Abstract
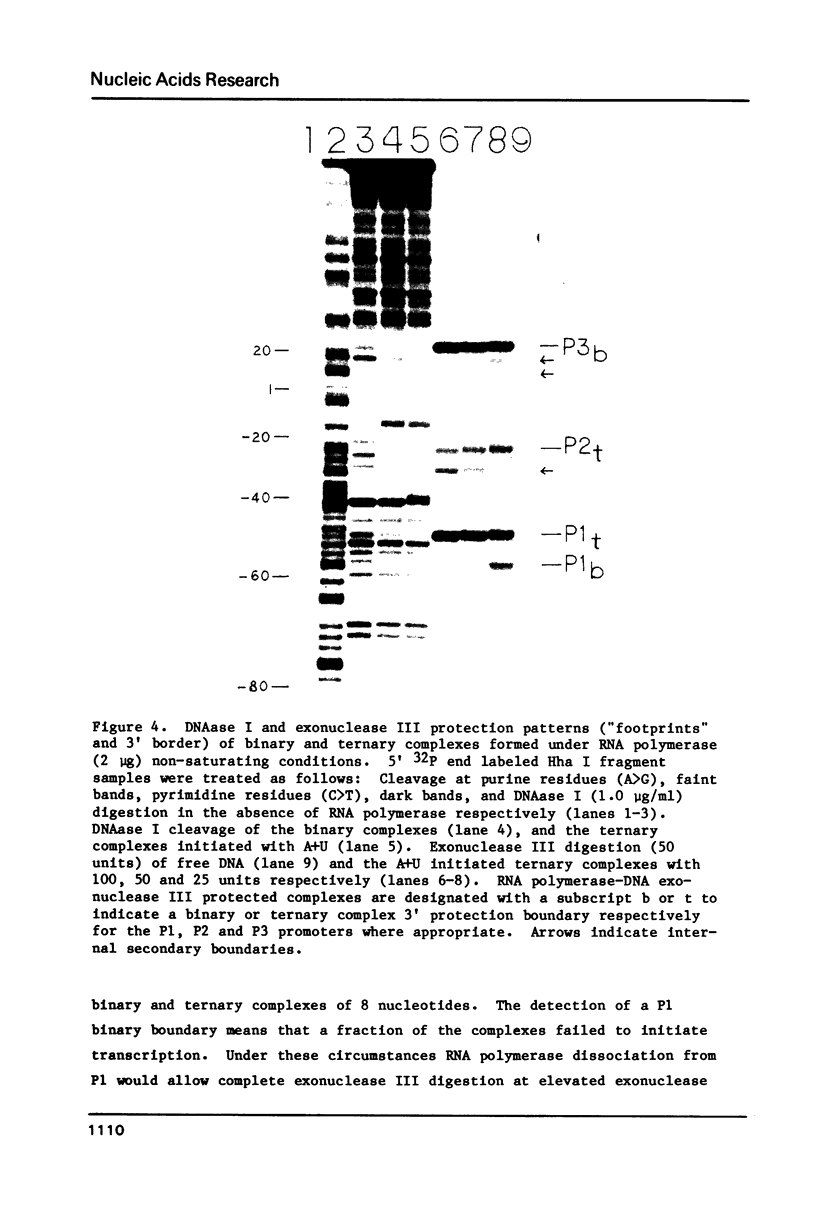
In order to determine the initiation site for three promoters P1, P2 and P3 (5' to 3') in close proximity in the colicin E1 control region we developed a new methodology that couples ternary complex formation and the analysis of the 3' border protected from exonuclease III digestion. The initiation of transcription could be detected by measuring the shift in the position of the 3' protected border when RNA polymerase moved from its binary complex position to its ternary complex position. The latter stops at a specific nucleotide because transcription is initiated with one or more NTPs missing. This approach, coupled with "footprinting", can also be used to decide whether the formation of an RNA polymerase binary or ternary complex at one site excludes or weakens binding at neighboring sites. The location of 3' protected borders reveals the formation of respective binary and ternary complexes at non-saturating RNA polymerase conditions, whereas at saturating conditions only the distal 3' boundary is seen and exonuclease cannot penetrate further. However, if "footprinting" reveals proximal 5' patterns this establishes that simultaneous binding has occurred on the same DNA fragment. The data showed that this was true for P1 and P3 which are only 8 nucleotides apart. P2 could only be detected at non-saturating conditions since it overlaps both P1 and P3. The evidence from the literature and this study establishes P1 as the true colicin E1 promoter with the possibility that supercoiling may eliminate any role for P2 and P3.
Full text
PDF
















Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brent R., Ptashne M. Mechanism of action of the lexA gene product. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4204–4208. doi: 10.1073/pnas.78.7.4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan P. T., Lebowitz J., Bastia D. Nucleotide sequence determination of a strong promoter of the colicin E 1 plasmid. Analysis of restriction sites protected by RNA polymerase interactions before and after limited transcription. Nucleic Acids Res. 1979 Nov 10;7(5):1247–1262. doi: 10.1093/nar/7.5.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebina Y., Kishi F., Miki T., Kagamiyama H., Nakazawa T., Nakazawa A. The nucleotide sequence surrounding the promoter region of colicin E1 gene. Gene. 1981 Nov;15(2-3):119–126. doi: 10.1016/0378-1119(81)90121-9. [DOI] [PubMed] [Google Scholar]
- Ebina Y., Kishi F., Nakazawa A. Direct participation of lexA protein in repression of colicin E1 synthesis. J Bacteriol. 1982 Jun;150(3):1479–1481. doi: 10.1128/jb.150.3.1479-1481.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horii T., Ogawa T., Ogawa H. Nucleotide sequence of the lexA gene of E. coli. Cell. 1981 Mar;23(3):689–697. doi: 10.1016/0092-8674(81)90432-3. [DOI] [PubMed] [Google Scholar]
- Little J. W., Mount D. W., Yanisch-Perron C. R. Purified lexA protein is a repressor of the recA and lexA genes. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4199–4203. doi: 10.1073/pnas.78.7.4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki T., Ebina Y., Kishi F., Nakazawa A. Organization of the lexA gene of Escherichia coli and nucleotide sequence of the regulatory region. Nucleic Acids Res. 1981 Feb 11;9(3):529–543. doi: 10.1093/nar/9.3.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overbye K. M., Margolin P. Role of the supX gene in ultraviolet light-induced mutagenesis in Salmonella typhimurium. J Bacteriol. 1981 Apr;146(1):170–178. doi: 10.1128/jb.146.1.170-178.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Sancar A., Stachelek C., Konigsberg W., Rupp W. D. Sequences of the recA gene and protein. Proc Natl Acad Sci U S A. 1980 May;77(5):2611–2615. doi: 10.1073/pnas.77.5.2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz A., Galas D. J. The interaction of RNA polymerase and lac repressor with the lac control region. Nucleic Acids Res. 1979 Jan;6(1):111–137. doi: 10.1093/nar/6.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalloway D., Kleinberger T., Livingston D. M. Mapping of SV40 DNA replication origin region binding sites for the SV40 T antigen by protection against exonuclease III digestion. Cell. 1980 Jun;20(2):411–422. doi: 10.1016/0092-8674(80)90627-3. [DOI] [PubMed] [Google Scholar]
- Sternglanz R., DiNardo S., Voelkel K. A., Nishimura Y., Hirota Y., Becherer K., Zumstein L., Wang J. C. Mutations in the gene coding for Escherichia coli DNA topoisomerase I affect transcription and transposition. Proc Natl Acad Sci U S A. 1981 May;78(5):2747–2751. doi: 10.1073/pnas.78.5.2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trucksis M., Depew R. E. Identification and localization of a gene that specifies production of Escherichia coli DNA topoisomerase I. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2164–2168. doi: 10.1073/pnas.78.4.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada M., Ebina Y., Miyata T., Nakazawa T., Nakazawa A. Nucleotide sequence of the structural gene for colicin E1 and predicted structure of the protein. Proc Natl Acad Sci U S A. 1982 May;79(9):2827–2831. doi: 10.1073/pnas.79.9.2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H. L., Heller K., Gellert M., Zubay G. Differential sensitivity of gene expression in vitro to inhibitors of DNA gyrase. Proc Natl Acad Sci U S A. 1979 Jul;76(7):3304–3308. doi: 10.1073/pnas.76.7.3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg E., Zwetsloot J., Noordermeer I., Pannekoek H., Dekker B., Dijkema R., van Ormondt H. The structure and function of the regulatory elements of the Escherichia coli uvrB gene. Nucleic Acids Res. 1981 Nov 11;9(21):5623–5643. doi: 10.1093/nar/9.21.5623. [DOI] [PMC free article] [PubMed] [Google Scholar]