Replication forks stalled by excess DNA supercoiling can be resolved by DNA cleavage by the Mus81 endonuclease.
Abstract
Deoxyribonucleic acid (DNA) topoisomerases are essential for removing the supercoiling that normally builds up ahead of replication forks. The camptothecin (CPT) Top1 (topoisomerase I) inhibitors exert their anticancer activity by reversibly trapping Top1–DNA cleavage complexes (Top1cc’s) and inducing replication-associated DNA double-strand breaks (DSBs). In this paper, we propose a new mechanism by which cells avoid Top1-induced replication-dependent DNA damage. We show that the structure-specific endonuclease Mus81-Eme1 is responsible for generating DSBs in response to Top1 inhibition and for allowing cell survival. We provide evidence that Mus81 cleaves replication forks rather than excises Top1cc’s. DNA combing demonstrated that Mus81 also allows efficient replication fork progression after CPT treatment. We propose that Mus81 cleaves stalled replication forks, which allows dissipation of the excessive supercoiling resulting from Top1 inhibition, spontaneous reversal of Top1cc, and replication fork progression.
Introduction
Top1 (topoisomerase I) regulates DNA topology during replication, transcription, and chromatin remodeling (Champoux, 2001; Wang, 2002). It removes DNA torsional stress (supercoiling) by forming cleavage complexes (Top1–DNA cleavage complexes [Top1cc’s]) in which one DNA strand is cleaved by covalent binding of Top1 to a 3′ DNA phosphate. After DNA relaxation, Top1cc’s reverse rapidly, and Top1 is released as the DNA religates. The plant alkaloid camptothecin (CPT) and its clinical derivatives, topotecan and irinotecan, are highly selective Top1 inhibitors that reversibly trap Top1cc by binding at the enzyme–DNA interface (Hsiang et al., 1985; Pommier et al., 2010). At pharmacological concentrations, they kill cancer cells in a replication-dependent manner (Holm et al., 1989; Hsiang et al., 1989), as replication forks collide with the stabilized Top1cc’s. Such collisions are because Top1cc’s do not reverse fast enough ahead of moving replication forks (Pommier et al., 2006), resulting in DNA double-strand ends (DSEs; Hsiang et al., 1989; Strumberg et al., 2000).
The repair of Top1-mediated DNA damage requires both the excision of the covalently attached Top1 from the DNA and the repair of the DSE resulting from replication. A specialized pathway for the excision of Top1cc hinges on TDP1 (tyrosyl-DNA phosphodiesterase I), an enzyme conserved from yeast to humans, which hydrolyzes the covalent bond between Top1 and the 3′ DNA end (Yang et al., 1996; Pouliot et al., 1999; Interthal et al., 2005; Dexheimer et al., 2008a). Genetic studies also indicate that Top1cc can be repaired by other TDP1-independent pathways involving the endonucleases Rad1XPF-Rad10ERCC1 and Mus81-Mms4EME1 (Liu et al., 2002; Vance and Wilson, 2002; Deng et al., 2005; Ciccia et al., 2008; Zhang et al., 2011).
Mus81-Eme1 is a heterodimeric endonuclease that acts preferentially on DNA substrates mimicking stalled replication forks and nicked Holliday junctions in vitro (Interthal and Heyer, 2000; Fricke et al., 2005; Ciccia et al., 2008; Ehmsen and Heyer, 2008). It cleaves such structures in the duplex region adjacent to the branched point (Ehmsen and Heyer, 2009). Mus81-Eme1 has been shown to play a critical role in both replication fork rescue and homologous recombination. It converts collapsed replication forks into DNA double-strand breaks (DSBs; Hanada et al., 2006, 2007; Franchitto et al., 2008; Froget et al., 2008; Shimura et al., 2008) and is considered a back-up system for the resolution of replication fork stalling in the absence of RecQ helicases (Kaliraman et al., 2001; Mullen et al., 2001; Doe et al., 2002; Trowbridge et al., 2007; Franchitto et al., 2008; Shimura et al., 2008). Mus81 is also required for the proper completion of homologous recombination, both during meiosis (Smith et al., 2003; Jessop and Lichten, 2008; Oh et al., 2008) and mitosis (Blais et al., 2004; Roseaulin et al., 2008).
In the present study, we explore the role of Mus81 in the cellular response of human cells to Top1 inhibitors. We show that Mus81 is not involved in the direct excision of Top1cc but rather participates in the repair and recovery of damaged replication forks by incision of the stalled replication forks.
Results
Mus81-deficient cells are hypersensitive to CPT
To assess the potential role of Mus81 in the cellular responses to Top1 inhibitors, we compared survival of wild-type (WT) and Mus81-deficient (Mus81−/−) human HCT116 colon carcinoma cells (Fig. 1 A) after CPT treatment (Holm et al., 1989). Cells were exposed to CPT for 1 h, and CPT sensitivity was assessed by clonogenic survival assays. Fig. 1 B shows that Mus81-deficient cells are significantly more sensitive than WT cells to pharmacological concentrations (0.01, 0.1, and 1 µM) of CPT, which selectively target replicating cells (Huang et al., 2010). At high concentrations of CPT (10 µM), which exceed those achieved in cancer therapy and for which cell lethality is primarily related to transcription-induced damage (Sordet et al., 2009; Zhang et al., 2011), >98% of the WT cells were killed, and the impact of Mus81 knockdown was minimal (Fig. 1 B). These results demonstrate the involvement of Mus81 in the cellular response to Top1 inhibitors under conditions in which Top1 inhibitors primarily target S-phase cells.
Figure 1.
Mus81-deficient cells are hypersensitive to CPT. (A) Western blotting analysis of Mus81 expression in WT and Mus81−/− HCT116 cells. Actin was used as a loading control. (B) Mus81 deficiency decreases survival of HCT116 cells after CPT exposure. Cells were treated with the indicated concentrations of CPT for 1 h and cultured 13 d to allow colony formation. The surviving fraction of untreated cells was defined as 100%. Mean values ± SEM from six independent experiments are shown. *, P < 0.05; **, P < 0.01; Mann–Whitney test. (C) Cell cycle profiles of untreated or CPT-treated WT and Mus81−/− cells. Cells were incubated with 1 µM CPT for 1 h and were pulse labeled with BrdU during the last 10 min. The x axis shows DNA content, and the y axis shows BrdU incorporation. Boxed dotted lines and percentages indicate S-phase cell subpopulations.
To investigate whether the hypersensitivity of Mus81−/− cells resulted from an enrichment in S-phase cells, we determined the cell cycle distributions of WT and Mus81−/− cells by BrdU incorporation assays. WT and Mus81−/− cells showed similar cell cycle profiles, both in the absence and presence of 1 µM CPT for 1 h (Fig. 1 C). Thus, Mus81 deficiency did not result in a higher proportion of S-phase cells, which excludes the possibility that more cells in S phase could account for the increased sensitivity to CPT.
Mus81 does not excise Top1cc’s
Because Mus81-Eme1 is a 3′ flap endonuclease, it has been postulated that it would cleave 5′ from the Top1cc and participate in the excision of trapped Top1 from the DNA (Liu et al., 2002; Vance and Wilson, 2002; Deng et al., 2005). To determine whether Mus81 is involved in Top1cc removal, we analyzed the formation of Top1cc’s in WT and Mus81−/− cells treated with various CPT concentrations. Top1cc’s were detected by Western blotting of cellular DNA fractions with an anti-Top1 antibody (Miao et al., 2006; Zhang et al., 2011). Top1cc levels were similar in WT and Mus81−/− cells upon CPT treatment (Fig. 2, A and B). In addition, Top1cc reversed efficiently in both WT and Mus81−/− cells after CPT removal (Fig. 2 B). Consistent with these results, Mus81 silencing in MDA-MB-231 breast cancer cells did not impair the formation or reversal of Top1cc in response to CPT (Fig. S1, A and B). Under the same experimental conditions, TDP1 silencing resulted in higher Top1cc levels, both during and after CPT treatment (Fig. S2; Miao et al., 2006).
Figure 2.
Mus81-deficient cells are not defective in the removal of Top1cc. (A–C) Top1 trapping and reversal are similar in WT and Mus81−/− cells. (A) Top1cc levels in WT and Mus81−/− HCT116 cells treated with the indicated concentrations of CPT for 1 h. Data from three independent experiments are shown. (B) Detection of Top1cc in WT and Mus81−/− HCT116 cells treated with 1 µM CPT for 1 h. R30’, cells were harvested 30 min after CPT removal. Different concentrations of genomic DNA (5, 2.5, and 1.25 µg) were probed with an anti-Top1 antibody. (left) Representative experiment. Dashed lines indicate that intervening wells have been spliced out. (right) Quantification of Top1cc (n = 3). (A and B, right) Dashed lines indicate Top1cc levels in WT untreated cells. (C) Measurement of DNA–Top1 cross-links by alkaline elution in WT and Mus81−/− cells treated with 1 µM CPT for 1 h. R30’ and R60’, cells were harvested 30 and 60 min after CPT removal. (left) Representative alkaline elution experiment. (right) Quantification of DNA–Top1 cross-linking (in rad equivalents [rad-eq]; Covey et al., 1989). Data from four independent experiments are shown. Mean values ± SEM are shown. A.U., arbitrary unit; Ctrl, control.
Alkaline elution assays were used to detect Top1cc’s as DNA–protein cross-links (DPCs; Miao et al., 2006) in WT and Mus81−/− cells treated with CPT. DPCs formed similarly in WT and Mus81−/− cells (Fig. 2 C, left, top curves). 30 min after CPT removal, reversal of DPC was partial but was not significantly different in WT and Mus81−/− cells (Fig. 2 C). DPCs fully reversed after 60 min in both WT and Mus81−/− cells (Fig. 2 C, right). Together, these results demonstrate that Mus81 is not involved in the excision of Top1 from the DNA.
Mus81 generates DNA DSBs in CPT-treated cells
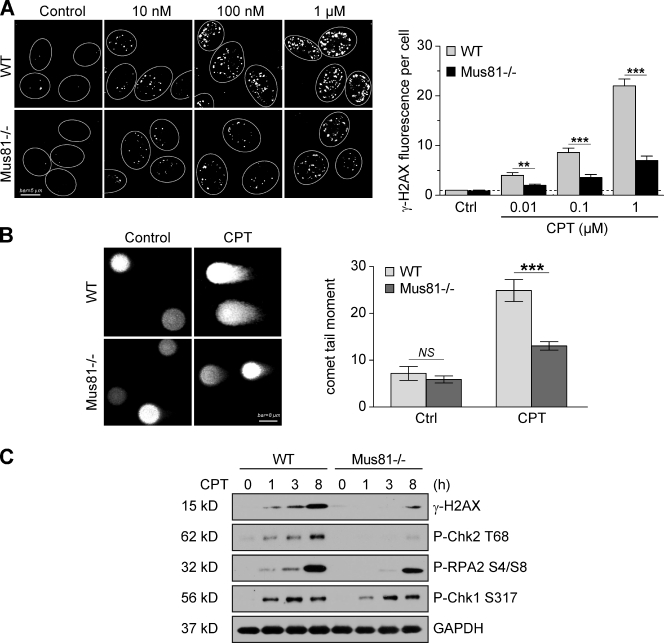
To elucidate the role of Mus81 in the cellular response to Top1-induced DNA damage, we next analyzed the influence of Mus81 on the formation of Top1-induced DNA DSBs. WT and Mus81−/− cells were exposed to various concentrations of CPT for 1 h, and the DSB marker γ-H2AX (Rogakou et al., 1998; Furuta et al., 2003) was examined by immunofluorescence. CPT-induced γ-H2AX levels were greatly reduced in Mus81-deficient cells (Fig. 3 A), indicating that Mus81 is involved in the formation of DSBs in CPT-treated cells. This result was further supported by neutral comet assays showing that Mus81−/− cells accumulated less DSBs than WT cells in response to CPT (Fig. 3 B). Time-course analysis showed reduced γ-H2AX induction in Mus81-deficient cells at both short and long time points after CPT treatment (Fig. 3 C).
Figure 3.
Mus81-dependent induction of DNA DSBs and DNA damage response in CPT-treated cells. (A) Immunostaining analysis of γ-H2AX induction in WT and Mus81−/− cells treated with the indicated concentrations of CPT for 1 h. (left) Representative microscopy images. Circles indicate nuclear contours. (right) Quantification of γ-H2AX fluorescence per cell. Fluorescence signals were scored in ∼60 cells per sample. (B) DNA DSBs measured by neutral comet assay in WT and Mus81−/− HCT116 cells treated with 1 µM CPT for 1 h. (left) Representative comet images. (right) Quantification of comet tail moments. At least 50 cells per sample were scored. (C) DNA damage response (phospho-H2AX (γ-H2AX), phospho [P]-Chk2, -RPA2, and -Chk1) in WT and Mus81−/− HCT116 cells treated with 1 µM CPT. GAPDH was used as a loading control. Mean values ± SEM are shown. **, P < 0.001; ***, P < 0.0001; Mann–Whitney test. Ctrl, control.
Mus81−/− cells also demonstrated a lower and delayed induction of other DNA damage response signals in response to CPT, including phosphorylation of Chk2 at threonine 68 (ATM [ataxia telangiectasia mutated] substrate) and phosphorylation of RPA2 (Fig. 3 C). However, phosphorylation of Chk1 at serine 317 (ATR [ataxia telangiectasia and Rad3 related] substrate) was induced similarly in Mus81−/− and WT cells upon CPT treatment, indicating reduced DSBs but efficient activation of the replication checkpoint in Mus81−/− cells (Fig. 3 C).
siRNA-mediated depletion of Mus81 in MDA-MB-231 cells also resulted in a marked reduction of CPT-induced γ-H2AX (Fig. S1, C and D). As Mus81 functions as a heterodimer with Eme1 (Ciccia et al., 2008), we measured γ-H2AX induction upon CPT exposure in HCT116 cells transfected with Eme1-targeting siRNAs. Eme1 silencing resulted in a marked reduction in CPT-induced γ-H2AX, recapitulating the phenotype of Mus81−/− cells (Fig. S1 E). Collectively, these results demonstrate that Mus81 is involved in the formation of DSBs in CPT-treated cells.
Mus81-dependent DNA DSBs form at replication foci
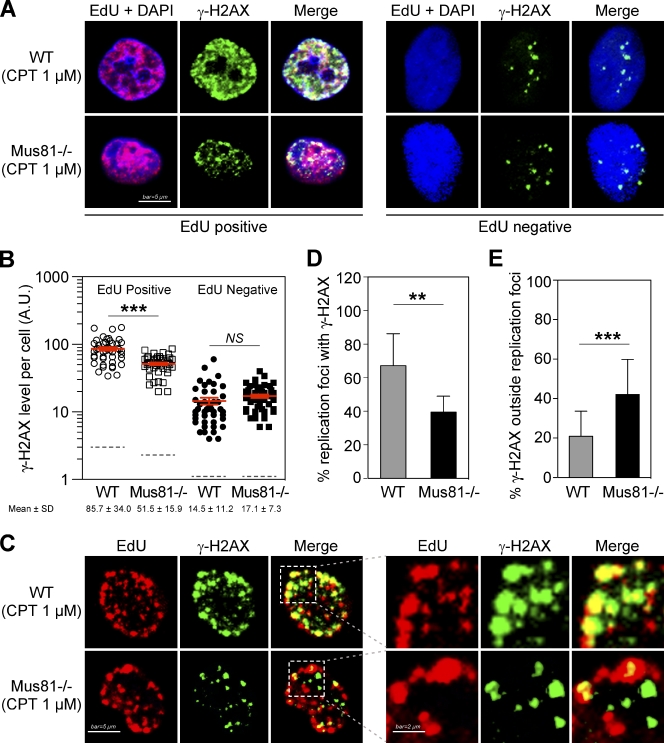
CPT-induced DSBs result from interference of stabilized Top1cc with either replication or transcription machineries (Hsiang et al., 1989; Ryan et al., 1991; Strumberg et al., 2000; Pommier et al., 2006; Sordet et al., 2009). To investigate whether Mus81-dependent DNA breaks are related to replication (Seiler et al., 2007) or transcription (Sordet et al., 2009; Zhang et al., 2011) in CPT-treated cells, we next analyzed γ-H2AX induction in replicating and nonreplicating cell subpopulations. WT and Mus81−/− cells were labeled with the thymidine analogue 5-ethynyl-2′-deoxyuridine (EdU; to detect replicating cells) before and during CPT treatment. EdU and γ-H2AX costaining showed that γ-H2AX was induced in both replicating (EdU positive) and nonreplicating (EdU negative) cells, with higher γ-H2AX levels being observed in the replicating cells (Fig. 4 A). Mus81 deficiency was associated with significantly lower γ-H2AX levels in replicating cells but did not affect γ-H2AX induction in nonreplicating cells (Fig. 4, A and B).
Figure 4.
Mus81-dependent DNA DSBs form at replication foci. Replication factories were labeled with EdU for 90 min. 1 µM CPT was added during the last 60 min. (A and B) Analysis of CPT-induced γ-H2AX in replicating (EdU positive) or nonreplicating (EdU negative) WT or Mus81−/− cells. (A) Representative microscopy images showing Mus81-dependent γ-H2AX response in replicating cells. (B) Quantification of γ-H2AX fluorescence signals in individual cells. 90–100 cells were scored per sample. Each dot represents a single cell. Mean values ± SEM are shown in red. Dotted lines indicate γ-H2AX mean levels in untreated cells. (C) Representative microscopy images showing the colocalization of EdU and γ-H2AX foci in EdU-positive cells. (right) Magnified images of squared areas from left images. (D and E) Detailed analysis of EdU and γ-H2AX colocalization in EdU-positive cells demonstrating that Mus81 is involved in γ-H2AX formation at replication foci. (D) Percentages of replication foci colocalized with γ-H2AX (defined as 100% x [number of EdU foci colocalizing with γ-H2AX foci divided by the total number of EdU foci]). (E) Percentages of γ-H2AX foci outside replication foci (defined as 100% x [number of γ-H2AX foci that do not colocalize with EdU foci divided by the total number of γ-H2AX foci]). 20 individual cells were scored per sample. Mean percentages ± SD are shown. **, P < 0.005; ***, P < 0.0001; Student’s t test. Blue, DAPI nuclear staining; red, EdU; green, γ-H2AX. A.U., arbitrary unit.
The occurrence of Mus81-dependent DNA breaks in replicating cells is consistent with the cleavage of stalled DNA replication forks by Mus81 (see Introduction). To address this possibility, we performed single-cell analyses of EdU and γ-H2AX foci. In WT cells treated with CPT, 68% of the replication foci (labeled with EdU) colocalized with γ-H2AX foci (Fig. 4, C and D; and Fig. S3, top), which is consistent with the preferential induction of DSBs by CPT in replication foci (Seiler et al., 2007). On the other hand, in Mus81-deficient cells, the fraction of damaged replication foci (i.e., those EdU foci colocalizing with γ-H2AX foci) was significantly decreased (39%; Fig. 4, C and D; and Fig. S3, bottom), and the fraction of γ-H2AX foci outside the EdU foci was significantly increased (Fig. 4, C and E; and Fig. S3). These results indicate the selective induction of Mus81-dependent DSBs at replication foci.
Mus81 promotes replication fork progression after Top1 inhibition
To study the effects of Mus81 on replication fork progression, we analyzed replication in single DNA molecules using DNA combing (Conti et al., 2007; Seiler et al., 2007). Incorporation of the thymidine analogues iododeoxyuridine (IdU) and chlorodeoxyuridine (CldU) was visualized on stretched DNA fibers prepared from untreated or CPT-treated WT and Mus81−/− cells. Cells were labeled with IdU and CldU for 30 min each, and CPT was added during the last 20 min of the IdU pulse (Fig. 5 A). This protocol allows the analysis of both replication fork slow down (by comparing replication fork velocity in the presence and in the absence of CPT) and replication fork recovery (by comparing fork velocity during CPT treatment and after CPT removal). In both WT and Mus81−/− cells, CPT induced a marked reduction in replication fork velocity (Fig. 5, B and C; and Fig. S4). After CPT removal, CldU incorporation was measured in both WT and Mus81−/− cells (Fig. 5, B and C). In WT cells, replication fork velocity resumed to ∼50% of its mean value before CPT (Fig. 5 B, left), whereas, in Mus81−/− cells, there was no detectable recovery upon CPT removal (Fig. 5 B, right). These results show that Mus81 is associated with recovery of replication fork velocity after CPT removal.
Figure 5.
Mus81 promotes efficient replication fork progression. (A) Experimental protocol. Cells were labeled sequentially with IdU and CldU for 30 min each. 1 µM CPT was added during the last 20 min of the IdU pulse. IdU and CldU were detected on combed DNA fibers using specific antibodies in green and red, respectively. W, wash. (B) Analysis of replication forks during and after CPT exposure, demonstrating efficient replication fork recovery in WT but not in Mus81−/− cells. At least 160 green/red replication signals were measured per sample. Each dot represents a single replication signal. Mean values ± SEM are shown in black. P-values were calculated using the Student’s t test. n, number of replication signals measured. (C) Representative images and schematic drawings of replication signals detected on DNA fibers. Ctrl, control.
Discussion
CPT and its clinically used derivatives topotecan and irinotecan are primarily cytotoxic to S-phase cells at pharmacological concentrations (Holm et al., 1989; Hsiang et al., 1989), as they induce replication-associated DSBs that are primarily processed by homologous recombination (Eng et al., 1988; Nitiss and Wang, 1988; Vance and Wilson, 2002; Hochegger et al., 2006; Nakamura et al., 2010). At least some of those DSBs have been shown to result from the conversion of Top1-associated single-strand breaks into DSBs by replication runoff (Fig. 6 A; Hsiang et al., 1989; Tsao et al., 1993; Shao et al., 1999; Strumberg et al., 2000).
Figure 6.
Model for the processing of stalled replication forks by Mus81-Eme1. (A) Classical model illustrating Mus81-independent induction of replication-associated DNA DSE by replication fork runoff (Strumberg et al., 2000). (B–H) Novel model showing Mus81-mediated DSBs. A Top1cc on the leading strand is shown, but the same would apply if the Top1cc was on the lagging strand. (B) Top1 trapping by CPT (green rectangle) results in the accumulation of positive supercoiling (Sc+), which stalls the replication fork. Purple triangle, DNA polymerase complex. (C) Cleavage of the stalled fork by Mus81-Eme1. (D) Cleavage of the template DNA allows supercoiling relaxation and 5′-end exonucleolytic resection. (E) After spontaneous dissociation of CPT, Top1cc’s reverse, and the newly synthesized strands hybridize, forming a “chicken foot”; alternatively, the fork cleaved by Mus81 can be processed by homologous recombination. (F) Annealing of the template strands. (G) Gap filling by DNA polymerase. (H) Fork reestablishment and replication restart.
Our study proposes an alternative by showing that the structure-specific endonuclease Mus81-Eme1 is responsible for the generation of DSBs at stalled replication forks in response to Top1 trapping. This conclusion is based on our findings that Mus81 inactivation either by stable knockout (Hiyama et al., 2006) or siRNA reduces CPT-induced DSBs selectively in replicating cells and at replication foci (Fig. 4). Our conclusion is consistent with the known biochemical activities of Mus81-Eme1, which preferentially processes substrates mimicking replication forks (Kaliraman et al., 2001; Doe et al., 2002; Fricke et al., 2005; Ehmsen and Heyer, 2008). The Mus81-dependent DSBs are not lethal but are associated with DNA repair. Indeed, inactivation of Mus81 results in more cell killing (Fig. 1 B) and less DSBs (Figs. 3 and 4). Mus81-dependent DNA breakage has also been observed in mammalian cells upon treatment with cisplatin (Hanada et al., 2006) and with the replication fork-blocking drugs aphidicolin and hydroxyurea (Hanada et al., 2007; Franchitto et al., 2008; Froget et al., 2008; Shimura et al., 2008). These studies have established that Mus81 can convert a stalled replication fork into a DSB. Here, we show the involvement of Mus81 in the formation of replication-associated DSBs after CPT treatment rather than in the direct excision of Top1 from the DNA. Our findings are best explained by the possibility that Top1 inhibition leads to replication fork stalling, which is resolved by Mus81-dependent DNA cleavage (Fig. 6, B–H). This interpretation is consistent with an independent publication proposing that Top1 inhibition by CPT in yeast leads to supercoiling accumulation ahead of replication forks, leading to replication fork stalling (Koster et al., 2007).
Because Mus81 is responsible for only a fraction of the replication-associated DSBs after Top1 inhibition (Fig. 4, C and D), it is plausible that Top1-mediated replication-associated DSBs are produced both by Mus81-dependent and Mus81-independent mechanisms. Thus, it is likely that Top1 trapping induces replication-associated DSBs either by replication runoff (Fig. 6 A) or by Mus81-dependent cleavage of replication forks stalled by positive supercoils (Fig. 6 C). Replication fork cleavage by Mus81-Eme1 could allow the dissipation of the excessive superhelical tension (Fig. 6, C and D), which could resolve the topological block resulting from Top1 deficiency ahead of the fork. This hypothesis is supported by genetic experiments in yeast, showing that mutations in both Mus81 and Mms4Eme1 results in growth defects in top1Δ strains (Mullen et al., 2001). It is also supported by the recent findings that Top1 deficiency in yeast, murine, and human cells is associated with genomic instability (Christman et al., 1993; Miao et al., 2007; Tuduri et al., 2009). Top1 deficiency in yeast is associated with enhanced genomic instability in the ribosomal DNA cluster, suggesting that Top1 may be particularly important in highly transcribed regions (Christman et al., 1993). In mammalian cells, Top1 deficiency leads to an accumulation of stalled replication forks and DNA breakage at replication sites (Miao et al., 2007; Tuduri et al., 2009). Because Top1 is required for efficient DNA relaxation during replication (Kim and Wang, 1989; Koster et al., 2007), Top1-deficient cells probably accumulate unresolved supercoiling in replicating DNA, which further leads to replication fork stalling and breakage.
We also show that Mus81 is required for efficient replication fork progression after CPT removal. Parallel observations have been made in cells treated with hydroxyurea or aphidicolin (Hanada et al., 2007; Shimura et al., 2008), indicating that Mus81-induced DSBs act as resolution intermediates for replication fork recovery. DSBs arising during replication are repaired primarily by homologous recombination (Takata et al., 1998; Arnaudeau et al., 2001; Rothkamm et al., 2003; Sonoda et al., 2006). Thus, Mus81-dependent DSBs could support strand invasion and initiate sister chromatid exchange. Mus81 is also required for the resolution of Holliday junctions (Blais et al., 2004; Fricke et al., 2005; Ehmsen and Heyer, 2008; Jessop and Lichten, 2008; Oh et al., 2008; Roseaulin et al., 2008) and could therefore participate in both initiation and completion of homologous recombination during S phase (Fig. 6 E). Alternatively, Mus81-dependent fork cleavage could enable replication progression by allowing retraction of the replication machinery (Fig. 6, D–H), possibly in association with the Bloom helicase. After nucleolytic resection of the 5′ end of the DNA, annealing of the two newly synthesized DNA strands would initiate replication fork regression, allowing reannealing of the two parental DNA strands and gap filling by DNA polymerase.
In conclusion, our study demonstrates that the Mus81-Eme1 endonuclease is involved in the repair of Top1-mediated DNA damage by promoting the cleavage and restoration of stalled replication forks. This mechanism may not be limited to CPT-induced DNA damage, as Top1 can be trapped by a variety of endogenous DNA lesions, such as oxidized bases, mismatches, abasic sites, adducts, and strand breaks (Pourquier and Pommier, 2001; Pommier et al., 2006; Dexheimer et al., 2008b). Mus81 is therefore a novel and important determinant of the cellular response of cancer cells to Top1 inhibitors and replication fork stalling by Top1 deficiency and excessive supercoiling.
Materials and methods
Cell lines and drugs
Human MDA-MB-231 breast adenocarcinoma cells were obtained from the Developmental Therapeutics Program (National Cancer Institute) and were maintained in RPMI 1640 supplemented with 10% fetal bovine serum. Human HCT116 colorectal carcinoma cells and HCT116 Mus81−/− cells (Hiyama et al., 2006) were grown in DME supplemented with 10% fetal bovine serum. CPT was obtained from the Drug Synthesis and Chemistry Branch (National Cancer Institute).
Clonogenic survival assay
After drug treatment, cells were seeded in 6-well plates at a density of 50, 250, or 500 cells/well and incubated for 10–13 d to allow colony formation. Colonies were fixed with methanol and stained with 0.05% methylene blue (Sigma-Aldrich) for 30 min. The surviving fraction was calculated by dividing the number of colonies in treated wells by the number of colonies in untreated wells.
Alkaline elution assays
DPCs were detected using alkaline elution as previously described (Covey et al., 1989). In brief, cells were radiolabeled overnight with 0.02 µCi/ml [14C]thymidine and chased with radioisotope-free medium 4 h before drug treatment. Cell aliquots were placed in ice-cold HBSS and irradiated with 30 grays to break the DNA. Cells were layered onto polyvinylchloride-acrylic copolymer (protein adsorbing) filters and lysed with LS-10 (2 M NaCl, 0.2% sarkosyl, and 0.04 M disodium EDTA, pH 10). DNA was eluted from the filters with tetrapropylammonium hydroxide-EDTA. After elution, filters were incubated for 1 h at 65°C with 1 M HCl and an additional hour at room temperature in the presence of 0.3 M NaCl. Radioactivity in fractions and filters was measured with a liquid scintillation analyzer (2200A Tri Carb Scintillation Analyzer; Packard Instruments).
BrdU incorporation assay
Cells were pulse labeled with 50 µM BrdU (EMD) during the last 10 min of CPT treatment. Cells were harvested by trypsinization, fixed in 70% ice-cold ethanol, and stored at −20°C. Cells were incubated for 30 min at room temperature in 2 N HCl–0.5% Triton X-100 to allow DNA denaturation. The medium was neutralized in 0.1 M sodium borate, pH 8.5, and cells were washed twice in PBS containing 0.5% Tween 20 and 0.5% bovine serum albumin. Cells were incubated for 1 h at room temperature with an FITC-conjugated anti-BrdU antibody (BD) and were treated with 0.5 mg/ml RNase A and 50 µg/ml propidium iodide. Samples were analyzed with a flow cytometer (FACScan; BD) using the CellQuest software (BD).
Detection of Top1cc’s
After drug treatment, cells were lysed in a reagent (DNAzol; Invitrogen), and genomic DNA was prepared according to the manufacturer’s instructions. Samples were sonicated briefly to shear the DNA. Varying concentrations of DNA were spotted on transfer membranes (Immobilon-FL; Millipore) using a slot-blot manifold (GE Healthcare). Membranes were blocked in blocking buffer (Odyssey; LI-COR Biosciences) and incubated successively with a C21 anti-Top1 antibody (gift from Y.-C. Cheng, Yale University, New Haven, CT) and with the goat anti–mouse secondary antibody (IRDye 800CW; LI-COR Biosciences). Both antibodies were diluted in blocking buffer. Membranes were imaged with the Odyssey Infrared Imaging System (LI-COR Biosciences).
Immunofluorescence
For immunofluorescence assays, cells were grown in chamber slides (Lab-Tek; Thermo Fisher Scientific). The staining for γ-H2AX was performed as previously described (Zhang et al., 2011). After drug treatment, cells were fixed and permeabilized by a 20-min incubation at room temperature in 2% paraformaldehyde and an overnight incubation at 4°C in 70% ethanol. Slides were blocked in 8% bovine serum albumin and stained successively with anti–γ-H2AX antibody (Abcam) and with a fluorescent secondary antibody (anti–mouse Alexa Fluor 488; Invitrogen). Both antibodies were diluted in 1% bovine serum albumin. Slides were mounted in mounting medium containing DAPI (Vectashield; Vector Laboratories) and visualized using a confocal microscope (Eclipse TE300; Nikon). Fluorescent signals in individual cells were quantified using Photoshop CS5 (Adobe).
For the simultaneous detection of γ-H2AX and replication foci, cells were labeled with 30 µM EdU for 90 min. 1 µM CPT was added during the last 60 min of the EdU pulse. γ-H2AX staining was performed as described in the previous paragraph, and EdU was detected with a flow cytometry assay kit (Click-iT EdU Alexa Fluor 647 Flow Cytometry Assay; Invitrogen) according to the manufacturer’s instructions. Confocal images were sequentially acquired with ZEN (2009, SP1; Carl Zeiss) software on a confocal system (LSM 510; Carl Zeiss) with an inverted microscope (Axio Observer.Z1; Carl Zeiss) and a UV laser tuned to 364 nm, a 25-mW argon visible laser tuned to 488 nm, and a 5-mW HeNe laser tuned to 633 nm. A 63× Plan Apochromat 1.4 NA oil immersion objective was used. Emission signals after sequential excitation of DAPI, Alexa Fluor 488, and Alexa Fluor 633 by the 364-, 488-, or 633-nm laser lines were collected with a band pass 385–470, band pass 505–550, or long pass 650 filter, respectively, using individual photomultipliers. Images were acquired at room temperature, and the mounting medium was Vectashield with DAPI. Images were adjusted using Photoshop and combined using Illustrator (Adobe).
Neutral comet assay
DNA DSBs were detected using the neutral comet assay according to the gel electrophoresis kit protocol (CometAssay; Trevigen; Zhang et al., 2008). Comet tail moments were measured with CometScore 1.5 software (TriTek Corporation).
Western blotting
Whole-cell extracts were obtained by homogenization of cell pellets in lysis buffer (1% SDS and 10 mM Tris-HCl, pH 7.4) supplemented with proteases and phosphatases inhibitors (Roche). Proteins were separated by SDS-PAGE electrophoresis and immunoblotted with the following antibodies: anti-Mus81 (Abcam), anti–γ-H2AX, antiphospho-RPA2 (S4/8; Novus Biologicals), antiphospho-Chk1 (S317), antiphospho-Chk2 (T68), anti–glyceraldehyde 3-phosphate dehydrogenase (GAPDH; Cell Signaling Technology), and anti–β-actin (Sigma-Aldrich).
siRNA transfection
Cells seeded in 6-well plates or LabTek chambers were transfected with targeting or nontargeting siRNAs using transfection reagent (DharmaFECT; Thermo Fisher Scientific). Mus81-targeting, Eme1-targeting, and nontargeting siRNAs were products of Thermo Fisher Scientific. TDP1-targeting siRNAs were purchased from QIAGEN.
DNA replication profiling by molecular combing
Molecular combing was performed as previously described (Conti et al., 2007; Seiler et al., 2007). In brief, asynchronous exponentially growing cells were labeled sequentially with the thymidine analogues IdU and CldU for 30 min each. To study replication fork arrest and restart, 1 µM CPT was added during the last 20 min of the IdU pulse. At the end of the CldU pulse, cells were harvested by trypsinization and embedded in low-melting agarose plugs. Agarose plugs were treated with proteinase K and then melted at 70°C in the presence of 4-morpholinepropanesulfonic acid, pH 6.5. After β-agarase digestion, DNA was combed on silanized surfaces (MicroSurfaces, Inc.). DNA fibers were denatured in 0.5 M NaOH and probed with the following primary antibodies: mouse anti-BrdU fluorescein isothiocyanate (IdU specific; BD) and rat anti-BrdU (CldU specific; Accurate Chemical and Scientific Corp.). After labeling with fluorescent secondary antibodies (anti–mouse Alexa Fluor 488 and anti–rat Alexa Fluor 594; Invitrogen), slides were mounted in Vectashield mounting medium. Images were captured with the AttoVision software (BD) using an epifluorescence microscope (Pathway; BD). Replication signals were measured using ImageJ (National Institutes of Health) with custom-made modifications. Measurements were converted from micrometers to kilobases according to a constant stretching factor (1 µm = 2 kb). Fork velocities were calculated by dividing the length of a replication signal (in kilobases) by the labeling time (in minutes).
Statistical analyses
All results are presented as means ± SEM or SD. Differences between samples were assessed using the Mann–Whitney test or the Student’s t test, depending on the distribution of the sample. The normal distribution of a sample was tested with the Kolmogorov–Smirnov test. All analyses were conducted with Prism 5.0 (GraphPad Software). P-values were two sided and considered statistically significant when <0.05.
Online supplemental material
Fig. S1 shows the effect of Mus81 and Eme1 silencing on the cellular response to CPT. Fig. S2 shows the effect of TDP1 silencing on the formation and reversal of CPT-induced Top1cc’s. Fig. S3 shows additional microscopy images of replication foci (EdU foci) and γ-H2AX foci in CPT-treated WT and Mus81−/− cells. Fig. S4 shows the frequency distribution of replication fork velocities in WT and Mus81−/− cells. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201104003/DC1.
Acknowledgments
We thank Drs. Shar-Yin Huang, Elisabetta Leo, and Benu Brata Das for helpful discussions during the conduction of the study. We thank Drs. Céline Douarre, Mathieu Métifiot, Christophe Redon, and Asako Nakamura for technical assistance. We also thank Dr. Susan Garfield for providing the information of image acquisition.
This work was supported by the Center for Cancer Research (National Cancer Institute Intramural Research Program, National Institutes of Health).
Footnotes
Abbreviations used in this paper:
- CldU
- chlorodeoxyuridine
- CPT
- camptothecin
- DPC
- DNA–protein cross-link
- DSB
- double-strand break
- DSE
- double-strand end
- EdU
- 5-ethynyl-2′-deoxyuridine
- GAPDH
- glyceraldehyde 3-phosphate dehydrogenase
- IdU
- iododeoxyuridine
- Top1cc
- Top1–DNA cleavage complex
- WT
- wild type
References
- Arnaudeau C., Lundin C., Helleday T. 2001. DNA double-strand breaks associated with replication forks are predominantly repaired by homologous recombination involving an exchange mechanism in mammalian cells. J. Mol. Biol. 307:1235–1245 [DOI] [PubMed] [Google Scholar]
- Blais V., Gao H., Elwell C.A., Boddy M.N., Gaillard P.H., Russell P., McGowan C.H. 2004. RNA interference inhibition of Mus81 reduces mitotic recombination in human cells. Mol. Biol. Cell. 15:552–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champoux J.J. 2001. DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 70:369–413 [DOI] [PubMed] [Google Scholar]
- Christman M.F., Dietrich F.S., Levin N.A., Sadoff B.U., Fink G.R. 1993. The rRNA-encoding DNA array has an altered structure in topoisomerase I mutants of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 90:7637–7641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A., McDonald N., West S.C. 2008. Structural and functional relationships of the XPF/MUS81 family of proteins. Annu. Rev. Biochem. 77:259–287 [DOI] [PubMed] [Google Scholar]
- Conti C., Saccà B., Herrick J., Lalou C., Pommier Y., Bensimon A. 2007. Replication fork velocities at adjacent replication origins are coordinately modified during DNA replication in human cells. Mol. Biol. Cell. 18:3059–3067 10.1091/mbc.E06-08-0689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covey J.M., Jaxel C., Kohn K.W., Pommier Y. 1989. Protein-linked DNA strand breaks induced in mammalian cells by camptothecin, an inhibitor of topoisomerase I. Cancer Res. 49:5016–5022 [PubMed] [Google Scholar]
- Deng C., Brown J.A., You D., Brown J.M. 2005. Multiple endonucleases function to repair covalent topoisomerase I complexes in Saccharomyces cerevisiae. Genetics. 170:591–600 10.1534/genetics.104.028795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dexheimer T.S., Antony S., Marchand C., Pommier Y. 2008a. Tyrosyl-DNA phosphodiesterase as a target for anticancer therapy. Anticancer. Agents Med. Chem. 8:381–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dexheimer T.S., Kozekova A., Rizzo C.J., Stone M.P., Pommier Y. 2008b. The modulation of topoisomerase I-mediated DNA cleavage and the induction of DNA-topoisomerase I crosslinks by crotonaldehyde-derived DNA adducts. Nucleic Acids Res. 36:4128–4136 10.1093/nar/gkn334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doe C.L., Ahn J.S., Dixon J., Whitby M.C. 2002. Mus81-Eme1 and Rqh1 involvement in processing stalled and collapsed replication forks. J. Biol. Chem. 277:32753–32759 10.1074/jbc.M202120200 [DOI] [PubMed] [Google Scholar]
- Ehmsen K.T., Heyer W.D. 2008. Saccharomyces cerevisiae Mus81-Mms4 is a catalytic, DNA structure-selective endonuclease. Nucleic Acids Res. 36:2182–2195 10.1093/nar/gkm1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehmsen K.T., Heyer W.D. 2009. A junction branch point adjacent to a DNA backbone nick directs substrate cleavage by Saccharomyces cerevisiae Mus81-Mms4. Nucleic Acids Res. 37:2026–2036 10.1093/nar/gkp038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng W.K., Faucette L., Johnson R.K., Sternglanz R. 1988. Evidence that DNA topoisomerase I is necessary for the cytotoxic effects of camptothecin. Mol. Pharmacol. 34:755–760 [PubMed] [Google Scholar]
- Franchitto A., Pirzio L.M., Prosperi E., Sapora O., Bignami M., Pichierri P. 2008. Replication fork stalling in WRN-deficient cells is overcome by prompt activation of a MUS81-dependent pathway. J. Cell Biol. 183:241–252 10.1083/jcb.200803173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricke W.M., Bastin-Shanower S.A., Brill S.J. 2005. Substrate specificity of the Saccharomyces cerevisiae Mus81-Mms4 endonuclease. DNA Repair (Amst.). 4:243–251 10.1016/j.dnarep.2004.10.001 [DOI] [PubMed] [Google Scholar]
- Froget B., Blaisonneau J., Lambert S., Baldacci G. 2008. Cleavage of stalled forks by fission yeast Mus81/Eme1 in absence of DNA replication checkpoint. Mol. Biol. Cell. 19:445–456 10.1091/mbc.E07-07-0728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta T., Takemura H., Liao Z.Y., Aune G.J., Redon C., Sedelnikova O.A., Pilch D.R., Rogakou E.P., Celeste A., Chen H.T., et al. 2003. Phosphorylation of histone H2AX and activation of Mre11, Rad50, and Nbs1 in response to replication-dependent DNA double-strand breaks induced by mammalian DNA topoisomerase I cleavage complexes. J. Biol. Chem. 278:20303–20312 10.1074/jbc.M300198200 [DOI] [PubMed] [Google Scholar]
- Hanada K., Budzowska M., Modesti M., Maas A., Wyman C., Essers J., Kanaar R. 2006. The structure-specific endonuclease Mus81-Eme1 promotes conversion of interstrand DNA crosslinks into double-strands breaks. EMBO J. 25:4921–4932 10.1038/sj.emboj.7601344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada K., Budzowska M., Davies S.L., van Drunen E., Onizawa H., Beverloo H.B., Maas A., Essers J., Hickson I.D., Kanaar R. 2007. The structure-specific endonuclease Mus81 contributes to replication restart by generating double-strand DNA breaks. Nat. Struct. Mol. Biol. 14:1096–1104 10.1038/nsmb1313 [DOI] [PubMed] [Google Scholar]
- Hiyama T., Katsura M., Yoshihara T., Ishida M., Kinomura A., Tonda T., Asahara T., Miyagawa K. 2006. Haploinsufficiency of the Mus81-Eme1 endonuclease activates the intra-S-phase and G2/M checkpoints and promotes rereplication in human cells. Nucleic Acids Res. 34:880–892 10.1093/nar/gkj495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochegger H., Dejsuphong D., Fukushima T., Morrison C., Sonoda E., Schreiber V., Zhao G.Y., Saberi A., Masutani M., Adachi N., et al. 2006. Parp-1 protects homologous recombination from interference by Ku and Ligase IV in vertebrate cells. EMBO J. 25:1305–1314 10.1038/sj.emboj.7601015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm C., Covey J.M., Kerrigan D., Pommier Y. 1989. Differential requirement of DNA replication for the cytotoxicity of DNA topoisomerase I and II inhibitors in Chinese hamster DC3F cells. Cancer Res. 49:6365–6368 [PubMed] [Google Scholar]
- Hsiang Y.-H., Hertzberg R., Hecht S., Liu L.F. 1985. Camptothecin induces protein-linked DNA breaks via mammalian DNA topoisomerase I. J. Biol. Chem. 260:14873–14878 [PubMed] [Google Scholar]
- Hsiang Y.-H., Lihou M.G., Liu L.F. 1989. Arrest of replication forks by drug-stabilized topoisomerase I-DNA cleavable complexes as a mechanism of cell killing by camptothecin. Cancer Res. 49:5077–5082 [PubMed] [Google Scholar]
- Huang T.H., Chen H.C., Chou S.M., Yang Y.C., Fan J.R., Li T.K. 2010. Cellular processing determinants for the activation of damage signals in response to topoisomerase I-linked DNA breakage. Cell Res. 20:1060–1075 10.1038/cr.2010.95 [DOI] [PubMed] [Google Scholar]
- Interthal H., Heyer W.D. 2000. MUS81 encodes a novel helix-hairpin-helix protein involved in the response to UV- and methylation-induced DNA damage in Saccharomyces cerevisiae. Mol. Gen. Genet. 263:812–827 10.1007/s004380000241 [DOI] [PubMed] [Google Scholar]
- Interthal H., Chen H.J., Champoux J.J. 2005. Human Tdp1 cleaves a broad spectrum of substrates, including phosphoamide linkages. J. Biol. Chem. 280:36518–36528 10.1074/jbc.M508898200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessop L., Lichten M. 2008. Mus81/Mms4 endonuclease and Sgs1 helicase collaborate to ensure proper recombination intermediate metabolism during meiosis. Mol. Cell. 31:313–323 10.1016/j.molcel.2008.05.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaliraman V., Mullen J.R., Fricke W.M., Bastin-Shanower S.A., Brill S.J. 2001. Functional overlap between Sgs1-Top3 and the Mms4-Mus81 endonuclease. Genes Dev. 15:2730–2740 10.1101/gad.932201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim R.A., Wang J.C. 1989. Function of DNA topoisomerases as replication swivels in Saccharomyces cerevisiae. J. Mol. Biol. 208:257–267 10.1016/0022-2836(89)90387-2 [DOI] [PubMed] [Google Scholar]
- Koster D.A., Palle K., Bot E.S., Bjornsti M.A., Dekker N.H. 2007. Antitumour drugs impede DNA uncoiling by topoisomerase I. Nature. 448:213–217 10.1038/nature05938 [DOI] [PubMed] [Google Scholar]
- Liu C., Pouliot J.J., Nash H.A. 2002. Repair of topoisomerase I covalent complexes in the absence of the tyrosyl-DNA phosphodiesterase Tdp1. Proc. Natl. Acad. Sci. USA. 99:14970–14975 10.1073/pnas.182557199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Z.H., Agama K., Sordet O., Povirk L., Kohn K.W., Pommier Y. 2006. Hereditary ataxia SCAN1 cells are defective for the repair of transcription-dependent topoisomerase I cleavage complexes. DNA Repair (Amst.). 5:1489–1494 10.1016/j.dnarep.2006.07.004 [DOI] [PubMed] [Google Scholar]
- Miao Z.H., Player A., Shankavaram U., Wang Y.H., Zimonjic D.B., Lorenzi P.L., Liao Z.Y., Liu H., Shimura T., Zhang H.L., et al. 2007. Nonclassic functions of human topoisomerase I: genome-wide and pharmacologic analyses. Cancer Res. 67:8752–8761 10.1158/0008-5472.CAN-06-4554 [DOI] [PubMed] [Google Scholar]
- Mullen J.R., Kaliraman V., Ibrahim S.S., Brill S.J. 2001. Requirement for three novel protein complexes in the absence of the Sgs1 DNA helicase in Saccharomyces cerevisiae. Genetics. 157:103–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K., Kogame T., Oshiumi H., Shinohara A., Sumitomo Y., Agama K., Pommier Y., Tsutsui K.M., Tsutsui K., Hartsuiker E., et al. 2010. Collaborative action of Brca1 and CtIP in elimination of covalent modifications from double-strand breaks to facilitate subsequent break repair. PLoS Genet. 6:e1000828 10.1371/journal.pgen.1000828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitiss J., Wang J.C. 1988. DNA topoisomerase-targeting antitumor drugs can be studied in yeast. Proc. Natl. Acad. Sci. USA. 85:7501–7505 10.1073/pnas.85.20.7501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S.D., Lao J.P., Taylor A.F., Smith G.R., Hunter N. 2008. RecQ helicase, Sgs1, and XPF family endonuclease, Mus81-Mms4, resolve aberrant joint molecules during meiotic recombination. Mol. Cell. 31:324–336 10.1016/j.molcel.2008.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pommier Y., Barcelo J.M., Rao V.A., Sordet O., Jobson A.G., Thibaut L., Miao Z.H., Seiler J.A., Zhang H., Marchand C., et al. 2006. Repair of topoisomerase I-mediated DNA damage. Prog. Nucleic Acid Res. Mol. Biol. 81:179–229 10.1016/S0079-6603(06)81005-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pommier Y., Leo E., Zhang H., Marchand C. 2010. DNA topoisomerases and their poisoning by anticancer and antibacterial drugs. Chem. Biol. 17:421–433 10.1016/j.chembiol.2010.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouliot J.J., Yao K.C., Robertson C.A., Nash H.A. 1999. Yeast gene for a Tyr-DNA phosphodiesterase that repairs topoisomerase I complexes. Science. 286:552–555 10.1126/science.286.5439.552 [DOI] [PubMed] [Google Scholar]
- Pourquier P., Pommier Y. 2001. Topoisomerase I-mediated DNA damage. Adv. Cancer Res. 80:189–216 10.1016/S0065-230X(01)80016-6 [DOI] [PubMed] [Google Scholar]
- Rogakou E.P., Pilch D.R., Orr A.H., Ivanova V.S., Bonner W.M. 1998. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 273:5858–5868 10.1074/jbc.273.10.5858 [DOI] [PubMed] [Google Scholar]
- Roseaulin L., Yamada Y., Tsutsui Y., Russell P., Iwasaki H., Arcangioli B. 2008. Mus81 is essential for sister chromatid recombination at broken replication forks. EMBO J. 27:1378–1387 10.1038/emboj.2008.65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothkamm K., Krüger I., Thompson L.H., Löbrich M. 2003. Pathways of DNA double-strand break repair during the mammalian cell cycle. Mol. Cell. Biol. 23:5706–5715 10.1128/MCB.23.16.5706-5715.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan A.J., Squires S., Strutt H.L., Johnson R.T. 1991. Camptothecin cytotoxicity in mammalian cells is associated with the induction of persistent double strand breaks in replicating DNA. Nucleic Acids Res. 19:3295–3300 10.1093/nar/19.12.3295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler J.A., Conti C., Syed A., Aladjem M.I., Pommier Y. 2007. The intra-S-phase checkpoint affects both DNA replication initiation and elongation: single-cell and -DNA fiber analyses. Mol. Cell. Biol. 27:5806–5818 10.1128/MCB.02278-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao R.-G., Cao C.-X., Zhang H., Kohn K.W., Wold M.S., Pommier Y. 1999. Replication-mediated DNA damage by camptothecin induces phosphorylation of RPA by DNA-dependent protein kinase and dissociates RPA:DNA-PK complexes. EMBO J. 18:1397–1406 10.1093/emboj/18.5.1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimura T., Torres M.J., Martin M.M., Rao V.A., Pommier Y., Katsura M., Miyagawa K., Aladjem M.I. 2008. Bloom’s syndrome helicase and Mus81 are required to induce transient double-strand DNA breaks in response to DNA replication stress. J. Mol. Biol. 375:1152–1164 10.1016/j.jmb.2007.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G.R., Boddy M.N., Shanahan P., Russell P. 2003. Fission yeast Mus81.Eme1 Holliday junction resolvase is required for meiotic crossing over but not for gene conversion. Genetics. 165:2289–2293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda E., Hochegger H., Saberi A., Taniguchi Y., Takeda S. 2006. Differential usage of non-homologous end-joining and homologous recombination in double strand break repair. DNA Repair (Amst.). 5:1021–1029 10.1016/j.dnarep.2006.05.022 [DOI] [PubMed] [Google Scholar]
- Sordet O., Redon C.E., Guirouilh-Barbat J., Smith S., Solier S., Douarre C., Conti C., Nakamura A.J., Das B.B., Nicolas E., et al. 2009. Ataxia telangiectasia mutated activation by transcription- and topoisomerase I-induced DNA double-strand breaks. EMBO Rep. 10:887–893 10.1038/embor.2009.97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strumberg D., Pilon A.A., Smith M., Hickey R., Malkas L., Pommier Y. 2000. Conversion of topoisomerase I cleavage complexes on the leading strand of ribosomal DNA into 5′-phosphorylated DNA double-strand breaks by replication runoff. Mol. Cell. Biol. 20:3977–3987 10.1128/MCB.20.11.3977-3987.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata M., Sasaki M.S., Sonoda E., Morrison C., Hashimoto M., Utsumi H., Yamaguchi-Iwai Y., Shinohara A., Takeda S. 1998. Homologous recombination and non-homologous end-joining pathways of DNA double-strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. EMBO J. 17:5497–5508 10.1093/emboj/17.18.5497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trowbridge K., McKim K., Brill S.J., Sekelsky J. 2007. Synthetic lethality of Drosophila in the absence of the MUS81 endonuclease and the DmBlm helicase is associated with elevated apoptosis. Genetics. 176:1993–2001 10.1534/genetics.106.070060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsao Y.P., Russo A., Nyamuswa G., Silber R., Liu L.F. 1993. Interaction between replication forks and topoisomerase I-DNA cleavable complexes: studies in a cell-free SV40 DNA replication system. Cancer Res. 53:5908–5914 [PubMed] [Google Scholar]
- Tuduri S., Crabbé L., Conti C., Tourrière H., Holtgreve-Grez H., Jauch A., Pantesco V., De Vos J., Thomas A., Theillet C., et al. 2009. Topoisomerase I suppresses genomic instability by preventing interference between replication and transcription. Nat. Cell Biol. 11:1315–1324 10.1038/ncb1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance J.R., Wilson T.E. 2002. Yeast Tdp1 and Rad1-Rad10 function as redundant pathways for repairing Top1 replicative damage. Proc. Natl. Acad. Sci. USA. 99:13669–13674 10.1073/pnas.202242599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.C. 2002. Cellular roles of DNA topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell Biol. 3:430–440 10.1038/nrm831 [DOI] [PubMed] [Google Scholar]
- Yang S.W., Burgin A.B., Jr, Huizenga B.N., Robertson C.A., Yao K.C., Nash H.A. 1996. A eukaryotic enzyme that can disjoin dead-end covalent complexes between DNA and type I topoisomerases. Proc. Natl. Acad. Sci. USA. 93:11534–11539 10.1073/pnas.93.21.11534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.W., Zhang Z.X., Miao Z.H., Ding J. 2008. The telomeric protein TRF2 is critical for the protection of A549 cells from both telomere erosion and DNA double-strand breaks driven by salvicine. Mol. Pharmacol. 73:824–832 10.1124/mol.107.039081 [DOI] [PubMed] [Google Scholar]
- Zhang Y.W., Regairaz M., Seiler J.A., Agama K.K., Doroshow J.H., Pommier Y. 2011. Poly(ADP-ribose) polymerase and XPF-ERCC1 participate in distinct pathways for the repair of topoisomerase I-induced DNA damage in mammalian cells. Nucleic Acids Res. 39:3607–3620 10.1093/nar/gkq1304 [DOI] [PMC free article] [PubMed] [Google Scholar]