Abstract
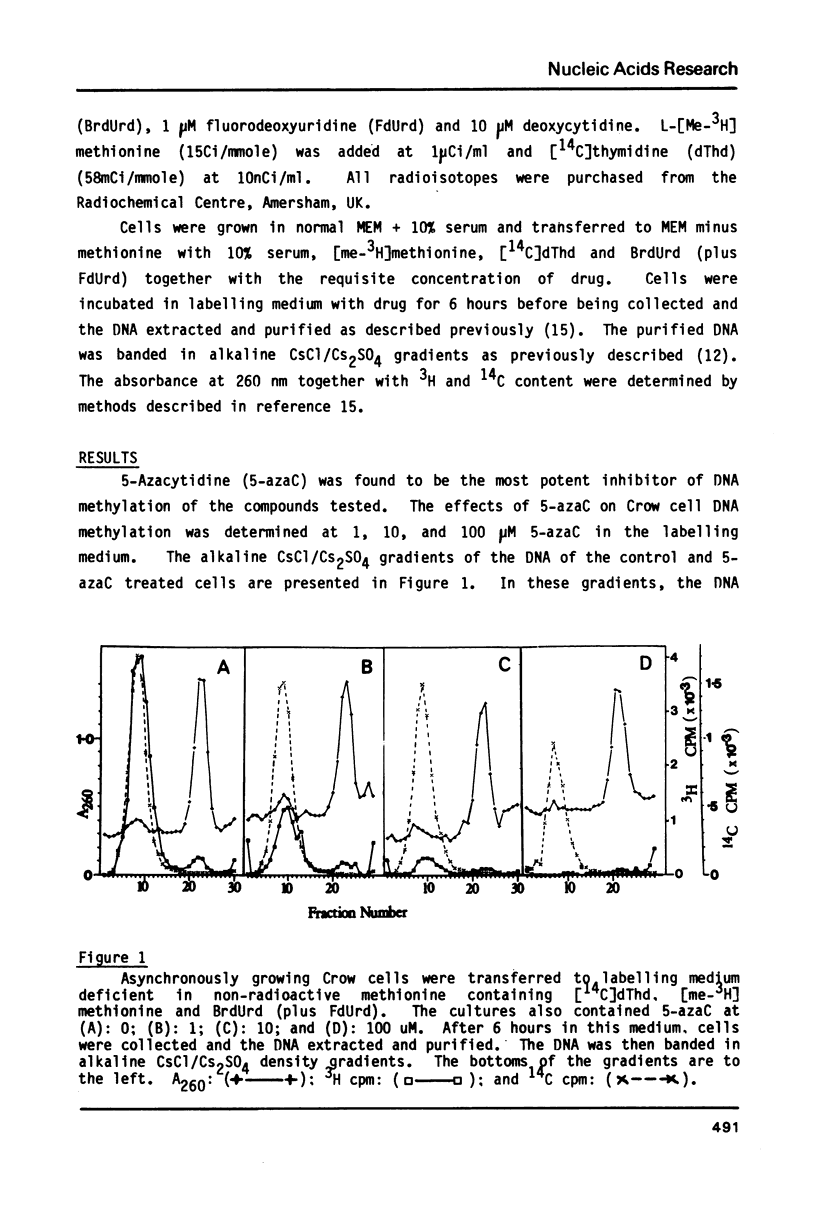
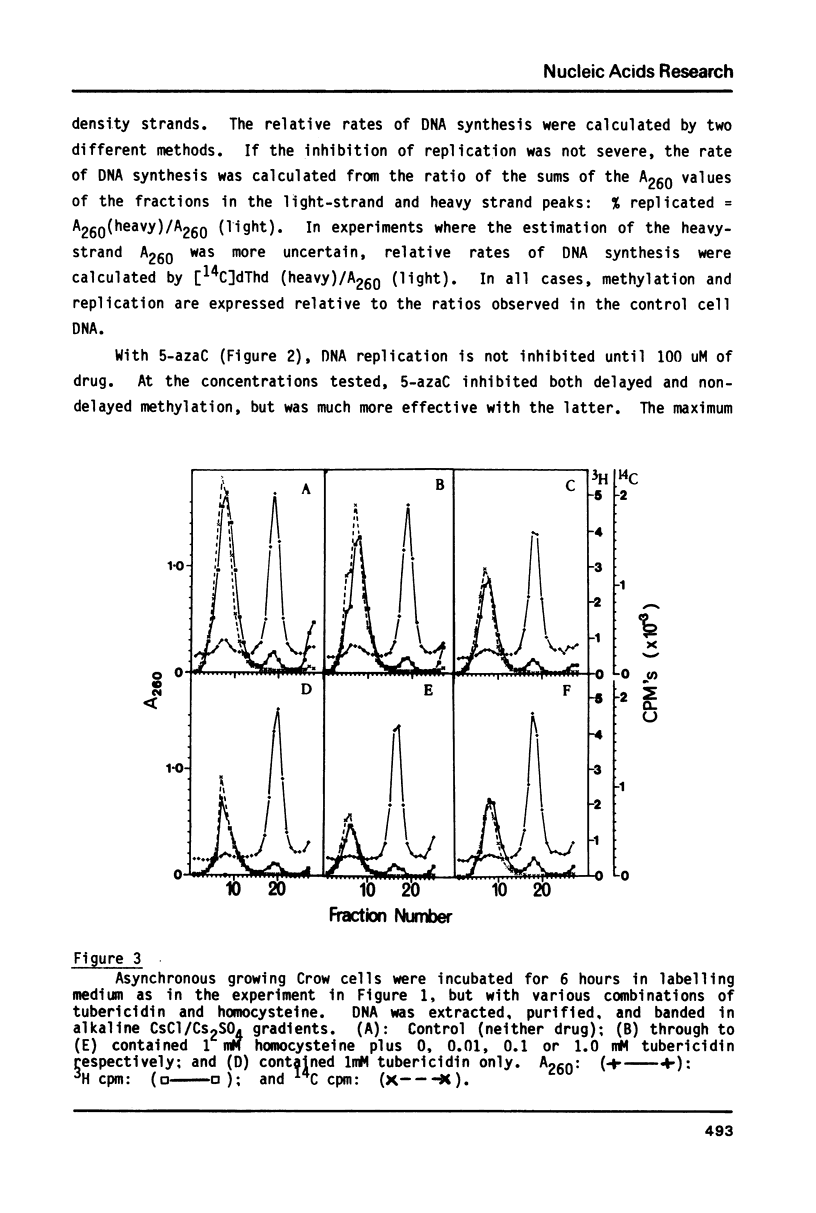
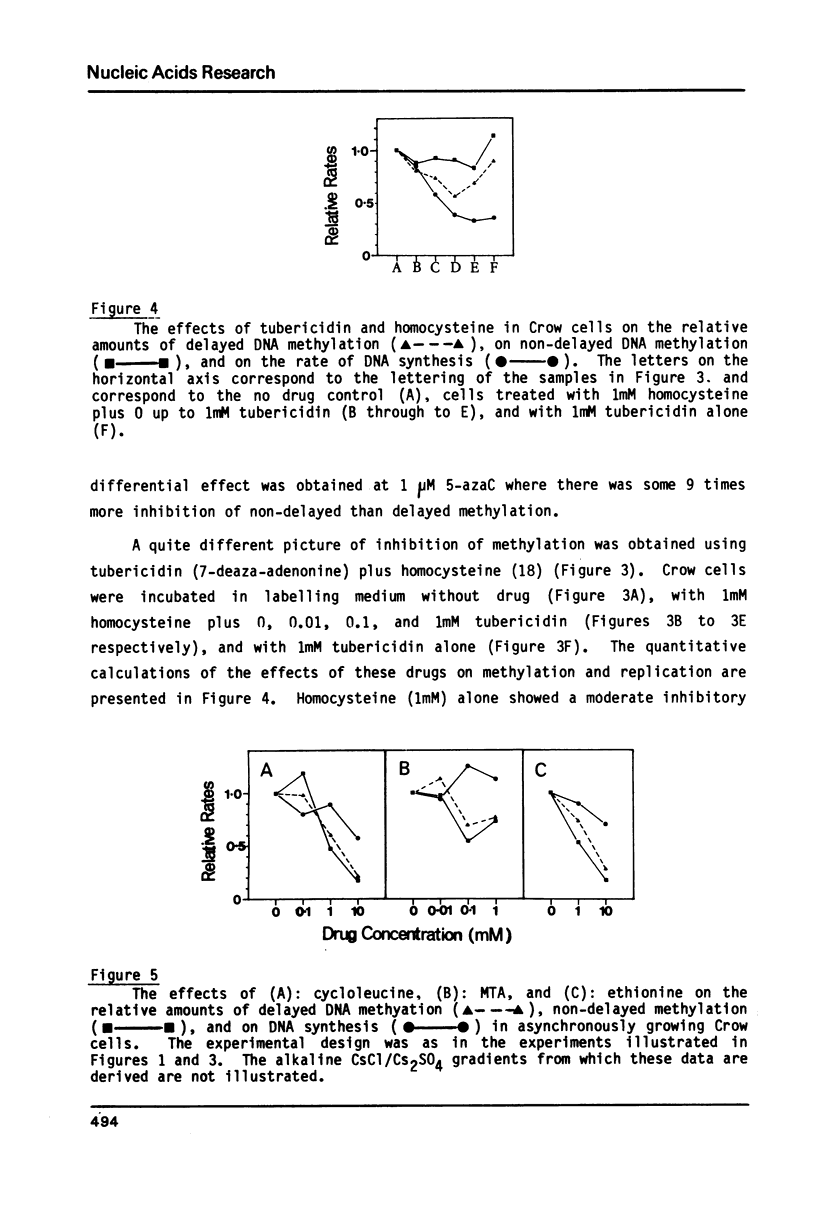
We have previously demonstrated that, while most enzymatic formation of 5-methylcytosine in the DNA of mammalian cells occurs very shortly after strand synthesis, there is also a minor fraction of methylation which occurs in some DNA sequences up to at least several hours after strand synthesis. Using a human cell line, we have examined the effects on these two classes of enzymatic DNA methylation of several compounds which have been reported to be inhibitors of methylation reactions. We have found that cycloleucine, ethionine, and 5'-deoxy-5'-methylthioadenosine (MTA) are all effective as inhibitors of enzymatic DNA methylation, but that there is no differential effect between the delayed and non-delayed methylation reactions. Tubericidin (7-deaza-adenosine) plus homocysteine inhibited delayed DNA methylation much more than non-delayed methylation (by up to 4 times). By contrast, 5-azacytidine produced a higher level of inhibition of DNA methylation at sites in the DNA in which the methylation occurred very shortly after strand synthesis. Also 5-azacytidine was by far the most potent inhibitor of DNA methylation of the compounds tested. S-Adenosyl-homocysteine and caffeine were found to have no effect on DNA methylation. These results are discussed in relation to the number and specificity of DNA methylases in these cells and to the cellular functions of those DNA sequences in which methylation is delayed for some hours after strand synthesis.
Full text
PDF







Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Christman J. K., Price P., Pedrinan L., Acs G. Correlation between hypomethylation of DNA and expression of globin genes in Friend erythroleukemia cells. Eur J Biochem. 1977 Nov 15;81(1):53–61. doi: 10.1111/j.1432-1033.1977.tb11926.x. [DOI] [PubMed] [Google Scholar]
- Creusot F., Acs G., Christman J. K. Inhibition of DNA methyltransferase and induction of Friend erythroleukemia cell differentiation by 5-azacytidine and 5-aza-2'-deoxycytidine. J Biol Chem. 1982 Feb 25;257(4):2041–2048. [PubMed] [Google Scholar]
- Crooks P. A., Dreyer R. N., Coward J. K. Metabolism of S-adenosylhomocysteine and S-tubercidinylhomocysteine in neuroblastoma cells. Biochemistry. 1979 Jun 12;18(12):2601–2609. doi: 10.1021/bi00579a026. [DOI] [PubMed] [Google Scholar]
- Enouf J., Lawrence F., Tempete C., Robert-Gero M., Lederer E. Relationship between inhibition of protein methylase I and inhibition of Rous sarcoma virus-induced cell transformation. Cancer Res. 1979 Nov;39(11):4497–4502. [PubMed] [Google Scholar]
- Friedman S. The inhibition of DNA(cytosine-5)methylases by 5-azacytidine. The effect of azacytosine-containing DNA. Mol Pharmacol. 1981 Mar;19(2):314–320. [PubMed] [Google Scholar]
- Groudine M., Eisenman R., Weintraub H. Chromatin structure of endogenous retroviral genes and activation by an inhibitor of DNA methylation. Nature. 1981 Jul 23;292(5821):311–317. doi: 10.1038/292311a0. [DOI] [PubMed] [Google Scholar]
- Jones P. A., Taylor S. M. Cellular differentiation, cytidine analogs and DNA methylation. Cell. 1980 May;20(1):85–93. doi: 10.1016/0092-8674(80)90237-8. [DOI] [PubMed] [Google Scholar]
- Kiryanov G. I., Kirnos M. D., Demidkina N. P., Alexandrushkina N. I., Vanyushin B. F. Methylation of DNA in L cells on replication. FEBS Lett. 1980 Apr 7;112(2):225–228. doi: 10.1016/0014-5793(80)80185-2. [DOI] [PubMed] [Google Scholar]
- Landolph J. R., Jones P. A. Mutagenicity of 5-azacytidine and related nucleosides in C3H/10T 1/2 clone 8 and V79 cells. Cancer Res. 1982 Mar;42(3):817–823. [PubMed] [Google Scholar]
- Lombardini J. B., Chou T. C., Talalay P. Regulatory properties of adenosine triphosphate-L-methionine S-adenosyltransferase of rat liver. Biochem J. 1973 Sep;135(1):43–57. doi: 10.1042/bj1350043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L. J., Randerath K. Mechanism of 5-azacytidine-induced transfer RNA cytosine-5-methyltransferase deficiency. Cancer Res. 1980 Aug;40(8 Pt 1):2701–2705. [PubMed] [Google Scholar]
- McGhee J. D., Ginder G. D. Specific DNA methylation sites in the vicinity of the chicken beta-globin genes. Nature. 1979 Aug 2;280(5721):419–420. doi: 10.1038/280419a0. [DOI] [PubMed] [Google Scholar]
- McKeon C., Ohkubo H., Pastan I., de Crombrugghe B. Unusual methylation pattern of the alpha 2 (l) collagen gene. Cell. 1982 May;29(1):203–210. doi: 10.1016/0092-8674(82)90104-0. [DOI] [PubMed] [Google Scholar]
- Mohandas T., Sparkes R. S., Shapiro L. J. Reactivation of an inactive human X chromosome: evidence for X inactivation by DNA methylation. Science. 1981 Jan 23;211(4480):393–396. doi: 10.1126/science.6164095. [DOI] [PubMed] [Google Scholar]
- Razin A., Riggs A. D. DNA methylation and gene function. Science. 1980 Nov 7;210(4470):604–610. doi: 10.1126/science.6254144. [DOI] [PubMed] [Google Scholar]
- Sano H., Sager R. Deoxyribonucleic acid methyltransferase from the eukaryote, Chlamydomonas reinhardi. Eur J Biochem. 1980 Apr;105(3):471–480. doi: 10.1111/j.1432-1033.1980.tb04522.x. [DOI] [PubMed] [Google Scholar]
- Shen C. K., Maniatis T. Tissue-specific DNA methylation in a cluster of rabbit beta-like globin genes. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6634–6638. doi: 10.1073/pnas.77.11.6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomkins G. A., MacGregor A., Pye D., Atkinson M. Rapid detection and isolation of mycoplasmas from cell cultures. Aust J Exp Biol Med Sci. 1975 Aug;53(4):257–263. doi: 10.1038/icb.1975.28. [DOI] [PubMed] [Google Scholar]
- Vedel M., Robert-Géro M. Comparative effect of S-adenosyl-homocysteine (SAH) and Sinefungin on tRNA-base methylation in whole cells and in vitro. FEBS Lett. 1981 Jun 1;128(1):87–89. doi: 10.1016/0014-5793(81)81086-1. [DOI] [PubMed] [Google Scholar]
- Viegas-Péquignot E., Dutrillaux B. Segmentation of human chromosomes induced by 5-ACR (5-azacytidine). Hum Genet. 1976 Dec 15;34(3):247–254. doi: 10.1007/BF00295287. [DOI] [PubMed] [Google Scholar]
- Woodcock D. M., Adams J. K., Cooper I. A. Characteristics of enzymatic DNA methylation in cultured cells of human and hamster origin, and the effect of DNA replication inhibition. Biochim Biophys Acta. 1982 Jan 26;696(1):15–22. doi: 10.1016/0167-4781(82)90004-5. [DOI] [PubMed] [Google Scholar]
- Woodcock D. M., Cooper I. A. Evidence for double replication of chromosomal DNA segments as a general consequence of DNA replication inhibition. Cancer Res. 1981 Jun;41(6):2483–2490. [PubMed] [Google Scholar]
- Woodcock D. M., Fox R. M., Cooper I. A. Evidence for a new mechanism of cytotoxicity of 1-beta-D arabinofuranosylcytosine. Cancer Res. 1979 Apr;39(4):1418–1424. [PubMed] [Google Scholar]
- van der Ploeg L. H., Flavell R. A. DNA methylation in the human gamma delta beta-globin locus in erythroid and nonerythroid tissues. Cell. 1980 Apr;19(4):947–958. doi: 10.1016/0092-8674(80)90086-0. [DOI] [PubMed] [Google Scholar]


