SUMMARY
It is proposed that caloric restriction (CR) increases mitochondrial biogenesis. However, it is not clear why CR increases an energetically costly biosynthetic process. We hypothesized that 40% CR would decrease mitochondrial protein synthesis and would be regulated by translational rather than transcriptional mechanisms. We assessed cumulative mitochondrial protein synthesis over 6 weeks and its transcriptional and translational regulation in the liver, heart, and skeletal muscle of young (6 mo), middle (12 mo), and old (24 mo) male B6D2F1 mice that were lifelong CR or ad lib (AL) controls. Mitochondrial protein synthesis was not different between AL and CR (Fractional synthesis over 6-weeks (range): liver 91 – 100%, heart 74–85% skeletal muscle 53–72%) despite a decreased cellular proliferation in liver and heart with CR. With CR there was an increase in AMP activated protein kinase (AMPK) phosphorylation:total (P:T) in heart and liver, and an increase in peroxisome proliferator-activated receptor gamma coactivator 1-α (PGC-1α) mRNA in all tissues, but not protein. Ribosomal protein S6 (RpS6) was decreased with CR. In conclusion, CR maintained mitochondrial protein synthesis while decreasing cellular proliferation during a time of energetic stress, which is consistent with the concept that CR increases somatic maintenance. Alternative mechanisms to global translation initiation may be responsible for selective translation of mitochondrial proteins.
Keywords: Biogenesis, stable isotope, aging, deuterium oxide, long-term, proliferation
Introduction
The protein-turnover theory of aging posits that an increase in the number of damaged proteins with age leads to decreased protein turnover and is thought to be a major mechanism underlying senescence during aging. By some estimations protein damage in older individuals is at minimum 20% and may be as high as 50% of total protein (Stadtman 1992) and this damage is accompanied by a decrease in protein turnover of 20–80% compared to younger controls (Rattan 1996). The ability to maintain functional proteins has been termed proteostasis and is thought to be key to the aging process. Mitochondrial proteins are particularly important to aging and their synthesis has been shown to decrease by middle age in humans (Rooyackers et al. 1996).
It has been proposed that the healthspan extension effects of caloric restriction (CR) are at least partially mediated through increased mitochondrial biogenesis (Civitarese et al. 2007). Although mitochondrial biogenesis is ill defined, it can be assumed that the making of new mitochondria requires protein synthesis. The idea that CR increases mitochondrial biogenesis is perplexing because increasing an energetically expensive process (protein synthesis) in a time of energy shortage is counterintuitive. Since CR mice eat once every 24 hrs upon food provision, the majority of the day is spent in an energy-restricted state. While some evidence suggests that mitochondrial biogenesis and synthesis of mitochondrial proteins is maintained or increased during caloric restriction (Nisoli et al. 2005; Zid et al. 2009), very recently others have questioned whether CR increases mitochondrial biogenesis (Hancock et al. 2011).
Reports documenting an increase in protein synthesis with CR (Goldspink & Kelly 1984; Lewis et al. 1985; el Haj et al. 1986; Merry et al. 1987; Zangarelli et al. 2006) have incorporated a flooding dose of an essential amino acid, which may stimulate a feeding response. It is clear that the acute response measured by the flooding dose of amino acids likely differs from long-term patterns especially when considering tissue specificity. An alternative method to measure synthesis involves using a labeled precursor that has free access to all pools of the body, namely stable isotopically labeled water (2H2O) (Hellerstein 2004). The 2H2O equilibrates throughout all tissues within an hour and decays with a half-life of one week (Raman et al. 2004). It is therefore easy to achieve constant body water enrichment over an extended period of time. Since hydrogen from water is nearly universal in biosynthetic processes the equilibrated water pool is a constant and easily accessible precursor pool for the measurement of a variety of tissue and cellular proteins. The long-term labeling design allows for the determination of average effects over time, rather than acute changes (e.g. response to recent feeding), which is an advantage when studying changes with aging or CR.
Many measurements of “mitochondrial biogenesis” assess either mRNA or mitochondrial DNA (mtDNA) content and it is possible that these results are misleading. Studies that report increases in mRNA as evidence of increased mitochondrial biogenesis (Nisoli et al. 2005; Civitarese et al. 2007) must consider that transcripts are only adaptive potentials and proteomic outcomes are ultimately dependent on the provision of energy, and amino acids. For example, it was demonstrated that in a skeletal muscle cell line that overexpression of transcription factor A of mitochondria (Tfam) was sufficient to increase mtDNA transcription, but not mtDNA number (Maniura-Weber et al. 2004) and proteomic analysis of mitochondria have demonstrated that mRNA content does not equal protein abundance (Mootha et al. 2003). Just because mRNA content of mitochondrial proteins increase, it does not necessarily mean that translation does too since protein translation is sensitive to feeding and energy status through the mTOR pathway.
Finally, there is an important distinction between protein content and mitochondrial biogenesis. Mitochondrial biogenesis is the making of new mitochondrial proteins since mitochondria are not made de novo but rather incorporate new proteins while expanding its reticulum (Ryan & Hoogenraad 2007). Content on the other hand is the balance of synthesis and breakdown. In that regard, an increase in content can result from an increased synthesis or decreased breakdown. Therefore, to properly assess mitochondrial biogenesis, the making of new mitochondrial proteins, mitochondrial protein synthesis is the appropriate measurement since the measurement of mitochondrial protein content does not indicate which of those proteins are new.
It is possible that the regulation of mitochondrial protein synthesis is different between tissues. A proteomic analysis of skeletal muscle, heart, and liver demonstrated that one-third of the mitochondrial proteins identified were associated predominantly to only one tissue but not the other two (Forner et al. 2006) demonstrating tissue-specificity of proteins. Further, mtDNA point mutations and deletions are non-uniformly distributed between tissues (Trifunovic et al. 2004) and it has been observed that some tissues contribute more to the aging phenotype than others. Therefore, mitochondrial protein synthesis likely varies from tissue-to-tissue and must be considered when exploring aging-related treatments.
It has also been hypothesized that CR might slow the aging process through reduced cell division leading to decreased replicative senescence. Decreased cell division has been measured with CR in tissues of the digestive tract (Lok et al. 1990), keratinocytes, mammary epithelial cells, and splenic T cells (Bruss et al. 2011), and hepatic cells (Bruss et al. 2011). The decrease in cell proliferation may allow for energetic allocation of somatic maintenance. The examination of skeletal muscle, heart, and liver allows a comparison of tissues representative of post-mitotic, semi-mitotic, and mitotic to more fully evaluate the effect of CR on cellular proliferation.
The goal of the current study was to comprehensively assess mitochondrial-specific protein synthesis during the aging process, with and without CR. We have focused on mitochondrial protein synthesis because these proteins are important in the aging process. We hypothesized that mitochondrial protein synthesis would decrease both because of age and CR. By obtaining a more accurate picture of long-term tissue-specific mitochondria synthesis and the role of nutrients and energy in regulating the translation of the proteins of mitochondria, it may be possible to target mitochondrial-specific translation initiation factors as a potential treatment for aging-related senescence.
Results
Across ages, AL mice weighed significantly more than CR mice (Supplementary Data Figure 1). Mice that were housed for 6 weeks at the CSU animal facilities (Cohort 2) did not gain weight over that period of time.
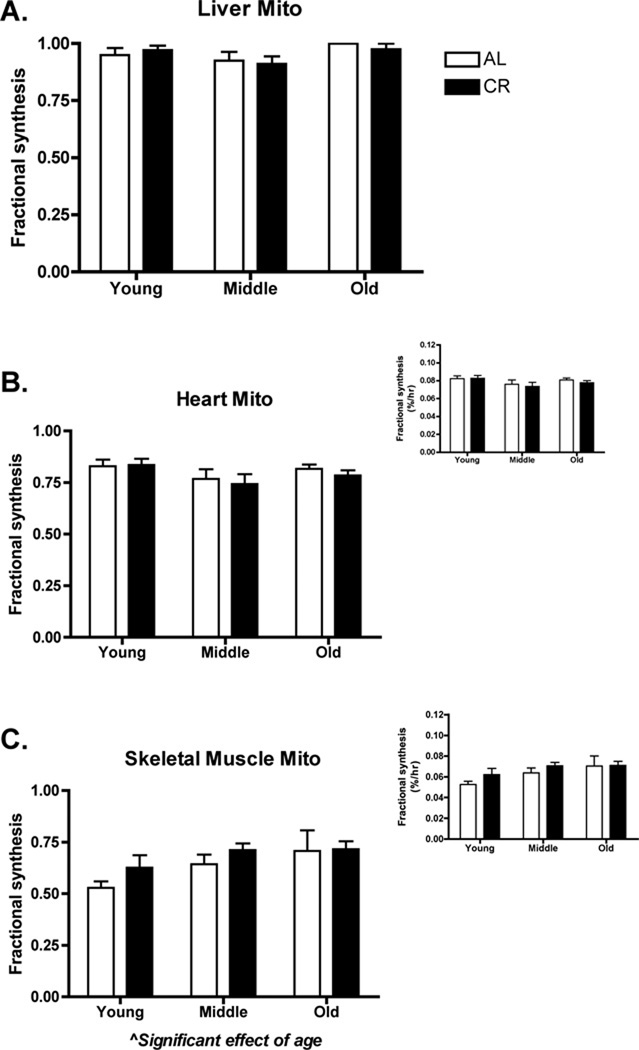
Liver mitochondrial protein synthesis (Figure 1A) was qualitatively greater than heart mitochondrial protein synthesis (Figure 1B), which was greater than skeletal muscle mitochondrial protein synthesis (Figure 1C). There was no effect of CR or age on liver (CR: p=0.81, F= 0.056; Age: p=0.06, F=2.99) or heart (CR: p=0.57, F=0.32; Age: p=0.10, F=2.39) mitochondrial protein synthesis. There was a significant effect of age on skeletal muscle mitochondrial protein synthesis with an increase observed with advancing age (p=0.03, F=3.75) (Figure 1C). As with heart and liver, there was no effect of CR on skeletal muscle protein synthesis (p=0.18, F=1.90). As a determination of mitochondrial content, we performed western blot analysis of COX4. In liver, there was a trend (p=0.06, F=3.86) toward increased COX4 with CR, whereas there were no differences (treatment or age) in heart and skeletal muscle (Supplementary Data Figure 2). In sum, the synthesis of mitochondrial proteins was not different between AL and CR conditions.
Figure 1.
Mitochondrial protein synthesis over a 6-week period in young, middle, and old AL or CR mice. Liver mitochondrial protein was fully turned over in the 6-week period (A). Heart mitochondrial protein synthesis was less than liver and there was no difference because of age or CR (B). Skeletal muscle mitochondrial protein synthesis was less than heart and had a significant effect of age (p ≤ 0.05), but not because of CR (C). Figure insets for heart and skeletal muscle are in units of %/hr. It was not possible to calculate %/hr for liver since proteins were fully turned over. n = 6–8 per group.
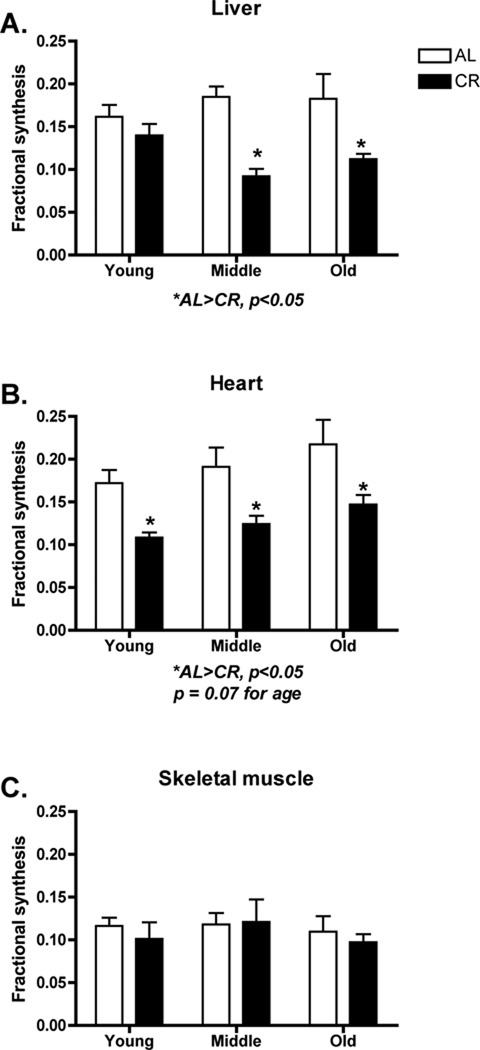
In contrast to mitochondrial protein synthesis, DNA synthesis showed a CR effect and tissue specificity. In liver, DNA synthesis was significantly decreased in CR compared to AL (p<0.0001, F=22.74) at middle (p<0.001) and old (p<0.01) age (Figure 2A). In heart CR caused a significant decrease compared to AL (p<0.0001, F=21.79) in young (p<0.05), middle (p<0.05), and old age (p<0.05) (Figure 2B). In contrast, in skeletal muscle there were no significant differences in DNA synthesis due to CR (p<0.57, F=0.33) at any age (p<0.64, F=0.46) (Figure 2C). Of note is that there was measurable DNA synthesis in skeletal muscle, a post-mitotic tissue.
Figure 2.
Cellular proliferation as measured by DNA synthesis over a 6-week period in young, middle, and old AL or CR mice. In liver (A) and heart (B) there were significant decreases in cellular proliferation in CR mice. In heart, there was a trend (p = 0.07) for increased cellular proliferation with age. In skeletal muscle there were no differences in cellular proliferation (C) although it is worth noting that there was measureable DNA synthesis in skeletal muscle. n = 6–8 per group.
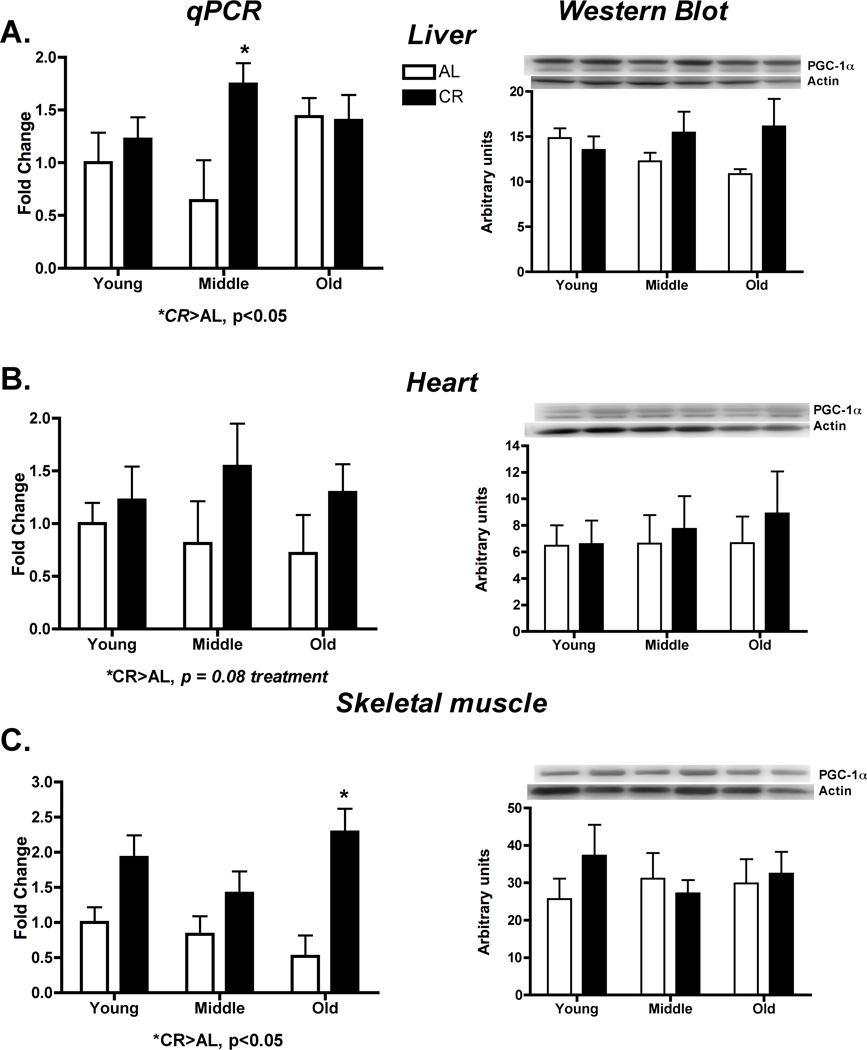
We hypothesized that any potential differences in mitochondrial protein synthesis would more likely be the result of changes translational control rather than transcriptional control. Therefore, we assessed changes in PGC-1α and its downstream effectors and changes in translation initiation of the mTOR pathway. CR caused a significant increase in the mRNA of PGC-1α in liver (p=0.05, F=4.22) (Figure 3A) and skeletal muscle (p=0.001, F=21.56) (Figure 3C), although this did not quite reach significance in the heart (p=0.08, F=3.41)(Figure 3B). These changes in mRNA did not result in differences in PGC-1α protein (Liver: p=0.11, F=2.69; Heart: p = 0.53, F=0.40; Skeletal muscle: p=0.50, F=0.46) (Figure 3). Downstream of PGC-1α there were no differences in mRNA of NRF-1, Tfam, or mitochondrial proteins (Supplementary Data Figure 3).
Figure 3.
PGC-1α was assessed by qPCR and western blot for potential transcriptional regulation of mitochondrial protein synthesis in liver (A), heart (B) and skeletal muscle (C). CR increased skeletal muscle PGC-1α at old age. There were no other differences in any tissue or at any age. *Significant difference between AL and CR, p ≤ 0.05, #Significant effect of age, p ≤ 0.05. n = 4–5 per group.
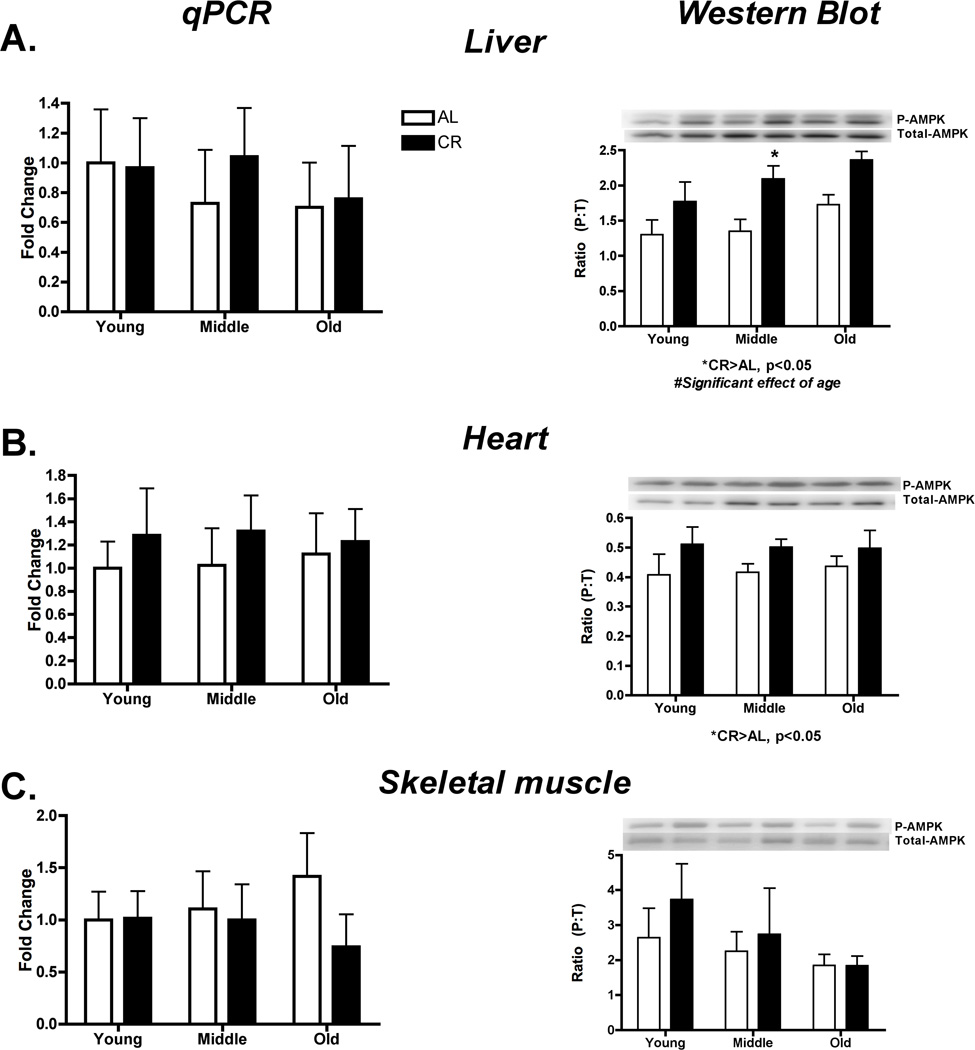
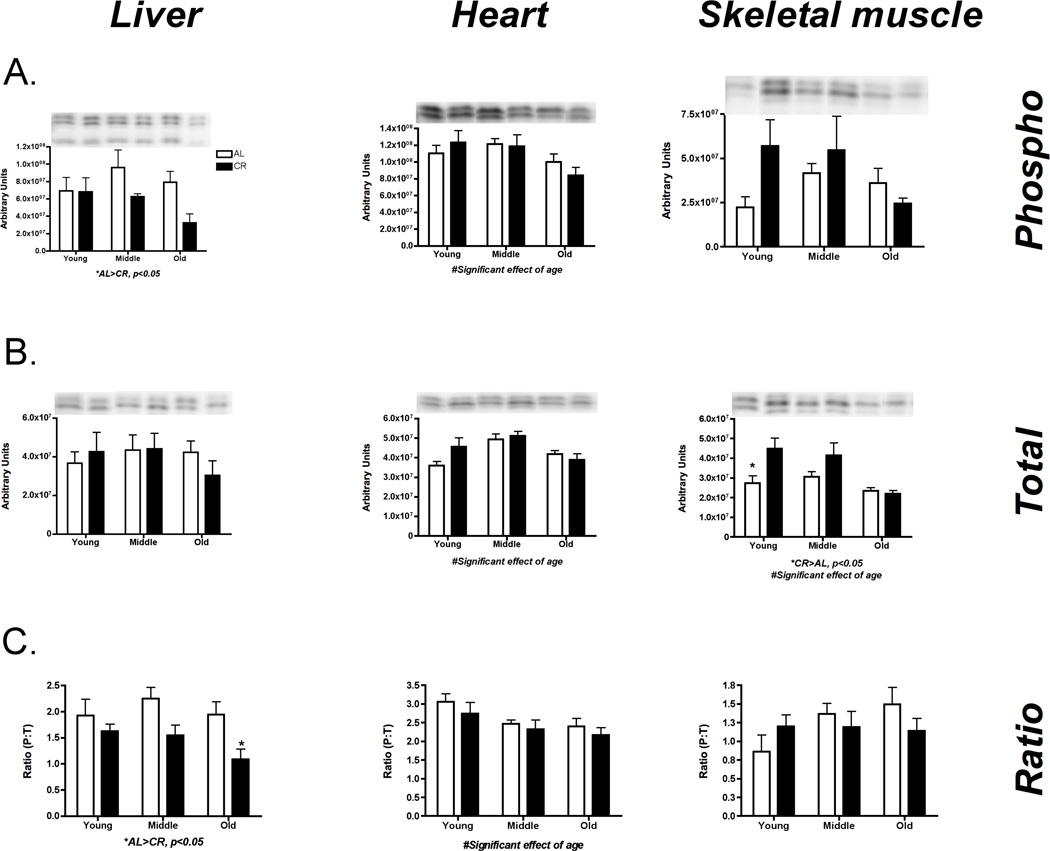
Long-term changes in energy may affect AMPK phosphorylation upstream of PGC-1α. In the current study, there was no significant effect of CR on AMPK mRNA (Liver: p=0.69, F=0.16; Heart: p = 0.39, F=0.78; Skeletal muscle: p=0.35, F=0.92) (Figure 4 A, B, C). However, AMPK phosphorylation to total ratio (P:T) was significantly greater in CR versus AL in liver (p=0.0006, F=15.78) and heart (p=0.05, F=4.27), while in liver only there was also a significant increase with age (p=0.04, F=3.63). Conversely, in skeletal muscle there was no change in AMPK P:T (p=0.41, F=0.69) (Figure 4C). Importantly, total AMPK did not change indicating that changes in P:T were not obscured by a change in content (Supplementary Data Figure 4).
Figure 4.
AMPK was assessed by qPCR and western blot for potential transcriptional regulation of mitochondrial protein synthesis in liver (A), heart (B) and skeletal muscle (C). P:T of AMPK was significantly greater in CR versus AL in liver and heart and in liver there was also a significant increase with age. In skeletal muscle there were no changes noted for AMPK. *Significant difference between AL and CR, p ≤ 0.05. n = 4–5 per group.
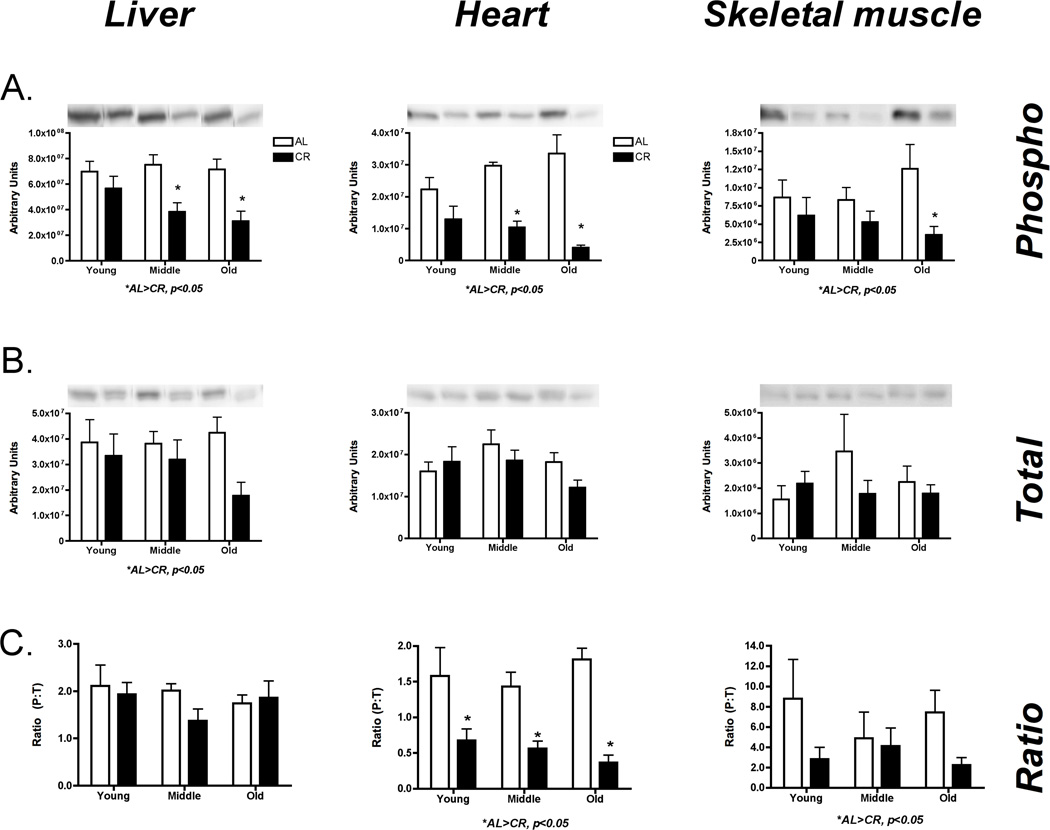
To assess mTOR activity, we examined downstream proteins RpS6 and 4e-BP. In liver there was a significant decrease in phosphorylated 4eBP-1 with CR (p=0.03, F=5.50), which resulted in decreased P:T (p=0.002, F=11.57) (Figure 5A). In heart there was a significant decrease in phospho (p=0.03, F=4.07), total (p=0.04, F=7.21), and P:T (p=0.02, F=4.53) with age for 4e-BP (Figure 5B). In skeletal muscle, there was an increase in total 4e-BP with CR (p=0.01, F=7.46) with no change in P:T (p=0.69, F=0.16) (Figure 5C). Across liver (Figure 6A), heart (Figure 6B), and skeletal muscle (Figure 6C) there was a significant decrease in RpS6 phosphorylation with CR (Liver: p=0.0001, F=21.05; Heart: p < 0.0001, F=47.53; Skeletal muscle: p=0.01, F=7.15). These decreases in phosphorylation resulted in an overall decrease in P:T for heart (p< 0.0001, F=38.09) and skeletal muscle (p=0.04, F=4.52). In sum, there is an overall decrease in global protein synthesis initiation through the mTOR pathway.
Figure 5.
Assessment of global translation initiation by changes in 4e-BP-1 activation in liver (A), heart (B), and skeletal muscle (C). There was a significant increased phosphorylation in liver with CR, which indicates decreased translation initiation. Skeletal muscle appears to have variable results since total 4e-BP decreases with CR and age, but not phosphorylation. In heart phopshorylation decreased with age. *Significant difference between AL and CR, p ≤ 0.05. n = 5 per group.
Figure 6.
Assessment of global translation initiation by changes in RpS6 activation in liver (A), heart (B), and skeletal muscle (C). In general, there is a decrease in global translation initiation in all three tissues. These changes were reflected in total, phosphorylation or the ratio. *Significant difference between AL and CR, p ≤ 0.05, #Significant effect of age, p ≤ 0.05. n = 5 per group.
Discussion
It has been hypothesized that one of the lifespan extension effects of CR is an increase in mitochondrial biogenesis. To date, an increase in mitochondrial biogenesis with CR has been poorly documented. Here we present a tissue-specific assessment of the synthesis of mitochondrial proteins across three ages of lifelong AL and CR animals over 6 weeks. We demonstrate that CR does not increase mitochondrial protein synthesis, but it does maintain it equal to synthesis rates during AL. The maintenance of mitochondrial protein synthesis occurs in liver and heart despite decreased cellular proliferation. Further, we demonstrate that the maintenance of mitochondrial protein synthesis is in the face of an energetic challenge, decreased activation of global protein synthesis, and small changes in the transcriptional regulation. These findings lead us to believe that other post-transcriptional mechanisms are maintaining mitochondrial protein synthesis during CR.
Assessment of mitochondrial protein synthesis and cellular proliferation
Previous studies examining CR and mitochondrial biogenesis have provided an incomplete picture since they have focused on mRNA content (Nisoli et al. 2005; Civitarese et al. 2007; Hancock et al. 2011), short-term synthesis (Lewis et al. 1985; el Haj et al. 1986; Merry et al. 1987; Zangarelli et al. 2006), or short-term restriction (Yuan et al. 2008; Bruss et al. 2011). Although recent data has lead to speculation that CR does not increase mitochondrial biogenesis (Hancock et al. 2011), we felt that a tissue-specific, long-term assessment of mitochondrial protein synthesis during CR was warranted. We chose to use deuterium oxide (2H2O, heavy water) to simultaneously assess mitochondrial protein synthesis and cellular proliferation. There are many advantages to this technique for our study design. First, it determines the sum changes over our 6-week period of study and thus is the sum total of protein synthesis to activity, feeding, and recovery. All these factors can change protein synthesis and the short-term nature of most isotope techniques usually captures only one of these factors. Second, mice were free-living and undisturbed throughout the labeling period. Last, it allowed simultaneous assessment of multiple synthetic processes.
Two important previous studies differ in their conclusions of mitochondrial biogenesis (Nisoli et al. 2005; Hancock et al. 2011). Some have speculated that these results could be the result of the different models used (mice versus rats and feeding paradigm). Our study differs slightly from these in that it used life-long CR (compared to 14 or 60 weeks) and three different ages, including an old group. However, most important is that the studies of Hancock et al. and Nisoli et al. make inferences of mitochondrial biogenesis based on mRNA and protein contents while ours directly measures the rate of mitochondrial protein synthesis. There is an important distinction between these measurements. Mitochondrial biogenesis is the result of mitochondrial protein synthesis since mitochondria are not made de novo (Ryan & Hoogenraad 2007). Nisoli et al. published an increase in the mtDNA:nDNA, which is an indirect determination of mitochondrial content, and mitochondrial protein content by Western Blot (Nisoli et al. 2005). Hancock et al. also measured protein content by Western Blot (Hancock et al. 2011). Protein content is the net result of synthesis and breakdown. Therefore, in the strictest sense, mitochondrial biogenesis is the making of new mitochondria and is reflective of synthesis while content is the end result of the process of synthesis and breakdown. Our study is the first to directly measure mitochondrial protein synthesis in CR mice over an extended period of time.
Previous studies have also used 2H2O during CR (Yuan et al. 2008; Bruss et al. 2011). Yuan et al., demonstrated that acute fasting (20 hr) did not change mitochondrial protein synthesis in the heart (Yuan et al. 2008). Although the authors also calorically restricted the rats for 7 days, they only present data for mixed protein synthesis, rather than mitochondrial, for liver, heart and skeletal muscle. Nevertheless, the authors demonstrated a decreased mixed protein synthesis in liver and skeletal muscle, but preserved protein synthesis in the heart, thus illustrating tissue specificity (Yuan et al. 2008). More recently, Bruss and others showed that short-term (4 week) CR decreased cellular proliferation in a variety of tissues including liver (Bruss et al. 2011). As opposed to the previously mentioned studies, our study examines life-long CR. Further, we examine mitochondrial protein synthesis and cellular proliferation in three tissues with varied proliferative ability.
Our primary finding was that mitochondrial protein synthesis was not different between treatments in any tissue in AL compared to CR. Some have argued that the increased foraging behavior of CR mice may be responsible for potential increases in mitochondrial biogenesis. Recent results from our laboratory indicate that although exercise alone increases mitochondrial biogenesis in skeletal muscle, it does not do so in the heart (unpublished observations). A comparison of synthesis rates between tissues accurately reflects what is commonly believed about tissue synthesis rates (e.g. (Wilson et al. 2011), with liver mitochondrial protein fully turned-over in 6 weeks, heart slightly less, and skeletal muscle having roughly 50–70% new protein. Because the liver was completely turned over in the time period of our study, it is possible that differences between treatments were missed since one may have fully turned over prior to the other. A novel observation was that in skeletal muscle, mitochondrial protein synthesis increased with age, which is counter to what has been previously published in short-term assessment of older human subjects (Rooyackers et al. 1996). We are confident in our rates over the six-week period because when compared to short-term measures, our rates compare to previously published values (approximately 0.04 – 0.08%/hr, inset Figure 1) when extrapolated to 6 weeks (approximately 40–80%) (Rooyackers et al. 1996). In addition, our time period of measurement (6 weeks) is within the linear phase of incorporation as previously demonstrated (Busch et al. 2006).
Although others have described decreases in content of skeletal muscle mitochondrial proteins with age (Short et al. 2005) our results are not at odds with these findings. Content is the result of changes in synthesis and breakdown. It is equally likely that a decrease in content is due to an increase in breakdown as a decrease in synthesis. In the current study we measure the synthesis of mitochondrial proteins. Measurements of mitochondrial protein breakdown over time, a measurement that has significant technical challenges, may shed more light on the determination of content.
It is also important to consider the mitochondrial protein synthesis in the context of cellular proliferation. CR decreased cellular proliferation in liver and heart with no change in skeletal muscle. Therefore, liver and heart are maintaining mitochondrial protein synthesis in light of an overall decrease in cellular proliferation. With less cellular proliferation in CR mice, it is likely that a greater proportion of the mitochondrial protein synthesis is directed at existing mitochondrial reticulum. This idea falls in line with the idea that CR increases somatic maintenance and that CR animals maintain existing cellular structures during energetic stress.
Of note is that skeletal muscle cellular proliferation did not follow the pattern of the liver and heart. Skeletal muscle is a post-mitotic tissue and is therefore assumed to have no cellular proliferation. Notably, we measured cellular proliferation in skeletal muscle albeit at relatively low rates. We recently demonstrated a similar finding in human subjects (Robinson et al. 2011). In our study on human subjects, we attributed the proliferation to the recruitment of satellite cells. Although the role of the satellite cell in skeletal muscle cellular repair has been well established (Kadi & Ponsot 2010), the decline in satellite cell function with aging or CR is less clear. Some have described a decrease in satellite cell number as an organism ages (Verdijk et al. 2007) and decreased ability to create reserve cells (Day et al. 2010). However, there seems to be little evidence on actual rate of recruitment and thus we present novel information in this regard. If our measure of DNA synthesis in skeletal muscle represents satellite cell recruitment, the finding of no decrease with aging could provide additional insight into satellite cell depletion (maintained recruitment with limited repletion). At this point we cannot definitely rule out the contribution of cellular proliferation, a finding that would be interesting skeletal muscle that is thought to be post-mitotic. In this regard it is worth mentioning that mice have telomerase positive skeletal muscle cells (Gorbunova et al. 2008) that could provide a means for limited proliferation. Nonetheless the difference in proliferation between CR and AL mice and no difference in mitochondrial protein synthesis may represent a slightly different balance of growth versus repair in skeletal muscle compared to liver and heart.
Transcriptional versus translation regulation of mitochondrial protein synthesis
Mitochondrial remodeling is thought to be under transcriptional regulation (Hock & Kralli 2009), which has resulted in a focused investigation into transcriptional regulation by PGC-1α and the energetic sensor, AMPK. During energetic stress AMPK activates PGC-1α, increasing the transcription of mitochondrial protein mRNA presumably to increase aerobic energy production potential. However, activation of AMPK generally down-regulates biosynthetic processes through inhibition of mTOR and its downstream effectors (Inoki et al. 2003). An apparent question is how can the activation of AMPK, and consequent upregulation of PGC-1α by energetic stress, stimulate mitochondrial protein synthesis if AMPK is simultaneously down-regulating translation initiation through mTOR? It is for this reason that post-transcriptional mechanisms must be considered.
We approached our assessment hypothesizing that transcriptional regulation of mitochondrial biogenesis during CR may be less important than previously thought and that translation initiation was where regulation occurred. The rationale for translational regulation relies on cellular energetics. In the basal state protein synthesis is the largest consumer of ATP (Rolfe & Brown 1997). Due to the ATP demands of peptide recruitment and assembly, translation is ten times more energetically costly than transcription (Wagner 2005). Therefore, efficient translation is dependent on energy and nutrient factors. Even though it can be argued that CR mice are not energetically restricted because of their stable yet decreased body mass, CR mice eat once per upon provision of food rather than ad lib, thus creating prolonged periods of negative energy balance. As hypothesized, our study demonstrated that CR leads to an overall increased activation of AMPK in mice. Surprisingly, very few groups have demonstrated increased phosphorylation of AMPK with CR (Palacios et al. 2009; Edwards et al. 2010). In agreement with others (Civitarese et al. 2007) CR increased the transcription of PGC-1α. Notably, changes in skeletal muscle PGC-1α mRNA did not correspond to changes in PGC-1α protein or mitochondrial protein synthesis. Similarly, we found very few changes downstream of PGC-1α. Therefore, we question whether PGC-1α mRNA is indicative of “mitochondrial biogenesis”. Our contention is not at odds with the vast amount of data that demonstrates that PGC-α and its modifications and localization (e.g. (Anderson et al. 2008) are important to its mitochondrial biogenesis. We simply contend that in stressful conditions, such as CR, energetic constraints may limit actual biosynthesis through post-transcriptional mechanisms.
We explored global translation initiation through the mTOR pathway. Because of recent evidence of tissue-specific mTOR effects (Polak & Hall 2009), we did so in all three tissues. With CR there was a general global downregulation of translation initiation. These results are consistent with our hypothesis that energetic stress would result in a global decrease in translation initiation because of the energetically costly process of translation. Our results, particularly with RpS6, are consistent with previous studies examining lifespan extension (Harrison et al. 2009; Zid et al. 2009). Although we hypothesized that energetic stress would downregulate protein synthesis, it is puzzling why this has life extension effects. In accordance with the mitochondrial theory of aging and the protein turnover theory of aging, an increased turnover should result in the breakdown of damaged proteins and replacement with “new” better functioning proteins. Conversely a decrease in turnover would indicate decreased protein replacement. It is now our thought that the synthesis of key proteins, such as mitochondrial proteins, is maintained by evading the mTOR-mediated inhibition of cap-dependent translation initiation. Currently, there is evidence of mechanisms for selective translation initiation of mitochondrial proteins in yeast (Zid et al. 2009) and heat shock proteins in a mouse cell line (Sun et al. 2011), but limited data in mammalian species. We believe that the life-extension effects of mTOR inhibition are mediated through selective translation of key proteins at the cost of global protein synthesis and is thus in line with somatic maintenance theories of aging.
Perspective and conclusion
Protein stability, or proteostasis, contributes to healthy aging. It is fairly clear that, in general, protein turnover decreases with aging and mitochondrial dysfunction contributes to age-related senescence. CR is a robust and reproducible means to extend lifespan and healthspan in a variety of species. Less clear is how acute and chronic changes in cellular energetics affect mitochondrial protein turnover. Although it is speculated that the making of mitochondria contributes to healthspan extension, it is unclear how or why an energetically stressful environment activates the energetically expensive process of protein synthesis. Here we demonstrate that CR maintains mitochondrial protein synthesis while decreasing cellular proliferation during a time of energetic stress and is consistent with the hypothesized somatic maintenance of CR. We also demonstrate that alternative mechanisms may be responsible for selective translation of mitochondrial proteins, which represent key proteins for health and longevity. Further, because of subtle differences in tissue-specific responses in our study we believe tissue specificity warrants further investigation. Finally, we strongly caution against equating changes in mRNA and signaling to changes in proteomic outcomes as demonstrated by our actual measures of protein synthesis.
Experimental Procedures
Overall study design
Male B6D2F1 mice from the National Institute of Aging (NIA) calorically restricted (CR) colony and ad lib controls (AL) were used for all aspects of the study. Mice were purchased at 6 (young), 12 (middle), and 21 (old) months of age to assess both age and lifelong CR effects. CR animals were maintained on the same absolute quantity of NIH-31/NIA Fortified Diet as arrival while AL animals were maintained on NIH-31 diet. Two separate cohorts were used for the study. The first cohort was used after a one-week acclimatization to housing conditions, while the second cohort was used after a one-week acclimatization and six-week assessment of synthesis (described below). Body weights were recorded upon arrival and every two weeks thereafter. The animals were individually housed and continued the NIA diets while being maintained until experimentation at the CSU Laboratory Animal Resource Center, at 18–26°C (dry bulb), 30–70% humidity, and a 12-hr light/dark cycle. All procedures at the facility meet or exceed the standards for facilities housing animals as described in the Animal Welfare Act regulations, the Guide for the Care and Use of Laboratory Animals and the Guide for the Care and Use of Agricultural Animals in Agricultural Research and Teaching and was approved by the CSU Animal Care and Use Committee (protocol #09-022A).
Sixteen hours prior to sacrifice, all food, but not water, was removed from the animals’ cages. Animals were anesthetized with an intraperitoneal (i.p.) injection of sodium pentobarbital. Blood was then obtained by cardiac puncture (approximately 1 ml) followed by rapid excision and cryopreservation of the heart, liver, the posterior aspect of both the distal hind limbs (mixed skeletal muscle), and extraction of marrow from the tibia. All tissues were stored at −80°C until analysis.
Labeled water
The use of heavy water (2H2O) allows simultaneous assessment of multiple synthetic processes. In this case we assessed the synthesis of mitochondrial protein and DNA in three tissues (heart, liver, and skeletal muscle) according to procedures previously described (Neese et al. 2002; Busch et al. 2006). After acclimation (1 week) to the CSU housing environment, animals received an i.p. injection of 99% enriched 2H2O calculated to enrich the body water pool (assumed 60% of body weight) to 5% (Neese et al. 2002). Animals were then allowed to drink ad lib water enriched to 4% for the next 6 weeks.
Tissue isolation
Tissue from heart, liver, and skeletal muscle were fractionated according to our previously published procedures (Robinson et al. 2010). Tissue (50–70 mg) was homogenized 1:10 in isolation buffer (100 mM KCl, 40 mM Tris HCl, 10 mM Tris Base, 5 mM MgCl2, 1 mM EDTA, 1 mM ATP, pH=7.5) with phosphatase and protease inhibitors (HALT, Thermo Scientific) using a bead homogenizer (Next Advance Inc, Averill Park NY). After homogenization the samples were centrifuged at low speed (800g) for 10 min at 4°C (Eppendorf, model 5415R) from which the pellet was stored. The supernatant from the low speed spin was centrifuged at 9000g for 10 min. The pellet from the 9000g spin was resuspended in a buffer two (100 mM KCl, 10 mM Tris HCl, 10 mM Tris Base, 1 mM MgCl2, 0.1 mM EDTA, 0.02 mM ATP, 1.5% BSA, pH=7.5) and centrifuged at 8000g for 10 min at 4°C. The supernatant was discarded, the pellet was resuspended in buffer two, and centrifuged at 6000g for 10 min at 4°C. From this spin, the supernatant was discarded and the remaining mitochondrial pellet (Mito) was washed in ethanol and water.
Preparation of analytes for mass spectrometric analyses
Protein was hydrolyzed by incubation in 6 N HCl at 120°C for 24 hr. The hydrolysates were ion-exchanged, dried under vacuum and then suspended in 1 ml of 50% acetonitrile, 50 mM K2HPO4, pH 11. Twenty microliters of pentafluorobenzyl bromide (Pierce Scientific, Rockford, IL, USA) were added, and the sealed mixture was incubated at 100°C for 1 h. Derivatives were extracted into ethyl acetate and the organic layer was removed and dried by addition of solid Na2SO4 followed by vacuum centrifugation.
GC-MS analysis of derivatized amino acids
Using negative chemical ionization (NCI), derivatized amino acids (AA) were analyzed on a DB225 gas chromatograph column. The starting temperature was 100°C, increasing 10°C per minute to 220°C. The mass spectrometry used NCI with helium as the carrier gas and methane as the reagent gas. The mass-to-charge ratios of 448, 449, and 450 were monitored for the pentafluorobenzyl-N,N-di(pentafluorobenzyl)alaninate derivative. In all cases, these mass-to-charge ratios represented the primary daughter ions that included all of the original hydrocarbon bonds from the given amino acid. 2H enrichment was calculated as described previously (Fanara et al. 2004). The newly synthesized fraction (f) of muscle proteins was calculated from the true precursor enrichment (p) using mass isotopomer distribution analysis [MIDA (Hellerstein & Neese 1999; Busch et al. 2006)]. Protein synthesis was calculated as the change in enrichment of deuterium labeled alanine (Busch et al. 2006) bound in muscle proteins over the entire labeling period and expressed as the common unit for protein synthesis rates (%*hr−1).
DNA isolation
Total DNA (~8 µg) was extracted from approximately 30 mg tissue following manufacturer’s instructions (MiniDNA kit, Qiagen) and eluted in 200 µl TE buffer (10 mM Tris, 1 mM EDTA, pH 8.0). The remaining DNA was precipitated with cold ethanol, suspended into 200 µl nuclease free H2O and hydrolyzed to free deoxyribonucleic acids. The deoxyadenosine fraction was separated and analyzed for deuterium content by GC-MS as previously described (Busch et al. 2006).
DNA synthesis measurement
Determination of 2H incorporation into purine deoxyribose (dR) of DNA was performed as described previously (Busch et al. 2006). Briefly, isolated DNA was hydrolyzed overnight at 37 °C with nuclease S1 and potato acid phosphatase. Hydrolysates were reacted with pentafluorobenzyl hydroxylamine and acetic acid, then acetylated with acetic anhydride and 1-methylimidazole. Dichloromethane extracts were dried, resuspended in ethyl acetate, and analyzed by GC/MS on a DB-17 column with negative chemical ionization, using helium as carrier and methane as the reagent gas. The fractional molar isotope abundances at m/z 435 (M0 mass isotopomer) and 436 (M1) of the pentafluorobenzyl triacetyl derivative of purine dR were quantified using ChemStation software. Excess fractional M+1 enrichment (EM1) was calculated as:
where sample and standard (std) refer to the analyzed sample and an unenriched pentafluorobenzyl triacetyl purine dR derivative standard, respectively. The fractional synthesis rate (f) was calculated by a comparison to bone marrow cells in the same animal, which represents an essentially fully turned over population of cells.
Real-time PCR
Real-time PCR of nuclear gene transcripts related to mitochondrial biogenesis was performed using a custom PCR array (SABiosciences, Frederick MD). Each 384 well array included eight gene targets, two reference genes (for normalization), a mouse genomic DNA control (for potential contamination) and a reverse transcription control (to verify reverse transcription efficiency). Each plate included samples from 22 mice, an internal control and blank.
Total RNA (~4 µg) was extracted from ~30 mg of frozen tissue using standard Trizol® methodology (Invitrogen, Carlsbad CA) and treated with a column based DNAse kit (RNeasy, Qiagen, Valencia CA). RNA purity (260/280 and 260/230 > 1.9) and final concentration (80 ng/µl) were verified with a spectrophotometer (Nanodrop, Wilmington DE). RNA was considered intact by visualizing two distinct bands on a native 1% agarose gel following ethidium bromide staining. RNA was reverse transcribed to cDNA using an Rt2 First Strand Kit (SABiosciences, Frederick MD). Approximately 2 ng of cDNA and master mix were loaded into each well that contained the appropriate primers. Real-time PCR was performed using a hot-start (10 min at 95°C) followed by 45 cycles of denaturing (15 sec 95°C) and elongation (1 min 60°C, ramp 1°/sec) on a Lightcycler® 480 (Roche Diagnostic Corporation, Indianapolis IN). A melting curve was performed for each plate (60°C for 15 sec followed by 95°C with ramp at 4.8°/sec). The threshold cycle (Ct) for each sample was determined using a second-order derivative maximum and relative quantification calculated using the 2−ΔΔCt method (Livak & Schmittgen 2001) using two separate reference genes (Beta 2 microglobulin (B2M) and TaTa box binding protein (Tbp)). Data were not different between reference genes and are expressed as fold change increase following feeding normalized to B2M with the Young-AL group as the reference group.
Western blot
Approximately 30 mg of frozen tissue was homogenized (Next Advance Inc, Averill Park NY) in 500 µl of ice-cold buffer (100 mM KCl, 40 mM Tris HCl, 10 mM Tris Base, 5 mM MgCl2, 1 mM EDTA, 1 mM ATP, pH 7.4) and commercial protease with phosphate inhibitor (Halt, Thermo Fisher, Rockford IL). Samples were centrifuged (10 min, 10000g, 4°C), then the supernatant was removed and protein concentration determined using a bicinchoninic acid assay (Thermo Fisher, Rockford IL). Samples were diluted to the same concentration, boiled with Laemmlli buffer, then 20 µg (heart and skeletal muscle) or 30 µg (liver) were separated using 12% SDS-PAGE at 200V. A positive control for PGC-1α (SC-2394, Santa Cruz, CA, USA) was loaded on each gel. Proteins were transferred at 4°C (100 V for 60 minutes in 20% w/v methanol, 0.02 %w/v SDS, 25 mM Tris, 192 mM glycine, pH 8.3) to nitrocellulose paper and incubated in Superblock (Thermo Fisher, Rockford IL) for one hour. Antibodies were purchased from Cell Signaling Technologies (Boston, MA, USA; RpS6 #2217, RpS6 phospho-Ser[240/244] #2215, 4e-BP1 #9452, phospho-4e-BP1[Thr37/46] #9459, AMPK #2532, p-AMPK (Thr 172) #2531) or Santa Cruz Biotechnology (Santa Cruz, CA, USA; PGC-1α #SC-13067, COX4 #SC-58348). Blots were incubated overnight with primary antibodies diluted 1:500 (AMPK, phospho-AMPK), 1:250 (RpS6, phospho-RpS6, 4e-BP1, phospho-4eBP-1) or 1:200 (PGC-1α, COX4) in Superblock reagent. Blots were washed in tris-buffered saline with tween and incubated with anti-rabbit HRP conjugated secondary antibody diluted 1:2000 in Superblock with subsequent chemiluminescence detection (West Dura, Pierce, Rockford, IL, USA). Images were captured and densitometry analyzed using a UVP Bioimaging system (Upland CA, USA). Blots were probed for phosphorylated proteins first, then stripped and re-probed for total protein. Equal loading was verified using ponceau-s staining as well as actin antibodies (sc-8432, Santa Cruz Biotechnology, Santa Cruz CA).
Statistics
Statistical analysis was performed using PRISM v4.0c (GraphPad Software Inc, La Jolla CA). Differences between treatment (AL and CR) and age (young, middle, old) were compared using a two-way analysis of variance (ANOVA). When a significant difference was detected, post-hoc analysis was performed with Bonferroni's Multiple Comparison Test. Significance was set at p ≤ 0.05 and p values of < 0.10 are noted. Data are presented as means ± SEM.
Supplementary Material
Acknowledgements
Elise Donovan, Frederick Peelor, Dan Heusinger and Nellie Reuland are thanked for their help with the experiments. We thank Ryan Yee and Jon Bahn for their help with GC-MS analyses. We thank Gerald Bauma for equipment time. Finally, Michael Pagliassotti, Greg Cartee, and Manfred Diehl are acknowledged for their initial assistance and experimental advice. This projected was funded by NIH 1K01AG031829-01.
Footnotes
Author Contributions: BFM designed the study, performed the data collection, directed the study, analyzed the data, interpreted the data and wrote the manuscript. MMR developed the methods and collected and analyzed the data. MDB and MH developed methods and analyzed the data. KLH collected, analyzed, and interpreted the data. All authors contributed critical feedback to the manuscript.
Contributor Information
Benjamin F Miller, Email: Benjamin.f.miller@colostate.edu.
Matthew M Robinson, Email: Matthew.Robinson@ColoState.edu.
Matthew D. Bruss, Email: mattbruss@berkeley.edu.
Marc Hellerstein, Email: march@berkeley.edu.
Karyn L Hamilton, Email: Karyn.Hamilton@ColoState.edu.
References
- Anderson RM, Barger JL, Edwards MG, Braun KH, O'Connor CE, Prolla TA, Weindruch R. Dynamic regulation of PGC-1alpha localization and turnover implicates mitochondrial adaptation in calorie restriction and the stress response. Aging Cell. 2008;7:101–111. doi: 10.1111/j.1474-9726.2007.00357.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruss MD, Thompson AC, Aggarwal I, Khambatta CF, Hellerstein MK. The effects of physiological adaptations to calorie restriction on global cell proliferation rates. American journal of physiology. 2011;300:E735–E745. doi: 10.1152/ajpendo.00661.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch R, Kim Y-K, Neese RA, Schade-Serin V, Collins M, Awada M, Gardner JL, Beysen C, Marino ME, Misell LM, Hellerstein MK. Measurement of protein turnover rates by heavy water labeling of nonessential amino acids. Biochim Biophys Acta. 2006;1760:730–744. doi: 10.1016/j.bbagen.2005.12.023. [DOI] [PubMed] [Google Scholar]
- Civitarese AE, Carling S, Heilbronn LK, Hulver MH, Ukropcova B, Deutsch WA, Smith SR, Ravussin E. Calorie Restriction Increases Muscle Mitochondrial Biogenesis in Healthy Humans. PLoS Med. 2007;4:e76. doi: 10.1371/journal.pmed.0040076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day K, Shefer G, Shearer A, Yablonka-Reuveni Z. The depletion of skeletal muscle satellite cells with age is concomitant with reduced capacity of single progenitors to produce reserve progeny. Developmental biology. 2010;340:330–343. doi: 10.1016/j.ydbio.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards AG, Donato AJ, Lesniewski LA, Gioscia RA, Seals DR, Moore RL. Life-long caloric restriction elicits pronounced protection of the aged myocardium: A role for AMPK. Mechanisms of ageing and development. 2010 doi: 10.1016/j.mad.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- el Haj AJ, Lewis SE, Goldspink DF, Merry BJ, Holehan AM. The effect of chronic and acute dietary restriction on the growth and protein turnover of fast and slow types of rat skeletal muscle. Comparative biochemistry and physiology. 1986;85:281–287. doi: 10.1016/0300-9629(86)90251-3. [DOI] [PubMed] [Google Scholar]
- Fanara P, Turner S, Busch R, Killion S, Awada M, Turner H, Mahsut A, Laprade KL, Stark JM, Hellerstein MK. In vivo measurement of microtubule dynamics using stable isotope labeling with heavy water. Effect of taxanes. The Journal of biological chemistry. 2004;279:49940–49947. doi: 10.1074/jbc.M409660200. [DOI] [PubMed] [Google Scholar]
- Forner F, Foster LJ, Campanaro S, Valle G, Mann M. Quantitative proteomic comparison of rat mitochondria from muscle, heart, and liver. Mol Cell Proteomics. 2006;5:608–619. doi: 10.1074/mcp.M500298-MCP200. [DOI] [PubMed] [Google Scholar]
- Goldspink DF, Kelly FJ. Protein turnover and growth in the whole body, liver and kidney of the rat from the foetus to senility. The Biochemical journal. 1984;217:507–516. doi: 10.1042/bj2170507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbunova V, Bozzella MJ, Seluanov A. Rodents for comparative aging studies: from mice to beavers. Age (Dordr) 2008;30:111–119. doi: 10.1007/s11357-008-9053-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock CR, Han DH, Higashida K, Kim SH, Holloszy JO. Does calorie restriction induce mitochondrial biogenesis? A reevaluation. Faseb J. 2011;25:785–791. doi: 10.1096/fj.10-170415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison DE, Strong R, Sharp ZD, Nelson JF, Astle CM, Flurkey K, Nadon NL, Wilkinson JE, Frenkel K, Carter CS, Pahor M, Javors MA, Fernandez E, Miller RA. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–395. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellerstein MK. New stable isotope-mass spectrometric techniques for measuring fluxes through intact metabolic pathways in mammalian systems: introduction of moving pictures into functional genomics and biochemical phenotyping. Metabolic engineering. 2004;6:85–100. doi: 10.1016/j.ymben.2003.10.005. [DOI] [PubMed] [Google Scholar]
- Hellerstein MK, Neese RA. Mass isotopomer distribution analysis at eight years: theoretical, analytic, and experimental considerations. The American journal of physiology. 1999;276:E1146–E1170. doi: 10.1152/ajpendo.1999.276.6.E1146. [DOI] [PubMed] [Google Scholar]
- Hock MB, Kralli A. Transcriptional control of mitochondrial biogenesis and function. Annu Rev Physiol. 2009;71:177–203. doi: 10.1146/annurev.physiol.010908.163119. [DOI] [PubMed] [Google Scholar]
- Inoki K, Zhu T, Guan K-L. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003;115:577–590. doi: 10.1016/s0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- Kadi F, Ponsot E. The biology of satellite cells and telomeres in human skeletal muscle: effects of aging and physical activity. Scandinavian journal of medicine & science in sports. 2010;20:39–48. doi: 10.1111/j.1600-0838.2009.00966.x. [DOI] [PubMed] [Google Scholar]
- Lewis SE, Goldspink DF, Phillips JG, Merry BJ, Holehan AM. The effects of aging and chronic dietary restriction on whole body growth and protein turnover in the rat. Experimental gerontology. 1985;20:253–263. doi: 10.1016/0531-5565(85)90050-6. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lok E, Scott FW, Mongeau R, Nera EA, Malcolm S, Clayson DB. Calorie restriction and cellular proliferation in various tissues of the female Swiss Webster mouse. Cancer Lett. 1990;51:67–73. doi: 10.1016/0304-3835(90)90232-m. [DOI] [PubMed] [Google Scholar]
- Maniura-Weber K, Goffart S, Garstka HL, Montoya J, Wiesner RJ. Transient overexpression of mitochondrial transcription factor A (TFAM) is sufficient to stimulate mitochondrial DNA transcription, but not sufficient to increase mtDNA copy number in cultured cells. Nucleic acids research. 2004;32:6015–6027. doi: 10.1093/nar/gkh921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merry BJ, Holehan AM, Lewis SE, Goldspink DF. The effects of ageing and chronic dietary restriction on in vivo hepatic protein synthesis in the rat. Mechanisms of ageing and development. 1987;39:189–199. doi: 10.1016/0047-6374(87)90008-x. [DOI] [PubMed] [Google Scholar]
- Mootha VK, Bunkenborg J, Olsen JV, Hjerrild M, Wisniewski JR, Stahl E, Bolouri MS, Ray HN, Sihag S, Kamal M, Patterson N, Lander ES, Mann M. Integrated analysis of protein composition, tissue diversity, and gene regulation in mouse mitochondria. Cell. 2003;115:629–640. doi: 10.1016/s0092-8674(03)00926-7. [DOI] [PubMed] [Google Scholar]
- Neese RA, Misell LM, Turner S, Chu A, Kim J, Cesar D, Hoh R, Antelo F, Strawford A, McCune JM, Christiansen M, Hellerstein MK. Measurement in vivo of proliferation rates of slow turnover cells by 2H2O labeling of the deoxyribose moiety of DNA. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:15345–15350. doi: 10.1073/pnas.232551499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nisoli E, Tonello C, Cardile A, Cozzi V, Bracale R, Tedesco L, Falcone S, Valerio A, Cantoni O, Clementi E, Moncada S, Carruba MO. Calorie restriction promotes mitochondrial biogenesis by inducing the expression of eNOS. Science (New York, N.Y. 2005;310:314–317. doi: 10.1126/science.1117728. [DOI] [PubMed] [Google Scholar]
- Palacios OM, Carmona JJ, Michan S, Chen KY, Manabe Y, Ward JL, 3rd, Goodyear LJ, Tong Q. Diet and exercise signals regulate SIRT3 and activate AMPK and PGC-1alpha in skeletal muscle. Aging (Albany NY) 2009;1:771–783. doi: 10.18632/aging.100075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polak P, Hall MN. mTOR and the control of whole body metabolism. Curr Opin Cell Biol. 2009;21:209–218. doi: 10.1016/j.ceb.2009.01.024. [DOI] [PubMed] [Google Scholar]
- Raman A, Schoeller DA, Subar AF, Troiano RP, Schatzkin A, Harris T, Bauer D, Bingham SA, Everhart JE, Newman AB, Tylavsky FA. Water turnover in 458 American adults 40–79 yr of age. Am J Physiol Renal Physiol. 2004;286:F394–F401. doi: 10.1152/ajprenal.00295.2003. [DOI] [PubMed] [Google Scholar]
- Rattan SI. Synthesis, modifications, and turnover of proteins during aging. Experimental gerontology. 1996;31:33–47. doi: 10.1016/0531-5565(95)02022-5. [DOI] [PubMed] [Google Scholar]
- Robinson MM, Richards JC, Hickey MS, Moore DR, Phillips SM, Bell C, Miller BF. Acute {beta}-adrenergic stimulation does not alter mitochondrial protein synthesis or markers of mitochondrial biogenesis in adult men. Am J Physiol Regul Integr Comp Physiol. 2010;298:R25–R33. doi: 10.1152/ajpregu.00524.2009. [DOI] [PubMed] [Google Scholar]
- Robinson MM, Turner SM, Hellerstein MK, Hamilton KL, Miller BF. Long-term synthesis rates of skeletal muscle DNA and protein are higher during aerobic training in older humans than in sedentary young subjects but are not altered by protein supplementation. Faseb J. 2011 doi: 10.1096/fj.11-186437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolfe DF, Brown GC. Cellular energy utilization and molecular origin of standard metabolic rate in mammals. Physiological reviews. 1997;77:731–758. doi: 10.1152/physrev.1997.77.3.731. [DOI] [PubMed] [Google Scholar]
- Rooyackers OE, Adey DB, Ades PA, Nair KS. Effect of age on in vivo rates of mitochondrial protein synthesis in human skeletal muscle. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:15364–15369. doi: 10.1073/pnas.93.26.15364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan MT, Hoogenraad NJ. Mitochondrial-nucleaRcommunications. Annu Rev Biochem. 2007;76:701–722. doi: 10.1146/annurev.biochem.76.052305.091720. [DOI] [PubMed] [Google Scholar]
- Short KR, Bigelow ML, Kahl J, Singh R, Coenen-Schimke J, Raghavakaimal S, Nair KS. Decline in skeletal muscle mitochondrial function with aging in humans. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:5618–5623. doi: 10.1073/pnas.0501559102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtman ER. Protein oxidation and aging. Science (New York, N.Y. 1992;257:1220–1224. doi: 10.1126/science.1355616. [DOI] [PubMed] [Google Scholar]
- Sun J, Conn CS, Han Y, Yeung V, Qian SB. PI3K-mTORC1 attenuates stress response by inhibiting cap-independent Hsp70 translation. The Journal of biological chemistry. 2011;286:6791–6800. doi: 10.1074/jbc.M110.172882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trifunovic A, Wredenberg A, Falkenberg M, Spelbrink JN, Rovio AT, Bruder CE, Bohlooly YM, Gidlof S, Oldfors A, Wibom R, Tornell J, Jacobs HT, Larsson NG. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429:417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- Verdijk LB, Koopman R, Schaart G, Meijer K, Savelberg HH, van Loon LJ. Satellite cell content is specifically reduced in type II skeletal muscle fibers in the elderly. American journal of physiology. 2007;292:E151–E157. doi: 10.1152/ajpendo.00278.2006. [DOI] [PubMed] [Google Scholar]
- Wagner A. Energy Constraints on the Evolution of Gene Expression. Molecular Biology and Evolution. 2005;22:1365. doi: 10.1093/molbev/msi126. [DOI] [PubMed] [Google Scholar]
- Wilson FA, Suryawan A, Orellana RA, Gazzaneo MC, Nguyen HV, Davis TA. Differential effects of long-term leucine infusion on tissue protein synthesis in neonatal pigs. Amino Acids. 2011;40:157–165. doi: 10.1007/s00726-010-0629-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan CL, Sharma N, Gilge DA, Stanley WC, Li Y, Hatzoglou M, Previs SF. Preserved protein synthesis in the heart in response to acute fasting and chronic food restriction despite reductions in liver and skeletal muscle. American journal of physiology. 2008;295:E216–E222. doi: 10.1152/ajpendo.00545.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zangarelli A, Chanseaume E, Morio B, Brugere C, Mosoni L, Rousset P, Giraudet C, Patrac V, Gachon P, Boirie Y, Walrand S. Synergistic effects of caloric restriction with maintained protein intake on skeletal muscle performance in 21-month-old rats: a mitochondria-mediated pathway. Faseb J. 2006;20:2439–2450. doi: 10.1096/fj.05-4544com. [DOI] [PubMed] [Google Scholar]
- Zid BM, Rogers AN, Katewa SD, Vargas MA, Kolipinski MC, Lu TA, Benzer S, Kapahi P. 4E-BP extends lifespan upon dietary restriction by enhancing mitochondrial activity in Drosophila. Cell. 2009;139:149–160. doi: 10.1016/j.cell.2009.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.