Background: The Clostridium difficile cell wall protein CwpV is post-translationally processed by an unknown mechanism.
Results: CwpV undergoes enzyme-independent intramolecular cleavage most likely through Thr-413-mediated N-O acyl shift whereas neighboring residues appear to be important in maintaining a precise backbone conformation.
Conclusion: Intramolecular cleavage of CwpV is independent of enzymes and cofactors.
Significance: Our results provide insight into protein autoprocessing in diverse biological systems.
Keywords: Bacteria, Bacterial Pathogenesis, Cell Surface Protein, Protein Processing, Site-directed Mutagenesis, Autoproteolysis, Clostridium difficile
Abstract
Clostridium difficile infection is a leading cause of antibiotic-associated diarrhea, placing considerable economic pressure on healthcare systems and resulting in significant morbidity and mortality. The pathogen produces a proteinaceous array on its cell surface known as the S-layer, consisting primarily of the major S-layer protein SlpA and a family of SlpA homologs. CwpV is the largest member of this family and is expressed in a phase-variable manner. The protein is post-translationally processed into two fragments that form a noncovalent, heterodimeric complex. To date, no specific proteases capable of cleaving CwpV have been identified. Using site-directed mutagenesis we show that CwpV undergoes intramolecular autoproteolysis, most likely facilitated by a N-O acyl shift, with Thr-413 acting as the source of a nucleophile driving this rearrangement. We demonstrate that neighboring residues are also important for correct processing of CwpV. Based on protein structural predictions and analogy to the glycosylasparaginase family of proteins, it appears likely that these residues play key roles in determining the correct protein fold and interact directly with Thr-413 to promote nucleophilic attack. Furthermore, using a cell-free protein synthesis assay we show that CwpV maturation requires neither cofactors nor auxiliary enzymes.
Introduction
Clostridium difficile is a Gram-positive, spore-forming anaerobe and a leading cause of antibiotic-associated diarrhea (1). C. difficile infection typically occurs among hospitalized patients, whose natural intestinal microflora has been disrupted by prolonged treatment with broad spectrum antibiotics, allowing the pathogen to colonize the compromised gastrointestinal tract. Strains causing disease produce one or two related toxins, TcdA and TcdB, which modulate the activity of host cell Rho GTPases, destroying the integrity of the epithelial cell barrier and inducing a variety of effects on intestinal cells (2). Although both toxins have been studied extensively in recent years, and their contribution to C. difficile pathogenicity has been well characterized, relatively little is known about the early stages of infection that are critical for colonization of the human gut.
Factors expressed on the bacterial cell surface are likely to contribute to host colonization via interactions with host tissue, the immune system, and other bacterial cells. C. difficile produces a surface layer (S-layer),2 composed of the high molecular weight S-layer protein (SLP) and the low molecular weight SLP (3). These SLPs are produced from a common precursor SlpA (3) via post-translational cleavage by a specific, surface layer-associated cysteine protease Cwp84 (4, 5). Together, they form a heterodimeric complex that assembles into a two-dimensional array completely surrounding the bacterial cell (6). 28 SLP paralogs have been identified in C. difficile forming a cell wall protein (CWP) family (7). Each paralog contains a conserved cell wall-anchoring domain, and many contain a second unique domain that may specify a functional property.
In C. difficile 630 (7), CwpV is the largest member of the CWP family. The protein is surface expressed in a phase variable manner (8) and is post-translationally processed into two fragments that reassociate to form a stable, noncovalently associated complex (9). Mature CwpV therefore consists of two distinct domains: the N-terminal domain with cell wall anchoring activity and the C-terminal domain consisting of nine repeats of 120 amino acids each. In the precursor protein, these domains are separated by a region containing the cleavage site and putative interaction domains, terminating in a serine-glycine-rich flexible linker (see Fig. 1A). Although we have shown that cleavage occurs at a defined site upstream of the serine-glycine-rich region (9), no specific protease capable of cleaving CwpV has been identified. Furthermore, cleavage is unaffected by protease inhibitors.3 If an enzyme were involved in CwpV cleavage, oversaturation of the processing pathway would be expected to lead to accumulation of full-length CwpV. Yet, full-length CwpV is never observed within the S-layer, even when CwpV is overexpressed. Together, these observations suggest that CwpV cleavage is enzyme-independent and that the protein might undergo intramolecular autoprocessing. Given that a threonine residue is located directly downstream of the cleavage site, forming the N terminus of the C-terminal domain, a plausible mechanism of cleavage is a N-O acyl migration with the oxygen of the hydroxyl group of threonine acting as the nucleophile and attacking the α-carbonyl carbon of the preceding amino acid to generate a hydroxazolidine intermediate (10). This autoprocessing mechanism was first identified in bacterial glycosylasparaginase which belongs to a family of N-terminal nucleophile hydrolases that share similar three-dimensional structures and whose activity depends on an N-terminal serine, threonine, or cysteine residue that is generated by the autocleavage of a precursor protein (11).
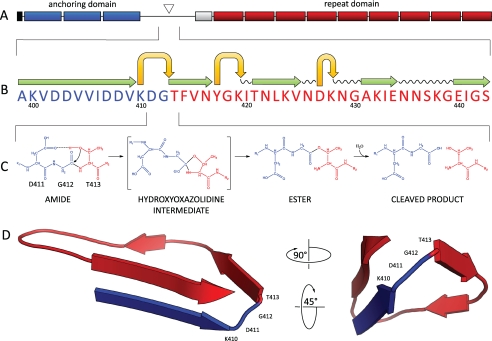
FIGURE 1.
A, domain structure of CwpV. The N terminus of CwpV contains a signal sequence (black), followed by three PF04122 cell wall anchoring motifs (blue), and a region containing the cleavage site (triangle). A serine-glycine rich region (white) precedes a number of repeats (red). B, predicted secondary structure of the region spanning the CwpV cleavage site generated using Phyre. A motif consisting of stacked β-strands (green) and tight turns (yellow) can be seen within the cleavage site, which has been previously identified and confirmed in this study by N-terminal sequencing. C, proposed mechanism of CwpV intramolecular autoprocessing. The reactive hydroxyl of Thr-413 is deprotonated by the carboxylate group of Asp-411 and launches a nucleophilic attack on the α-carbonyl carbon of Gly-412 to form a hydroxazolidine intermediate. After a proton is transferred to the leaving amino group of Thr-413, the α-carbonyl of Gly-412 is shifted to the hydroxyl of Thr-413, leading to the ester intermediate via an N-O shift. The ester is then hydrolyzed to form two peptides, the N-terminal anchoring domain and the C-terminal repeat domain. D, three-dimensional model of CwpV cleavage site (106–135 amino acids). The tertiary structure of CwpV cleavage region was predicted using Phyre with an estimated precision of 80%. A tight turn can be observed directly upstream of Thr-413 and consisting of Gly-412, Asp-411, and Lys-410.
In this study we use site-directed mutagenesis to investigate the post-translational processing of CwpV. We show that CwpV undergoes intramolecular autoproteolysis that requires neither cofactors nor auxiliary enzymes and that this reaction is most likely facilitated by a N-O acyl rearrangement with the oxygen of the hydroxyl group of Thr-413 acting as the nucleophile. Structural predictions of CwpV using the Protein homology analogy research engine (Phyre) suggest that this is a plausible mechanism, and our data suggest that the structural context of Thr-413 is important for cleavage. To our knowledge, this is the first reported case of such a protein maturation mechanism in a Gram-positive bacterium.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Growth Conditions
C. difficile was routinely cultured on blood-agar base II (Oxoid) supplemented with 7% horse blood (TCS Biosciences), brain-heart infusion agar or in brain-heart infusion broth (Oxoid). Cultures were grown in an anaerobic cabinet (Don Whitley Scientific) at 37 °C in an atmosphere of 10% CO2, 10% H2, and 80% N2. C. difficile strains containing recombinant plasmids (supplemental Table S1) were grown under thiamphenicol selection. Commercial, chemically competent NovaBlue Escherichia coli cells (Merck) were used for cloning and recombinant plasmid maintenance. E. coli strains were routinely grown at 37 °C on LB agar plates or in LB liquid culture in the presence of selective antibiotics where appropriate.
DNA Manipulations
DNA manipulations were carried out according to standard techniques. C. difficile genomic DNA for use in cloning and PCR analysis was prepared as described previously (8). PCRs used KOD (Merck), in accordance with the manufacturer's protocols using primers detailed in supplemental Table S2.
Conjugation
Plasmids were transformed into E. coli CA434 and then conjugated into C. difficile as described previously (12) using thiamphenicol (15 μg/ml) to select for pMTL960-based plasmids and cycloserine (250 μg/ml) to counterselect against E. coli.
S-layer Extraction, SDS-PAGE, and Western Blotting
C. difficile cultures were grown overnight and harvested by centrifugation, and S-layer extracts were prepared using low pH glycine incubation as described previously (3). Proteins in the S-layer extracts were subjected to SDS-PAGE and Western blotting according to standard protocols. Both rabbit primary anti-CwpV antibodies (anti-CwpV N-term and anti-CwpV rpt1) were used at 1/5,000 dilution, followed by anti-rabbit-HRP (Dako Cytomation) at 1/2,000 dilution. Anti-Strep-tag II antibody (Merck) was used at 1/1,000 dilution, followed by anti-mouse-HRP conjugate at 1/1,000 dilution. Signal was detected using SuperSignal West Pico Chemiluminescent Substrate (Pierce).
In Vitro Protein Synthesis
CwpV was synthesized in vitro using the PURExpress in vitro protein synthesis kit (NEB) in accordance with the manufacturer's protocols. 250 ng of plasmid DNA was used per reaction. Protein synthesis was carried out at 37 °C for 4 h.
StrepTactin Binding/Elution Assay
Strep-tagged CwpV fragments were purified on StrepTactin (Merck) in accordance with the manufacturer's protocols. Briefly, PURExpress reactions were diluted with wash buffer (150 mm NaCl, 100 mm Tris-HCl, 1 mm EDTA, pH 8.0) to 100 μl and mixed with pre-equilibrated resin (50-μl bed volume). The mixture was incubated overnight at 4 °C with rotation and then applied to a column. After washing with 5 volumes of wash buffer, the Strep-tagged proteins were eluted in 1 volume of elution buffer (150 mm NaCl, 100 mm Tris-HCl, 1 mm EDTA, 2.5 mm desthiobiotin, pH 8.0) and concentrated using a Microcon spin column (10 kDa; Millipore). Purified proteins were analyzed by SDS-PAGE and Western blotting.
N-terminal Sequencing
Purified CwpV fragments were separated on 12% SDS-polyacrylamide gels and transferred to a PVDF membrane. The membrane was washed in water for 10 min and stained in 0.1% Coomassie Blue in 50% MeOH for 5 min. After destaining in 50% MeOH, 10% acetic acid (3 × 5 min with rocking) and washing in water (3 × 5 min with rocking) the membrane was allowed to dry thoroughly. N-terminal sequencing by Edman degradation was carried out at the PNAC Facility, Department of Biochemistry, University of Cambridge.
Structural Predictions
Phyre structure predictions were conducted on a 142-amino acid sequence containing the CwpV cleavage site (residues 350–492). Models were rendered using PyMOL version 1.3 (13).
RESULTS
Hydroxyl Group of Thr-413 Is Involved in CwpV Cleavage
Autoproteolysis of bacterial glycosylasparaginase occurs adjacent to a threonine residue located in a highly conserved region. It has been shown previously that replacement of threonine with amino acids other than serine or cysteine prevents autoproteolysis, whereas replacement with cysteine or serine reduces its rate (14). In CwpV, a threonine residue (Thr-413) is located directly downstream of the cleavage site (see Fig. 1B). To test the hypothesis that Thr-413 is involved in the post-transcriptional processing of CwpV, site-directed mutagenesis was used to replace Thr-413 with serine, cysteine, alanine or valine. Mutations were created using inverse PCR in plasmid pCwpV carrying a wild-type copy of cwpV under the control of a constitutive promoter. The resulting plasmids were introduced by conjugation into C. difficile 630 Δerm ΔcwpV, a CwpV knock-out strain (designated ΔcwpV). Expression of CwpV in S-layer extracts from wild-type C. difficile 630 and ΔcwpV strains carrying plasmids pCwpV, CwpVT413S, pCwpVT413C, pCwpVT413A, and pCwpVT413V was analyzed by SDS-PAGE and Western blotting.
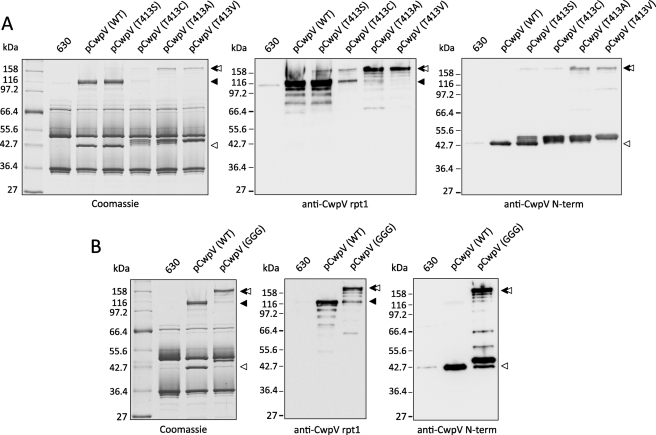
In 630 and ΔcwpV (pCwpV) extracts the two cleavage products of CwpV were seen: the ∼42-kDa N-terminal anchoring domain and the ∼116-kDa C-terminal repeat domain (Fig. 2A). Due to the phase variable expression of cwpV, the overall amount of CwpV in the 630 S-layer was low compared with the CwpV-overexpressing strain containing pCwpV. In all Thr-413 mutants a product of ∼158 kDa, corresponding to full-length protein and recognized by both the anti-CwpV N-term (detects the ∼42-kDa anchoring domain) and anti-CwpV rpt1 (detects the ∼116-kDa repeat domain) antibodies, could be seen. The amount of the ∼158-kDa product was higher in strains carrying nonconservative mutations (T413A and T413V) than those carrying conservative mutations (T413S and T413C), indicating that the nucleophile-carrying residues of serine and cysteine can partially mimic the activity of Thr-413 in CwpV cleavage. This is in agreement with the observations of Guan et al. (14) in their study of bacterial glycosylasparaginase autoprocessing. The ∼116-kDa repeat domain was present at similar levels in the ΔcwpV (pCwpVT413S) and ΔcwpV (pCwpV) strains. In contrast, its level in the ΔcwpV (pCwpVT413C) S-layer was significantly lower while being totally absent in the S-layer from ΔcwpV (pCwpVT413A) and ΔcwpV (pCwpVT413V) strains. This shows that although the conservative mutations disrupt and inhibit CwpV processing allowing the full-length precursor to be detected, they do not abolish it completely as the nonconservative mutations do.
FIGURE 2.
A, hydroxyl group of Thr-413 is involved in CwpV processing. S-layer extracts were prepared from overnight cultures of C. difficile 630, ΔcwpV (pCwpV (WT)), ΔcwpV (pCwpVT413S), ΔcwpV (pCwpVT413C), ΔcwpV (pCwpVT413A), and ΔcwpV (pCwpVT413V). Samples were separated on 10% SDS-polyacrylamide gels and analyzed via Coomassie Blue staining and Western blotting. Full-length CwpV was detected in all Thr-413 mutants. Nonconservative mutations T413A and T413V abolished cleavage of the precursor protein whereas conservative mutations T413S and T413C significantly reduced its rate. B, Lys-410 and Asp-411 play key roles in promoting CwpV autocleavage. S-layer extracts were prepared from overnight cultures of C. difficile 630, ΔcwpV (pCwpV (WT)), and ΔcwpV (pCwpVK410G/D411G). Samples were separated on 10% SDS-polyacrylamide gels and analyzed via Coomassie Blue staining and Western blotting. Full-length CwpV was detected in CwpVK410G/D411G. ◂, repeat domain; ◁, anchoring domain.
Interestingly, a slight increase in the apparent mass of the ∼42-kDa anchoring domain was also observed (Fig. 2A), with multiple bands corresponding to protein species of varying sizes being detected across all Thr-413 mutants. This was most apparent in T413A and T413V strains and was likely the result of proteolytic degradation of the full-length precursor protein. The resulting disruption of interaction with the cell wall anchoring domain would explain why the anchoring domain but not the repeat domain could be detected within the S-layer of these mutants. This was further suggested by analysis of culture supernatants revealing significant amounts of the repeat domain shed from the cells (supplemental Fig. S1). A small amount of the ∼158-kDa precursor could also be detected, indicating that the full-length protein is not incorporated into the S-layer as efficiently as its processed variant (supplemental Fig. S1).
Lys-410 and Asp-411 Play Key Roles in Promoting CwpV Autocleavage
N-O acyl rearrangements require extreme chemical conditions, unless catalyzed by the tertiary structure of the protein. It has been reported previously that a strained backbone conformation at the scissile peptide bond is a key structural feature critical for driving autoproteolysis through an N-O acyl shift (15–17). Qian et al. (18) have shown that an aspartic acid residue located directly upstream of the bacterial glycosylasparaginase cleavage site is responsible for maintaining the distorted backbone conformation by forming an unusually tight turn, allowing the carboxylate oxygen to deprotonate the hydroxyl oxygen of threonine, enhancing its nucleophility. Thus, aspartic acid plays a dual role in glycosylasparaginase autoproteolysis, acting as the general base to activate the nucleophile and holding the distorted backbone conformation that is critical for initiating an N-O acyl rearrangement.
Because no crystal structures of CwpV were available at the time of this study, we used Phyre (19) to predict the tertiary structure of the protein. This method relies on the observation that the structure of a protein is more conserved in evolution than its amino acid sequence, and therefore a protein sequence of interest can be modeled with reasonable accuracy on a very distantly related sequence of known structure, provided that the relationship between target and template can be discerned through sequence alignment. Our analysis revealed a predicted motif consisting of stacked β-sheets spanning the cleavage site (residues 372–425) (Fig. 1B) Furthermore, a small, tight turn consisting of three amino acids, one of which is an aspartic acid residue (Lys-410, Asp-411, Gly-412) was predicted to be located directly upstream of the cleavage site (Fig. 1D). In an attempt to reduce the strain within the cleavage region and elucidate the role of Lys-410 and Asp-411 in CwpV processing, both amino acids were replaced with glycine, resulting in the sequence G410G411G412 (GGG). These mutations were constructed in pCwpV and introduced into C. difficile ΔcwpV. Expression of CwpV in S-layer extracts from strains 630, ΔcwpV (pCwpV) and ΔcwpV (pCwpVK410G/D411G) was analyzed by SDS-PAGE and Western blotting.
The K410G/D411G mutation significantly reduced the efficiency of CwpV cleavage. As shown in Fig. 2B, a product of ∼158 kDa, corresponding to full-length protein and recognized by both the anti-CwpV N-term and anti-CwpV rpt1 antibodies, was detected in ΔcwpV (pCwpVK410G/D411G). This is likely due to a loss of the critical base required to activate the nucleophile Thr-413 and/or relaxation of the scissile peptide bond and thus loss of the precise backbone conformation necessary to initiate a nucleophilic attack and a N-O acyl rearrangement. Further structural analysis, beyond the scope of this study, would be necessary to gain mechanistic insight into how these mutations affect CwpV autocleavage.
CwpV Cleavage Is Enzyme-independent
Heterologous gene expression is a standard way of “uncoupling” the protein from the processing pathways of the host organism. To investigate further the possibility of CwpV autoprocessing, a Strep-tagged fragment of cwpV including the entire N-terminal anchoring domain and part of the C-terminal domain (including one repeat) was cloned into pET28a, forming pCwpVfr, and expressed in E. coli BL21 (DE3). Whole cell lysates were prepared and analyzed by SDS-PAGE and Western blotting. Extensive degradation of the protein of interest was observed (data not shown). In an attempt to reduce the effect of endogenous proteases, the PURExpress in vitro protein synthesis kit (NEB) was used. This cell-free transcription/translation system is free from exonucleases, RNases, and proteases, allowing for synthesis of undegraded proteins. Using pCwpVfr as a template, the CwpV derivative was synthesized, purified by affinity chromatography on StrepTactin resin, and analyzed by SDS-PAGE and Western blotting.
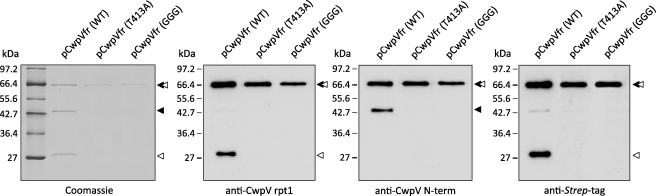
Three products were identified: a ∼62-kDa protein corresponding to the full-length CwpV fragment, the ∼42-kDa anchoring domain, and a ∼28-kDa fragment of the repeat domain (Fig. 3). The apparent mass of the observed products and the fact that no additional degradation products were observed suggest that the protein was cleaved within the previously identified cleavage site. N-terminal sequence analysis of the ∼28-kDa product confirmed that cleavage occurred on the N-terminal side of Thr-413. Because the protein was C-terminally Strep-tagged, the fact that the anchoring domain was co-purified would indicate that the interaction domains responsible for the formation of the heterodimeric complex remained intact after cleavage.
FIGURE 3.
CwpV undergoes enzyme-independent intramolecular autoprocessing. Plasmids encoding a Strep-tagged fragment of CwpV (pCwpVfr (WT), pCwpVfr(T413A), and pCwpVfr(GGG)) were used as templates in PURExpress in vitro protein synthesis reactions. Proteins of interest were purified on StrepTactin resin separated on 12% SDS-polyacrylamide gels and analyzed via Coomassie Blue staining and Western blotting. In the pCwpVfr (WT) sample, two products corresponding to the N-terminal domain and a fragment of the repeat domain could be seen aside from the full-length CwpV fragment. In both pCwpVfr(T413A) and pCwpVfr(GGG)samples, no cleavage was observed as only the full-length CwpV fragment could be seen. ◂, repeat domain; ◁, anchoring domain.
To determine whether the T413A and K410G/D411G mutations would prevent cleavage, inverse PCR was used to construct these mutations in pCwpVfr. The plasmids were used as templates in PURExpress reactions, and the synthesized proteins were purified and analyzed by SDS-PAGE and Western blotting. In both mutants only the ∼62-kDa full-length product was identified, indicating that no cleavage took place (Fig. 3).
Full-length CwpV Retains Its Aggregation-promoting Properties
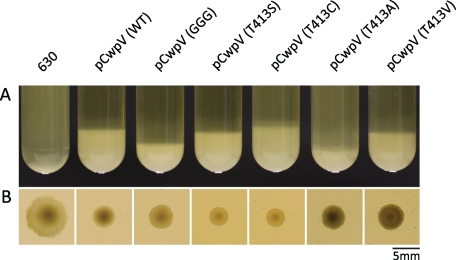
We have shown previously that the C-terminal repeat domain of CwpV is an aggregation-promoting factor and that overexpression of CwpV can cause C. difficile to aggregate from suspension and results in a smaller, smooth edged colony morphology compared with WT strains when grown on solid media (9). To investigate whether disruption of CwpV processing had any effect on this phenotype, the propensity of C. difficile to aggregate from suspension was assessed across the panel of strains used in this study. In all mutant strains, overexpression of CwpV induced autoaggregation from a liquid suspension with a starting A600 nm of 4 (Fig. 4). Aggregation was followed in real time and could be observed as soon as 1 h after suspension of bacteria. What is more, similar to the ΔcwpV (pCwpV) strain, all mutant strains had a smaller, smoother edged colony morphology than seen in the WT strain (Fig. 4). This is consistent with our expectation that the introduced mutations would not affect the repeat domain of the protein. The slightly larger colonies observed in ΔcwpV (pCwpVK410G/D411G), ΔcwpV (pCwpVT413A), and ΔcwpV (pCwpVT413V) could be explained by the fact that less CwpV is present in the S-layer of those strains.
FIGURE 4.
Full-length CwpV retains its aggregation-promoting properties. A, aggregation of C. difficile from liquid culture (starting A600 nm of 4) observed in suspensions of C. difficile ΔcwpV (pCwpV (WT)), ΔcwpV (pCwpVK410G/D411G), ΔcwpV (pCwpVT413S), ΔcwpV (pCwpVT413C), ΔcwpV (pCwpVT413A), and ΔcwpV (pCwpVT413V) but not C. difficile 630. B, colony morphology caused by CwpV overexpression in C. difficile. All strains overexpressing CwpV show a smaller, smoother edged morphology compared with the larger, ruffled edged colonies of C. difficile 630 (WT).
DISCUSSION
Here, we show for the first time that C. difficile cell wall protein CwpV undergoes intramolecular autoprocessing that requires neither cofactors nor auxiliary enzymes and that this reaction is most likely facilitated by a nucleophile-driven N-O acyl migration. Based on the data presented here and in other published studies (14, 18, 20), we propose the following model of autoproteolysis of C. difficile CwpV, illustrated in Fig. 1C. Following Sec-mediated translocation of the precursor polypeptide across the cell membrane (21), the signal peptide is removed, and the protein is allowed to fold. The reactive hydroxyl of Thr-413 is deprotonated by the carboxylate group of Asp-411 and launches a nucleophilic attack on the α-carbonyl carbon of Gly-412 to form a hydroxazolidine intermediate. After a proton is transferred to the leaving amino group of Thr-413, the α-carbonyl of Gly-412 is shifted to the hydroxyl of Thr-413, leading to the ester intermediate via an N-O shift. The ester is then hydrolyzed to form two peptides, the anchoring domain and the repeat domain, which assemble into a noncovalent, heterodimeric complex.
In C. difficile, the majority of the CWPs do not appear to undergo a second cleavage event following removal of the signal peptide. The known exceptions are the major SLP, SlpA, processed by the cysteine protease Cwp84, and CwpV, which undergoes enzyme-independent intramolecular autoprocessing. At least five antigenically distinct types of CwpV are found in C. difficile strains (9), and all contain a cleavage site identical to the one described here, implying that the cleavage mechanism is conserved among these antigenically distinct strains. Interestingly, both SlpA and CwpV are present in high quantities within the S-layer, and their processing appears to be highly efficient. In both cases, uncleaved protein can be incorporated into the S-layer, although the amount present is reduced, and protein is found in the culture supernatant. This suggests that cleavage of CwpV and SlpA is necessary for efficient incorporation into the bacterial S-layer. To our knowledge, CwpV is the first reported case of a protein undergoing intramolecular autoprocessing via a N-O acyl shift in a Gram-positive bacterium. It is plausible that further studies of the CWP family and cell wall proteins in other bacteria will yield further proteins that undergo autocleavage.
The mechanisms of secretion of the C. difficile S-layer and the CWPs are poorly understood, and we believe this study increases our understanding of how S-layer proteins are processed, secreted, and assembled. Translocation of proteins across the cytoplasmic membrane is an essential process in bacteria and is largely mediated by the Sec system consisting of the heteromeric SecYEG membrane channel and the SecA ATPase. Recently, a parallel accessory Sec system has been described in a small number of Gram-positive species (22–26), characterized by the presence of a second copy of SecA, termed SecA2. Bacteria that possess an accessory Sec system employ it for translocation of a defined subset of proteins, many of which appear to be virulence factors (27). C. difficile strain 630 encodes the core components of the canonical Sec secretion system (7), including the SecYEG membrane channel and also has a second copy of the SecA ATPase, indicative of an accessory Sec system. We have recently shown that knock-down of SecA2 expression results in significant accumulation of full-length CwpV and the major S-layer protein SlpA in the cell, demonstrating an absolute requirement for SecA2 for the translocation of both proteins (21). This indicates that CwpV autocleavage occurs after translocation and requires specific conditions to occur. It is possible that SecA2-dependent translocation is a feature of the entire CWP family, but confirmation of this will require additional study.
Autoproteolysis plays important roles in post-translational processing of gene products. A number of proteins undergoing autocleavage have now been identified, including Hedgehog proteins, the precursors of the β-subunits of proteosomes, and members of the N-terminal nucleophile hydrolase family mentioned above (11, 28, 29). All must undergo specific autoproteolysis to perform their biological function. Autoproteolytic cleavage is also involved in protein splicing (30). Understanding the basis of this unique post-translational process in CwpV may not only provide valuable information for studies related to C. difficile cell wall assembly, it may also provide insight into autoproteolytic processing of gene products in different biological systems.
Supplementary Material
Acknowledgment
We thank Robert Fagan for helpful discussions.
This work was supported by Wellcome Trust studentships (to M. D. and C. B. R.).

This article contains supplemental Fig. S1 and Tables S1 and S2.
M. Dembek, C. B. Reynolds, and N. F. Fairweather, unpublished observations.
- S-layer
- surface layer
- CWP
- cell wall protein
- SLP
- S-layer protein.
REFERENCES
- 1. McFarland L. V. (2008) Antibiotic-associated diarrhea: epidemiology, trends and treatment. Future Microbiol. 3, 563–578 [DOI] [PubMed] [Google Scholar]
- 2. Voth D. E., Ballard J. D. (2005) Clostridium difficile toxins: mechanism of action and role in disease. Clin. Microbiol. Rev. 18, 247–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Calabi E., Ward S., Wren B., Paxton T., Panico M., Morris H., Dell A., Dougan G., Fairweather N. (2001) Molecular characterization of the surface layer proteins from Clostridium difficile. Mol. Microbiol. 40, 1187–1199 [DOI] [PubMed] [Google Scholar]
- 4. Kirby J. M., Ahern H., Roberts A. K., Kumar V., Freeman Z., Acharya K. R., Shone C. C. (2009) Cwp84, a surface-associated cysteine protease, plays a role in the maturation of the surface layer of Clostridium difficile. J. Biol. Chem. 284, 34666–34673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dang T. H., de la Riva L., Fagan R. P., Storck E. M., Heal W. P., Janoir C., Fairweather N. F., Tate E. W. (2010) Chemical probes of surface layer biogenesis in Clostridium difficile. ACS Chem. Biol. 5, 279–285 [DOI] [PubMed] [Google Scholar]
- 6. Sleytr U. B., Egelseer E. M., Ilk N., Pum D., Schuster B. (2007) S-layers as a basic building block in a molecular construction kit. FEBS J. 274, 323–334 [DOI] [PubMed] [Google Scholar]
- 7. Sebaihia M., Wren B. W., Mullany P., Fairweather N. F., Minton N., Stabler R., Thomson N. R., Roberts A. P., Cerdeño-Tárraga A. M., Wang H., Holden M. T., Wright A., Churcher C., Quail M. A., Baker S., Bason N., Brooks K., Chillingworth T., Cronin A., Davis P., Dowd L., Fraser A., Feltwell T., Hance Z., Holroyd S., Jagels K., Moule S., Mungall K., Price C., Rabbinowitsch E., Sharp S., Simmonds M., Stevens K., Unwin L., Whithead S., Dupuy B., Dougan G., Barrell B., Parkhill J. (2006) The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat. Genet. 38, 779–786 [DOI] [PubMed] [Google Scholar]
- 8. Emerson J. E., Reynolds C. B., Fagan R. P., Shaw H. A., Goulding D., Fairweather N. F. (2009) A novel genetic switch controls phase variable expression of CwpV, a Clostridium difficile cell wall protein. Mol. Microbiol. 74, 541–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Reynolds C. B., Emerson J. E., de la Riva L., Fagan R. P., Fairweather N. F. (2011) The Clostridium difficile cell wall protein CwpV is antigenically variable between strains, but exhibits conserved aggregation-promoting function. PLoS Pathog. 7, e1002024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Perler F. B., Xu M. Q., Paulus H. (1997) Protein splicing and autoproteolysis mechanisms. Curr. Opin. Chem. Biol. 1, 292–299 [DOI] [PubMed] [Google Scholar]
- 11. Brannigan J. A., Dodson G., Duggleby H. J., Moody P. C., Smith J. L., Tomchick D. R., Murzin A. G. (1995) A protein catalytic framework with an N-terminal nucleophile is capable of self-activation. Nature 378, 416–419 [DOI] [PubMed] [Google Scholar]
- 12. Purdy D., O'Keeffe T. A., Elmore M., Herbert M., McLeod A., Bokori-Brown M., Ostrowski A., Minton N. P. (2002) Conjugative transfer of clostridial shuttle vectors from Escherichia coli to Clostridium difficile through circumvention of the restriction barrier. Mol. Microbiol. 46, 439–452 [DOI] [PubMed] [Google Scholar]
- 13. DeLano W. L. (2010) The PyMOL Molecular Graphics System, version 1.3r1, Schrödinger, LLC, New York [Google Scholar]
- 14. Guan C., Cui T., Rao V., Liao W., Benner J., Lin C. L., Comb D. (1996) Activation of glycosylasparaginase: formation of active N-terminal threonine by intramolecular autoproteolysis. J. Biol. Chem. 271, 1732–1737 [DOI] [PubMed] [Google Scholar]
- 15. Ditzel L., Huber R., Mann K., Heinemeyer W., Wolf D. H., Groll M. (1998) Conformational constraints for protein self-cleavage in the proteasome. J. Mol. Biol. 279, 1187–1191 [DOI] [PubMed] [Google Scholar]
- 16. Klabunde T., Sharma S., Telenti A., Jacobs W. R., Jr., Sacchettini J. C. (1998) Crystal structure of GyrA intein from Mycobacterium xenopi reveals structural basis of protein splicing. Nat. Struct. Biol. 5, 31–36 [DOI] [PubMed] [Google Scholar]
- 17. Xu Q., Buckley D., Guan C., Guo H. C. (1999) Structural insights into the mechanism of intramolecular proteolysis. Cell 98, 651–661 [DOI] [PubMed] [Google Scholar]
- 18. Qian X., Guan C., Guo H. C. (2003) A dual role for an aspartic acid in glycosylasparaginase autoproteolysis. Structure 11, 997–1003 [DOI] [PubMed] [Google Scholar]
- 19. Kelley L. A., Sternberg M. J. (2009) Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4, 363–371 [DOI] [PubMed] [Google Scholar]
- 20. Liu Y., Guan C., Aronson N. N., Jr. (1998) Site-directed mutagenesis of essential residues involved in the mechanism of bacterial glycosylasparaginase. J. Biol. Chem. 273, 9688–9694 [DOI] [PubMed] [Google Scholar]
- 21. Fagan R. P., Fairweather N. F. (2011) Clostridium difficile has two parallel and essential Sec secretion systems. J. Biol. Chem. 286, 27483–27493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen Q., Wu H., Fives-Taylor P. M. (2004) Investigating the role of secA2 in secretion and glycosylation of a fimbrial adhesin in Streptococcus parasanguis FW213. Mol. Microbiol. 53, 843–856 [DOI] [PubMed] [Google Scholar]
- 23. Lenz L. L., Portnoy D. A. (2002) Identification of a second Listeria secA gene associated with protein secretion and the rough phenotype. Mol. Microbiol. 45, 1043–1056 [DOI] [PubMed] [Google Scholar]
- 24. Bensing B. A., Sullam P. M. (2002) An accessory sec locus of Streptococcus gordonii is required for export of the surface protein GspB and for normal levels of binding to human platelets. Mol. Microbiol. 44, 1081–1094 [DOI] [PubMed] [Google Scholar]
- 25. Braunstein M., Brown A. M., Kurtz S., Jacobs W. R., Jr. (2001) Two nonredundant SecA homologues function in mycobacteria. J. Bacteriol. 183, 6979–6990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Siboo I. R., Chaffin D. O., Rubens C. E., Sullam P. M. (2008) Characterization of the accessory Sec system of Staphylococcus aureus. J. Bacteriol. 190, 6188–6196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rigel N. W., Braunstein M. (2008) A new twist on an old pathway: accessory Sec [corrected] systems. Mol. Microbiol. 69, 291–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Porter J. A., von Kessler D. P., Ekker S. C., Young K. E., Lee J. J., Moses K., Beachy P. A. (1995) The product of hedgehog autoproteolytic cleavage active in local and long-range signalling. Nature 374, 363–366 [DOI] [PubMed] [Google Scholar]
- 29. Zwickl P., Kleinz J., Baumeister W. (1994) Critical elements in proteasome assembly. Nat. Struct. Biol. 1, 765–770 [DOI] [PubMed] [Google Scholar]
- 30. Xu M. Q., Southworth M. W., Mersha F. B., Hornstra L. J., Perler F. B. (1993) In vitro protein splicing of purified precursor and the identification of a branched intermediate. Cell 75, 1371–1377 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.