Abstract
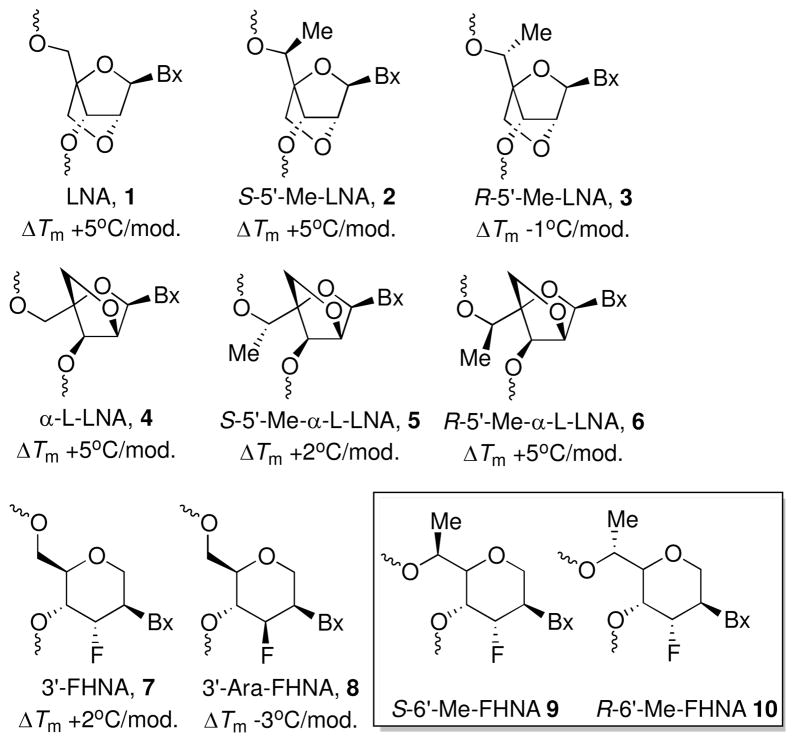
Locked nucleic acid (LNA) analogs with 2′,4′-bridged sugars show promise in antisense applications. S-5′-Me-LNA has high RNA affinity and modified oligonucleotides show reduced immune stimulation in vivo. Conversely, an R-5′-methyl group dramatically lowers RNA affinity. To test the effects of S- and R-6′-methyl groups on 3′-fluoro hexitol nucleic acid (FHNA) stability, we synthesized S- and R-6′-Me-FHNA thymidine and incorporated them into oligo-2′-deoxynucleotides. As with LNA, S-6′-Me is stabilizing whereas R-6′-Me is destabilizing. Crystal structures of 6′-Me-FHNA-modified DNAs explain the divergent consequences for stability and suggest convergent origins of these effects by S- and R-6′-Me (FHNA) [-5′-Me (LNA, RNA)] substituents.
Second generation antisense oligonucleotides (ASOs) are being evaluated for their therapeutic potential in the clinic.1,2 The most advanced ASOs are gapmers that combine the 2′-(2-methoxy)-ethyl (MOE) RNA modification3 in their flanks with a central DNA window and a fully modified phosphorothioate (PS4) backbone. Additional ASO modifications with enhanced RNA affinity and a signature 2′,4′-bridged nucleic acid (BNA) sugar framework have been found to exhibit promising properties for antisense applications (Figure 1). Among them, locked nucleic acid (LNA 15,6) constitutes the basic representative and recent research has demonstrated that ASOs carrying locked nucleotides allow modulation of gene expression via a variety of mechanisms.7–9
Figure 1.
Structures and duplex thermal stability properties of LNA, α-L-LNA, FHNA and 6′-Me-modified FHNAs.
As part of a comprehensive program aimed at elucidating the structure activity relationships (SAR) of gapmer ASOs containing high affinity modifications,10–15 we combined the LNA modification with a methyl substitution at the 5′-position of the bicyclic sugar.14 Introduction of S-5′-Me-LNA 2 residues into ASOs furnished high affinity recognition comparable to that seen with native LNA. Conversely, introduction of R-5′-Me-LNA 3 residues neutralized the gains afforded by the LNA modification and resulted in an unfavorable RNA affinity relative to native DNA. In animal experiments, gapmers with central DNA windows and S-5′-Me-LNA in their wings exhibited lower drug-induced increases in spleen weights, indicative of reduced immune stimulation, as compared to their LNA counterparts.
Recently, we also evaluated the effect of introducing a methyl group in the (R) and (S) configuration at the 5′-position of α-L-LNA 4, which also shows LNA-like high affinity recognition of complementary RNA. However, the consequences on RNA affinity were different from those observed in the β-D-LNA series with the R-5′-Me isomer 6 now displaying enhanced affinity as compared to the S-5′-Me analog 5.16
In view of the attractive antisense properties displayed by the S-5′-Me-LNA modification, and the configuration-dependent divergent effects on RNA affinity in the α-L versus the β-D series, we decided to evaluate the consequences for stability and structure of the methyl backbone modification in the context of a hexitol nucleic acid (HNA17) analog, 3′-fluoro hexitol nucleic acid (FHNA) 7. FHNA-modified ASOs (unlike those containing Ara-FHNA 8) showed comparable potency to LNA in animal trials without producing hepatotoxicity.15 Interestingly, the excellent in vivo activity observed with FHNA was achieved in the absence of elaborate fomulations to improve delivery and despite the lower RNA affinity of this modification relative to LNA.
Here we report the synthesis, biophysical evaluation and crystal structures of oligonucleotides containing S-6′-Me-FHNA 9 or R-6′-Me-FHNA 10 residues (Figure 1). The phosphoramidite T building blocks of 9 and 10 and the modified oligonucleotides were synthesized as outlined in Schemes S1 and S2 and Figure S1 (supporting information file). To establish the consequences of the two analogs for the stability of hybrids between modified DNA and RNA, we conducted UV melting experiments with duplexes containing either one or two modified nucleotides (Table 1). The S-6′-Me-FHNA-T enhances duplex thermal stability similar to FHNA-T (Figure 1). On the other hand, incorporation of R-6′-Me-FHNA-T has a destabilizing effect, amounting to ca. 4°C relative to FHNA-T.15
TABLE 1.
Thermal stabilities of the duplexes between S-and R-6′-Me-FHNA modified DNA and RNA.
| Oligonucleotidea | FHNA-T* | Tmb[°C] | ΔTm/mod[°C] |
|---|---|---|---|
| GCGTTTTTTGCT | DNA | 45.6 | — |
| GCGTT-T*-TTTGCT | S-6′-Me 9 | 46.5 | + 0.9 |
| GCGTT-T*T*-TTTGCT | S-6′-Me 9 | 48.2 | + 1.3 |
| GCGTT-T*-TTTGCT | R-6′-Me 10 | 42.3 | − 3.3 |
| GCGTT-T*T*-TTTGCT | R-6′-Me 10 | 40.7 | − 2.4 |
T* indicates a modified nucleotide.
Tm values (error ± 0.5°C) were measured at 4 μM oligo concentration in 10 mM sodium phosphate buffer (pH 7.2) containing 100 mM NaCl and 0.1 mM EDTA. The RNA complement was 5′-r(AGCAAAAAACGC)-3′.
To understand the opposite effects on stability triggered by a methyl substituent at C6′ with R or S configuration, we studied the crystal structures of A-form decamer duplexes [d(GCGTAT*ACGC)]2 (T*=S-6′-Me-FHNA-T 13 or R-6′-Me-FHNA-T 14) with a single modified nucleotide per strand. Both crystallize in the same space group (P212121) and are isomorphous. The structure of the duplex with S-6′-Me-FHNA Ts (S-6′-Me decamer) was refined to 1.55 Å resolution and that of the duplex with R-6′-Me-FHNA Ts (R-6′-Me decamer) was refined to 1.24 Å resolution. Experimental procedures are summarized in the supporting information, selected crystal data and refinement parameters are listed in Table S1, and examples of the quality of the final electron density are depicted in Figure S2 (si file).
In both duplexes all 2′-deoxyribose sugars adopt the C3′-endo conformation, consistent with the overall RNA-like A-form conformation (Figure S3; si file). In the region of the modified residues T*6 and T*16 (nucleotides in strands 1 and 2 are numbered 1–10 and 11–20, resp.), paired strands exhibit only minimal conformational deviations (Figure S4; si file).
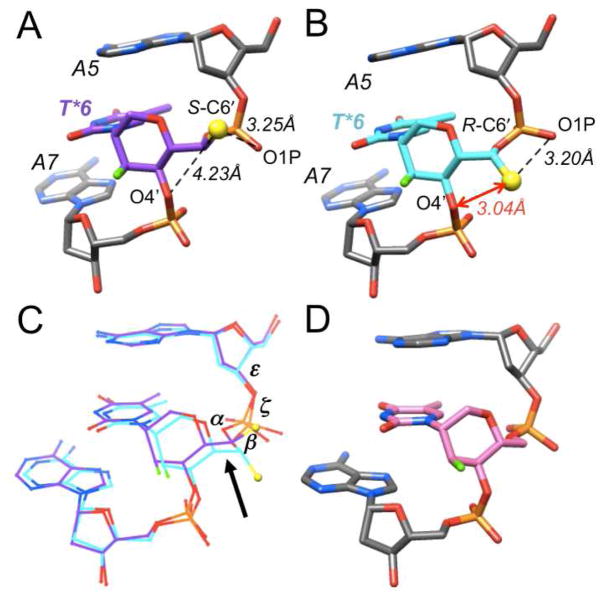
Inspection of the helical parameters and backbone torsion angles in the S- and R-6′-Me duplexes and comparing them to the structure of the decamer with FHNA T residues at positions 6 and 16 (Figure 2),15 reveals subtle changes in the torsion angles α (wider in S-6′-Me-FHNA and compressed in R-6′-Me-FHNA), β (expanded to pure ap in S-6′-Me-FHNA and compressed in R-6′-Me-FHNA), as well as in torsion angle ε of the preceding residues (A5 and A15; Figure 2C, arrow). However, in both 6′-Me-FHNA structures, the sugar-phosphate backbone geometries of modified residues conform to the standard sc−, ap, sc+, sc+ (60° in HNAs15,17), ap, sc− (α to ζ) genus of A-form duplexes. In both the S- and R-6′-Me decamers, residue A5 exhibits an extended backbone variant with α, β and γ in the ap conformations. In the latter duplex, this conformation is also observed for residue G13.
Figure 2.
Conformations of (A) S- and (B) R-6′-Me-FHNA (purple and cyan carbon atoms, resp.), (C) superimposition of the two, and (D) the conformation of FHNA (pink carbon atoms) for comparison. The methyl carbon is shown as a yellow sphere, F3′ is green, residues are labeled and the short 1···5 contact in R-6′-Me-FHNA T is highlighted with a red arrow.
The most obvious difference between the methyl group in the S and R configurations at C6′ (note the different atom numberings in FHNA and LNA) in the two structures is a short 1···5 intra-nucleoside contact between C7′ (Me) and O4′ in R-6′-Me-FHNA (Figure 2B). In the S-6′-Me decamer the spacing between the methyl group and O4′ is considerably larger (Figure 2A). Apart from the aforementioned minor deviations in the torsion angles α, β and ε in the region of the modified residue (Figure 2C), there are no obvious deviations between the conformations of the R- and S-6′-Me-FHNA nucleotides and the backbone of the latter appears unable to avoid the 1···5 contact.
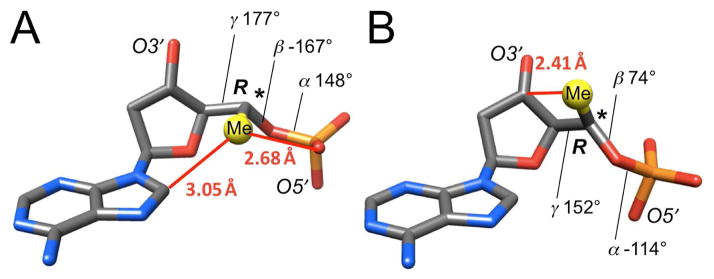
Because of the conformational similarities between FHNA, HNA, LNA and RNA,15 the above energetically unfavorable interaction involving O4′ (O3′ in LNA and RNA) as a result of an R-6′-Me (R-5′-Me in LNA and RNA) substituent will persist in all of these analogs as well as in A-form DNA duplexes. Even when alternative backbone conformations of DNA are considered,14 such as the above extended backbone variant with α, β and γ all in the ap range, or the tricyclo-DNA ac− (α), sc+ (β), sc+ (γ) backbone,18 an R-configured methyl group will cause energetically unfavorable interactions (Figure 3).
Figure 3.
An R-5′-methyl substituent (yellow) in RNA or A-DNA will cause energetically unfavorable, short contacts (red lines) even when sugar-phosphate backbone conformations other than the standard sc−, ap, sc+, sc+, ap, sc− (α to ζ) geometry are considered. (A) Extended backbone variant with α, β and γ in the ap range (seen for residue A5 in the S-6′-Me decamer structure). (B) Backbone conformation in tricyclo-DNA18 with a compensatory change in β and γ.
In addition to causing an unfavorable 1···5 backbone, contact, a 6′-methyl group (5′ in LNA and RNA) in the R configuration can also be expected to perturb the water structure around O2P phosphate oxygens (Figure 4). By comparison, the S-6′-methyl group is directed toward the minor groove (Figures 2, S3) and away from the negatively polarized environment around phosphates.
Figure 4.
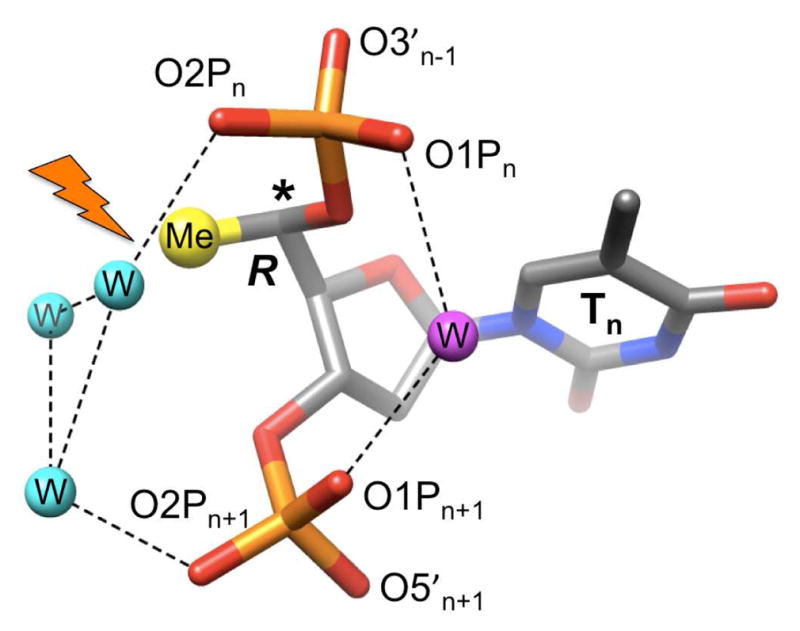
The R-5′-methyl group (modeled) juts into a hydrophilic environment and will interfere (flash) with phosphate hydration (water molecules are cyan and purple spheres) as observed in the 0.83 Å crystal structure of an A-form DNA.19 The shorter distance between O1P oxygens on the edge of the major groove can typically be bridged by a single water (purple), whereas the wider spacing between O2P oxygens requires 2-water bridges (cyan).
In summary, the structural data provide insight into the opposite effects on RNA affinity seen with the two 6′-Me-FHNA modifications described here and help rationalize the previous observations regarding the modulation of β-D-LNA’s duplex stability as a function of the configuration of the 5′-methyl substituent.14
Supplementary Material
Footnotes
Supported by NIH grant R01 GM55237 (to M.E.).
COORDINATE DEPOSITION
The PDB ID codes (http://www.rcsb.org) for the S- and R- 6′-Me decamers are 3V06 and 3V07, respectively.
Materials and methods, Schemes S1 and S2, Tables S1 and S2, Figures S1–S4. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Bennett CF, Swayze EE. Annu Rev Pharmacol Toxicol. 2010;50:259–293. doi: 10.1146/annurev.pharmtox.010909.105654. [DOI] [PubMed] [Google Scholar]
- 2.Raal FJ, Santos RD, Blom DJ, Marais AD, Charng MJ, Cromwell WC, Lachmann RH, Gaudet D, Tan JL, Chasan-Taber S, Tribble DL, Flaim JD, Crooke ST. Lancet. 2010;375:998–1006. doi: 10.1016/S0140-6736(10)60284-X. [DOI] [PubMed] [Google Scholar]
- 3.Teplova M, Minasov G, Tereshko V, Inamati GB, Cook PD, Manoharan M, Egli M. Nat Struct Biol. 1999;6:535–539. doi: 10.1038/9304. [DOI] [PubMed] [Google Scholar]
- 4.Eckstein F. Antisense Nucleic Acid Drug Dev. 2000;10:117–121. doi: 10.1089/oli.1.2000.10.117. [DOI] [PubMed] [Google Scholar]
- 5.Wengel J. Acc Chem Res. 1999;32:301–310. [Google Scholar]
- 6.Imanishi T, Obika S. Chem Commun. 2002:1653–1659. doi: 10.1039/b201557a. [DOI] [PubMed] [Google Scholar]
- 7.Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk ME, Kauppinen S, Orum H. Science. 2010;327:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Straarup EM, Fisker N, Hedtjarn M, Lindholm MW, Rosenbohm C, Aarup V, Hansen HF, Orum H, Hansen JB, Koch T. Nucleic Acids Res. 2010;38:7100–7111. doi: 10.1093/nar/gkq457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Graziewicz MA, Tarrant TK, Buckley B, Roberts J, Fulton L, Hansen H, Orum H, Kole R, Sazani P. Mol Ther. 2008;16:1316–1322. doi: 10.1038/mt.2008.85. [DOI] [PubMed] [Google Scholar]
- 10.Seth PP, Siwkowski A, Allerson CR, Vasquez G, Lee S, Prakash TP, Wancewicz EV, Witchell D, Swayze EE. J Med Chem. 2009;52:10–13. doi: 10.1021/jm801294h. [DOI] [PubMed] [Google Scholar]
- 11.Seth PP, Vasquez G, Allerson CA, Berdeja A, Gaus H, Kinberger GA, Prakash TP, Migawa MT, Bhat B, Swayze EE. J Org Chem. 2010;75:1569–1581. doi: 10.1021/jo902560f. [DOI] [PubMed] [Google Scholar]
- 12.Seth PP, Allerson CR, Berdeja A, Siwkowski A, Pallan PS, Gaus H, Prakash TP, Watt AT, Egli M, Swayze EE. J Am Chem Soc. 2010;132:14942–14950. doi: 10.1021/ja105875e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prakash TP, Siwkowski A, Allerson CR, Migawa MT, Lee S, Gaus HJ, Black C, Seth PP, Swayze EE, Bhat B. J Med Chem. 2010;53:1636–1650. doi: 10.1021/jm9013295. [DOI] [PubMed] [Google Scholar]
- 14.Seth PP, Allerson CR, Siwkowski A, Vasquez G, Berdeja A, Migawa MT, Gaus H, Prakash TP, Bhat B, Swayze EE. J Med Chem. 2010;53:8309–8318. doi: 10.1021/jm101207e. [DOI] [PubMed] [Google Scholar]
- 15.Egli M, Pallan PS, Allerson CR, Prakash TP, Bedeja A, Yu J, Lee S, Watt A, Gaus H, Bhat B, Swayze EE, Seth PP. J Am Chem Soc. 2011;133:16642–16649. doi: 10.1021/ja207086x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seth PP, Allerson CR, Østergaard ME, Swayze EE. Bioorg Med Chem Lett. doi: 10.1016/j.bmcl.2011.11.012. [DOI] [PubMed] [Google Scholar]
- 17.Herdewijn P. Chem Biodiv. 2010;7:1–59. doi: 10.1002/cbdv.200900185. [DOI] [PubMed] [Google Scholar]
- 18.Pallan PS, Ittig D, Heroux A, Wawrzak Z, Leumann CJ, Egli M. Chem Commun. 2008:883–885. doi: 10.1039/b716390h. [DOI] [PubMed] [Google Scholar]
- 19.Egli M, Tereshko V, Teplova M, Minasov G, Joachimiak A, Sanishvili R, Weeks CM, Miller R, Maier MA, An H, Cook PD, Manoharan M. Biopolymers (Nucleic Acid Sciences) 2000;48:234–252. doi: 10.1002/(SICI)1097-0282(1998)48:4<234::AID-BIP4>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.