling system that reveals distinct CD4/CCR5 usage patterns among human immunodeficiency virus type 1 and simian immunodeficiency virus strains. J. Virol. 83:016–11026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kirchherr JL, et al. 2007. High throughput functional analysis of HIV-1 env genes without cloning. J. Virol. Methods 143:4–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kuhmann SE, Platt EJ, Kozak SL, Kabat D. 2000. Cooperation of multiple CCR5 coreceptors is required for infections by human immunodeficiency virus type 1. J. Virol. 74:05–7015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kuhmann SE, et al. 2004. Genetic and phenotypic analyses of human immunodeficiency virus type 1 escape from a small-molecule CCR5 inhibitor. J. Virol. 78:90–2807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kwong PD, Wyatt R, Sattentau QJ, Sodroski J, Hendrickson WA. 2000. Oligomeric modeling and electrostatic analysis of the gp120 envelope glycoprotein of human immunodeficiency virus. J. Virol. 74:61–1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lassen KG, et al. 2009. Elite suppressor-derived HIV-1 envelope glycoproteins exhibit reduced entry efficiency and kinetics. PLoS Pathog. 5:e1000377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lin NH, et al. 2010. The design and validation of a novel phenotypic assay to determine HIV-1 coreceptor usage of clinical isolates. J. Virol. Methods 169:39–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. 2008. Molecular architecture of native HIV-1 gp120 trimers. Nature 455:9–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lu J, Sista P, Giguel F, Greenberg M, Kuritzkes DR. 2004. Relative replicative fitness of human immunodeficiency virus type 1 mutants resistant to enfuvirtide (T-20). J. Virol. 78:28–4637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Maeda K, et al. 2006. Structural and molecular interactions of CCR5 inhibitors with CCR5. J. Biol. Chem. 281:688–12698 [DOI] [PubMed] [Google Scholar]
- 24. Marozsan AJ, et al. 2005. Generation and properties of a human immunodeficiency virus type 1 isolate resistant to the small molecule CCR5 inhibitor, SCH-417690 (SCH-D). Virology 338:2–199 [DOI] [PubMed] [Google Scholar]
- 25. Marozsan AJ, et al. 2005. Differences in the fitness of two diverse wild-type human immunodeficiency virus type 1 isolates are related to the efficiency of cell binding and entry. J. Virol. 79:7121–7134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. McNicholas P, et al. 2010. Characterization of emergent HIV resistance in treatment-naive subjects enrolled in a vicriviroc phase 2 trial. J. Infect. Dis. 201:1470–1480 [DOI] [PubMed] [Google Scholar]
- 27. McNicholas PM, et al. 2011. Mapping and characterization of vicriviroc resistance mutations from human immunodeficiency virus type-1 isolated from treatment-experienced subjects enrolled in a phase II study (VICTOR-E1). J. Acquir. Immune Defic. Syndr. 56:222–229 [DOI] [PubMed] [Google Scholar]
- 28. Melikyan GB, et al. 2000. Evidence that the transition of HIV-1 gp41 into a six-helix bundle, not the bundle configuration, induces membrane fusion. J. Cell Biol. 151:413–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Miyauchi K, Kim Y, Latinovic O, Morozov V, Melikyan GB. 2009. HIV enters cells via endocytosis and dynamin-dependent fusion with endosomes. Cell 137:3–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Miyauchi K, Kozlov MM, Melikyan GB. 2009. Early steps of HIV-1 fusion define the sensitivity to inhibitory peptides that block 6-helix bundle formation. PLoS Pathog. 5:e1000585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mkrtchyan SR, et al. 2005. Ternary complex formation of human immunodeficiency virus type 1 Env, CD4, and chemokine receptor captured as an intermediate of membrane fusion. J. Virol. 79:161–11169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Moore JP, Kitchen SG, Pugach P, Zack JA. 2004. The CCR5 and CXCR4 coreceptors—central to understanding the transmission and pathogenesis of human immunodeficiency virus type 1 infection. AIDS Res. Hum. Retroviruses 20:1–126 [DOI] [PubMed] [Google Scholar]
- 33. Moore JP, McKeating JA, Weiss RA, Sattentau QJ. 1990. Dissociation of gp120 from HIV-1 virions induced by soluble CD4. Science 250:1139–1142 [DOI] [PubMed] [Google Scholar]
- 34. Ogert RA, et al. 2010. Clinical resistance to vicriviroc through adaptive V3 loop mutations in HIV-1 subtype D gp120 that alter interactions with the N-terminus and ECL2 of CCR5. Virology 400:5–155 [DOI] [PubMed] [Google Scholar]
- 35. Ogert RA, et al. 2008. Mapping resistance to the CCR5 co-receptor antagonist vicriviroc using heterologous chimeric HIV-1 envelope genes reveals key determinants in the C2-V5 domain of gp120. Virology 373:387–399 [DOI] [PubMed] [Google Scholar]
- 36. Pfaff JM, et al. 2010. HIV-1 resistance to CCR5 antagonists associated with highly efficient use of CCR5 and altered tropism on primary CD4+ T cells. J. Virol. 84:05–6514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Platt EJ, Durnin JP, Kabat D. 2005. Kinetic factors control efficiencies of cell entry, efficacies of entry inhibitors, and mechanisms of adaptation of human immunodeficiency virus. J. Virol. 79:4347–4356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Platt E. J., Kozak S. L., Durnin J. P., Hope T. J., Kabat D. 2010. Rapid dissociation of HIV-1 from cultured cells severely limits infectivity assays, causes the inactivation ascribed to entry inhibitors, and masks the inherently high level of infectivity of virions. J. Virol. 84:06–3110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Platt EJ, Wehrly K, Kuhmann SE, Chesebro B, Kabat D. 1998. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J. Virol. 72:55–2864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pugach P, et al. 2007. HIV-1 clones resistant to a small molecule CCR5 inhibitor use the inhibitor-bound form of CCR5 for entry. Virology 361:2–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pugach P, et al. 2009. Inefficient entry of vicriviroc-resistant HIV-1 via the inhibitor-CCR5 complex at low cell surface CCR5 densities. Virology 387:296–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rangel HR, et al. 2003. Role of the human immunodeficiency virus type 1 envelope gene in viral fitness. J. Virol. 77:9069–9073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ray N, Blackburn LA, Doms RW. 2009. HR-2 mutations in human immunodeficiency virus type 1 gp41 restore fusion kinetics delayed by HR-1 mutations that cause clinical resistance to enfuvirtide. J. Virol. 83:2989–2995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Reeves JD, et al. 2002. Sensitivity of HIV-1 to entry inhibitors correlates with envelope/coreceptor affinity, receptor density, and fusion kinetics. Proc. Natl. Acad. Sci. U. S. A. 99:16249–16254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Reeves JD, et al. 2005. Enfuvirtide resistance mutations: impact on human immunodeficiency virus envelope function, entry inhibitor sensitivity, and virus neutralization. J. Virol. 79:91–4999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Reeves JD, et al. 2004. Impact of mutations in the coreceptor binding site on human immunodeficiency virus type 1 fusion, infection, and entry inhibitor sensitivity. J. Virol. 78:76–5485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Strizki JM, et al. 2005. Discovery and characterization of vicriviroc (SCH 417690), a CCR5 antagonist with potent activity against human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 49:4911–4919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tilton JC, et al. 2010. HIV type 1 from a patient with baseline resistance to CCR5 antagonists uses drug-bound receptor for entry. AIDS Res. Hum. Retroviruses 26:13–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Trkola A, et al. 2002. HIV-1 escape from a small molecule, CCR5-specific entry inhibitor does not involve CXCR4 use. Proc. Natl. Acad. Sci. U. S. A. 99:395–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Troyer RM, et al. 2009. Variable fitness impact of HIV-1 escape mutations to cytotoxic T lymphocyte (CTL) response. PLoS Pathog. 5:e1000365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tsamis F, et al. 2003. Analysis of the mechanism by which the small-molecule CCR5 antagonists SCH-351125 and SCH-350581 inhibit human immunodeficiency virus type 1 entry. J. Virol. 77:01–5208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tsibris AM, et al. 2009. Quantitative deep sequencing reveals dynamic HIV-1 escape and large population shifts during CCR5 antagonist therapy in vivo. PLoS One 4:e5683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tsibris AM, Kuritzkes DR. 2007. Chemokine antagonists as therapeutics: focus on HIV-1. Annu. Rev. Med. 58:445–459 [DOI] [PubMed] [Google Scholar]
- 54. Tsibris AM, et al. 2008. In vivo emergence of vicriviroc resistance in a human immunodeficiency virus type 1 subtype C-infected subject. J. Virol. 82:8210–8214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. U.S. Food and Drug Administration 24 April 2007. Maraviroc tablets NDA 22-128: Antiviral Drugs Advisory Committee (AVDAC) briefing document, p 104 http://www.fda.gov/OHRMS/DOCKETS/AC/07/briefing/2007-4283b1-01-Pfizer.pdf U.S. Food and Drug Administration, Rockville, MD [Google Scholar]
- 56. Watson C, Jenkinson S, Kazmierski W, Kenakin T. 2005. The CCR5 receptor-based mechanism of action of 873140, a potent allosteric noncompetitive HIV entry inhibitor. Mol. Pharmacol. 67:68–1282 [DOI] [PubMed] [Google Scholar]
- 57. Weiss CD, Levy JA, White JM. 1990. Oligomeric organization of gp120 on infectious human immunodeficiency virus type 1 particles. J. Virol. 64:74–5677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Westby M, et al. 2006. Emergence of CXCR4-using human immunodeficiency virus type 1 (HIV-1) variants in a minority of HIV-1-infected patients following treatment with the CCR5 antagonist maraviroc is from a pretreatment CXCR4-using virus reservoir. J. Virol. 80:09–4920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Westby M, et al. 2007. Reduced maximal inhibition in phenotypic susceptibility assays indicates that viral strains resistant to the CCR5 antagonist maraviroc utilize inhibitor-bound receptor for entry. J. Virol. 81:59–2371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wu L, et al. 1996. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature 384:9–183 [DOI] [PubMed] [Google Scholar]
- 61. Wu L, et al. 1997. Interaction of chemokine receptor CCR5 with its ligands: multiple domains for HIV-1 gp120 binding and a single domain for chemokine binding. J. Exp. Med. 186:73–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Xu Y, Zhang X, Matsuoka M, Hattori T. 2000. The possible involvement of CXCR4 in the inhibition of HIV-1 infection mediated by DP178/gp41. FEBS Lett. 487:5–188 [DOI] [PubMed] [Google Scholar]
- 63. Zhu P, et al. 2006. Distribution and three-dimensional structure of AIDS virus envelope spikes. Nature 441:847–852 [DOI] [PubMed] [Google Scholar]