Abstract
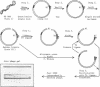
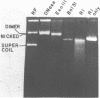
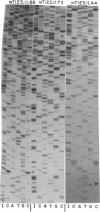
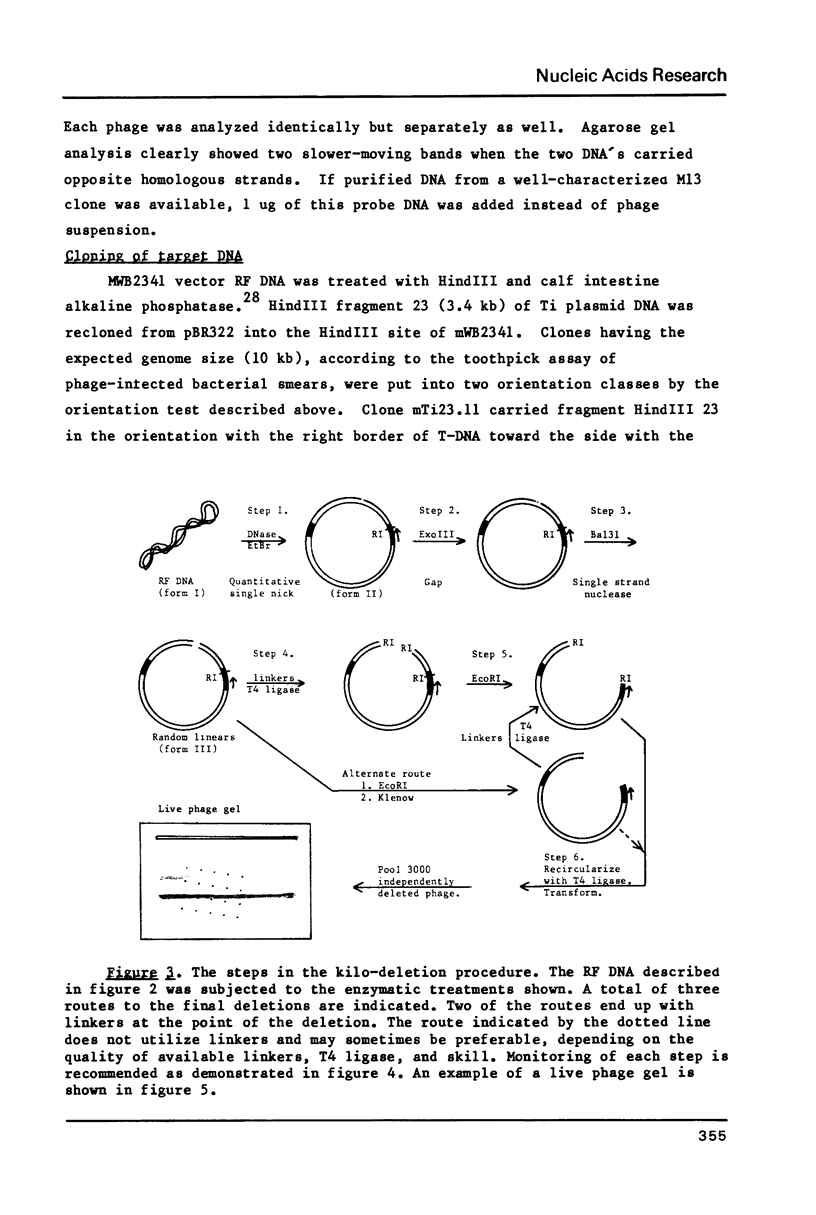
A strategy for rapid DNA sequence acquisition in an ordered, nonrandom manner, while retaining all of the conveniences of the dideoxy method with M13 transducing phage DNA template, is described. Target DNA 3 to 14 kb in size can be stably carried by our M13 vectors. Suitable targets are stretches of DNA which lack an enzyme recognition site which is unique on our cloning vectors and adjacent to the sequencing primer; current sites that are so useful when lacking are Pst, Xba, HindIII, BglII, EcoRI. By an in vitro procedure, we cut RF DNA once randomly and once specifically, to create thousands of deletions which start at the unique restriction site adjacent to the dideoxy sequencing primer and extend various distances across the target DNA. Phage carrying a desired size of deletions, whose DNA as template will give rise to DNA sequence data in a desired location along the target DNA, may be purified by electrophoresis alive on agarose gels. Phage running in the same location on the agarose gel thus conveniently give rise to nucleotide sequence data from the same kilobase of target DNA.
Full text
PDF

















Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barnes W. M. Construction of an M13 histidine-transducing phage: a single-stranded cloning vehicle with one EcoRI site. Gene. 1979 Feb;5(2):127–139. doi: 10.1016/0378-1119(79)90098-2. [DOI] [PubMed] [Google Scholar]
- Barnes W. M. Plasmid detection and sizing in single colony lysates. Science. 1977 Jan 28;195(4276):393–394. doi: 10.1126/science.318764. [DOI] [PubMed] [Google Scholar]
- Chan H. W., Wells R. D. Structural uniqueness of lactose operator. Nature. 1974 Nov 15;252(5480):205–209. doi: 10.1038/252205a0. [DOI] [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C., Hsu L. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc Natl Acad Sci U S A. 1972 Aug;69(8):2110–2114. doi: 10.1073/pnas.69.8.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dürwald H., Hoffmann-Berling H. Endonuclease-I-deficient and ribonuclease I-deficient Escherichia coli mutants. J Mol Biol. 1968 Jul 14;34(2):331–346. doi: 10.1016/0022-2836(68)90257-x. [DOI] [PubMed] [Google Scholar]
- Egner C., Berg D. E. Excision of transposon Tn5 is dependent on the inverted repeats but not on the transposase function of Tn5. Proc Natl Acad Sci U S A. 1981 Jan;78(1):459–463. doi: 10.1073/pnas.78.1.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frischauf A. M., Garoff H., Lehrach H. A subcloning strategy for DNA sequence analysis. Nucleic Acids Res. 1980 Dec 11;8(23):5541–5549. doi: 10.1093/nar/8.23.5541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingeras T. R., Milazzo J. P., Sciaky D., Roberts R. J. Computer programs for the assembly of DNA sequences. Nucleic Acids Res. 1979 Sep 25;7(2):529–545. doi: 10.1093/nar/7.2.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingeras T. R., Rice P., Roberts R. J. A semi-automated method for the reading of nucleic acid sequencing gels. Nucleic Acids Res. 1982 Jan 11;10(1):103–114. doi: 10.1093/nar/10.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenfield L., Simpson L., Kaplan D. Conversion of closed circular DNA molecules to single-nicked molecules by digestion with DNAase I in the presence of ethidium bromide. Biochim Biophys Acta. 1975 Oct 15;407(3):365–375. doi: 10.1016/0005-2787(75)90104-5. [DOI] [PubMed] [Google Scholar]
- Gronenborn B., Messing J. Methylation of single-stranded DNA in vitro introduces new restriction endonuclease cleavage sites. Nature. 1978 Mar 23;272(5651):375–377. doi: 10.1038/272375a0. [DOI] [PubMed] [Google Scholar]
- Guyer M. S., Reed R. R., Steitz J. A., Low K. B. Identification of a sex-factor-affinity site in E. coli as gamma delta. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):135–140. doi: 10.1101/sqb.1981.045.01.022. [DOI] [PubMed] [Google Scholar]
- Hartley J. L., Chen K. K., Donelson J. E. A mercury-thiol affinity system for rapid generation of overlapping labeled DNA fragments for DNA sequencing. Nucleic Acids Res. 1982 Jul 10;10(13):4009–4025. doi: 10.1093/nar/10.13.4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong G. F. A systemic DNA sequencing strategy. J Mol Biol. 1982 Jul 5;158(3):539–549. doi: 10.1016/0022-2836(82)90213-3. [DOI] [PubMed] [Google Scholar]
- Jorgensen R. A., Reznikoff W. S. Organization of structural and regulatory genes that mediate tetracycline resistance in transposon Tn10. J Bacteriol. 1979 Jun;138(3):705–714. doi: 10.1128/jb.138.3.705-714.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz L., Kingsbury D. T., Helinski D. R. Stimulation by cyclic adenosine monophosphate of plasmid deoxyribonucleic acid replication and catabolite repression of the plasmid deoxyribonucleic acid-protein relaxation complex. J Bacteriol. 1973 May;114(2):577–591. doi: 10.1128/jb.114.2.577-591.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M. H., Hines J. C., Ray D. S. Viable deletions of the M13 complementary strand origin. Proc Natl Acad Sci U S A. 1981 Nov;78(11):6784–6788. doi: 10.1073/pnas.78.11.6784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klenow H., Overgaard-Hansen K., Patkar S. A. Proteolytic cleavage fo native DNA polymerase into two different catalytic fragments. Influence of assay condtions on the change of exonuclease activity and polymerase activity accompanying cleavage. Eur J Biochem. 1971 Oct 14;22(3):371–381. doi: 10.1111/j.1432-1033.1971.tb01554.x. [DOI] [PubMed] [Google Scholar]
- Legerski R. J., Hodnett J. L., Gray H. B., Jr Extracellular nucleases of pseudomonas BAL 31. III. Use of the double-strand deoxyriboexonuclease activity as the basis of a convenient method for the mapping of fragments of DNA produced by cleavage with restriction enzymes. Nucleic Acids Res. 1978 May;5(5):1445–1464. doi: 10.1093/nar/5.5.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meselson M., Yuan R. DNA restriction enzyme from E. coli. Nature. 1968 Mar 23;217(5134):1110–1114. doi: 10.1038/2171110a0. [DOI] [PubMed] [Google Scholar]
- Messing J., Crea R., Seeburg P. H. A system for shotgun DNA sequencing. Nucleic Acids Res. 1981 Jan 24;9(2):309–321. doi: 10.1093/nar/9.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J., Gronenborn B., Müller-Hill B., Hans Hopschneider P. Filamentous coliphage M13 as a cloning vehicle: insertion of a HindII fragment of the lac regulatory region in M13 replicative form in vitro. Proc Natl Acad Sci U S A. 1977 Sep;74(9):3642–3646. doi: 10.1073/pnas.74.9.3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojica T. Transduction by phage P1CM clr-100 in Salmonella typhimurium. Mol Gen Genet. 1975;138(2):113–126. doi: 10.1007/BF02428116. [DOI] [PubMed] [Google Scholar]
- Poncz M., Solowiejczyk D., Ballantine M., Schwartz E., Surrey S. "Nonrandom" DNA sequence analysis in bacteriophage M13 by the dideoxy chain-termination method. Proc Natl Acad Sci U S A. 1982 Jul;79(14):4298–4302. doi: 10.1073/pnas.79.14.4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R. J., Lau L. F., Bahl C. P., Narang S. A., Wu R. Synthetic adaptors for cloning DNA. Methods Enzymol. 1979;68:98–109. doi: 10.1016/0076-6879(79)68009-6. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Barrell B. G., Smith A. J., Roe B. A. Cloning in single-stranded bacteriophage as an aid to rapid DNA sequencing. J Mol Biol. 1980 Oct 25;143(2):161–178. doi: 10.1016/0022-2836(80)90196-5. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R. The use of thin acrylamide gels for DNA sequencing. FEBS Lett. 1978 Mar 1;87(1):107–110. doi: 10.1016/0014-5793(78)80145-8. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setlow P., Brutlag D., Kornberg A. Deoxyribonucleic acid polymerase: two distinct enzymes in one polypeptide. I. A proteolytic fragment containing the polymerase and 3' leads to 5' exonuclease functions. J Biol Chem. 1972 Jan 10;247(1):224–231. [PubMed] [Google Scholar]
- Shinnick T. M., Lund E., Smithies O., Blattner F. R. Hybridization of labeled RNA to DNA in agarose gels. Nucleic Acids Res. 1975 Oct;2(10):1911–1929. doi: 10.1093/nar/2.10.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. A new computer method for the storage and manipulation of DNA gel reading data. Nucleic Acids Res. 1980 Aug 25;8(16):3673–3694. doi: 10.1093/nar/8.16.3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. A strategy of DNA sequencing employing computer programs. Nucleic Acids Res. 1979 Jun 11;6(7):2601–2610. doi: 10.1093/nar/6.7.2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugino A., Goodman H. M., Heyneker H. L., Shine J., Boyer H. W., Cozzarelli N. R. Interaction of bacteriophage T4 RNA and DNA ligases in joining of duplex DNA at base-paired ends. J Biol Chem. 1977 Jun 10;252(11):3987–3994. [PubMed] [Google Scholar]
- Sutcliffe J. G. Complete nucleotide sequence of the Escherichia coli plasmid pBR322. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):77–90. doi: 10.1101/sqb.1979.043.01.013. [DOI] [PubMed] [Google Scholar]
- Ullrich A., Shine J., Chirgwin J., Pictet R., Tischer E., Rutter W. J., Goodman H. M. Rat insulin genes: construction of plasmids containing the coding sequences. Science. 1977 Jun 17;196(4296):1313–1319. doi: 10.1126/science.325648. [DOI] [PubMed] [Google Scholar]
- Yadav N. S., Vanderleyden J., Bennett D. R., Barnes W. M., Chilton M. D. Short direct repeats flank the T-DNA on a nopaline Ti plasmid. Proc Natl Acad Sci U S A. 1982 Oct;79(20):6322–6326. doi: 10.1073/pnas.79.20.6322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wezenbeek P. M., Hulsebos T. J., Schoenmakers J. G. Nucleotide sequence of the filamentous bacteriophage M13 DNA genome: comparison with phage fd. Gene. 1980 Oct;11(1-2):129–148. doi: 10.1016/0378-1119(80)90093-1. [DOI] [PubMed] [Google Scholar]