Abstract
Acute lymphoblastic leukemia (ALL) likely has a multistep etiology, with initial genetic aberrations occurring early in life. An abnormal immune response to common infections has emerged as a plausible candidate for triggering the proliferation of pre-leukemic clones and the fixation of secondary genetic mutations and epigenetic alterations. We investigated whether evidence of infection with a specific common myelotropic childhood virus, parvovirus B19 (PVB19), relates to patterns of gene promoter DNA methylation in ALL patients. We serologically tested bone marrow samples at diagnosis of B-cell ALL for PVB19 infection and DNA methylation using a high-throughput bead array and found that 4.2% and 36.7% of samples were seroreactive to PVB19 IgM and IgG, respectively. Leukemia samples were grouped by DNA methylation pattern. Controlling for age and immunophenotype, unsupervised modeling confirmed that the DNA methylation pattern was associated with history of PVB19 (assessed by IgG, p = 0.02), but not recent infection (assessed by IgM). Replication assays on single genes were consistent with the association. The data indicate that a common viral illness may drive specific DNA methylation patterns in susceptible B-precursor cells, contributing to the leukemogenic potential of such cells. Infections may impact childhood leukemia by altering DNA methylation patterns and specific key genes in susceptible cells; these changes may be retained even after the clearance of infection.
Key words: childhood leukemia, DNA methylation, parvovirus B19, serology
Introduction
Many studies support a role for infections in the etiology of childhood acute lymphoblastic leukemia (ALL), leading to two prevailing hypotheses on the underlying mechanisms. Kinlen suggested that childhood leukemia could result from a specific infection introduced by infected persons into a population without herd immunity.1 Greaves proposed a related hypothesis: delayed exposure to common infections during infancy leads to an increased risk of common pre-B-cell ALL (cALL).2 Under this latter theory, children may harbor genetic mutations that create a pre-leukemic clone. These clones, in turn, would be susceptible to proliferative expansion and ALL if they underwent aberrant stimulation from infections acquired later.3 A rich epidemiologic literature supports both of these hypotheses; however, very little support for specific molecular mechanisms has been presented. A challenging problem in delineating the role of infections in the onset of c-ALL is the lack of consensus regarding the precise causal infections. Indeed, molecular screening for specific candidate viruses or virus families has produced negative results.3–7 These data seem to provide evidence against direct viral transformation. However, it is possible that a very small virus or proviral component might be involved or that a potentially transforming virus has operated through a “hit-and-run” mechanism. A provocative report8 noted an increased frequency of group-C Adenovirus (AdV) in Guthrie cards (GC) from children who developed ALL. We were unable to repeat these observations in a patient population in California—AdV DNA was not detected at a higher frequency among neonates who later developed leukemia, when compared with controls.9
Evidence for infectious etiologies supporting Greaves' hypothesis include a broad range of epidemiologic studies using surrogate measures of exposure to infections, including birth order, child's history of infections, child's day-care and play group attendance and parental social contacts in the workplace (reviewed in refs. 10 and 11). In western countries these studies indicate that reduced exposure very early in life to common infections of childhood increases risk to leukemia. It becomes more challenging in tropical countries where the exposures of endemic viral and non-viral infections are seasonal and geographically heterogeneous. Recently, Reis and collaborators observed that substantial regional differences exist in the incidence of acute leukemia in Brazil12 and that such differences may, in part, be related to differences in prevalence of exposures to infectious agents and subsequent infection.
A plausible mechanism by which a viral infection might impact the process of leukemogenesis is through alteration of DNA methylation, leading to changes in the expression of hematopoietic genes, which could persist after viral clearance. A cell's response to viral infection can result in the activation of de novo DNA methylation processes in efforts to control viral gene expression.13 A question of interest is whether global gene promoter methylation changes might reflect the cells overall response to an infection. DNA methylation is sensitive to several factors: the availability of methyl groups, instructive histone modifications in chromatin, cell development and lineage properties, expression of methyltransferase genes and the interaction of environmental variables.
A candidate ALL-related infection is parvovirus B19 (PVB19), which is tropic to the bone marrow and specifically infects erythroblasts. PVB19 infection is the cause of “fifth disease” (also known as “Slapped Cheek Syndrome”). The vast majority of the population is infected early in life and tests seropositive by the age of 7 y, in both developed and developing countries including Brazil.14 While some reports describe an association between ALL and persistent parvovirus B19 (PVB19) infection, it was assumed that the infection was opportunistic, concurrent with the ALL.15–18 In the present study, we addressed the question whether gene promoter methylation patterns can provide insight into infection-mediated pathophysiology and etiology of childhood ALL in Brazil, specifically assessing whether DNA methylation patterns were associated with cytogenetic subtypes of ALL and evidence of a past history of PVB19 infection.
Results
The patients studied included a predominance of males with an age range of 0.08–14 y old (median 2.5 y) at diagnosis. “Common ALL” (CD19+, CD10+) represented the majority of cases. Serological analysis revealed an overall prevalence of 39.8% seroreactivity to PVB19, with 4.2% being IgM and 36.7% IgG seropositive (Table 1). Two cases were both IgM and IgG positive.
Table 1.
Demographic and diagnostic characteristics of acute lymphocytic leukemia cases used for Parvovirus B19/DNA methylation analysis
| n | % | |
| Sex | ||
| Female | 43 | 41 |
| Male | 61 | 59 |
| Age (years) | ||
| <1 | 16 | 15 |
| 1–9 | 70 | 67 |
| ≥10 | 18 | 18 |
| WBC (x109/L) | ||
| <50 | 60 | 58 |
| ≥50 | 44 | 42 |
| Immunophenotype | ||
| pro-B ALL | 17 | 16 |
| c-ALL | 76 | 73 |
| pre-B ALL | 11 | 15 |
| Cytogenetic/Molecular abnormalities | ||
| TEL-AML1 | 22 | 21 |
| E2A-PBX1 | 4 | 3.8 |
| MLL rearranged | 14 | 13.5 |
| Hyperdiploidy | 24 | 23 |
| Other | 3 | 3 |
| None | 28 | 27 |
| Unknown | 9 | 8.7 |
| Seroprevalence PVB19 | ||
| IgM positive | 2 | 2 |
| IgM undetermined | 1 | 1 |
| IgG positive | 28 | 27 |
| IgG undetermined | 11 | 10.5 |
| Double IgM + IgG+ | 1 | 1 |
| IgM pos + IgG und | 1 | 1 |
Methylation profiling.
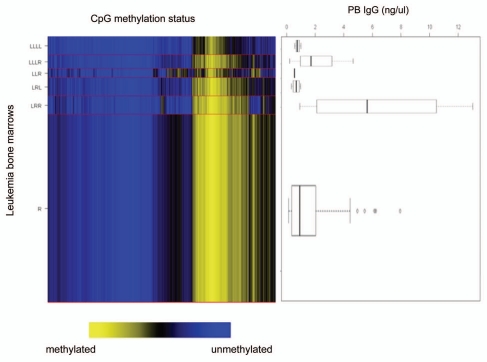
We determined methylation status of the promoter and first exon regions of 803 genes in bone marrow samples from 59 childhood ALL cases selected as a training set, with the same overall prevalence of leukemia subtypes as our available sample set. Array methylation data were assembled and analyzed applying recursively partitioned mixture modeling (RPMM) 24 resulting in 6 methylation classes (Fig. 2). DNA methylation class membership was significantly associated with the two B precursor cell leukemia immunophenotypes, c-ALL and pro-B ALL, as well as age, separately (Permutation p = 0.01 and p = 0.02, respectively). After controlling for age and immunophenotype, DNA methylation classes were associated with IgG levels, as a continuous variable, but not with IgM levels (Permutation test p = 0.02, p = 0.92, respectively; Fig. 3). In particular, the DNA methylation class “LRR” had a higher mean profile of PVB IgG seroreactivity (Fig. 2). Despite the relationship between immunophenotype and DNA methylation class, we did not find any further association between methylation status and individual leukemia molecular characteristics (MLL rearrangement, TEL-AML1, E2A-PBX1) or high hyperdiploidy (data not shown).
Figure 2.
Relationship of serum Parvovirus B19 levels to leukemia DNA methylation class. DNA methylation class determined by recursively-partitioned mixture modeling is shown on the left—six subtypes of leukemia are shown with each subclass in proportionate size to the numbers of leukemia samples within each. The right part consists of box and whisker plots for IgG levels in each class. Controlling for immunophenotype and age, Parvovirus B19 IgG levels were associated with DNA methylation class (p = 0.02) but not IgM (p = 0.92).
Figure 3.
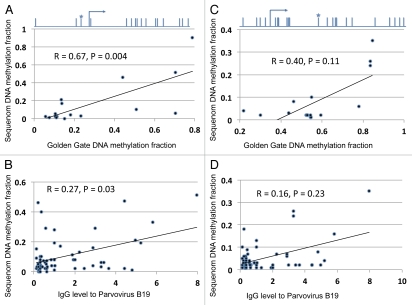
Validation/replication of DNA methylation marks in two genes using Sequenom analysis. (A and C) Samples assayed for DNA methylation status by GoldenGate and Sequenom (n = 18). Spearman correlation statistics are shown. (B and D) relation between DNA methylation and Parvovirus levels in a unique set of leukemia DNA samples than those used for GoldenGate analysis (n = 45), with Spearman correlation. The location of the CpG site assayed by both the GoldenGate and the Sequenom analysis is graphically illustrated in relation to the transcription start site for gene LTA above (A), and for LCK above the (C).
We individually analyzed the GoldenGate array CpG site β values, and found a number of sites (n = 30) associated with evidence of Parvovirus IgG seropositivity (Table S2). We chose genes for validation analysis based on their statistical significance (p < 0.05), the level of variation of β values (ranges >0.5), and successful Sequenom assay design. LCK and LTA were chosen after assessing their significant associations with IgG to PVB19, as well as having a high range of β values. We replicated the results in the same samples as used for the GoldenGate (n = 18), and an independent set of samples (n = 45) with the same immunophenotype profile as the parent sample set. The latter patients were not part of the cohort included in microarray analysis but were also serologically analyzed. The results obtained for LTA, with Golden Gate panel and Sequenom, are positively correlated (p = 0.004, Fig. 3A and B) and PVB19 IgG level is also correlated with methylation fraction as seen by microarray (R = 0.67, p = 0.03). However, the correlation between LCK results with microarray and Sequenom DNA methylation were not statistically significant (R = 0.40, p = 0.11; Fig. 3C and D).
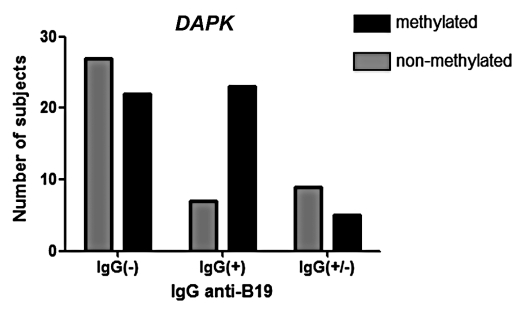
An existing methylation specific PCR assay for DAPK was also applied to these samples (n = 55 that were tested with Illumina array plus n = 45 independent samples), as this gene was more methylated with higher PVB19-IgG in the array results. We detected DAPK methylation in 52% of the total samples by MSP and a positive association was seen between methylation and the presence of PVB19 IgG antibodies (p = 0.008; Fig. 4). While DAPK was not one of the top genes on the Golden Gate array locus-by-locus results, there was a relationship between DAPK and IgG results (R = 0.2) and the qualitative, rather than quantitative, nature of the MSP reaction may be more sensitive to this difference than Sequenom, which provides a true ratio between methylated and unmethylated CpG sites.
Figure 4.
DAPK DNA methylation Methylation-specific PCR. DNA methylation is significantly more prevalent in leukemias, which came from patients with a history of Parvovirus B19 reaction. The X-axis indicates classification of leukemia patients into 3 categories based on the level of anti-PBV19 antibodies in serum as explained in Methods.
Discussion
Our data provide evidence of a potential link between history of a viral illness and a DNA methylation pattern in ALL cells. The DNA methylation pattern that was linked with higher levels of PVB19 IgG is a combination of both increased and decreased DNA methylation at specific CpGs compared with other ALLs of the same immunophenotype but different DNA methylation patterns. This observation was replicated for specific individual genes, but future replications using different populations and technologies will be necessary to further characterize and confirm this phenomenon. Kerr and colleagues25 proposed that infection with PVB19 was involved in the onset of ALL, although the virus was not in blood circulation at the time of diagnosis. These authors also showed a prominent presence of PVB19 DNA in the cerebrospinal fluid of patients with ALL when compared with with normal controls (p = 0.046).25 Heegaard et al. proposed that PVB19 infection could be the cause of cytopenic episodes occasionally preceding an ALL diagnosis, this relationship being possibly etiologic.26 Patients with weak erythropoiesis typically experience a transient aplastic crisis and anemia with PVB19 infection; viral clearance is associated with positive regulation and regrowth of the bone marrow.27 This presents a possible scenario where viral damage impacting a DNA methylation pattern is followed by enhanced proliferation and fixation of that pattern. An alternative explanation for the DNA methylation pattern associated with the virus could be the creation of a permissive environment for the growth of cells with a particular phenotype that is reflected in a common DNA methylation pattern. Either of these mechanisms could be considered a “bystander” effect, with the primary infected cells (erythroblasts) not the target cells for cancer. Precursor-B cells could be affected by a cytokine “storm” during an infection, as well as being exposed to toxic reactive oxygen species in the clearance of infection. Developing B cells have a capacity for rapid expansion and proliferation and exhibit a limited repertoire of genetic changes such as chromosomal translocations and gene deletions, which may be promoted by localized inflammation in the bone marrow following an infection.
Our result is consistent with a single prior study measuring the presence of PVB19 and DNA methylation in diagnostic samples. While “history” of PVB19 infection was not measured, Yalcin and colleagues noted a significant association between the concurrent presence of PVB19 DNA in diagnostic samples and methylation in the p15 gene promoter in adult leukemia patients.28 Future studies should incorporate measures of both current and historic infections.
The proposed role of viruses in childhood ALL can seem somewhat paradoxical. Epidemiological studies have long portrayed a protective role for childhood ALL concerning infections, as surrogate markers for infection such as early daycare seem to be protective.10 However, additional studies indicate that infections severe enough to result in a physician visit may be a risk factor.29,30 While exposure to a variety of antigens may be protective via a normal modulation and development of the immune system, a strong reaction to infections agents may induce widespread immune stimulation as well as specific anti-viral defenses that may impact leukemogenesis, in particular for a virus with bone marrow tropism like PVB19. Children who contract leukemia tend to have pre-leukemic mutations, present in many cases prior to birth.31–33 A strong reaction in the vicinity of a pre-cancerous cell may produce a characteristic immune reaction that results in the homogeneous DNA methylation pattern observed among some DNA methylation classes, in particular class “LRR” (Fig. 2) whose children harbor the highest PVB19 IgG levels. It should be noted that this methylation class does not harbor an overall “more methylated” phenotype, only a different pattern of DNA methylation compared with other childhood ALLs from this series.
Epigenetic traits (e.g., aberrant DNA methylation) are known to be related to ALL pathogenesis along with genetic variations such as translocation and altered chromosome number. There are few reports, however, analyzing methylation profile in childhood lymphocytic leukemias.34–38 Most of them analyze a small numbers of genes except for one, where the authors used a customized platform by Illumina with probes for 386 genes.35 Our current study is the only one we are aware of that shows any relation of DNA methylation in ALL to infections.
We chose genes to replicate based only on their strong associations in the GoldenGate panel, but do note that they are derived from a “Cancer Panel” and may have an impact on leukemogenesis. All three genes also have key roles in hematopoietic and immune function as well. LCK and LTA are both expressed in lymphocytes and are critical immune regulatory genes. LCK is a T-cell specific tyrosine kinase, and LTA encodes a cytokine produced by lymphocytes that is highly induced during viral infection. DAPK is a serine-threonine kinase that is a positive mediator of interferon-associated cell death. Future studies should explore a potential for mechanistic links between viral infection and DNA methylation of key immune genes with subsequent impact on leukemogenesis.
Finally, our analysis here does not implicate PVB19 as a “causative agent” in leukemogenesis nor does it satisfy Koch's postulates. We do provide evidence for a potential new role for a common virus in leukemogenesis; a virus may impact the epigenetic state of a leukemia clone in a specific manner. The findings have implications on infections impacting leukemia risk, and may impact current etiologic models of childhood leukemogenesis involving infection. Further research should examine the possible role of other infectious agents on DNA methylation patterns as well as tease out the mechanistic relationship between Parvovirus B19 and DNA methylation in leukemia cells. Such future investigations should also investigate the role of PVB19 infection on the DNA methylation patterns of normal cells, sorted into homogenous populations, including both the direct targets (erythroblasts) and bystander cells.
Materials and Methods
Patient samples.
Subjects were identified from 2001 to 2008 from Brazilian regions through population-based cancer registries, as described in reference 19. Peripheral blood (PB) and bone marrow (BM) aspirates were collected for diagnostic purposes (pre-treatment). Patients were classified according to standard immunophenotype and molecular techniques.20,21 The Ethics and Scientific Committees of Instituto Nacional de Câncer approved the study and children's parents provided written informed consent (CEP#070/07). The main clinical-pathological features of the ALL series including the distribution of precursor B ALL subtypes and cytogenetic/molecular abnormalities are shown in Table 1. There were a predominance of males and the age range of 0.08–14 y-old, median 2.5 y at the diagnosis of ALL. The distribution of precursor B-ALL demonstrated that common ALL (c-ALL, CD10+, CD19+) were the majority of the random selected B-cell cases.
Samples used in this study included patient serum for serology, and pre-treatment leukemia blast cells for DNA methylation analysis. Bone marrows were subjected to mononuclear cell purification through Ficoll gradient. All patients were tested for PVB19 seroreactivity. Fifty-nine subjects were tested on the Illumina DNA methylation array; 45 additional subjects were utilized for DNA methylation replication by Sequenom and methylation-specific PCR.
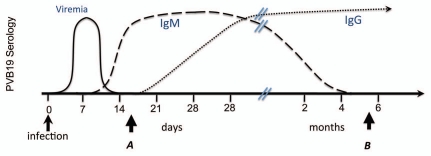
Anti-PVB19 IgM and IgG enzyme immunoassay.
Plasma anti-PVB19 antibodies (IgM and IgG) were detected by ELISA, according to the commercial manufacturer's instructions (Biotrin). For each plasma sample two separate assays were performed, one for a recent or current infection (IgM), and another as a marker of past history of infection (IgG) with PVB19 (Fig. 1). The presence or absence of anti-PVB19 antibodies for clinical purposes is determined in relation to a calculated Cut-Off Value (COV). Samples with a mean absorbance reading greater than the COV X 1.1 are considered reactive (positive) for anti-PVB19 antibodies. Samples with a mean absorbance reading less than the COV X 0.9 are considered non-reactive (negative) for anti-PVB19 Igs. Samples with a mean absorbance reading greater than or equal to the COV X 0.9 and less than or equal to the COV X 1.1 are equivocal. Mean absorbance was also used as a continuous variable in statistical analyses.
Figure 1.
History of infection as measured by antibody titers. A recent, non-etiologic infection will be characterized by an IgM reaction with the absence of IgG (time point A). An infection in an individual's past that might impact leukemogenesis will be characterized by an IgG reaction with the absence of IgM (time point B).
Methylation analysis.
DNA was extracted from purified bone marrow blast cells using the QIAamp DNA mini kit according to the manufacturer's protocol (Qiagen). DNA was modified by sodium bisulfite to convert unmethylated cytosines to uracil using the EZ DNA Methylation kit (Zymo Research) according to the manufacturer's protocol.
Illumina GoldenGate methylation bead arrays were used to simultaneously interrogate 1,505 CpG loci associated with 803 cancer-related genes. The Illumina array interrogates approximately two CpGs per gene, and although sequencing methods would provide additional details, CpGs were cultivated from reports that have shown the methylation-expression relationship in large part through sequencing experiments. Bead arrays have a similar sensitivity as quantitative methylation-specific PCR and were run at the University of California at San Francisco Institute for Human Genetics, Genomics Core Facility, according to the manufacturer's protocol and as described in reference 22.
Methylation specific PCR (MSP) and sequenom.
MSP was performed using two pairs of primers for DAPK gene, one for methylated sequence and other to unmethylated sequences. Primers (Table S1) were designed using MethyOligonucleotide Express software based on published sequences from the NCBI. Polymerase chain reactions were set up in a total volume of 20 µl, including 1x PCR buffer with magnesium, 200 µM of each DNTP, 1 U of Taq Gold enzyme, 25 pmoles of each primer and 5 ng of bisulfite converted DNA. After an initial denaturation step for 10 min at 95°C, each cycle of a total of 30, consisted of denaturation for 30 sec at 94°C, annealing for 30 sec at 58°C and primer extension for 50 sec at 72°C and a final step for 7 min at 72°C.
Sequenom reactions for replication/validation were designed to include the specific CpG site associated with PVB19 levels in the GoldenGate analysis. Assays were designed using Sequenom Epityper software (primers, Table S1). PCR assays were performed with Qiagen Hot-Start polymerase (Qiagen), and analyzed on a Sequenom MassArray using manufacturer's protocols. All DNA methylation assays were performed on randomized, coded samples by laboratory personnel who were blinded to PVB19 serological results.
Statistical analysis.
BeadStudio Methylation software from the array manufacturer Illumina was used for data set assembly. All array data points are represented by fluorescent signals from both methylated (M) and unmethylated (U) alleles, and methylation level is given by β = [max (M, 0)]/[|U| + |M| + 100], the average methylation (β) value is derived from the ∼30 replicate methylation measurements and a Cy3/Cy5 methylated/unmethylated ratio. At each locus for each sample, the detection p value was used to determine sample performance, no samples had >25% of loci having with a detection p value of >1e−5 and so all passed performance quality criteria. CpG loci with a median detection p value of >0.05 (n = 8, 0.5%), were eliminated from analysis. Subsequent analyses were performed using the R software (R Development CT, 2007). Associations between sample type or covariates such as age or gender and methylation at individual CpG loci were tested with a generalized linear model. The β distribution of average β values was accounted for with a quasi-binomial logit link with an estimated scale variable constraining the mean between 0 and 1, in a manner similar to that described by Hsuing and colleagues.23 Array-wide tests for association between CpG loci and covariates of interest used false discovery rate correction and Q values computed by the q value package in R (R Development CT). For inference, data were clustered using a recursively-partitioned mixture modeling (RPMM) as described previously in reference 24. RPMM is freely available as an R package. Permutation tests (running 10,000 permutations) were used to test for association with methylation class by generating a distribution of the test statistic for the null distribution for comparison to the observed distribution. For continuous variables, the permutation test was run with the statistic Kruskal-Wallis test. Significant associations from permutation tests were controlled for potential confounders where appropriate using logistic regression with methylation classes and potential confounders and a likelihood ratio test of the model with and without methylation classes.
Acknowledgments
Conceived and designed the experiments: G.M.V., M.P.O., J.L.W. Performed the experiments: G.M.V., J.X., M.P.O., J.L.W. Analyzed the data: G.M.V., B.C.C., E.A.H., M.P.O., J.L.W. Contributed to the analysis and interpretation of data: J.X., J.K.W., S.Z., M.R.K., H.H.N., M.R.W., K.T.K. Managed subject recruitment and diagnostic analysis: M.P.O. with G.M.V. Authored the manuscript: G.M.V., M.P.O., J.L.W. Read, edited and approved the manuscript: all authors. Funded the study: M.P.O., J.L.W.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Grant Support
G.M.V. was supported by CAPES no. BEX2663/07-4, Brazil. Also supported by grants from the Children with Leukaemia Foundation, the Leukemia and Lymphoma Society 6026-10, the Tobacco-Related Disease Research Program of California, and R01CA109745 and P01ES018172 (J.L.W.), and the Swiss Bridge Foundation and CNPq-Brazil (M.P.O.).
Supplementary Material
References
- 1.Kinlen L. Evidence for an infective cause of childhood leukaemia: comparison of a Scottish new town with nuclear reprocessing sites in Britain. Lancet. 1988;2:1323–1327. doi: 10.1016/S0140-6736(88)90867-7. [DOI] [PubMed] [Google Scholar]
- 2.Greaves MF. Speculations on the cause of childhood acute lymphoblastic leukemia. Leukemia. 1988;2:120–125. [PubMed] [Google Scholar]
- 3.Greaves M. Infection, immune responses and the aetiology of childhood leukaemia. Nat Rev Cancer. 2006;6:193–203. doi: 10.1038/nrc1816. [DOI] [PubMed] [Google Scholar]
- 4.Bender AP, Robison LL, Kashmiri SV, McClain KL, Woods WG, Smithson WA, et al. No involvement of bovine leukemia virus in childhood acute lymphoblastic leukemia and non-Hodgkin's lymphoma. Cancer Res. 1988;48:2919–2922. [PubMed] [Google Scholar]
- 5.MacKenzie J, Gallagher A, Clayton RA, Perry J, Eden OB, Ford AM, et al. Screening for herpesvirus genomes in common acute lymphoblastic leukemia. Leukemia. 2001;15:415–421. doi: 10.1038/sj.leu.2402049. [DOI] [PubMed] [Google Scholar]
- 6.MacKenzie J, Perry J, Ford AM, Jarrett RF, Greaves M. JC and BK virus sequences are not detectable in leukaemic samples from children with common acute lymphoblastic leukaemia. Br J Cancer. 1999;81:898–899. doi: 10.1038/sj.bjc.6690783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Priftakis P, Dalianis T, Carstensen J, Samuelsson U, Lewensohn-Fuchs I, Bogdanovic G, et al. Human polyomavirus DNA is not detected in Guthrie cards (dried blood spots) from children who developed acute lymphoblastic leukemia. Med Pediatr Oncol. 2003;40:219–223. doi: 10.1002/mpo.10246. [DOI] [PubMed] [Google Scholar]
- 8.Gustafsson B, Huang W, Bogdanovic G, Gauffin F, Nordgren A, Talekar G, et al. Adenovirus DNA is detected at increased frequency in Guthrie cards from children who develop acute lymphoblastic leukaemia. Br J Cancer. 2007;97:992–994. doi: 10.1038/sj.bjc.6603983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vasconcelos GM, Kang M, Pombo-de-Oliveira MS, Schiffman JD, Lorey F, Buffler P, et al. Adenovirus detection in Guthrie cards from paediatric leukaemia cases and controls. Br J Cancer. 2008;99:1668–1672. doi: 10.1038/sj.bjc.6604714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Urayama KY, Buffler PA, Gallagher ER, Ayoob JM, Ma X. A meta-analysis of the association between day-care attendance and childhood acute lymphoblastic leukaemia. Int J Epidemiol. 2010;39:718–732. doi: 10.1093/ije/dyp378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McNally RJ, Eden TO. An infectious aetiology for childhood acute leukaemia: a review of the evidence. Br J Haematol. 2004;127:243–263. doi: 10.1111/j.1365-2141.2004.05166.x. [DOI] [PubMed] [Google Scholar]
- 12.de Souza Reis RSr, de Camargo B, de Oliveira Santos M, de Oliveira JM, Azevedo Silva F, Pombo-de-Oliveira MS. Childhood leukemia incidence in Brazil according to different geographical regions. Pediatr Blood Cancer. 2011;56:58–64. doi: 10.1002/pbc.22736. [DOI] [PubMed] [Google Scholar]
- 13.Fernandez AF, Esteller M. Viral epigenomes in human tumorigenesis. Oncogene. 2010;29:1405–1420. doi: 10.1038/onc.2009.517. [DOI] [PubMed] [Google Scholar]
- 14.Nascimento JP, Buckley MM, Brown KE, Cohen BJ. The prevalence of antibody to human parvovirus B19 in Rio de Janeiro, Brazil. Rev Inst Med Trop Sao Paulo. 1990;32:41–45. doi: 10.1590/S0036-46651990000100007. [DOI] [PubMed] [Google Scholar]
- 15.Heegaard ED, Madsen HO, Schmiegelow K. Transient pancytopenia preceding acute lymphoblastic leukaemia (pre-ALL) precipitated by parvovirus B19. Br J Haematol. 2001;114:810–813. doi: 10.1046/j.1365-2141.2001.03021.x. [DOI] [PubMed] [Google Scholar]
- 16.Lee SM, Kim DG, Bang D. Persistent erythema infectiosum-like rash as a prodrome of acute lymphocytic leukemia. Pediatr Dermatol. 1994;11:156–159. doi: 10.1111/j.1525-470.1994.tb00571.x. [DOI] [PubMed] [Google Scholar]
- 17.Petrella T, Bailly F, Mugneret F, Caillot D, Chavanet P, Guy H, et al. Bone marrow necrosis and human parvovirus associated infection preceding an Ph1+ acute lymphoblastic leukemia. Leuk Lymphoma. 1992;8:415–419. doi: 10.3109/10428199209051023. [DOI] [PubMed] [Google Scholar]
- 18.Sava an S, Ozdemir O. Parvovirus B19 infection and acute lymphoblastic leukaemia. Br J Haematol. 2003;120:168–169. doi: 10.1046/j.1365-2141.2003.03983_3.x. [DOI] [PubMed] [Google Scholar]
- 19.de Camargo B, de Oliveira Santos M, Rebelo MS, de Souza Reis R, Ferman S, Noronha CP, et al. Cancer incidence among children and adolescents in Brazil: first report of 14 population-based cancer registries. Int J Cancer. 2010;126:715–720. doi: 10.1002/ijc.24799. [DOI] [PubMed] [Google Scholar]
- 20.Bene MC, Castoldi G, Knapp W, Ludwig WD, Matutes E, Orfao A, et al. Proposals for the immunological classification of acute leukemias. European Group for the Immunological Characterization of Leukemias (EGIL) Leukemia. 1995;9:1783–1786. [PubMed] [Google Scholar]
- 21.van Dongen JJ, Macintyre EA, Gabert JA, Delabesse E, Rossi V, Saglio G, et al. Standardized RT-PCR analysis of fusion gene transcripts from chromosome aberrations in acute leukemia for detection of minimal residual disease. Report of the BIOMED-1 Concerted Action: investigation of minimal residual disease in acute leukemia. Leukemia. 1999;13:1901–1928. doi: 10.1038/sj.leu.2401592. [DOI] [PubMed] [Google Scholar]
- 22.Bibikova M, Lin Z, Zhou L, Chudin E, Garcia EW, Wu B, et al. High-throughput DNA methylation profiling using universal bead arrays. Genome Res. 2006;16:383–393. doi: 10.1101/gr.4410706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hsiung DT, Marsit CJ, Houseman EA, Eddy K, Furniss CS, McClean MD, et al. Global DNA methylation level in whole blood as a biomarker in head and neck squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev. 2007;16:108–114. doi: 10.1158/1055-9965.EPI-06-0636. [DOI] [PubMed] [Google Scholar]
- 24.Houseman EA, Christensen BC, Yeh RF, Marsit CJ, Karagas MR, Wrensch M, et al. Model-based clustering of DNA methylation array data: a recursive-partitioning algorithm for high-dimensional data arising as a mixture of beta distributions. BMC Bioinformatics. 2008;9:365. doi: 10.1186/1471-2105-9-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kerr JR, Barah F, Cunniffe VS, Smith J, Vallely PJ, Will AM, et al. Association of acute parvovirus B19 infection with new onset of acute lymphoblastic and myeloblastic leukaemia. J Clin Pathol. 2003;56:873–875. doi: 10.1136/jcp.56.11.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heegaard ED, Jensen L, Hornsleth A, Schmiegelow K. The role of parvovirus B19 infection in childhood acute lymphoblastic leukemia. Pediatr Hematol Oncol. 1999;16:329–334. doi: 10.1080/088800199277155. [DOI] [PubMed] [Google Scholar]
- 27.Brown KE, Hibbs JR, Gallinella G, Anderson SM, Lehman ED, McCarthy P, et al. Resistance to parvovirus B19 infection due to lack of virus receptor (erythrocyte P antigen) N Engl J Med. 1994;330:1192–1196. doi: 10.1056/NEJM199404283301704. [DOI] [PubMed] [Google Scholar]
- 28.Yalcin A, Serin MS, Emekdas G, Tiftik N, Aslan G, Eskandari G, et al. Promoter methylation of P15(INK4B) gene is possibly associated with parvovirus B19 infection in adult acute leukemias. Int J Lab Hematol. 2009;31:407–419. doi: 10.1111/j.1751-553X.2008.01052.x. [DOI] [PubMed] [Google Scholar]
- 29.Roman E, Simpson J, Ansell P, Kinsey S, Mitchell CD, McKinney PA, et al. Childhood acute lymphoblastic leukemia and infections in the first year of life: a report from the United Kingdom Childhood Cancer Study. Am J Epidemiol. 2007;165:496–504. doi: 10.1093/aje/kwk039. [DOI] [PubMed] [Google Scholar]
- 30.Chan LC, Lam TH, Li CK, Lau YL, Yuen HL, Lee CW, et al. Is the timing of exposure to infection a major determinant of acute lymphoblastic leukaemia in Hong Kong? Paediatr Perinat Epidemiol. 2002;16:154–165. doi: 10.1046/j.1365-3016.2002.00406.x. [DOI] [PubMed] [Google Scholar]
- 31.McHale CM, Wiemels JL, Zhang L, Ma X, Buffler PA, Feusner J, et al. Prenatal origin of childhood acute myeloid leukemias harboring chromosomal rearrangements t(15;17) and inv(16) Blood. 2003;101:4640–4641. doi: 10.1182/blood-2003-01-0313. [DOI] [PubMed] [Google Scholar]
- 32.McHale CM, Wiemels JL, Zhang L, Ma X, Buffler PA, Guo W, et al. Prenatal origin of TEL-AML1-positive acute lymphoblastic leukemia in children born in California. Genes Chromosomes Cancer. 2003;37:36–43. doi: 10.1002/gcc.10199. [DOI] [PubMed] [Google Scholar]
- 33.Wiemels J, Kang M, Greaves M. Backtracking of leukemic clones to birth. Methods Mol Biol. 2009;538:7–27. doi: 10.1007/978-1-59745-418-6_2. [DOI] [PubMed] [Google Scholar]
- 34.Nakamura M, Sugita K, Inukai T, Goi K, Iijima K, Tezuka T, et al. p16/MTS1/INK4A gene is frequently inactivated by hypermethylation in childhood acute lymphoblastic leukemia with 11q23 translocation. Leukemia. 1999;13:884–890. doi: 10.1038/sj.leu.2401437. [DOI] [PubMed] [Google Scholar]
- 35.Milani L, Lundmark A, Kiialainen A, Nordlund J, Flaegstad T, Forestier E, et al. DNA methylation for subtype classification and prediction of treatment outcome in patients with childhood acute lymphoblastic leukemia. Blood. 2010;115:1214–1225. doi: 10.1182/blood-2009-04-214668. [DOI] [PubMed] [Google Scholar]
- 36.Davidsson J, Lilljebjorn H, Andersson A, Veerla S, Heldrup J, Behrendtz M, et al. The DNA methylome of pediatric acute lymphoblastic leukemia. Hum Mol Genet. 2009;18:4054–4065. doi: 10.1093/hmg/ddp354. [DOI] [PubMed] [Google Scholar]
- 37.Dunwell T, Hesson L, Rauch TA, Wang L, Clark RE, Dallol A, et al. A genome-wide screen identifies frequently methylated genes in haematological and epithelial cancers. Mol Cancer. 2010;9:44. doi: 10.1186/1476-4598-9-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weeks RJ, Kees UR, Song S, Morison IM. Silencing of TESTIN by dense biallelic promoter methylation is the most common molecular event in childhood acute lymphoblastic leukaemia. Mol Cancer. 2010;9:163. doi: 10.1186/1476-4598-9-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.