Abstract
Virus gene sequencing and phylogenetics can be used to study the epidemiological dynamics of rapidly evolving viruses. With complete genome data, it becomes possible to identify and trace individual transmission chains of viruses such as influenza virus during the course of an epidemic. Here we sequenced 153 pandemic influenza H1N1/09 virus genomes from United Kingdom isolates from the first (127 isolates) and second (26 isolates) waves of the 2009 pandemic and used their sequences, dates of isolation, and geographical locations to infer the genetic epidemiology of the epidemic in the United Kingdom. We demonstrate that the epidemic in the United Kingdom was composed of many cocirculating lineages, among which at least 13 were exclusively or predominantly United Kingdom clusters. The estimated divergence times of two of the clusters predate the detection of pandemic H1N1/09 virus in the United Kingdom, suggesting that the pandemic H1N1/09 virus was already circulating in the United Kingdom before the first clinical case. Crucially, three clusters contain isolates from the second wave of infections in the United Kingdom, two of which represent chains of transmission that appear to have persisted within the United Kingdom between the first and second waves. This demonstrates that whole-genome analysis can track in fine detail the behavior of individual influenza virus lineages during the course of a single epidemic or pandemic.
INTRODUCTION
Molecular phylogenies can reveal many aspects of the transmission, epidemiology, and evolution of rapidly evolving pathogens (17). Analysis of influenza virus genomes during the emergence of pandemic H1N1/09 virus, causing the first influenza pandemic in 40 years, provides a unique opportunity to track the transmission dynamics of a new influenza virus in an immunologically naïve population. The application of whole-virus genome sequencing and analysis has already provided detailed insights into seasonal influenza virus infections (5, 18). The evolutionary dynamics of seasonal influenza A arise from a complex combination of rapid mutation and genome segment reassortment, generating viral diversity that is modulated by natural selection, global patterns of virus circulation, and host population biology. The import of influenza A virus strains from tropical regions such as Southeast Asia, where different virus lineages cocirculate, may account for the genetic relationships among seasonal epidemics in the northern and southern hemispheres (18, 19). However, the evolutionary dynamics of pandemic influenza virus lineages during their initial emergence and establishment in the human population are not well understood.
The detection of a novel influenza A virus of swine origin (pandemic H1N1/09 virus) in Mexico and the United States in April 2009 and its subsequent rapid global spread provide a unique opportunity to observe such dynamics, particularly in regions, such as the United Kingdom, where virological surveillance is comprehensive, closely matched to the well-defined chronology of epidemic waves, and linked to disease surveillance. The first two laboratory-confirmed cases in the United Kingdom were detected on 27 April 2009, in travellers returning from Mexico, and were followed by further cases in travellers returning from different parts of Mexico and the United States. Virus introductions into the United Kingdom gave rise to secondary and tertiary cases in chains of transmission and clusters in May and early June (16). During this time, the United Kingdom initiated a containment strategy whereby individual cases were investigated and laboratory confirmed, contacts traced, and antiviral treatment initiated, including administration of prophylaxis to close contacts. Gradually, sporadic cases emerged in the community that could not be linked to specific epidemiological clusters, leading to sustained community transmission and the first pandemic wave. The first pandemic wave peaked in July, during which up to 42% of school children aged 5 to 14 years became infected in high-incidence areas of the United Kingdom, with clear regional differences in infection rates (12). Following a decline of pandemic H1N1/09 virus infections over the summer, a second wave of infections began in mid-September, coincident with the opening of schools, with an extended peak of infection between the middle of October and the middle of November. Characteristics of virus transmission and disease incidence in the second wave were similar to those of the first wave, with an estimated total cumulative number of symptomatic patients seeking health care of 788,000 (range, 375,000 to 1,644,000).
Analysis of whole-genome pandemic H1N1/09 virus sequences demonstrated that distinct lineages arose early during the pandemic and rapidly disseminated globally (4, 13). Here we have sequenced the complete genomes of 153 pandemic H1N1/09 virus isolates from the first and second epidemiological waves in the United Kingdom and the Republic of Ireland. These data demonstrate that the first wave of infections in the United Kingdom was genetically complex, comprising multiple cocirculating and genetically distinct lineages that had each been introduced independently to the United Kingdom from elsewhere. We show that at least two pandemic H1N1/09 virus lineages were circulating before the first case in the United Kingdom was laboratory confirmed. Crucially, our data suggest that at least two United Kingdom transmission chains persisted in the country between the first and second waves of infections.
MATERIALS AND METHODS
Sample ethics, origin, and isolation.
This was an observational study undertaken as part of management of a national outbreak. The samples were taken during routine diagnostic treatment by hospital physicians. It was carried out under United Kingdom legislation NHS Act 2006 (section 251), which provides statutory support for disclosure of data by the NHS and their processing by the Health Protection Agency (HPA) for communicable disease control. Health Protection Scotland is also embedded as part of the NHS, in which the sharing of outbreak and investigation data is undertaken as part of its role in the coordination of national outbreaks. Viruses were obtained by isolation from respiratory specimens provided by sentinel general practitioners (GPs), obtained through self-sampling schemes, or submitted by hospital diagnostic laboratories. Sentinel samples were analyzed at the Health Protection Agency Centre for Infections (CfI) by real-time reverse transcription-PCR (RT-PCR) for detection of influenza A virus, and viruses were subtyped as seasonal H1 and H3 and pandemic H1N1 2009 viruses. A range of specimens were selected for cell culture, including samples representing a broad temporal and geographical distribution, and also different clinical manifestations, including hospitalized and fatal cases. First-wave sampling attempted to capture a very large proportion of all infected individuals in the first 8 weeks of the epidemic in the United Kingdom, whereas the second-wave sampling represented a return to more normal national virological surveillance. Representative virology samples were obtained from ill patients in the community, but there was a focus on hospitalized cases to ensure sampling of severe cases in case of emergence of more virulent strains. The mean age of first-wave patients was 23.96 years, and that of second-wave patients was 14.88 years. Viruses were passaged in either Madin-Darby canine kidney (MDCK) cells or the SIAT1 derivative (10) in the presence of 2.5 μg/ml of trypsin and were stored at −80°C. A total of 153 viruses were selected for further whole-genome amplification and sequencing: 127 viruses were randomly selected from the first wave (24 April to 10 July 2009), and 26 were selected from the second wave (9 September to 2 December 2009) (see Table S1 in the supplemental material).
RNA isolation, PCR, and sequencing.
Virus RNA extraction, PCR, and sequencing are described in detail in the supplemental material. Briefly, virus RNA was extracted either from aliquots of original respiratory material or from cell-grown viral isolates. Amplification and sequencing of influenza virus whole genomes were performed with original respiratory material or cell-grown isolates. A number of methods were used. Clinical samples were sequenced by performing two-step RT-PCR amplification of full or half segments (see Table S2 in the supplemental material), followed by direct sequencing of the products. Cultured viruses were sequenced using a two-step RT-PCR approach employing pandemic H1N1/09 virus-specific PCR primers containing 5′ extensions identical to the M13F and M13R sequencing primers (see Table S3), similar to the method described by Ghedin et al. (5). For the second-wave samples, RNAs from clinical samples were amplified by RT-PCR by the 8-segment PCR method of Zhou et al. (25), with some modifications. Products were used as templates for PCRs with M13F/M13R-tailed primers, as described above, or were sequenced on an Illumina Genome Analyzer IIe sequencer (see Table S1).
Sequence characterization and phylogenetics.
Alignments for all segments were created individually using CLUSTAL W v1.83 (23), manually inspected, and trimmed to include coding regions only. The coding regions of all segments for each isolate were concatenated, in the order PB2-PB1-PA-HA-NP-NA-M-NS. The final concatenated alignments were used for all subsequent evolutionary analyses. Global pandemic H1N1/09 virus sequences were downloaded from the NCBI influenza virus database. Only those sequences satisfying the following criteria were used: (i) genomes must have had complete coverage over all reading frames; (ii) the year, month, and day of isolate sampling must be known; and (iii) sequences must have had ≤20% nucleotide ambiguities. The complete global pandemic H1N1/09 virus sequence set, including our United Kingdom and Irish sequences, comprised 1,523 sequences. A maximum likelihood phylogeny of the unfiltered global H1N1/09 sequence set, comprising 1,523 genomes, was estimated using RAxML-VI-HPC (22) with the GTRGAMMA substitution model and 100 bootstrap replicates.
The resulting data set was further filtered such that for heavily sampled geographical regions (except the United Kingdom), only one isolate per location per day was used. The final number of sequences in the filtered set was 595 (153 United Kingdom/Republic of Ireland sequences and 442 global sequences). We estimated the phylogeny and divergence times of the filtered pandemic H1N1/09 virus sequences by using a Bayesian Markov chain Monte Carlo (MCMC) approach as implemented in the BEAST package (2). This approach has been validated extensively in the context of human influenza (18). Evolutionary model specification followed that employed for pandemic H1N1/09 virus (3): we used an HKY+Γ substitution model with nucleotide frequencies and substitution rates estimated from the data (an exponential population growth model). Statistical support informed the use of the relaxed molecular clock as described previously (3). MCMC chain lengths were 50,000,000 generations, with sampling every 2,500 generations. Effective sample sizes were estimated using Tracer (version 1.4; http://beast.bio.ed.ac.uk/Tracer), and multiple chains were run to check chain convergence. Map analysis of the spatial locations of United Kingdom viruses was achieved using MapInfo Professional (version 10.5; PitneyBowes).
BaTS (Bayesian tip significance testing) (15) was used to test for spatial phylogenetic structure within the United Kingdom and Republic of Ireland data set. BaTS calculates two statistics (association index and parsimony score) for a given data set and compares these to their null distributions under the null hypothesis of no correlation between phylogenetic position and isolate location. A set of 200 trees representing the evolutionary history of the United Kingdom and Republic of Ireland data set was obtained using BEAST (using an HKY+Γ substitution model; 50 million MCMC states were computed, with the first 20% discarded as burn-in, and trees were sampled every 200,000 generations thereafter). Each isolate was labeled with its corresponding United Kingdom region of sampling (see Fig. S1 in the supplemental material). We used BaTS to calculate the association index and parsimony score statistics from these 200 trees and compared them with appropriate null distributions (obtained using 1,000 replicates).
Nucleotide sequence accession numbers.
All sequences were submitted to GenBank under accession numbers GQ166654 to GQ166661 (A/England/195/2009), CY065139 to CY065746, and HM567541 to HM568156 (see Table S1 in the supplemental material).
RESULTS
The first pandemic H1N1/09 wave in the United Kingdom consisted of multiple independent introductions.
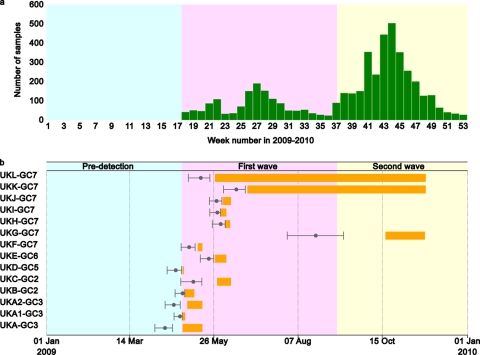
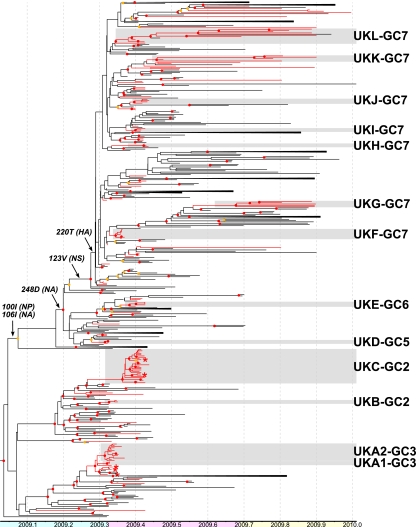
A total of 153 whole-genome sequences were obtained from original pretreatment clinical samples or virus isolates during the first (n = 127) and second (n = 26) waves of United Kingdom infections (Fig. 1a). The coding regions of all gene segments of these isolates were aligned with a filtered set of global pandemic H1N1/09 virus whole-genome sequences and used to derive a molecular clock phylogeny using Bayesian methods as employed in the BEAST software package (Fig. 2; see Fig. S1 in the supplemental material). This phylogeny demonstrates the existence of multiple independent introductions of pandemic H1N1/09 virus into the United Kingdom during the first wave of infections, as opposed to a single introduction (or a few introductions) followed by clonal expansion.
Fig 1.
(a) Numbers of laboratory-confirmed cases of pandemic H1N1/09 influenza during 2009. (b) Divergence dates and life spans of United Kingdom clusters. Gray dots represent divergence times of clusters, with bars showing 95% confidence intervals. Orange bars represent ranges of sampling dates of viruses within each cluster.
Fig 2.
Simplified BEAST tree showing distribution of United Kingdom isolates in the context of global pandemic H1N1/09 virus isolates. United Kingdom lineages are shown with red lines, and all other lineages are shown with black lines. United Kingdom-specific clusters are indicated by gray boxes with adjacent cluster labels. Red dots indicate nodes with ≥95% posterior support, and orange dots indicate nodes with posterior support of <95% but ≥80%. Amino acid changes which characterize global cluster 7 are shown adjacent to the branches on which they likely occurred. Known (or suspected) importations are indicated with an asterisk adjacent to the terminal branch. Some groups with high posterior support are collapsed for clarity. Blue, pink, and yellow bars represent predetection, first-wave, and second-wave phases of the pandemic in the United Kingdom. The full tree with taxon names is shown in Fig. S1 in the supplemental material.
We sought to identify United Kingdom-specific transmission chains by using three previously published criteria (7): (i) the cluster must be significantly supported, with a phylogenetic posterior probability of 95%; (ii) the cluster must contain more than 2 isolates; and (iii) more than 80% of isolates within the cluster must be sampled in the United Kingdom or Republic of Ireland. The Republic of Ireland samples were included due to the close geographical proximity of and large population movements between the Republic of Ireland, Northern Ireland, and the rest of the United Kingdom. By these criteria, we identified 13 United Kingdom clusters, each containing 3 to 27 isolates, representing 94 (61%) isolates in total (Fig. 1b and 2; Table 1). Clusters were named to reflect their placement within global clades 1 to 7, defined previously (13); hence, UKC-GC2 indicates United Kingdom cluster C, which falls within global clade 2. To ensure that United Kingdom clusters were robust to the method of filtering global pandemic H1N1/09 virus sequences, we confirmed that all clusters were also intact in a maximum likelihood phylogeny of the unfiltered sequence set (see Fig. S2 in the supplemental material). Clusters UKA1-GC3 and UKA2-GC3 were initially joined in a larger cluster (cluster UKA-GC3), which also included four additional United Kingdom isolates and four non-United Kingdom isolates. However, three of these United Kingdom isolates were independent importations, and a maximum likelihood phylogeny including all global isolates suggested that more non-United Kingdom isolates grouped with these sequences. For these reasons, the larger cluster UKA-GC3, with >95% phylogenetic posterior probability support, was divided into clusters UKA1-GC3 and UKA2-GC3, with the same support.
Table 1.
Divergence times and detection ranges of United Kingdom clusters
| Cluster | No. of isolates | % of isolates from United Kingdom/Republic of Ireland | TMRCA | TMRCA |
Date of isolation |
Maximum duration (days)a | Summaryb | Pre-UK detection (days)c | Precluster detection (days)d | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Low 95% HPD | High 95% HPD | Earliest UK isolate | Latest UK isolate | ||||||||
| UKA1-GC3 | 3 | 100.00 | 26 Apr 2009 | 21 Apr 2009 | 28 Apr 2009 | 28 Apr 2009 | 30 Apr 2009 | 9 | 1st wave | 1 | 2 |
| UKA2-GC3 | 14 | 100.00 | 20 Apr 2009 | 13 Apr 2009 | 26 Apr 2009 | 2 May 2009 | 15 May 2009 | 32 | 1st wave | 7 | 12 |
| UKB-GC2 | 3 | 100.00 | 27 Apr 2009 | 21 Apr 2009 | 30 Apr 2009 | 30 Apr 2009 | 8 May 2009 | 17 | 1st wave | 0 | 3 |
| UKC-GC2 | 27 | 100.00 | 7 May 2009 | 26 Apr 2009 | 15 May 2009 | 27 May 2009 | 8 Jun 2009 | 43 | 1st wave | 0 | 20 |
| UKD-GC5 | 3 | 100.00 | 22 Apr 2009 | 14 Apr 2009 | 27 Apr 2009 | 27 Apr 2009 | 28 Apr 2009 | 14 | 1st wave | 5 | 5 |
| UKE-GC6 | 4 | 100.00 | 20 May 2009 | 13 May 2009 | 25 May 2009 | 26 May 2009 | 4 Jun 2009 | 22 | 1st wave | 0 | 6 |
| UKF-GC7 | 8 | 100.00 | 3 May 2009 | 26 Apr 2009 | 9 May 2009 | 11 May 2009 | 15 May 2009 | 19 | 1st wave | 0 | 8 |
| UKG-GC7 | 5 | 100.00 | 21 Aug 2009 | 27 Jul 2009 | 15 Sep 2009 | 20 Oct 2009 | 24 Nov 2009 | 120 | 2nd wave | 0 | 60 |
| UKH-GC7 | 3 | 100.00 | 31 May 2009 | 23 May 2009 | 4 Jun 2009 | 4 Jun 2009 | 8 Jun 2009 | 16 | 1st wave | 0 | 4 |
| UKI-GC7 | 3 | 100.00 | 28 May 2009 | 21 May 2009 | 31 May 2009 | 31 May 2009 | 4 Jun 2009 | 14 | 1st wave | 0 | 3 |
| UKJ-GC7 | 4 | 100.00 | 27 May 2009 | 21 May 2009 | 1 Jun 2009 | 1 Jun 2009 | 8 Jun 2009 | 18 | 1st wave | 0 | 5 |
| UKK-GC7 | 5 | 100.00 | 13 Jun 2009 | 2 Jun 2009 | 21 Jun 2009 | 23 Jun 2009 | 24 Nov 2009 | 175 | Persistent | 0 | 10 |
| UKL-GC7 | 12 | 83.33 | 14 May 2009 | 3 May 2009 | 22 May 2009 | 26 May 2009 | 25 Nov 2009 | 205 | Persistent | 0 | 12 |
Time between TMRCA (low 95% HPD) and latest United Kingdom isolate.
1st wave, cluster contains first-wave isolates only; 2nd wave, cluster contains second-wave isolates only; persistent, cluster contains isolates from both first and second waves.
Time between TMRCA and first detected pandemic H1N1/09 case in the United Kingdom (27 April 2009).
Time between TMRCA and earliest detection date within cluster.
The United Kingdom-specific transmission clusters were interspersed with isolates from other countries. The remaining United Kingdom isolates appeared as single branches or as pairs of United Kingdom isolates within groups of isolates from other countries. The first reported pandemic H1N1/09 virus isolate in the United Kingdom was sampled from a traveler returning from Mexico on 27 April 2009. However, our estimates indicate that pandemic H1N1/09 virus was circulating in the United Kingdom for up to 1 week prior to this date (Table 1). The inferred date of common ancestry of all first-wave United Kingdom clusters predates the first documented sampling of the respective cluster, in some cases by as much as 20 days (cluster UKC-GC2) (Fig. 1b; Table 1). The estimated date of origin of one predominantly United Kingdom cluster (cluster UKA2-GC3) (Fig. 1b and 2) was 20 April 2009 (95% highest posterior density [HPD] range, 13 April 2009 to 26 April 2009), 12 days before the earliest sampling date from this cluster and 7 days (95% HPD range, 1 to 14 days) before any pandemic H1N1/09 virus-infected individual was detected in the United Kingdom. Similarly, cluster UKD-GC5 (Fig. 1b and 2; Table 1) arose on or around 22 April 2009, 5 days (95% HPD range, 0 to 13 days) before the earliest confirmed United Kingdom isolate.
Isolates within clusters UKA1-GC3/UKA2-GC3 and UKC-GC2 were associated predominantly (but not exclusively) with early, large, individual educational establishment outbreaks in the United Kingdom. Within cluster UKC-GC2, however, isolate A/England/361/2009 was a reported new importation, A/England/374/2009 was suspected to be a new importation, and A/England/399/2009 was a community-acquired virus epidemiologically unconnected to the outbreak in this cluster (Fig. 2; see Fig. S1 in the supplemental material). However, our sequence data suggest that all of the isolates in cluster UKC-GC2 were closely linked.
Based on our samples, it appears that the majority of pandemic H1N1/09 virus transmission chains that comprised the first epidemic wave were not sustained in the United Kingdom. There were no known epidemiological linkages between clusters.
Persistence of first-wave United Kingdom pandemic H1N1/09 virus strains into the second wave.
Most significantly, it is clear that two United Kingdom clusters (UKK-GC7 and UKL-GC7) contained isolates from both the first and second waves of infections. Cluster UKK-GC7 was estimated to have arisen on 13 June 2009 and persisted for at least 175 days, to 24 November. Similarly, cluster UKI-GC7 was estimated to have arisen on 14 May 2009 and persisted for at least 205 days, to 25 November (Table 1). This provides the first evidence that individual chains of pandemic H1N1/09 virus transmission can persist between pandemic infection waves in a specific location. In addition, four second-wave United Kingdom isolates that do not belong to a defined global cluster also group with United Kingdom sequences related to cluster UKK-GC7 from the first wave, but statistical support for this grouping is not significant (Fig. 2). In contrast, the remaining United Kingdom clusters appeared only in the first wave of infection (Fig. 1b and 2).
Cluster UKG-GC7 comprised only isolates from the second wave of infection in the United Kingdom. This transmission chain was placed phylogenetically within a large and persistent lineage of U.S. isolates (from New York State, California, District of Columbia, and Texas) (Fig. 2). Therefore, it is highly likely that cluster UKG-GC7 originated in the United States. Cluster UKG-GC7 appears to have been present for some time (60 days) before first being detected, which most likely reflects less intense sampling of isolates in both the United Kingdom and the United States as the pandemic progressed during 2009.
Of the remaining second-wave United Kingdom samples, five grouped together with other second-wave isolates from Russia, Norway, Greece, Poland, and Denmark, and six were distributed throughout the tree. Two viruses from Russia and Denmark grouped with United Kingdom isolates within cluster UKL-GC7. The identical genetic composition of the Russian virus and its earlier sampling date, which took place during the first United Kingdom wave, suggest that a similar virus may have given rise to these second-wave United Kingdom viruses. Viruses within cluster UKL-GC7 may therefore have been undetected in sampling in the United Kingdom during the first wave or introduced into the United Kingdom prior to the second wave.
Signature amino acid substitutions define United Kingdom pandemic H1N1/09 virus clusters.
We next sought to identify amino acid changes in the United Kingdom isolates and clusters with possible biological or antigenic relevance. Based on signature nonsynonymous changes, we concluded that all of the United Kingdom lineages are found within clades defined previously as circulating worldwide (13). Clusters UKA1-GC3 and UKA2-GC3 fall within global clade 3, clusters UKB-GC2 and UKC-GC2 fall within global clade 2 (changes in PA [M581L] and nucleoprotein [NP] [T373I] relative to clade 3), cluster UKD-GC5 falls within global clade 5 (changes in NP [V100I] and neuraminidase [NA] [V106I and N248D]), cluster UKE-GC6 falls within global clade 6 (changes in hemagglutinin [HA] [K2E and Q310H], NP [V100I], and NA [V106I and N248D]), and the remaining United Kingdom clusters (UKF-GC7 to UKL-GC7) fall within global clade 7 (changes in HA [S220T], NP [V100I], NA [V106I and N248D], and NS1 [I123V]). Our estimated divergence time for global clade 7 is 2 to 3 April 2009 (95% HPD range, 22 March to 12 April 2009), which is slightly earlier but not significantly different from that estimated previously (95% HPD range, 28 March to 18 April 2009) (13). Interestingly, the signature amino acid changes seen in viruses from clusters UKF-GC7, UKG-GC7, UKH-GC7, UKI-GC7, UKK-GC7, and UKL-GC7, and which define global clade 7, predominated in the most recent pandemic H1N1/09 virus isolates worldwide (11).
Some United Kingdom clusters possessed signature coding changes which further distinguished them from the global isolates (see Fig. S3 and Table S4 in the supplemental material), although none of the United Kingdom cluster-specific changes were at positions likely to affect pathogenicity, drug resistance, or HA receptor binding (as previously described for other influenza A virus human subtypes). Some United Kingdom isolates and clusters showed amino acid changes of possible antigenic significance (see the supplemental material). However, none of the United Kingdom viruses with or without these changes had significant reductions in hemagglutination inhibition (HI) titers against A/California/07/2009 (pandemic vaccine strain) or A/England/195/2009 (United Kingdom prototype virus) ferret antisera. One United Kingdom isolate (A/Scotland/10/2009) possessed a glycine (G) at position 222 and was isolated from a sample taken late during the course of a fatal illness. This change has been noted in some severe and fatal cases of pandemic H1N1 influenza (8, 9, 24).
Geographical locations of clusters and isolates.
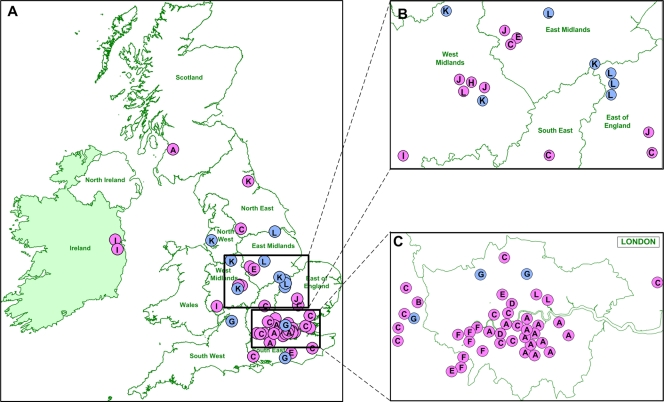
We used the geographical location of the individual from whom each sample was obtained in the context of the genetically defined transmission chains to examine the spatial pattern of pandemic H1N1/09 virus spread within the United Kingdom. The initial period of the first wave (global clades 2, 3, 5, and 6; United Kingdom clusters A1, A2, B, C, D, and E) was dominated by isolates from London and the Southeast and East of England (Fig. 3; see Fig. S1 in the supplemental material). UKC-GC2, which contains many isolates from a single educational establishment outbreak, is complex, with isolate locations spread geographically within the East, the Southeast, and the London area. Isolates that were not within defined United Kingdom clusters also tended to be located predominantly in the East, the Southeast, and the London area during this time. While this may reflect a sampling bias for the larger clusters, the similar trend for noncluster isolates (12 of 18 isolates located in the East, the Southeast, and the London area) supports a degree of geographical restriction early in the first wave. With the arrival of global clade 7 isolates (UKF-GC7 to UKL-GC7), more extensive spread to most United Kingdom regions was observed. Interestingly, UKK-GC7 and UKL-GC7, which persisted from the first to the second wave, contained isolates from many United Kingdom regions, suggesting that persistence was not constrained geographically (Fig. 3; see Fig. S1). Complex transmission dynamics are more evident with ultrafine spatial mapping (Fig. 3). From genome sequences alone, United Kingdom clusters are clearly distinct, but they are otherwise similar in their phylogenetic profiles. However, clusters A and F were geographically constrained in two distinct regions of London (Fig. 3C), whereas cluster C isolates occurred in and around distinct parts of London as well as other United Kingdom regions (Fig. 3A to C). Clusters E and G, in contrast, showed wide spatial distributions within the East, the Southeast, and the London area, indicating that different transmission chain dynamics were clearly visible early in pandemic H1N1/09 virus introduction into the United Kingdom. The BaTS analysis of the spatial phylogenetic structure within the United Kingdom and Republic of Ireland data set showed that there was a significant (P < 0.001) association between the regions from which isolates originated and their phylogenetic relationships. However, we note that the statistical significance of this result is perhaps raised artificially as a result of the intense sampling of some local outbreaks, particularly those in London (UKA1-GC3 and UKA2-GC3) and the Southeast (UKC-GC2).
Fig 3.
(A) Geographical distribution of viruses belonging to global clade-United Kingdom phylogenetic clusters. Two viruses isolated in the Republic of Ireland were also included. Magnified views of the Midlands (B) and the London area (C) are shown. Virus isolates are colored according to whether they were isolated in the first (pink) or second (blue) infection wave, with the United Kingdom cluster for each isolate identified by the cluster letter shown in Fig. 2.
DISCUSSION
We have used influenza virus whole-genome sequences combined with sophisticated evolutionary analysis to estimate the molecular epidemiological characteristics of pandemic H1N1 influenza in the United Kingdom. The distribution of United Kingdom-specific clusters and individual United Kingdom isolates throughout the global pandemic H1N1/09 virus phylogeny proves that the epidemic in the United Kingdom resulted from multiple independent introductions of the virus into the country during the first wave of infection. None of the viruses analyzed in this study, more broadly as part of United Kingdom surveillance, or worldwide showed any evidence of significant antigenic divergence from A/California/07/2009, although they are genetically distinct.
Epidemiological tracking of cases during this period suggested that there were laboratory-confirmed cases from at least 60 distinct introductions, grouping into at least 9 different epidemiologically defined clusters. In addition to the 13 United Kingdom clusters we defined here (containing 94 of the 153 isolates sequenced), there were 52 isolates, widespread geographically in the United Kingdom, distributed throughout our phylogeny. This suggests that our sampling of the lineages that arrived in the United Kingdom at the start of the outbreak was reasonably complete and that the 13 clusters and 52 dispersed lineages account for much of the United Kingdom pandemic H1N1/09 virus genetic diversity. Standard power calculations to add support to this observation are not possible, however, as sequences are not statistically independent observations but are in fact highly correlated due to the presence of shared ancestry.
Based on our analysis, at least one United Kingdom cluster (cluster UKA2-GC3) began to diverge 1 week (95% HPD range, 1 to 14 days) before the first clinically diagnosed United Kingdom case. The node which connects clusters UKA1-GC3 and UKA2-GC3, although poorly supported (posterior probability = 0.3542), has a time of most recent common ancestor (TMRCA) of 13 April 2009, suggesting that pandemic H1N1/09 virus was already spreading undetected in the United Kingdom around the same time that it was identified in Mexico, presumably as a result of unidentified infections in returning travelers. Note that there were approximately 5,000 travelers per week to and from Mexico in the weeks immediately preceding the first confirmed introduction to the United Kingdom.
The reasons for the apparent lag between the introductions of pandemic H1N1/09 virus into the United Kingdom and the peak of first-wave infections are not known. The spread of A/H1N1/09 in mainland Europe during the first wave in the United Kingdom was characterized by sporadic cases and isolated self-limiting outbreaks linked to importations. In the United Kingdom, however, the major generalized epidemic occurred in June and July and declined only once schools closed for the summer holidays at the end of July (6). No equivalent epidemic wave was reported in Europe until the autumn, during which time the United Kingdom had its second wave of infection. The beginning of the second wave in the United Kingdom also coincided with the reopening of schools after the summer holiday period. It is possible that the United Kingdom public health response, which was comprehensive and applied consistently during the early phases and which consisted of laboratory confirmation of suspected cases followed by antiviral prophylaxis of contacts, may have contributed to this delay. Antiviral prophylaxis appears to be effective at reducing household transmission (R. G. Pebody et al., unpublished data), suggesting that the strict prophylaxis policy acted to extinguish some of the early viral introductions, possibly slowing the rate of rise of the pandemic.
We were able to show clear evidence of virus lineage persistence in the United Kingdom between the first and second epidemic waves. A prominent characteristic of influenza epidemics in temperate regions is their seasonality, with the majority of infections occurring during the winter months in the northern and southern hemispheres, in contrast to year-round influenza virus circulation across tropical regions. This has led to the hypothesis that influenza virus genetic diversity is generated continually in tropical regions, primarily Southeast Asia, from where viral lineages migrate to temperate regions each year, founding the next seasonal epidemic (18, 19). Here we found two United Kingdom-specific clusters that appeared in the first wave of infection and persisted into the second wave of infections. The density of our sampling and phylogenetic inference suggest that this reflects true persistence rather than reintroduction within the United Kingdom between the epidemic waves. Additional HA gene sequencing of influenza virus isolates between the first and second waves showed that viruses of cluster UKK-GC7, which arose in the United Kingdom in June and possessed the signature amino acid substitution D222E, were present and circulating in the United Kingdom between July and September 2009. The likelihood of persistence is further supported by intensive surveillance as a consequence of the national response to the pandemic, which clearly indicates that there were low levels of infection sustained in the community in individuals without any history of travel abroad, as well as sporadic travel-associated cases. In 2009, persistence may have been facilitated by the timing of the waves, as the majority of first-wave infections occurred during the summer months in the United Kingdom (with a peak in late July), whereas the second wave of infections started in September and peaked earlier (October-November) than usual for seasonal influenza epidemics. Consequently, the time between waves was much shorter than that for typical seasonal influenza, where the length of time between the end of one winter wave and the onset of an autumn wave may be as much as 8 to 9 months. Furthermore, a recent study also suggested that the occurrence of two consecutive waves in the United Kingdom, as opposed to only sporadic cases in the rest of Europe during the summer, was a consequence of a relatively large number of early importations (as demonstrated here) combined with a low level of absolute humidity, which has been shown to affect both transmissibility and the survival of influenza virus (20). The relative immunological naïvety of the human population to the pandemic H1N1/09 variant may also have enabled higher-than-usual infection rates during the interwave period. Whatever the underlying mechanism, it seems that influenza virus continued to circulate in the United Kingdom between infection peaks without causing a sustained level of clinical disease.
It is important to consider the benefits that are brought by real-time monitoring of virus genome diversity during a pandemic or epidemic. The very-high-resolution tracking of individual lineages is achievable only with full genome sequences. Coupled with spatial information, we show that detailed insight into transmission chains and epidemiology becomes apparent, extending what can be observed by use of sequences alone (1, 14, 21).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Wellcome Trust and the Health Protection Agency.
Footnotes
Published ahead of print 19 October 2011
Supplemental material for this article may be found at http://jvi.asm.org/.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1. Barrero PR, Viegas M, Valinotto LE, Mistchenko AS. 2011. Genetic and phylogenetic analyses of influenza A H1N1pdm virus in Buenos Aires, Argentina. J. Virol. 85:1058–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Drummond AJ, Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fraser C, et al. 2009. Pandemic potential of a strain of influenza A (H1N1): early findings. Science 324:1557–1561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Garten RJ, et al. 2009. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science 325:197–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ghedin E, et al. 2005. Large-scale sequencing of human influenza reveals the dynamic nature of viral genome evolution. Nature 437:1162–1166 [DOI] [PubMed] [Google Scholar]
- 6. House T, et al. 2011. Modelling the impact of local reactive school closures on critical care provision during an influenza pandemic. Proc. Biol. Sci. 278:2753–2760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hué S, et al. 2009. Demonstration of sustained drug-resistant human immunodeficiency virus type 1 lineages circulating among treatment-naïve individuals. J. Virol. 83:2645–2654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kilander A, Rykkvin R, Dudman SG, Hungnes O. 2010. Observed association between the HA1 mutation D222G in the 2009 pandemic influenza A(H1N1) virus and severe clinical outcome, Norway 2009–2010. Euro Surveill. 15:19498. [DOI] [PubMed] [Google Scholar]
- 9. Mak GC, et al. 2010. Association of D222G substitution in haemagglutinin of 2009 pandemic influenza A (H1N1) with severe disease. Euro Surveill. 15:19534. [PubMed] [Google Scholar]
- 10. Matrosovich M, Matrosovich T, Carr J, Roberts NA, Klenk HD. 2003. Overexpression of the α-2,6-sialyltransferase in MDCK cells increases influenza virus sensitivity to neuraminidase inhibitors. J. Virol. 77:8418–8425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Maurer-Stroh S, et al. 2010. A new common mutation in the hemagglutinin of the 2009 (H1N1) influenza A virus. PLoS Curr. 2:RRN1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Miller E, et al. 2010. Incidence of 2009 pandemic influenza A H1N1 infection in England: a cross-sectional serological study. Lancet 375:1100–1108 [DOI] [PubMed] [Google Scholar]
- 13. Nelson M, et al. 2009. The early diversification of influenza A/H1N1pdm. PLoS Curr. 1:RRN1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nelson MI, et al. 2011. Phylogeography of the spring and fall waves of the H1N1/09 pandemic influenza virus in the United States. J. Virol. 85:828–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Parker J, Rambaut A, Pybus OG. 2008. Correlating viral phenotypes with phylogeny: accounting for phylogenetic uncertainty. Infect. Genet. Evol. 8:239–246 [DOI] [PubMed] [Google Scholar]
- 16. Pebody RG, et al. 2010. Pandemic influenza A (H1N1) 2009 and mortality in the United Kingdom: risk factors for death, April 2009 to March 2010. Euro Surveill. 15:19571. [PubMed] [Google Scholar]
- 17. Pybus OG, Rambaut A. 2009. Evolutionary analysis of the dynamics of viral infectious disease. Nat. Rev. Genet. 10:540–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rambaut A, et al. 2008. The genomic and epidemiological dynamics of human influenza A virus. Nature 453:615–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Russell CA, et al. 2008. The global circulation of seasonal influenza A (H3N2) viruses. Science 320:340–346 [DOI] [PubMed] [Google Scholar]
- 20. Shaman J, Pitzer VE, Viboud C, Grenfell BT, Lipsitch M. 2010. Absolute humidity and the seasonal onset of influenza in the continental United States. PLoS Biol. 8:e1000316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shiino T, et al. 2010. Molecular evolutionary analysis of the influenza A(H1N1)pdm, May-September, 2009: temporal and spatial spreading profile of the viruses in Japan. PLoS One 5:e11057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Stamatakis A, Ludwig T, Meier H. 2005. RAxML-III: a fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics 21:456–463 [DOI] [PubMed] [Google Scholar]
- 23. Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. World Health Organization 2010. Preliminary review of D222G amino acid substitution in the haemagglutinin of pandemic influenza A(H1N1) 2009 viruses. Wkly. Epidemiol. Rec. 85:21–22 [PubMed] [Google Scholar]
- 25. Zhou B, et al. 2009. Single-reaction genomic amplification accelerates sequencing and vaccine production for classical and Swine origin human influenza A viruses. J. Virol. 83:10309–10313 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.