Abstract
Foamy viruses (FV) are nonpathogenic retroviruses that have cospeciated with primates for millions of years. FV can be transmitted through severe bites from monkeys to humans. Viral loads remain generally low in infected humans, and no secondary transmission has been reported. Very little is known about the ability of FV to trigger an innate immune response in human cells. A few previous reports suggested that FV do not induce type I interferon (IFN) in nonhematopoietic cells. Here, we examined how human hematopoietic cells sense FV particles and FV-infected cells. We show that peripheral blood mononuclear cells (PBMCs), plasmacytoid dendritic cells (pDCs), and the pDC-like cell line Gen2.2 detect FV, produce high levels of type I IFN, and express the IFN-stimulated gene MxA. Fewer than 20 FV-infected cells are sufficient to trigger an IFN response. Both prototypic and primary viruses stimulated IFN release. Donor cells expressing a replication-defective virus, carrying a mutated reverse transcriptase, induced IFN production by target cells as potently as wild-type virus. In contrast, an FV strain with env deleted, which does not produce viral particles, was inactive. IFN production was blocked by an inhibitor of endosomal acidification (bafilomycin A1) and by an endosomal Toll-like receptor (TLR) antagonist (A151). Silencing experiments in Gen2.2 further demonstrated that TLR7 is involved in FV recognition. Therefore, FV are potent inducers of type I IFN by pDCs and by PBMCs. This previously underestimated activation of the innate immune response may be involved in the control of viral replication in humans.
INTRODUCTION
Foamy viruses (FV) or spumaretroviruses are a large family of retroviruses found in various mammals (for reviews, see references 13, 19, and 57). They are highly prevalent in nonhuman primates, with at least 16 different simian viral subtypes (6, 19, 49, 51, 52, 69). FV are particularly well adapted to their natural hosts. Simian FV (SFV) have cospeciated with Old World primates for 30 to 100 million years, making them the oldest known vertebrate RNA viruses (34, 69). These viruses are considered innocuous in naturally or experimentally infected animals, in which they induce life-long persistent infections (39, 49, 57). SFV are readily transmitted via saliva, and seroprevalence exceeds rates of 70% in some species (6, 19, 22, 36, 46, 51). In nonhuman primates, active replication seems to be restricted to superficial epithelial cells of the oral mucosa (50).
Numerous cases of simian to human transmissions have been reported, generally after severe bites or scratches (4, 5, 7, 22, 27, 28, 75). From 2 to 24% of humans in contact with monkeys harbor anti-SFV antibodies or are positive by PCR analysis (5, 22, 27, 32, 36, 68, 75). As for monkeys, human infection is apparently nonpathogenic. There is no evidence for secondary human transmission. Viral loads are low in the infected host (5, 6, 22, 28), suggesting that the immune system controls the infection.
In cell culture, FV generally cause characteristic foam-like cytopathic effects and large syncytia and display a wide tropism (30, 39, 46, 57). FV establish persistent productive infection in human hematopoietic cell lines, as well as acute infection in primary human lymphocytes (47, 57, 66, 80). In infected monkeys and humans, various hematopoietic cell types harbor viral sequences. It was initially reported that CD8+ T cells may represent a viral reservoir in monkeys (African green monkeys [AGM] and chimpanzees) and in some humans (73), but this remains controversial (7, 19). The replication strategy of FV differs in some aspects from that of other retroviruses, presenting similarities with the life cycles of pararetroviruses (i.e., hepatitis B virus) and endogenous retroviruses (13, 25, 40, 57). For instance, reverse transcription occurs to a large extent in the producer cell, leading to the presence of both RNA and double-stranded viral DNA in the extracellular virion (14, 58, 78). Other properties include the formation of a specific pol mRNA, the budding of virions into the endoplasmic reticulum rather than at the cell surface, and the requirement of Env to ensure viral budding (18, 40, 57). Interestingly, in culture systems, FV replication is sensitive to type I interferons (IFNs) (56, 59, 62) due to the induction of cellular proteins with antiviral activity. As for HIV and other retroviruses, APOBEC3 proteins act during FV reverse transcription and induce lethal mutations in the viral genome (12, 41, 53, 61), whereas tetherin inhibits viral release without affecting FV cell-to-cell spread (76). Other antiviral proteins include PML and IFP35 (56, 70). These cellular restriction factors probably limit or modulate viral spread in vivo.
The interaction of FV with the innate immune system remains poorly characterized. Sensing viruses is an essential step in the generation of a host response to infection. There are two main types of sensors that detect viral nucleic acids within cells. The cytosolic RLRs (RIG-I-like receptors) include DExD/H box-containing RNA helicase retinoic acid inducible gene I (RIG-I), melanoma differentiation antigen 5 (MDA5), and LGP2 and are activated by various RNA species (35). Some DNA viruses are also sensed by RIG-I after transcription of viral DNA by RNA polymerase III (1) (9). The main other type of sensors are the endosomal Toll-like receptors (TLRs). TLR3 senses double-stranded RNA, TLR7, and TLR8 are activated by single-stranded RNA, whereas TLR9 recognizes CpG-containing DNA. Activation of cytosolic or endosomal sensors leads to the production of IFN and inflammatory cytokines (reviewed in references 35 and 45). These cytokines in turn induce the expression of a wide array of proteins, with direct antiviral properties or which promote adaptive immune responses.
How HIV and other retroviruses are sensed by the innate immune system is the current subject of an intense scrutiny. For instance, in plasmacytoid dendritic cells (pDCs), the main IFN-producing cell in the organism, HIV is sensed in large part by TLR7. pDCs efficiently detect HIV-infected cells (3, 21, 29, 38, 67). In other cell types, detection of cell-free HIV particles is considered suboptimal. In macrophages and lymphocytes, TREX1, a host DNase, degrades HIV DNA generated during HIV infection, providing a mechanism for the virus to avoid detection by nucleic acid sensors (77). Monocyte-derived DCs are poorly sensitive to HIV-1 infection due to the presence of the SAMHD1 restriction factor (31, 37). Monocyte-derived DCs produce low levels of IFN when encountering HIV (21). However, when resistance to infection is circumvented, HIV-1 induces DC maturation and type I IFN production (44). HTLV-1 is also sensed by pDCs in culture experiments (11). In vivo, some murine retroviruses (MMTV and MLV), triggers immune activation through TLR7, as demonstrated using TLR7-KO mice (33). Very little is known about the sensing of FV. A few reports, published almost 2 decades ago, described an absence of type I IFN production by human and primate cell lines (such as U373-MG glioblastoma cells and AV3 embryonic amniotic cells) upon FV infection (10, 60, 62). However, how pDCs and other hematopoietic cells react when they encounter FV has not been investigated thus far.
We show here that FV particles and FV-infected cells are potent inducers of type I IFN. A few FV-infected cells are sufficient to trigger release of the cytokines by pDCs and PBMCs. Both the prototypic FV strain (PFV), and two primary viruses that we previously isolated from humans bitten by monkeys (5) were detected by hematopoietic cells. We characterized further the mechanism of FV sensing and report that TLR7 is a main sensor of FV in pDCs.
MATERIALS AND METHODS
Cells, viruses, plasmids, and reagents.
Cell lines were grown in Dulbecco modified essential medium containing GlutaMAX I and sodium pyruvate (Invitrogen) and supplemented with 10% fetal bovine serum (Sigma) and antibiotics. The following cell lines were used: BHK21 (baby hamster kidney cells), 293T (human epithelial kidney cell line), HeLa (human epithelial carcinoma cell line), FAB cells (BHK21-derived indicator cells containing a β-galactosidase gene under the control of the FV long terminal repeat [LTR]) (12, 79). The pDC cell line Gen2.2 has been described elsewhere (8). Gen2.2 cells were grown in RPMI medium supplemented with 10% fetal bovine serum (FBS), 1 mM sodium pyruvate and nonessential amino acids. PBMCs were isolated from the blood of healthy donors by Ficoll centrifugation. The blood was provided by the EFS (Etablissement Frana̧is du Sang, the French Official Blood Blank). pDCs were isolated by positive selection using anti-BDCA-4 magnetic beads (Miltenyi Biotec). The negative fraction was collected and constituted PBMCs depleted from pDCs. Influenza virus (FLUAV, A/PR/8/34; Charles River Laboratories) at 40 units of hemagglutination (UHA)/ml was used to stimulate PBMCs and Gen2.2 cells.
The following plasmids encoding for FV clones were kindly provided by Axel Rethwilm: pcHSRV2, termed here FV WT, an infectious FV provirus containing an heterologous cytomegalovirus promoter (48), and the isogenic mutants pcHSRV2M68, termed FVΔEnv (with a functional deletion of the env gene starting 160 aa downstream of the start codon), and pcHSRV2M69, termed FVΔRT (in which the reverse transcriptase [RT] active site has been mutated) (48, 65). The FV clone 13 (pFVcl13) (42), an infectious FV provirus, was used in the indicated experiments with results similar to those seen with pcHSRV2. Lysates from infected and control cells were prepared as described previously (79). Briefly, ∼107 infected cells in their supernatants were subjected to three freeze-thaw cycles. The cell mixtures were then harvested with a cell scraper. Cellular debris were removed by centrifugation at 2,000 × g for 5 min, followed by filtration (0.45-μm pore size; Millipore). The supernatants were then concentrated ×500 by ultracentrifugation for 2 h 30 at 76,200 × g on a 12% iodixanol cushion. Concentrated cell lysates were then divided into aliquotes and stored at −80°C. Mock-infected cells were similarly treated to generate negative controls. Titration of infectivity on FAB reporter cells was performed as described previously (12, 79).
A151, a TLR7/9 antagonist (24, 38), was synthesized by MWG Operon. Bafilomycin A1 was purchased from Sigma-Aldrich. CpG-2216 (Invivogen) was used at 2 μM.
FV infection and provirus transfection of donor cells.
Isolation and propagation of PFV, FV15, and FV16 has been previously described (5, 12). “Chronically” infected BHK21 cells were maintained by the regular addition (twice a week) of noninfected BHK21 cells in the cultures. For the production of WT and mutant viruses, BHK21 cells (106 cells) were transfected with the indicated FV proviral vectors (1 μg) by lipofection (MetafectenPRO; Biontex, Germany) according to the manufacturer's instructions. The levels of infected cells were assessed by flow cytometry for PFV or FV15 and by immunofluorescence or visual examination of the viral CPE for FV16. When stated, 293T and HeLa cells were transfected with FV proviruses as described for BHK21 cells.
Stimulation of PBMCs and Gen2.2 cells by coculture with FV-positive BHK21 cells or by FV virions.
Cocultures of infected donor cells and target PBMCs were performed as described previously (38). Briefly, the indicated number of donor cells (ranging from 2 to 20,000 cells) were mixed with 125,000 (PBMCs, PBMC-pDCs, Gen2.2) or 25,000 (pDCs) target cells in 96-well plates in a final volume of 250 μl. To distinguish donor from target cells, the latter were stained with 2.5 μM carboxyfluorescein diacetate succinimidyl ester (CFSE; Molecular Probes) for 10 min at 37°C before coculture. When stated, the indicated inhibitors were added to target cells 1 h before and maintained during the coculture.
Exposure of PBMCs or Gen2.2 cells to purified lysates from infected cells was performed by adding the indicated amount of virus (in 10 μl) in a final volume of 250 μl.
Flow cytometry.
Assessment of the level of FV-infected donor cells was performed by flow cytometry. Briefly, cells were fixed for 10 min with phosphate-buffered saline (PBS) and 4% paraformaldehyde, washed, permeabilized, and stained for 45 min with a rabbit anti-FV polyclonal serum (kindly provided by Ali Saïb) (63) in PBS containing 1% bovine serum albumin and 0.05% saponin. MxA expression was determined as described previously (38) using an anti-MxA monoclonal antibody (kindly provided by O. Haller). Characterization of pDCs among PBMCs was performed as described previously (38) using anti-BDCA2-APC and anti-BDCA4-PE antibodies (Miltenyi Biotech). Samples were analyzed by flow cytometry using a FACSCalibur (Becton Dickinson) with FlowJo software.
Immunofluorescence.
For immunofluorescence, BHK21cells infected or not with PFV, FV15, and FV16 were stained as previously described (5). Briefly, cells were fixed with 4% paraformaldehyde, rinsed with PBS–0.1% Tween, and permeabilized with methanol at 4°C for 5 min. The cells were then stained with rabbit anti-FV serum in PBS–0.1% Tween overnight at 4°C, rinsed, and stained with goat anti-rabbit IgG(H+L)-Alexa 488 (Invitrogen). Images were obtained with a Nikon Microphot-FXA microscope.
Titration of PFV and FV15.
Titration of FV was achieved using FAB cells as previously described (12, 79). Briefly, indicator FAB cells were incubated with the virus stock for 2 h at 37°C. Virus was then removed and replaced with growth medium. Two days later, the monolayer was fixed with 0.5% glutaraldehyde, washed, and incubated 30 min with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) staining solution. Blue cells were counted under a light microscope.
Gen2.2 cell silencing experiments.
Gen2.2 cell were transduced using lentiviral vector (LV) particles containing shRNA targeting TLR7 (NM_016562; clone ID TRCN0000056974 [Open Biosystems]) or control shRNA. The LV also expresses the puroR gene. At 2 days after transduction, Gen2.2 cells were selected in the presence of 1 μg of puromycin/ml. Resistant populations grew in a few days and were used without further subcloning. Measurement of TLR7 mRNA levels was performed as described previously (38).
Type I IFN detection.
Type I IFN secretion was quantified using the reporter cell line HL116, which carries the luciferase gene under the control of the IFN-inducible 6-16 promoter (kindly provided by Sandra Pellegrini, Institut Pasteur, France) (71). HL116 cells were grown in Dulbecco modified Eagle medium supplemented with 10% FBS and HAT (hypoxanthine, 20 μg/ml; thymidine, 20 μg/ml; aminopterin, 0.2 μg/ml). A total of 2 × 104 HL116 cells, plated 16 h prior the assay in 96-well plate, were incubated for 7 h with the desired culture supernatants or standards containing a titration of human IFN-α2a (Immunotools). The cells were then lysed (luciferase cell culture lysis, 5× reagent; Promega), and the luciferase activity was measured using luciferase assay reagent (Promega). Samples were analyzed using the Perkin-Elmer Wallac 1420. IFN levels are expressed as equivalent of IFN-α2a concentration in U/ml.
Data and statistical analysis.
The results of the experiments are expressed as means ± the standard deviations. Comparisons between groups were performed using the Student t test. Differences with a P value of <0.05 were considered statistically significant.
RESULTS
Description of FV strains used in the present study.
We selected three different viral strains to study the innate sensing of FV. We first used the prototypic viral clone PFV derived from a chimpanzee virus (2, 42) that we produced by transfecting the corresponding provirus in BHK21 cells. We recently isolated two novel primary FV strains (FV15 and FV16) from two FV-seropositive persons (AG15 and AG16) that have been bitten by a chimpanzee and an African green monkey (AGM), respectively (5). FV15 and FV16 were initially isolated by coculture of PBMCs with BHK21 target cells. After 3 to 4 weeks, typical giant multinucleated cells were observed. Sequence analysis of the integrase gene indicated that FV15 and FV16 corresponded to the chimpanzee and AGM viruses, respectively (5). The viruses produced in the cocultures were infectious. Reinfection of naive BHK21 cells generated a typical cytopathic effect (CPE) in 2 to 5 days, as observed with PFV (Fig. 1A). The syncytia expressed FV antigens, detected by immunofluorescence with a rabbit anti-PFV serum, raised against a chimpanzee virus (63) (Fig. 1B).
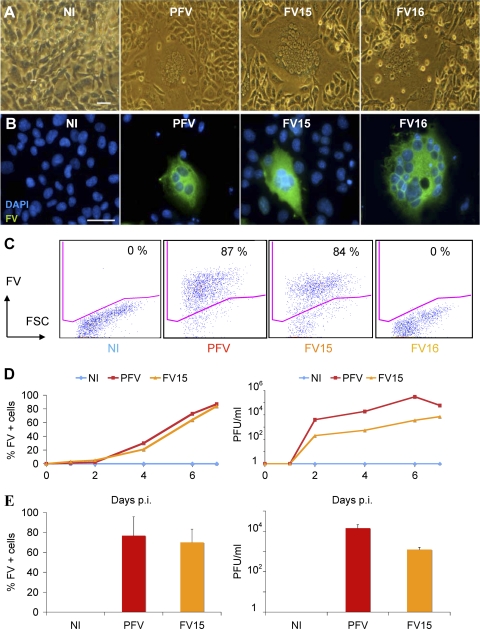
Fig 1.
Description of FV strains used in the present study. (A and B) Visualization of FV-infected cells. BHK21 cells were noninfected (NI) or infected with prototypic PFV, FV15, or FV16 primary strains. After 3 to 5 days of infection, large syncytia were visible. (A) Light microscopy analysis. (B) Immunofluorescence analysis. FV-infected cells were stained with an anti-FV polyclonal serum (green). Nuclei were stained with DAPI. Representative fields are shown. Scale bar, 5 μm. (C) Flow cytometry analysis of infected BHK21cells. After infection, cells were stained with an anti-FV polyclonal serum, which recognizes PFV- and FV15-infected cells, but not cells infected with the genetically distant FV16 strain. The percentage of FV+ cells is indicated. Staining was performed at day 3 postinfection for PFV and day 5 postinfection for FV15 and FV16. (D) Propagation of PFV and FV15 on BHK21cells. BHK21 cells were noninfected (NI) or infected with PFV (multiplicity of infection [MOI] = 0.05) or FV15 (MOI = 0.5) and then analyzed by flow cytometry at the indicated days postinfection (p.i.) (left panel). Cells were lysed by freeze-thaw cycles to harvest viruses. Viral infectivity (in PFU/ml) was determined using FAB-reporter cells (right panel). The results of a representative experiment are shown. The FV16 strain does not activate the PFV promoter present in FAB cells (data not shown). (E) Analysis of BHK21 cells chronically infected with PFV or FV15. BHK-infected cell cultures were maintained by addition of noninfected cells twice a week. Cells were analyzed by flow cytometry (left panel) and lysates were titrated on FAB-reporter cells (right panel). Means ± the standard deviations (SD) of three independent experiments are shown.
We further characterized FV infectivity by assessing viral spread using flow cytometry. The rabbit anti-PFV serum recognized PFV- and FV15-infected cells but not cells infected with the genetically distant FV16 strain (Fig. 1C). A kinetic analysis indicated that both PFV and FV15 efficiently spread in BHK21 cells, reaching >80% of Gag-expressing cells at day 7 postinfection (Fig. 1D), before destruction of the cell monolayer by the viral CPE. FV buds intracellularly and viral particles can be collected by lysis of infected cell cultures by freeze-thaw cycles, followed by an ultracentrifugation step (79). We generated and titrated such lysates using the FAB reporter cell line, which carries the β-galactosidase gene under the PFV LTR (12, 79). Upon infection, the viral protein Tas transactivates the LTR promoter. Virus titers reached 105 to 106 PFU/ml with PFV and 103 to 104 PFU/ml with FV15 (Fig. 1D). The lower titers observed with FV15 may reflect a lower infectivity, and/or a reduced efficiency of FV15 Tas, compared to PFV Tas, to transactivate the PFV LTR. The AGM-derived strain FV16 did not transactivate the PFV promoter, precluding titration on FAB cells (not shown). We also maintained long-term cultures of PFV-, FV15-, and FV16-infected BHK21 cells by adding noninfected cells twice a week in the cultures. The resulting “chronically” infected cell lines carried up to 60 to 80% FV+ cells, as assessed by flow cytometry and produced infectious virus (103 to 104 PFU/ml) (Fig. 1E). The “chronically” infected cells represent a convenient tool and were used, along with acutely infected cells (day 3 to 5 postinfection), in further experiments.
Thus, PFV, FV15, and FV16 are infectious in cell cultures. For the three strains, a typical CPE associated with the presence of FV-positive cells is observed within a few days of infection in BHK21 cells, allowing visual estimation of the level of infection. The three viruses also infect the human cell lines HeLa and 293T (not shown). PFV and FV15 infection can also be assessed by flow cytometry or by using FAB reported cells.
Primary hematopoietic cells efficiently sense FV particles and FV-infected cells.
We examined whether human PBMCs produce type I IFN when they encounter PFV-infected cells and virions. The experimental procedure is outlined in Fig. 2A. BHK21 donor cells were infected with FV or transfected with PFV proviruses. With PFV and FV15, after a few days of infection, when 60 to 90% of the population was FV positive by flow cytometry, BHK21 cells were cocultivated with PBMCs isolated from healthy donors. Since FV16-infected BHK21 cells were not detected by our flow cytometry assay, we roughly assessed infection by visual examination of the CPE, before coculture. “Chronically” PFV-, FV15-, or FV16-infected cells were also used as donor cells. In parallel, PBMCs were exposed to PFV particles. After 24 h of exposure to viral particles or coculture with FV-infected donor cells, type I IFN was measured in supernatants using a biological activity assay.
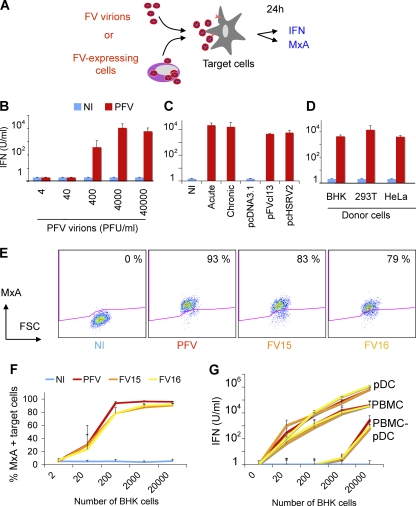
Fig 2.
Sensing of FV particles and FV-infected cells by hematopoietic cells. (A) Schematic representation of the experimental procedure. Target PBMCs were exposed to FV particles or cocultivated with FV-infected cells. After 24 h, type I IFN production was measured in supernatants, and expression of the IFN-stimulated gene MxA was monitored by flow cytometry. (B) IFN release by PBMCs exposed to FV particles. Whole PBMCs were exposed to the indicated doses (in PFU/ml) of PFV virions, obtained by ultracentrifugation of lysates from PFV-infected (PFV) BHK21 cells. Similar amounts of lysates from NI cells were used as a control. Type I IFN was measured after 24 h. Means ± the SD of at least three independent experiments are shown. (C) PBMCs recognize BHK21 cells infected with FV or transfected with PFV proviruses. Different types of FV-expressing BHK21 cells were used as donors. Acutely infected BHK21 cells (acute) correspond to cells used 3 to 5 days postinfection with FV. “Chronically” infected BHK21 cells (chronic) are maintained in long-term cultures by the addition of fresh BHK21 cells twice a week. BHK21 cells were also transfected with two reference PFV proviruses (pFVcl13 and pcHSRV2) or with a control plasmid (pcDNA3.1). These different donor cells were cocultivated with PBMCs and type I IFN was measured after 24 h. Means ± the SD of at least three independent experiments are shown. (D) IFN release by PBMCs exposed to various FV-expressing donor cells. PBMCs were also cocultivated with BHK21, 293T, or HeLa cells that had been transfected with PFV-encoding (pFVcl13) and control plasmid (pcDNA3.1). Type I IFN production was measured in the supernatants. Means ± the SD of three to five independent experiments are shown. (E) MxA expression in PBMCs. PBMCs were (i) cocultivated with noninfected BHK21 cells or with cells infected with PFV, FV15, or FV16, (ii) stained for MxA, and (iii) analyzed by flow cytometry. A representative experiment is shown. The percentage of MxA+ cells is indicated. (F) Dose-response analysis of MxA expression in PBMCs. PBMCs (1.25 × 105/well) were cocultivated with the indicated number BHK21cells infected with PFV, FV15, or FV16 (60 to 90% of the BHK21 cells were FV positive at the beginning of the coculture). Means ± the SD of MxA expression of three to five independent experiments are shown. (G) pDCs are the main hematopoietic cells producing type I IFN. Whole PBMCs, PBMCs depleted of pDCs (PBMC-pDCs; 1.25 × 105/well), or purified pDCs (0.25 × 105/well) were cocultivated with the indicated numbers of BHK21cells infected with PFV, FV15, or FV16. Type I IFN production was measured in the supernatants. Means ± the SD of three independent experiments are shown.
PFV particles efficiently induced type I IFN production by PBMCs, up to 103 to 104 U/ml (Fig. 2B). A dose-response analysis of the viral inoculum (40 to 40,000 PFU/ml) indicated that 400 PFU/ml is sufficient to trigger type I IFN release, whereas lysates from noninfected cells did not activate PBMCs (Fig. 2B). FV-expressing BHK21 cells also potently induced type I IFN release in cocultures, with levels reaching up to 104 U/ml (Fig. 2C). Similar levels of type I IFN were obtained in cocultures of PBMCs with BHK21 cells that were either “acutely” or “chronically” infected with FV or transiently transfected with two reference PFV proviral clones (pFVcl13 or pcHSRV2) (42, 48) (Fig. 2C). Noninfected donor cells were inactive (Fig. 2C). Other types of FV-expressing donor cells, such as 293T and HeLa cells (Fig. 2D), also promoted IFN production by PBMCs, demonstrating that this phenomenon was not an artifact due to the use of BHK21 cells as donors. Of note, 293T and HeLa cells did not produce detectable levels of type I IFN, upon FV expression (not shown).
To confirm the activation of PBMCs by either FV virions and FV-infected cells, we examined the induction of MxA, an IFN-stimulated gene (23). Flow cytometry indicated that MxA was expressed by PBMCs cocultivated with PFV-infected cells or exposed to cell-free PFV particles (Fig. 2E and F).
We then compared the recognition of the three FV strains. BHK21 cells were infected with PFV, FV15, or FV16 isolates. An increasing number of PFV-, FV15-, or FV16-infected BHK21 cells (from 2 to 20,000 donor cells/well) were cocultivated with 1.25 × 105 PBMCs. Infected BHK21 cells induced type I IFN release and MxA expression by PBMCs (Fig. 2E to G). As few as 20 FV-infected cells (corresponding to <0.02% of infected donor cells among target PBMCs) were sufficient to induce the cytokines, at levels which were similar for the three types of FV tested.
We determined which cells, among PBMCs, were the main producers of type I IFN. In PBMCs, pDCs, identified as BDCA4/BDCA2 double-positive cells, represented 0.2 to 1% of whole PBMCs (38; data not shown). We thus used as targets whole PBMCs, PBMCs depleted of pDCs (after removal of >80% of pDCs), or purified pDCs (corresponding to >85% of purity based on BDCA2/BDCA4+ double labeling) (38; data not shown). Upon coculture with FV-infected cells or exposure to FV particles, PBMCs depleted of pDCs produced very low amounts of type I IFN compared to total PBMCs (both PBMCs and PBMCs depleted of pDCs were at 1.25 × 105 cells/well) (Fig. 2G and data not shown). Conversely, purified pDCs (0.25 × 105 cells/well) produced huge amounts of type I IFN (above 105 U/ml) (Fig. 2G). Thus, among circulating hematopoietic cells, the early production of type I IFN induced by FV-infected cells and FV viral particles mainly originates from pDCs.
Altogether, these results indicate that FV virions and FV-infected cells are efficiently sensed by PBMCs, inducing high levels of type I IFN. Among PBMCs, pDCs are the main cells producing the antiviral cytokines. Donor cells infected with the three distinct viral strains PFV, FV15, and FV16 similarly trigger an IFN response.
Pathways of recognition of FV viral particles and FV-infected cells by PBMCs.
We characterized which step of the viral life cycle is required to trigger type I IFN release by PBMCs. We examined the role of Env and RT in this process. We used as donors BHK21 cells producing an Env-deleted (FVΔEnv) or an RT-defective virus (FVΔRT). Env is required for the budding of FV capsids and infectivity (20, 55). The FVΔRT strain carries a mutation that abrogates the enzymatic activity of RT (48). Since these mutants are not infectious, we transfected the corresponding proviral constructs in BHK21 cells. We also transfected the FV WT provirus as a positive control. At day 2 posttransfection, BHK21 cells displayed similar levels of Gag+ cells (10 to 20%) with FV WT, FVΔEnv, and FVΔRT constructs (Fig. 3A). As expected, only the WT virus was infectious, as assessed by the increase of FV+ cells over time in the culture (Fig. 3A) and by the measurement of infectivity of cell lysates on FAB reporter cells (data not shown). We then cocultivated BHK21 cells at day 1 posttransfection with PBMCs for 24 h. BHK21 cells expressing FVWT or FVΔRT induced IFN production by PBMCs (up to 6 × 103 U/ml [Fig. 3B]). Interestingly, FVΔEnv-expressing cells did not stimulate target cells.
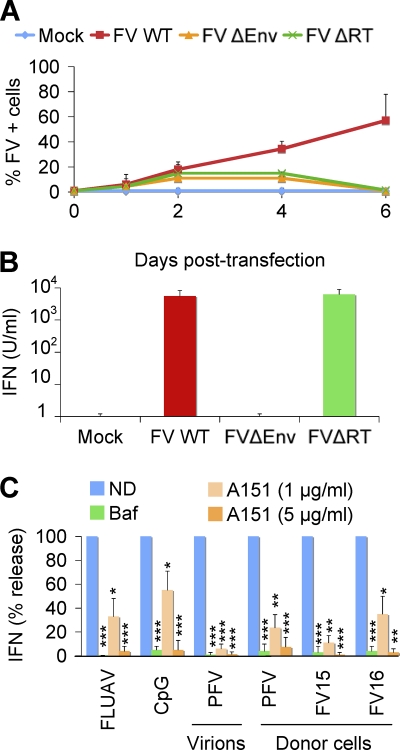
Fig 3.
Analysis of the recognition of FV infected cells by PBMCs. (A and B) Role of envelope (Env) and reverse transcriptase (RT) in type I IFN production by PBMCs. (A) Replication of FV WT, FVΔEnv, or FVΔRT proviruses in BHK21 cells. BHK21cells were transfected with FV WT, FVΔEnv, or FVΔRT proviruses. Viral propagation was studied at the indicated time points by flow cytometry. Means ± the SD of three independent experiments are shown. (B) Induction of type I IFN by BHK21 cells expressing FV WT, FVΔEnv, or FVΔRT in coculture with PBMCs. At day 1 posttransfection, BHK21 cells (2 × 104/well) were cocultivated with PBMCs (1.25 × 105/well) for 24 h. The supernatants were harvested, and the type I IFN production was measured. Means ± the SD of four independent experiments are shown. (C) Effect of drugs impairing endosomal TLR signaling. Whole PBMCs were preincubated for 1 h with bafilomycin A1 (Baf), an inhibitor of vesicular acidification (25 nM), or with A151, a TLR7/9 antagonist (1 or 5 μg/ml). The PBMCs were then stimulated with FLUAV(TLR7 agonist), CpG (TLR9 agonist, 2 μM), PFV virions, and PFV-, FV15-, or FV16-infected BHK21 cells. The supernatants were harvested after 24 h, and the type I IFN release was measured. The results are expressed as the precentage of the signal observed in the absence of drug (ND). Means ± the SD of three independent experiments are shown. *, P < 0.05; **, P < 0.005; ***, P < 0.001.
Therefore, induction of type I IFN release by PBMCs necessitates the Env protein. This highlights the requirement of assembled FV particles and/or fusion between virions or infected cells and PBMCs to trigger FV recognition by PBMCs. In contrast, reverse transcription is not required to trigger type I IFN production. Since FV reverse transcription occurs partly in the producer cell, leading to the presence of both RNA and double-stranded viral DNA in virions (14, 58, 78), our results suggest that viral DNA is not the main molecule recognized by cellular sensors.
To determine further the mechanisms by which PBMCs sense FV viral particles and FV-infected cells, we evaluated the effect of bafilomycin A1, an inhibitor of vesicular acidification. As controls, we used influenza virus (FLUAV) and CpG, stimuli for TLR7 and TLR9, respectively (16, 72). Bafilomycin A1 at 25 nM, a concentration that did not detectably affect cell viability (data not shown), inhibited type I IFN induction by FV particles and infected cells (Fig. 3C), which is consistent with the requirement for an acidic endosome and viral degradation to achieve TLR signaling (3, 24, 38). As expected, FLUAV and CpG stimulation were strongly inhibited by bafilomycin A1. Since bafilomycin A1 may also interfere with viral entry (54), we tested the effect of A151, an oligonucleotide described as an endosomal TLR antagonist (inhibiting TLR7 and, less efficiently, TLR9) (3, 24, 38). A151 at 1 and 5 μg/ml significantly decreased IFN production by PBMCs cocultivated with FV-infected cells (Fig. 3C). At 1 μg/ml, A151 inhibited FLUAV and, to a lesser extent, CpG stimulation of PBMCs. At a higher concentration (5 μg/ml), A151 inhibited CpG stimulation (Fig. 3C), confirming that this compound may antagonize both TLR7 and TLR9. Of note, bafilomycin A1 and A151 inhibited the activation of PBMCs by PFV-, FV15-, and FV16-infected BHK21 cells (Fig. 3C). Altogether, these results suggest that detection of FV-infected cells and FV particles by pDCs and PBMCs requires an acidic environment and is in large part mediated by endosomal TLRs.
Sensing of FV-infected cells by the Gen2.2 pDC-like cell line.
We used the Gen2.2 pDC-like cell line to dissect the pathways of recognition of FV particles and infected cells. This cell line was derived from a patient with pDC leukemia and possesses phenotypic and functional features of primary pDCs (8). Gen2.2 cells express TLR7, TLR9, and other pDC markers. They produce type I IFN when exposed to FLUAV, HIV-infected cells, or other TLR agonists, although at lower levels than primary pDCs (38, 43). These cells can be transduced with lentiviral vectors encoding shRNAs (15). We examined how Gen2.2 cells reacted when encountering FV. To this end, viral particles or FV-infected cells were mixed with Gen2.2 cells, and the IFN levels were measured after 6 and 24 h. The cytokine was barely detected at 6 h (data not shown). At 24 h, 3 × 103 to 104 U of type I IFN/ml were released by Gen2.2 cells exposed to FV particles or FV-infected cells (Fig. 4A), which is 7- to 20-fold less than in primary pDCs. Donor cells infected with PFV, FV15, and FV16 induced production of the antiviral cytokines by Gen2.2 cells (Fig. 4A). We also cocultivated Gen2.2 cells with increasing numbers (from 2 to 20,000 donor cells/well) of infected donor cells. As few as 20 PFV-infected cells were sufficient to induce type I IFN release and MxA expression by Gen2.2 cells (Fig. 4B). Therefore, although they produce less IFN, Gen2.2 cells behave like primary pDCs and efficiently sense cell-free FV particles and material from FV-infected cells.
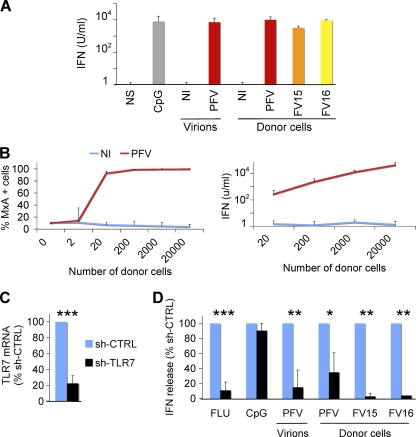
Fig 4.
FV Sensing by the Gen2.2 pDC cell line. (A) Type I IFN release by Gen2.2 cells in contact with FV particles or FV-infected cells. Gen2.2 cells (1.25 × 105/well) were either (i) not stimulated (NS), (ii) incubated with CpG (2 μM), lysates from noninfected (NI), or FV-infected (PFV) cells (4,000 PFU/ml), or (iii) cultivated with BHK21 cells (2 × 103/well) either not infected (NI) or infected with PFV, FV15, or FV16. Type I IFN production in supernatants was measured after 24 h. Means ± the SD of IFN release of four independent experiments are shown. (B) Dose-response analysis of type I IFN production. Gen2.2 cells were cocultivated for 24 h with the indicated number of BHK21cells and stained for MxA (left panel). The type I IFN production in supernatants was measured in supernatants (right panel). Means ± the SD of two (left panel) and four (right panel) independent experiments are shown. (C and D) Role of TLR7 in FV sensing. (C) Silencing of TLR7. Gen2.2 cells were transduced with lentiviral vectors expressing shRNAs against TLR7 (shTLR7) or an irrelevant target (shCTRL). The levels of TLR7 mRNA in transduced cells were measured by RT-PCR. The data are normalized to GAPDH mRNA and expressed as relative levels of mRNA compared to shCTRL cells. (D) Type I production in control and TLR7-silenced Gen2.2 cells. shCTRL and shTLR7 Gen2.2 cells were exposed for 24 h to FLUAV, CpG (2μM), PFV particles, or cocultivated with PFV-, FV15-, or FV16-infected BHK21cells (2 × 103/well). The type I IFN levels are expressed as the percentage of the signal obtained with shCTRL Gen2.2 cells. Means ± the SD of two to three independent experiments are shown. *, P < 0.05; **, P < 0.005; ***, P < 0.001.
We next assessed the involvement of TLR7 in FV sensing. TLR7 was silenced by transduction of Gen2.2 cells with a lentiviral vector coding for an anti-TLR7 shRNA (38). The vector also expressed a puromycin-resistant gene, allowing selection of a population of transduced cells (termed Gen2.2-shTLR7). Silencing decreased TLR7 mRNA levels by 80% compared to a control shRNA (Fig. 4C). Gen2.2-shTLR7 cells produced ca. 90% less IFN than did control cells when incubated with FV particles and 70 to 97% less IFN when cocultivated with donor cells infected with PFV, FV15, and FV16 (Fig. 4D). As expected, the response to FLUAV was impaired when TLR7 was downregulated, whereas CpG stimulation was not affected. These experiments directly demonstrate that TLR7 is a cellular receptor mediating recognition of FV by pDC-like cells.
DISCUSSION
There are numerous cases of simian FV infections in humans who have been in contact with monkeys or great apes (4, 5, 7, 27, 28, 75). FV does not replicate well in humans. Infection is apparently not pathogenic, the viral loads are low (5, 28), and there is no evidence of virus transmission between humans. However, in culture systems, FV replicates easily and displays a wide tropism for human cells (30, 39, 57). This suggests that the host controls viral replication through mechanisms that are not fully understood. In particular, the interaction of FV with the human immune system remains poorly characterized. It has been suggested that FV does not activate an innate response (10, 62). Previous experiments were performed mostly with human or simian epithelial cell lines, and the response of human hematopoietic cells to the virus is not known. We show here that FV triggers a rapid and intense type I IFN response in human hematopoietic cells. Among PBMCs, pDCs are the main cells producing the antiviral cytokines, since their removal strongly decreases type I IFN release. We have further studied the mechanisms involved. We demonstrate here that in pDCs, the TLR7 pathway detects FV. We have also tested the ability of various FV strains to induce type I IFN production. The reference strain PFV, as well as two primary viruses, FV15 and FV16, isolated in two independent FV-infected individuals (5) induced similar levels of type I IFN in PBMC targets, indicating that this phenomenon is not restricted to a laboratory-adapted strain and is observed with human FV originating from different simian species.
We have examined the recognition of cell-free viral particles and of FV-infected cells, since this retrovirus is barely released in supernatants and thus likely spreads mostly through cell-to-cell contacts (26, 57). Both virions and infected cells induced type I IFN production by PBMCs and pDCs. Of note, viral particles released in the supernatants and concentrated by ultracentrifugation also stimulated type I IFN production (not shown). Interestingly, as few as 20 FV-infected cells, added to 2.5 × 104 pDCs or 1.25 × 105 PBMCs, are sufficient to trigger IFN release. The recognition of FV-infected cells is therefore particularly efficient. This situation is reminiscent of that observed with another retrovirus, HIV, which easily spreads from lymphocytes to lymphocytes through virological synapses (64). HIV-infected lymphocytes are efficiently sensed by pDCs (38, 67). Contact with HIV-infected cells promotes a massive and rapid transfer of viral material to target cells (68). It is likely that similar events are occurring with FV-infected cells. It will be worth examining whether virological synapses are formed between FV-infected cells and targets and determining the nature of viral proteins and nucleic acids transmitted through cellular contacts. Using viral mutants, we have initiated an analysis of the components of FV-infected cells that are necessary to trigger an IFN response. Detection of FV-infected cells by PBMCs required the presence of viral envelope glycoproteins. In viruses from which Env has been deleted, FV particles are not released (18), indicating that the expression of other structural and nonstructural viral proteins in the absence of viral release is not sufficient to activate an innate immune response.
The presence of infectious virus was not necessary to generate a response to FV-positive cells. This was demonstrated using an RT-defective virus (FVΔRT) (48) that did not spread in cultures but stimulated IFN release in PBMC cocultures as efficiently as did the wild-type virus. Our results suggest that cells expressing defective viruses can be sensed by the host and may participate to the immune antiviral response. Moreover, with FVΔRT, viral DNA is not synthesized and is thus not involved in the recognition of FV by PBMCs and pDCs. It is more likely that the viral RNA itself is detected and promotes type I IFN secretion. Our results also strongly suggest that the productive infection of target pDCs and other PBMCs is not a prerequisite to trigger recognition of FV.
Several lines of evidence strongly suggest that the TLR pathway is involved in FV sensing. First, in PBMCs, bafilomycin A1 inhibited type I IFN production, indicating that endosomal acidification is required. However, FV-mediated viral entry has been reported to involve a pH-dependent fusion process (54, 57). Therefore, bafilomycin A1 may not only impair TLR activity but also the access of incoming viral material to the cytosol and hence detection by cytosolic sensors. Second, A151, a modified oligonucleotide with anti-TLR7/9 activity (3, 29, 38), blocked type I IFN release in PBMCs exposed to FV. Third, a direct demonstration of the involvement of TLR7 was achieved using the Gen2.2 pDC-like cell line (8). We show here that Gen2.2 cells, like primary pDCs, release IFN upon exposure to FV. We generated TLR7-negative Gen2.2 cells by RNA silencing. These cells were impaired in type I IFN production when cocultivated with FV-infected cells or incubated with FV virions, whereas CpG-induced TLR9 signaling was normal.
Therefore, as previously described for pathogenic retroviruses such as HIV-1, MMTV, and MLV (33, 38), our results show that TLR7 is able to sense nonpathogenic FV. The pathways of retrovirus sensing are known to be diverse and cell type dependent (38). It will be worth determining whether TLR7-independent, endosomal, or cytosolic mechanisms may also be operative in other immune and nonimmune cells. It will be also of interest following the fate of incoming FV in target cells to visualize precisely how captured viral material encounters TLR7 molecules or other putative sensors in various cell types.
Our observations suggest that human hosts can efficiently detect FV upon accidental infections and generate a type I IFN-induced antiviral response. In culture systems, the addition of type I IFN (56, 59, 62) or the expression of various IFN-stimulated genes, including APOBEC3 and tetherin, impair FV replication (12, 17, 41, 53, 61, 76). It is tempting to speculate that the type I IFN response described here is in large part responsible for the control of viral replication in infected individuals. In addition, neutralizing antibodies are known to inhibit SFV transmission and infection (74), indicating that the adaptive immune response may control further the virus in vivo.
In summary, we have shown that FV efficiently induces an innate immune response. We have focused our analysis on the production of type I IFN by human hematopoietic cells. It may be of interest to extend this observation to other cytokines and chemokines that may be released, by either human and primate cells, upon FV encounter. Experimental infections in primate models may further allow deciphering the breadth and duration of antiviral and inflammatory responses and their role in the innocuousness of the virus in its natural hosts. Moreover, FV are currently being evaluated as vectors for gene therapy purposes (18). Our results indicate that these vectors may generate a previously underestimated innate immune response.
ACKNOWLEDGMENTS
We thank Diana Ayinde, Nicoletta Casartelli, and members of the Virus and Immunity Unit for discussions and critical reading of the manuscript. We thank Sara Calattini and Edouard Betsem for discussions and sharing of viral reagents. We thank Ali Saïb, Axel Rethwilm, Sandra Pellegrini, Otto Haler, Joël Plumas, and Laurence Chaperot for kindly providing reagents.
This study was supported by grants from the Agence Nationale de Recherche sur le SIDA (ANRS), the Agence Nationale de Recherche (ANR), SIDACTION, the CNRS, AREVA, and the Institut Pasteur. We thank the Centre d'Immunologie Humaine at the Institut Pasteur for support.
The authors have no conflicting financial interests.
Footnotes
Published ahead of print 16 November 2011
REFERENCES
- 1. Ablasser A, et al. 2009. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat. Immunol. 10:1065–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Achong BG, Mansell PW, Epstein MA, Clifford P. 1971. An unusual virus in cultures from a human nasopharyngeal carcinoma. J. Natl. Cancer Inst. 46:299–307 [PubMed] [Google Scholar]
- 3. Beignon AS, et al. 2005. Endocytosis of HIV-1 activates plasmacytoid dendritic cells via Toll-like receptor-viral RNA interactions. J. Clin. Invest. 115:3265–3275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Calattini S, et al. 2011. Multiple retroviral infection by HTLV type 1, 2, 3 and simian foamy virus in a family of Pygmies from Cameroon. Virology 410:48–55 [DOI] [PubMed] [Google Scholar]
- 5. Calattini S, et al. 2007. Simian foamy virus transmission from apes to humans, rural Cameroon. Emerg. Infect. Dis. 13:1314–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Calattini S, et al. 2004. Natural simian foamy virus infection in wild-caught gorillas, mandrills and drills from Cameroon and Gabon. J. Gen. Virol. 85:3313–3317 [DOI] [PubMed] [Google Scholar]
- 7. Callahan ME, et al. 1999. Persistent zoonotic infection of a human with simian foamy virus in the absence of an intact orf-2 accessory gene. J. Virol. 73:9619–9624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chaperot L, et al. 2006. Virus or TLR agonists induce TRAIL-mediated cytotoxic activity of plasmacytoid dendritic cells. J. Immunol. 176:248–255 [DOI] [PubMed] [Google Scholar]
- 9. Chiu YH, Macmillan JB, Chen ZJ. 2009. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell 138:576–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Colas S, et al. 1995. Human foamy virus infection activates class I major histocompatibility complex antigen expression. J. Gen. Virol. 76(Pt 3):661–667 [DOI] [PubMed] [Google Scholar]
- 11. Colisson R, et al. 2010. Free HTLV-1 induces TLR7-dependent innate immune response and TRAIL relocalization in killer plasmacytoid dendritic cells. Blood 115:2177–2185 [DOI] [PubMed] [Google Scholar]
- 12. Delebecque F, et al. 2006. Restriction of foamy viruses by APOBEC cytidine deaminases. J. Virol. 80:605–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Delelis O, Lehmann-Che J, Saib A. 2004. Foamy viruses-a world apart. Curr. Opin. Microbiol. 7:400–406 [DOI] [PubMed] [Google Scholar]
- 14. Delelis O, Saib A, Sonigo P. 2003. Biphasic DNA synthesis in spumaviruses. J. Virol. 77:8141–8146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Di Domizio J, et al. 2009. TLR7 stimulation in human plasmacytoid dendritic cells leads to the induction of early IFN-inducible genes in the absence of type I IFN. Blood 114:1794–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Diebold SS, Kaisho T, Hemmi H, Akira S, Reis e Sousa C. 2004. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science 303:1529–1531 [DOI] [PubMed] [Google Scholar]
- 17. Dietrich I, et al. 2011. Feline tetherin efficiently restricts release of feline immunodeficiency virus but not spreading of infection. J. Virol. 85:5840–5852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Erlwein O, McClure MO. 2010. Progress and prospects: foamy virus vectors enter a new age. Gene Ther. 17:1423–1429 [DOI] [PubMed] [Google Scholar]
- 19. Falcone V, Schweizer M, Neumann-Haefelin D. 2003. Replication of primate foamy viruses in natural and experimental hosts. Curr. Top. Microbiol. Immunol. 277:161–180 [DOI] [PubMed] [Google Scholar]
- 20. Fischer N, et al. 1998. Foamy virus particle formation. J. Virol. 72:1610–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fonteneau JF, et al. 2004. Human immunodeficiency virus type 1 activates plasmacytoid dendritic cells and concomitantly induces the bystander maturation of myeloid dendritic cells. J. Virol. 78:5223–5232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gessain A, Calattini S. 2008. Emergence of simian foamy viruses in humans: facts and unanswered questions. Future Virol. 3:71–81 [Google Scholar]
- 23. Haller O, Stertz S, Kochs G. 2007. The Mx GTPase family of interferon-induced antiviral proteins. Microbes Infect. 9:1636–1643 [DOI] [PubMed] [Google Scholar]
- 24. Hardy AW, Graham DR, Shearer GM, Herbeuval JP. 2007. HIV turns plasmacytoid dendritic cells (pDC) into TRAIL-expressing killer pDC and downregulates HIV coreceptors by Toll-like receptor 7-induced IFN-alpha. Proc. Natl. Acad. Sci. U. S. A. 104:17453–17458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Heinkelein M, et al. 2000. Efficient intracellular retrotransposition of an exogenous primate retrovirus genome. EMBO J. 19:3436–3445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Heinkelein M, Rammling M, Juretzek T, Lindemann D, Rethwilm A. 2003. Retrotransposition and cell-to-cell transfer of foamy viruses. J. Virol. 77:11855–11858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Heneine W, Schweizer M, Sandstrom P, Folks T. 2003. Human infection with foamy viruses. Curr. Top. Microbiol. Immunol. 277:181–196 [DOI] [PubMed] [Google Scholar]
- 28. Heneine W, et al. 1998. Identification of a human population infected with simian foamy viruses. Nat. Med. 4:403–407 [DOI] [PubMed] [Google Scholar]
- 29. Herbeuval JP, et al. 2005. Regulation of TNF-related apoptosis-inducing ligand on primary CD4+ T cells by HIV-1: role of type I IFN-producing plasmacytoid dendritic cells. Proc. Natl. Acad. Sci. U. S. A. 102:13974–13979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hill CL, Bieniasz PD, McClure MO. 1999. Properties of human foamy virus relevant to its development as a vector for gene therapy. J. Gen. Virol. 80((Pt 8):2003–2009 [DOI] [PubMed] [Google Scholar]
- 31. Hrecka K, et al. 2011. Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature 474:658–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jones-Engel L, et al. 2008. Diverse contexts of zoonotic transmission of simian foamy viruses in Asia. Emerg. Infect. Dis. 14:1200–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kane M, et al. 2011. Innate immune sensing of retroviral infection via Toll-like receptor 7 occurs upon viral entry. Immunity 35:135–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Katzourakis A, Gifford RJ, Tristem M, Gilbert MT, Pybus OG. 2009. Macroevolution of complex retroviruses. Science 325:1512. [DOI] [PubMed] [Google Scholar]
- 35. Kawai T, Akira S. 2011. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity 34:637–650 [DOI] [PubMed] [Google Scholar]
- 36. Khan AS. 2009. Simian foamy virus infection in humans: prevalence and management. Expert Rev. Anti-Infect. Ther. 7:569–580 [DOI] [PubMed] [Google Scholar]
- 37. Laguette N, et al. 2011. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature 474:654–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lepelley A, et al. 2011. Innate sensing of HIV-infected cells. PLoS Pathog. 7:e1001284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Linial M. 2000. Why aren't foamy viruses pathogenic? Trends Microbiol. 8:284–289 [DOI] [PubMed] [Google Scholar]
- 40. Linial ML. 1999. Foamy viruses are unconventional retroviruses. J. Virol. 73:1747–1755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lochelt M, et al. 2005. The antiretroviral activity of APOBEC3 is inhibited by the foamy virus accessory Bet protein. Proc. Natl. Acad. Sci. U. S. A. 102:7982–7987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lochelt M, Zentgraf H, Flugel RM. 1991. Construction of an infectious DNA clone of the full-length human spumaretrovirus genome and mutagenesis of the bel1 gene. Virology 184:43–54 [DOI] [PubMed] [Google Scholar]
- 43. Manches O, et al. 2008. HIV-activated human plasmacytoid DCs induce Tregs through an indoleamine 2,3-dioxygenase-dependent mechanism. J. Clin. Invest. 118:3431–3439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Manel N, et al. 2010. A cryptic sensor for HIV-1 activates antiviral innate immunity in dendritic cells. Nature 467:214–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. McCartney SA, Colonna M. 2009. Viral sensors: diversity in pathogen recognition. Immunol. Rev. 227:87–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Meiering CD, Linial ML. 2001. Historical perspective of foamy virus epidemiology and infection. Clin. Microbiol. Rev. 14:165–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mikovits JA, Hoffman PM, Rethwilm A, Ruscetti FW. 1996. In vitro infection of primary and retrovirus-infected human leukocytes by human foamy virus. J. Virol. 70:2774–2780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Moebes A, et al. 1997. Human foamy virus reverse transcription that occurs late in the viral replication cycle. J. Virol. 71:7305–7311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Murray SM, Linial ML. 2006. Foamy virus infection in primates. J. Med. Primatol 35:225–235 [DOI] [PubMed] [Google Scholar]
- 50. Murray SM, et al. 2008. Replication in a superficial epithelial cell niche explains the lack of pathogenicity of primate foamy virus infections. J. Virol. 82:5981–5985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Neumann-Haefelin D, Fleps U, Renne R, Schweizer M. 1993. Foamy viruses. Intervirology 35:196–207 [DOI] [PubMed] [Google Scholar]
- 52. Pacheco B, Finzi A, McGee-Estrada K, Sodroski J. 2010. Species-specific inhibition of foamy viruses from South American monkeys by New World monkey TRIM5α proteins. J. Virol. 84:4095–4099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Perkovic M, et al. 2009. Species-specific inhibition of APOBEC3C by the prototype foamy virus protein bet. J. Biol. Chem. 284:5819–5826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Picard-Maureau M, Jarmy G, Berg A, Rethwilm A, Lindemann D. 2003. Foamy virus envelope glycoprotein-mediated entry involves a pH-dependent fusion process. J. Virol. 77:4722–4730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Pietschmann T, et al. 1999. Foamy virus capsids require the cognate envelope protein for particle export. J. Virol. 73:2613–2621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Regad T, et al. 2001. PML mediates the interferon-induced antiviral state against a complex retrovirus via its association with the viral transactivator. EMBO J. 20:3495–3505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Rethwilm A. 2010. Molecular biology of foamy viruses. Med. Microbiol. Immunol. 199:197–207 [DOI] [PubMed] [Google Scholar]
- 58. Rethwilm A. 2003. The replication strategy of foamy viruses. Curr. Top. Microbiol. Immunol. 277:1–26 [DOI] [PubMed] [Google Scholar]
- 59. Rhodes-Feuillette A, et al. 1990. Effects of human recombinant alpha and gamma and of highly purified natural beta interferons on simian Spumavirinae prototype (simian foamy virus 1) multiplication in human cells. Res. Virol. 141:31–43 [DOI] [PubMed] [Google Scholar]
- 60. Rhodes-Feuillette A, Mahouy G, Lasneret J, Flandrin G, Peries J. 1987. Characterization of a human lymphoblastoid cell line permanently modified by simian foamy virus type 10. J. Med. Primatol 16:277–289 [PubMed] [Google Scholar]
- 61. Russell RA, et al. 2005. Foamy virus Bet proteins function as novel inhibitors of the APOBEC3 family of innate antiretroviral defense factors. J. Virol. 79:8724–8731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sabile A, et al. 1996. In vitro studies on interferon-inducing capacity and sensitivity to IFN of human foamy virus. Res. Virol. 147:29–37 [DOI] [PubMed] [Google Scholar]
- 63. Saib A, Peries J, de The H. 1993. A defective human foamy provirus generated by pregenome splicing. EMBO J. 12:4439–4444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Sattentau Q. 2008. Avoiding the void: cell-to-cell spread of human viruses. Nat. Rev. Microbiol. 6:815–826 [DOI] [PubMed] [Google Scholar]
- 65. Schenk T, Enssle J, Fischer N, Rethwilm A. 1999. Replication of a foamy virus mutant with a constitutively active U3 promoter and deleted accessory genes. J. Gen. Virol. 80(Pt 7):1591–1598 [DOI] [PubMed] [Google Scholar]
- 66. Schiffer C, et al. 2004. Persistent infection with primate foamy virus type 1 increases human immunodeficiency virus type 1 cell binding via a Bet-independent mechanism. J. Virol. 78:11405–11410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Schmidt B, Ashlock BM, Foster H, Fujimura SH, Levy JA. 2005. HIV-infected cells are major inducers of plasmacytoid dendritic cell interferon production, maturation, and migration. Virology 343:256–266 [DOI] [PubMed] [Google Scholar]
- 68. Sourisseau M, Sol-Foulon N, Porrot F, Blanchet F, Schwartz O. 2007. Inefficient human immunodeficiency virus replication in mobile lymphocytes. J. Virol. 81:1000–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Switzer WM, et al. 2005. Ancient co-speciation of simian foamy viruses and primates. Nature 434:376–380 [DOI] [PubMed] [Google Scholar]
- 70. Tan J, et al. 2008. IFP35 is involved in the antiviral function of interferon by association with the viral Tas transactivator of bovine foamy virus. J. Virol. 82:4275–4283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Uze G, et al. 1994. Domains of interaction between alpha interferon and its receptor components. J. Mol. Biol. 243:245–257 [DOI] [PubMed] [Google Scholar]
- 72. Vollmer J, Krieg AM. 2009. Immunotherapeutic applications of CpG oligodeoxynucleotide TLR9 agonists. Adv. Drug Deliv. Rev. 61:195–204 [DOI] [PubMed] [Google Scholar]
- 73. von Laer D, Neumann-Haefelin D, Heeney JL, Schweizer M. 1996. Lymphocytes are the major reservoir for foamy viruses in peripheral blood. Virology 221:240–244 [DOI] [PubMed] [Google Scholar]
- 74. Williams DK, Khan AS. 2010. Role of neutralizing antibodies in controlling simian foamy virus transmission and infection. Transfusion 50:200–207 [DOI] [PubMed] [Google Scholar]
- 75. Wolfe ND, et al. 2004. Naturally acquired simian retrovirus infections in central African hunters. Lancet 363:932–937 [DOI] [PubMed] [Google Scholar]
- 76. Xu F, et al. 2011. Tetherin inhibits prototypic foamy virus release. Virol. J. 8:198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Yan N, Regalado-Magdos AD, Stiggelbout B, Lee-Kirsch MA, Lieberman J. 2010. The cytosolic exonuclease TREX1 inhibits the innate immune response to human immunodeficiency virus type 1. Nat. Immunol. 11:1005–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Yu SF, Baldwin DN, Gwynn SR, Yendapalli S, Linial ML. 1996. Human foamy virus replication: a pathway distinct from that of retroviruses and hepadnaviruses. Science 271:1579–1582 [DOI] [PubMed] [Google Scholar]
- 79. Yu SF, Linial ML. 1993. Analysis of the role of the bel and bet open reading frames of human foamy virus by using a new quantitative assay. J. Virol. 67:6618–6624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Yu SF, Stone J, Linial ML. 1996. Productive persistent infection of hematopoietic cells by human foamy virus. J. Virol. 70:1250–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]