Abstract
Elucidation of the relationship between targeting molecule binding properties and the adhesive behavior of therapeutic or diagnostic nanocarriers would aid in the design of optimized vectors and lead to improved efficacy. We measured the adhesion of 200 nm diameter particles under fluid flow that was mediated by a diverse array of molecular interactions, including recombinant single-chain antibodies (scFvs), full antibodies, and the avidin/biotin interaction. Within the panel of scFvs, we used a family of mutants that display a spectrum of binding kinetics, allowing us to compare nanoparticle adhesion to bond chemistry. In addition, we explored the effect of molecular size by inserting a protein linker into the scFv fusion construct and by employing scFvs that are specific for targets with vastly different sizes. Using computational models we extracted multivalent kinetic rate constants for particle attachment and detachment from the adhesion data and correlated the results to molecular binding properties. Our results indicate that the factors that increase encounter probability, such as adhesion molecule valency and size, directly enhance the rate of nanoparticle attachment. Bond kinetics had no influence on scFv-mediated nanoparticle attachment within the kinetic range tested however, but did appear to effect antibody/antigen and avidin/biotin mediated adhesion. We attribute this finding to a combination of multivalent binding and differences in bond mechanical strength between recombinant scFvs and the other adhesion molecules. Nanoparticle detachment probability correlated directly with adhesion molecule valency and size, as well as the logarithm of the affinity for all molecules tested. Based on this work, scFvs can serve as viable targeting receptors for nanoparticles, but improvements to their bond mechanical strength would likely be required to fully exploit their tunable kinetic properties and maximize the adhesion efficiency of nanoparticles that bear them.
Keywords: nanoparticles, multivalent binding, molecular targeting, single-chain antibody (scFv), avidin-biotin, VCAM-1
INTRODUCTION
A major goal of the field of drug delivery is to develop targeted therapeutic delivery systems that can reduce or eliminate the adverse side-effects associated with traditional systemic administration. Nanoparticles have emerged as important materials for this application because they can carry large drug cargos, can readily be modified with affinity molecules for targeted binding, and can make use of multivalency for improved binding properties.1
In previous work, we established fundamental principles to quantify the adhesion of multivalent nanoparticles under fluid flow, and elucidated fundamental scaling laws that related the particle adhesion rate constant and particle dissociation rate constant to molecular surface densities, flow rate, and nanoparticle size.2, 3 However, to date very little is known regarding the relationship between targeting molecule binding properties and nanoparticle adhesion. Here we provide insight into the influence of molecular binding properties by attaching a spectrum of different targeting molecules to nanoparticles and quantifying adhesion. A particular focus of this work is on recombinant antibody fragments (single-chain antibodies, scFvs), which have been widely used as nanocarrier targeting moieties both in vitro4–6 and in vivo.7–11 scFvs are particularly attractive for our purposes because their binding properties can readily be engineered using directed evolution.12 For example, the 4-4-20 anti-fluorescein scFv was evolved through multiple rounds of directed evolution to yield a library of mutants displaying greater than 10,000-fold differences in kr.13, 14 This family of scFv mutant proteins thus provides an excellent experimental model system to study the effect of bond kinetics on nanoparticle adhesion.
Molecular binding interactions are often quantified by the kinetic reaction rates (forward, kf, and reverse, kr) or the thermodynamic equilibrium affinity (KA = kf/kr). Hammer and Lauffenburger suggested that the adhesion of cells under fluid flow can be controlled by either kinetics or affinity depending on the time-scale of the force driven process; for fast flows, kinetics should dominate.15 However, bond mechanical strength can also influence the dynamics of adhesion under shear force. Bell first suggested a relationship between bond dissociation rate and force that depends on the unstressed kr and an internal property that set the sensitivity to force.16 Evans17 and later Dembo and coworkers18 suggested that detachment is driven by the logarithm of the affinity constant, a relationship that was later corroborated by Kuo and Lauffenburger experimentally19 and computationally using Adhesive Dynamics simulations.20 Our laboratory has also used Adhesive Dynamics simulations to elucidate the quantitative relationship between bond mechanical strength and the dynamics of cell adhesion, suggesting mechanical properties that lead to diverse phenomena such as rolling, firm, or weak adhesion.21 Thus it is expected that bond mechanical strength will vary as a weak function of the bond affinity, as well as an intrinsic property (reactive compliance) of the bond that establishes the sensitivity to force. Finally, bond length defines the spatial constraints over which adhesion molecules can locate binding partners. Israelachvili and coworkers firmly established that adhesion can be enhanced when ligands are placed on long, flexible tethers.22 Molecular length has also been shown to directly affect encounter frequency and bond formation rate in micropipette-based binding assays.23 While we expect that bond kinetics, thermodynamics, mechanics, and length may all play significant roles in dictating nanocarrier adhesion dynamics under fluid flow, detailed experiments aimed at quantifying these relationships have yet to be performed.
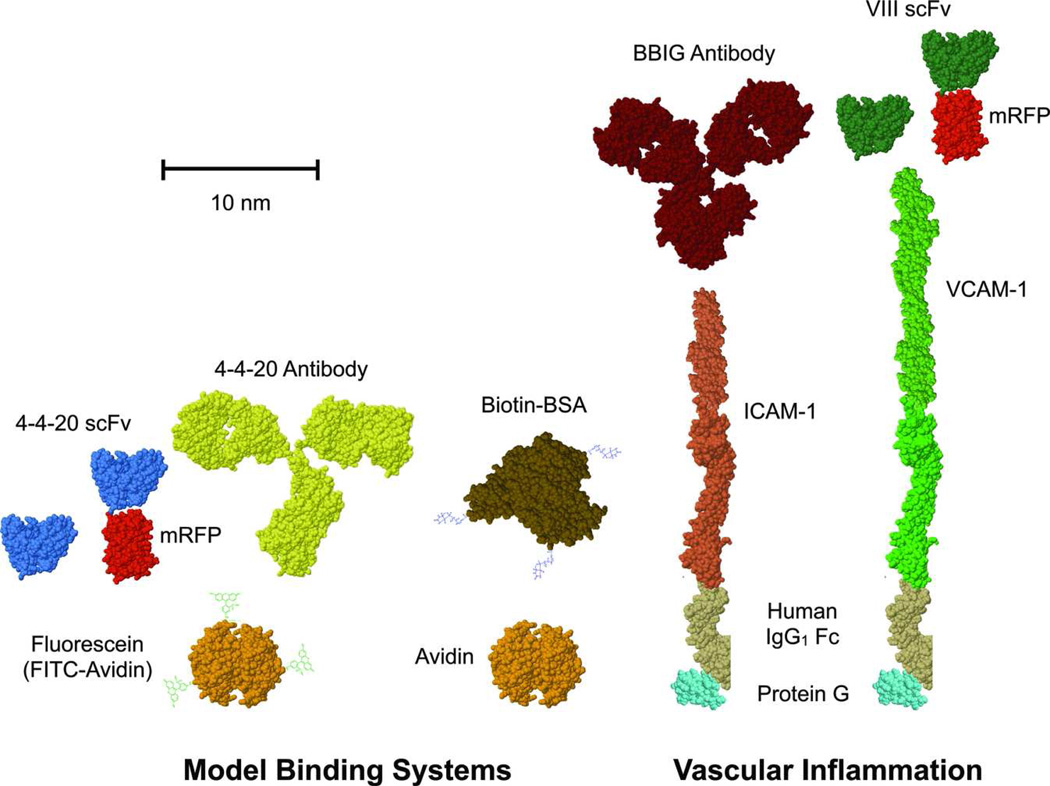
In this paper, we use a spectrum of molecular tools to explore the effects of chemical kinetics, bond length, and bond mechanical strength on the adhesion of 200 nm particles under fluid flow. This was accomplished using a diverse panel of molecular binding interactions including the 4-4-20 scFv family of mutants that bind to their ligand fluorescein with different kinetic rates. We also test nanoparticle binding mediated by the full 4-4-20 antibody and avidin/biotin, the latter widely considered the gold standard for non-covalent, high affinity biological binding24 We explore the direct effect of molecular size on adhesion by inserting monomeric red fluorescent protein (mRFP) into the scFv fusion construct. This was performed for both the 4-4-20 scFv and the novel VIII scFv that is specific for a much larger ligand, VCAM-1. Molecular sizes are based on measurements of the precise molecules or equivalent proxies obtained from the Protein Data Bank. The structures are depicted in Figure 1, and size measurements are listed along with published kinetic reaction rates in Table 1.
Figure 1.
Schematic representation of the receptor/ligand binding pairs employed in this study. Molecular sizes are depicted to scale, with the exception of fluorescein and biotin, based on measurements of actual or similar structures published in the Protein Data Bank (Table 1). The upper species were used as targeting receptors for particles and the lower species were used as ligands attached to glass coverslip substrates. From left to right, the receptor/ligand pairs are 4-4-20 scFv (wild-type and kinetic mutants)/fluorescein, 4-4-20 scFv-mRFP/fluorescein, 4-4-20 antibody/fluorescein, avidin/biotin, BBIG antibody/ICAM-1, VIII scFv/VCAM-1, and VIII scFv-mRFP/VCAM-1. The protein structures that are depicted and their Protein Data Bank codes include: 4M5.3 scFv (for all scFv species, 1X9Q), mRFP (2VAD), mouse IgG2a antibody (for 4-4-20 antibody, 1IGT), avidin (3FDC), human serum albumin (for BSA, 1AO6), ICAM-1 (combination of 1IAM and 1P53), VCAM-1 (combination of 1VSC and the 2 sequences used for ICAM-1), human IgG1 Fc (3D03), and protein G (3GB1).
Table 1.
Molecular size and kinetic properties for the various receptor/ligand pairs used in this study.
| Receptor | Ligand | Receptor | Ligand* | Kinetics | |||||
|---|---|---|---|---|---|---|---|---|---|
|
MWr (kDa) |
Lr (nm) |
MWl (kDa) |
Ll (nm) |
kf (M−1s−1) |
kr (s−1) |
KA (M−1) |
Ref | ||
| 4-4-20 scFv | Fluorescein | 37 | 4.4 | 61 | 5.9 | 8.0×107 | 2.5×10−2 | 3.2×109 | 14 |
| 4M2.4 scFv | Fluorescein | 37 | 4.4 | 61 | 5.9 | NM** | 4.9×10−4 | 1.2×1010 | 14 |
| 4M5.3 scFv | Fluorescein | 37 | 4.4 | 61 | 5.9 | 6.0×106 | 1.4×10−6 | 4.3×1012 | 14 |
| 4-4-20 scFv-mRFP | Fluorescein | 66 | 8.9 | 61 | 5.9 | 8.0×107 | 2.5×10−2 | 3.2×109 | 14 |
| 4-4-20 Ab | Fluorescein | 146 | 11.4 | 61 | 5.9 | 6.3×106 | 3.7×10−4 | 1.7×1010 | 37 |
| Biotin | Neutravidin | 67 | 7.3 | 60 | 5.9 | 7.5×107 | 7.5×10−5 | 1×1015 | 38 |
| VIII scFv | VCAM-1 | 36 | 4.4 | 152 | 38.0 | NM | NM | NM | NA |
| VIII scFv-mRFP | VCAM-1 | 65 | 8.9 | 152 | 38.0 | NM | NM | NM | NA |
| BBIG Ab | ICAM-1 | 146 | 11.4 | 142 | 29.7 | 1.6×105 | 1.1×10−4 | 1.5×109 | 39 |
Ligand size estimates are based on additive contributions from Neutravidin (60 kDa, 5.9 nm), fluorecein-biotin (1 kDa, 0.1 nm), protein G (22 kDa, 2.7 nm), human IgG1 Fc (50 kDa, 6.2 nm), ICAM-1 (70 kDa, 20.8 nm), and VCAM-1 (80 kDa, 29.1 nm).
NM, not measured.
Our results show substantial differences in nanoparticle binding dynamics for the various targeting molecules studied. Using transport-reaction modeling and stochastic binding simulations to calculate multivalent nanoparticle attachment and detachment rates,2, 3 we show that differences in nanoparticle binding correlate with molecular valency and size. We also find that the wild-type and mutant 4-4-20 scFvs mediate the same rate of nanoparticle attachment despite vast differences in kinetic reaction rates, and after correcting for molecular size effects all scFvs (4-4-20 and VIII) yielded identical attachment results. These findings demonstrate that nanoparticle attachment under fluid flow that is mediated by scFvs is independent of kinetic rates. This suggests that above a certain kinetic threshold, the influence of bond kinetics is mitigated by a factor such as multivalency or poor mechanical strength, leaving nanoparticle adhesion unchanged. In contrast, detachment assays with microbeads indicate that bond strengths for scFvs correlate exclusively with equilibrium affinity. We feel that this disparity is related to differences in context between nanoparticle attachment (dynamic, initial capture at low valency) and microbead detachment (equilibrium, large valency) assays, and suggest that we have uncovered a unique phenomenon that influences adhesion at the nano-scale. Nanoparticle attachment efficiencies are greater for the other molecular interactions (antibody/antigen, avidin/biotin) relative to the scFvs, and appear to be influenced by kinetics. For nanoparticle detachment, we observe a dependence on molecular size and kinetics for all species. In sum, our findings indicate that bond valency and length directly control both nanoparticle attachment and detachment. However, bond kinetics does not necessarily affect nanoparticle attachment due to possible influences from multivalency and insufficient bond mechanical strength. Conversely, bond kinetics universally controls the stability of nanoparticle adhesion after binding. We conclude that scFvs can serve as viable targeting receptors for nanoparticles, but attempts to tune kinetic properties will likely not improve nanoparticle adhesion efficiency in the absence of increased bond mechanical strength. This interdependence of bond kinetics and mechanical strength may also be relevant for other recombinant protein or synthetic construct (peptide, aptamer, affibody, avimer) targeting receptors.
MATERIALS AND METHODS
Reagents
VCAM-1/human IgG1 Fc chimera and control human IgG1 Fc were purchased from R&D Systems (Minneapolis, MN). Anti-fluorescein antibody (clone 4-4-20), fluorescein-biotin (FITC-biotin), HRP-conjugated Neutravidin, HRP-conjugated rat anti-mouse κ-light chain monoclonal antibody, R-phycoerithryn (RPE)-conjugated Neutravidin, RPE-conjugated anti-mouse IgG1 antibody, and Alexa 488-conjugated Protein G were purchased from Invitrogen. Biotinylated BSA, Neutravidin, D-biotin, and protein G were obtained from Thermo Fisher. Mouse anti-fluorescein monoclonal IgG1 antibody (clone FIT-22) was from BioLegend (San Diego, CA). Anti-c-myc antibody (clone 9E10) was purchased in purified form from Covance (Berkeley, CA) and conjugated to HRP from Roche (Indianapolis, IN). Mouse anti-human VCAM-1 monoclonal IgG1 antibody (clone 1.G11B1) was from Ancell (Bayport, MN). HRP-conjugated rat anti-mouse IgG1 monoclonal antibody (clone X56) was from BD Biosciences. IRDye 680-conjugated goat α-mouse IgG antibody was purchased from LI-COR Biotechnology (Lincoln, NB). Bovine serum albumin (BSA) was from Sigma (St. Loius, MO).
Strains and Media
Saccharomyces cerevisiae strain EBY100 containing an integrated copy of Aga1p under GAL1-10 promoter control13 was used for cell surface display of the VIII scFv. Strain BJ5464 (BJα) (α ura3-52 trp1 leu2Δ1 his3Δ200 pep4::HIS3 prb1Δ1.6R can1 GAL) was used for secretion of the 4-4-20 and VIII scFvs into culture supernatants. Escherichia coli strain DH5α (Invitrogen) was used for scFv gene cloning. Luria-Bertani (LB) medium (10.0 g/L tryptone, 5.0 g/L yeast extract, 10.0 g/L NaCl, pH 7.5, supplemented with 50 µg/ml ampicillin) was used for bacteria growth and plasmid amplification. EBY100 yeast were grown in minimal SD-CAA medium (20.0 g/L dextrose, 6.7 g/L yeast nitrogen base, 5.0 g/L casamino acids, 5.4 g/L Na2HPO4, 7.46 g/L NaH2PO4). BJ5464 were grown in similar medium with supplementation of uracil (40 µg/ml). Expression of scFv protein was induced using SG-CAA medium (20.0 g/L galactose instead of dextrose), which was supplemented with bovine serum albumin (BSA) at 1 mg/L as a nonspecific carrier. Hybridoma clone VIII-6G10 (HB-10519, American Type Culture Collection) was grown in RPMI 1640 including 2mM L-glutamine, 1 mM sodium pyruvate, 25 mM HEPES and 10% FBS.
Construction of VIII scFv
VIII-6G10 hybridoma cells were expanded in RPMI 1640 media at 37°C and passaged every 2 days. Total messenger RNA was extracted from 107 cells using OligoTex Direct mRNA kit (Qiagen). First-strand cDNA was generated and amplified by polymerase chain reaction (PCR) using TITAN One-Tube RT-PCR System (Roche) and primers based on those described by Barbas.25 PCR product was resolved by electrophoresis on a 1% agarose gel and all products approximately 350 bp in size were extracted, ligated into pDrive cloning vector (Qiagen), and transformed into E. coli strain DH5α. The products were then sequenced (University of Pennsylvania Cell Center), and those that were consistent with antibody light and heavy variable regions were combined by overlap extension PCR through introduction of a peptide linker (GGGGSGGGGSGGGGS), yielding the VIII scFv.
Plasmid Cloning
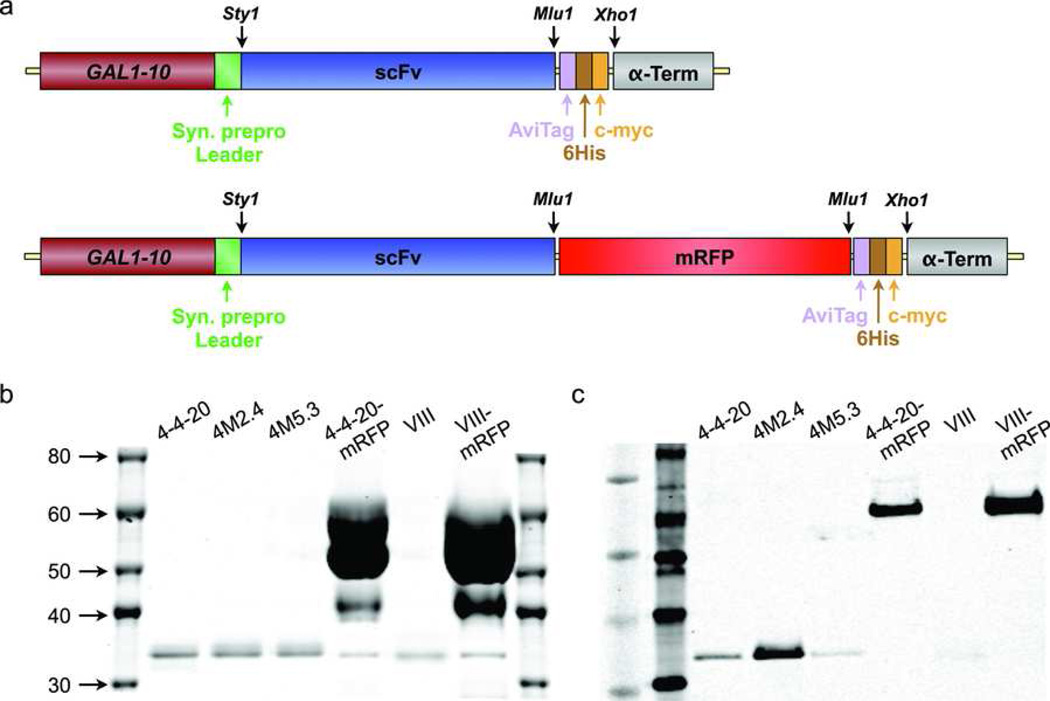
VIII scFv was amplified by PCR using sense (GAGGAGGCTAGCGAGCTCGATATTCAGATGACA) and anti-sense (CTCCTCACGCGTACTAGTGACAGATG GGGGTGT) primers, which introduced Nhe1 and Mlu1 restriction sites (underlined) at the 5’ and 3’ ends, respectively, to facilitate subcloning. The wild-type 4-4-20 and mutant 4M2.4 and 4M5.3 scFv sequences were similarly amplified from a pCT302-based plasmid used for yeast surface display13, 14 using sense (TGCCATTGGCCTTGGCTGACGTCGTTATGACTCAA) and anti-sense (GAGGAGACGCGTTGAGGAGACGGTGACTGAGGTTCC) primers. In this case the forward primer introduced a Sty1 restriction sites at the 5’ end. The VIII scFv was then inserted into pCT302 to create pCTVIII, which was transformed into EBY100 yeast to confirm specificity for VCAM-1 using surface display (see Supplemental Information). Likewise, the VIII, 4-4-20, 4M2.4 and 4M5.3 scFvs were inserted into a pRS314-based expression vector (GAL1-10 promoter, CEN-TRP1) similar to previous descriptions26, 27 to create pRSVIII, pRS4420, pRS4M24 and pRS4M53. The final expression cassette is depicted in Figure 2a, and was comprised of a GAL1-10 promoter, synthetic prepro leader sequence (MKVLIVLLAIFAALPLALA),28 scFv gene, biotin acceptor protein (AviTag, GLNDIFEAQKIEWHE), six-histidine tag (6His), c-myc epitope (EQKLISEEDL), and the α-factor 3’ untranslated region. In addition, mRFP was cloned by PCR using sense (GAGGAGACGCGTATGGCCTCCTCCGAGGACGTC) and anti-sense (GAGGAGACGCGTGGCGCCGGTGGAGTGGCGGCC) primers that added Mlu1 restriction sites at both 5’ and 3’ ends for insertion between the scFv and AviTag sequences (Figure 2a). After sequencing to confirm correct insertion of the scFv and mRFP open reading frames, yeast strain EBY100 was transformed with pCTVIII and strain BJ5464 was transformed with pRSVIII, pRSVIIImRFP, pRS4420, pRS4420mRFP, pRS4M24 and pRS4M53 plasmids using the lithium acetate method29 and Trp+ transformants were selected.
Figure 2.
Production of soluble scFv and scFv-mRFP fusions. (a) Schematic representation of the soluble expression cassette including GAL1-10 promoter, synthetic prepro leader sequence to direct the protein through the secretory machinery, scFv gene, AviTag biotin acceptor protein for site-specific biotinylation, six-histidine tag for nickel-chelate purification, c-myc eptiope tag, and α-factor 3’ untranslated region. To increase the size of the scFv fusion construct, mRFP was also inserted between the scFv and AviTag sequences. (b and c) Detection of scFv and scFv-mRFP protein following purification and biotinylation. Samples were resolved by SDS-PAGE and visualized using (b) Coomassie staining and (c) c-myc specific Western blot. The scFvs migrated at approximately 35 kDa and the scFv-mRFP fusions migrated at approximately 65 kDa, similar to the predicted molecular weights (Table 1). Protein samples were used directly for Coommassie stains and diluted 1000-fold for Western blots.
Production, Processing and Characterization of Soluble scFv Protein
BJ5464 yeast was grown in 1 liter SD-CAA cultures for 3 days at 30°C, centrifuged at 3000 × g and induced in 1 liter SG-CAA for 3 days at 20°C, as previously described.27 After induction, culture supernatants were recovered by centrifugation (10 minutes at 3000 × g) to remove cellular debris and concentrated by tangential flow filtration using 10,000 molecular-weight cut-off Pellicon XL cassettes (Millipore). A peristaltic pump was used to induce flow at 60 ml/min and the retentate backpressure was set to 20 psi using a tubing clamp and pressure gauge. Once the concentrate volume was reduced to approximately 10 ml, the buffer was exchanged to HisTrap binding buffer (50 mM NaH2PO4, 500 mM NaCl, 20 mM imidazole, pH 7) through three rounds dilution and filtration. The sample was then recovered, added to a 10 ml SuperLoop mounted on an ÄKTA Basic HPLC and pumped at 1 ml/min through a 1 ml HisTrap affinity column (all from GE Healthcare). After sample addition, the column was washed with 40 mM imidazole solution for 10 min before elution with 500 mM imidazole solution. Elution was monitored by absorbance at 280 nm and 0.5 ml fractions were collected.
Peak elution fractions from HisTrap column purifications were pooled in an Amicon Ultra-4 centrifugal concentrator, centrifuged for 10 min at 3000 × g and buffer exchanged to 100 mM Tris-HCl (pH 8) using two additional centrifugations. Recovered samples were then biotinylated using 10 µg/ml BirA biotin protein ligase (Avidity, Denver, CO) in a 0.5 ml reaction volume for 3 hours at 30°C. Following biotinylation, free biotin was removed by size-exclusion chromatography with Superdex 75 pg media (GE Healthcare) at 1 ml/min flow rate. Protein elution was monitored by absorbance at 280 nm and 1 ml fractions were collected once the absorbance signal increased above baseline. Peak scFv elution fractions were pooled and concentrated to approximately 250 µl using an Amicon Ultra-4 centrifugal concentrator. Protein concentration was estimated using the micro-BCA assay (Thermo Fisher) with BSA as a standard.
Processed scFv samples were resolved using SDS-polyacrylamide gel electrophoresis under reducing conditions with NuPAGE 4–12% gradient Bis-Tris gels and MOPS running buffer (Invitrogen). Gels were directly visualized using Coomassie Brilliant Blue staining (Thermo Fisher) or were transferred to PVDF membrane for Western blotting using an Xcell II Electrophoresis Cell (Invitrogen). Blots were blocked with 5% dried milk, probed sequentially with anti c-myc monoclonal antibody (1:1000 dilution) and IRDye 680-conjugated goat α-mouse IgG (1:5000 dilution), and imaged with an Odyssey Infrared Imaging System (LI-COR Biotechnology).
Particle Functionalization
Neutravidin-coated polystyrene particles with diameters of 210 nm (yellow-green fluorescent; Invitrogen) and 9.95 µm (non-fluorescent; Bangs Labs, Fishers, IN) were directly functionalized with biotinylated receptor protein or controls. Prior to reaction, nanoparticles (108 particles per sample) were diluted with 20 µl Block-Aid (Invitrogen), sonicated for 5 minutes, and diluted with PBS+ to 0.9 ml. Micron-sized particles (2×105 particles per sample) were washed twice with PBS+ by centrifugation for 1 min at 13,000 RPM, and diluted to 0.9 ml. Biotinylated protein was then added to 100 µl PBS+ to yield final protein concentrations as follows: 50 nM for scFv, 200 nM for 4-4-20 antibody and 500 nM for biotinylated BSA. Biotinylated BSA had approximately 8 biotin molecules per BSA, and therefore imparts biotin functionality. The 4-4-20 antibody (biotinylated using sulfo-NHS-LC-biotin as described in the Supplemental Information) and biotinylated BSA were used at much higher concentration than scFv protein due to biotin multivalency and the resulting possibility for particle cross-linking. In all cases, receptor site-density was modulated by mixing biotinylated ligands in different ratios with D-biotin. Reaction mixtures were incubated for at least 1 hour at room temperature on an end-to-end rotator.
Unbound protein was removed from 210 nm particles by size-exclusion chromatography using Sephacryl S-500 HR gel filtration media. Sample was added to the column at 1 ml/min using PBS as the running buffer while monitoring absorbance at 280 nm. For flow experiments, peak fractions were collected, analyzed for concentration using a fluorescence plate reader at 485 nm excitation/527 nm emission, pooled, and diluted with PBS+ to 108/ml concentration. Particle concentration was determined based on a calibration curve of stock particles. For the 9.95 µm particles, unbound protein was removed by centrifugation using three PBS+ washes as described above. Samples were then resuspended to 1 ml and used directly. Receptor site densities (nr) were determined by ELISA for 210 nm particles and flow cytometry for the 9.95 µm particles as described in the Supplemental Information, and values are listed in Table 2.
Table 2.
Receptor and ligand densities measured by ELISA (receptors on nanoparticles and ligands on flow chamber substrates) or flow cytometry (receptors on microparticles).
| Type | Molecule | Molecular Density (•m−2) | |||
|---|---|---|---|---|---|
| Low | Med | High | Max | ||
| Receptor (Nanoparticle) | 4-4-20 scFv | 6,900 ± 900 | 14,100 ± 700 | 19,800 ± 700 | |
| 4M2.4 scFv | 4,800 ± 400 | 16,400 ± 1000 | 20,700 ± 1300 | ||
| 4M5.3 scFv | 3,600 ± 300 | 14,800 ± 600 | 20,000 ± 700 | ||
| 4-4-20 scFv-mRFP | 6,700 ± 500 | 20,400 ± 900 | |||
| 4-4-20 Ab | 480 ± 30 | 960 ± 130 | 2,500 ± 60 | ||
| Biotin | 680 ± 50 | 1,100 ± 150 | 1,500 ± 350 | 5,800 ± 100 | |
| VIII scFv | 390 ± 80 | 1,900 ± 200 | 19,600 ± 1200 | ||
| VIII scFv-mRFP | 400 ± 80 | 980 ± 200 | |||
| Receptor (Microparticle) | 4-4-20 scFv | 1200 ± 10 | 4600 ± 40 | ||
| 4M2.4 scFv | 630 ± 10 | 4600 ± 90 | |||
| 4M5.3 scFv | 640 ± 10 | 4600 ± 80 | |||
| Biotin | 2100 ± 170 | 5,600 ± 140 | |||
| Ligand | Fluorescein | 47 ± 4 | 76 ± 7 | 120 ± 8 | 195 ± 3 |
| Neutravidin | 29 ± 4 | 41 ± 7 | 154 ± 8 | 195 ± 3 | |
| VCAM-1 | 142 ± 9 | 457 ± 6 | |||
Substrate Preparation
Substrates for parallel-plate flow experiments were prepared using (3-aminopropyl)trimethoxysilane (Sigma) treated glass coverslips that were fitted with modified FlexiPerm silicone gaskets, as described elsewhere.2 Silanized coverslips were washed with adsorption buffer (0.1 M NaHCO3, pH 9.2) and incubated for 2 hours at room temperature with a saturating concentration of Neutravidin or protein G in adsorption buffer (100 µg/ml, 0.45 ml). After three PBS washes, Neutravidin substrates were incubated for 1 hr with one of the following treatments: (1) 0.5 ml D-biotin at 0, 20, 50 or 500 nM in PBS for experiments using biotinylated BSA or (2) 0.5 ml FITC-biotin at 100 nM in PBS or mixtures of FITC-biotin and D-biotin for the 4-4-20 scFv and antibody cases. For experiments using VIII scFv, protein G substrates were incubated with 0.25 ml VCAM-1/Fc at 100 nM in PBS, or ratios of VCAM-1/Fc and human IgG1 Fc control. Substrates were washed three times immediately prior to use with SuperBlock (Thermo Fisher). Ligand site densities (nl) were measured by ELISA for all substrates as described in the Supplemental Information, and values are listed in Table 2.
Nanoparticle Binding Assays
Binding and detachment experiments were performed for 210 nm particles using a parallel plate flow chamber as described previously.2 The flow chamber was assembled with the fluorescein, Neutravidin, VCAM-1, or control coverslip acting as the bottom surface, and then was positioned on an inverted Nikon Diaphot microscope equipped with cooled CCD camera, motorized stage, and FITC filter cube. Fluorescent images were captured at 40× magnification with an exposure time of 500 milliseconds, with the camera and motorized stage controlled using custom Labview (National Instruments, Austin, TX) programs. A total of eight stage positions were monitored at the centerline of the chamber, starting 5 mm from the inlet and spaced 1 mm apart down the axis, and complete imaging cycles were completed each minute during the binding period and with at less frequent intervals (2–4 minutes) during the detachment period. Imaging was initiated once particles reached the chamber, however data was only used after the binding rate reached steady state. Binding experiments were conducted for 15 min at 100 s−1 shear rate, followed directly by detachment experiments at the same shear rate utilizing the flow of buffer. Flow was induced using a syringe pump (Harvard Apparatus, Natick, MA), and wall shear rate (γ̇w) was calculated from the volumetric flow rate Q as follows
| (1) |
where U is the average fluid velocity, H is the chamber height and W is the chamber width.
Binding Profile Analysis
Particle binding was assessed by manually overlaying consecutive fluorescence microscopy images using a custom Labview program. Binding and detachment profiles were constructed by tracking the instantaneous number of particles bound at each time point during binding and detachment experiments, respectively. Attachment profiles were constructed by summing the cumulative number of particles bound by each time point during the binding period, and thus detachment was ignored. Bound particle numbers were converted to bound densities using the known field of view area. Both time and bound particle number were initialized to the beginning of the steady state period, and the results from each of the stage positions were averaged.
Since nanoparticle attachment can be tracked independently from detachment, nanoparticle adhesion can be modeled using the following rate equation
| (2) |
where B is the bound particle density, t is time, kA is the multivalent kinetic attachment rate, and Cw is the unbound particle concentration at the wall of the reactive substrate. The attachment rate (kACw) can then be obtained directly from binding experiment attachment data by integration of equation 2
| (3) |
where BTotal is the total number of particles that bound per area. Isolation of kA from Cw requires knowledge of the particle concentration throughout the chamber, which is accomplished using a transport-reaction model described previously.2
Monte Carlo Simulation of Particle Detachment
Multivalent nanoparticle detachment is history dependent, with the probability of detachment decreasing with the time bound due to adhesion strengthening. Therefore we used a simulation technique to track bound lifetimes for each nanoparticle throughout both experimental binding and detachment periods and stochastically sample for detachment events, as described elsewhere.2 To capture the time-variant detachment dynamics, the multivalent detachment rate constant was modeled using constant (detachment rate magnitude, •D0) and functional power law (•) components. These parameters are used to calculate the detachment probability for each bound particle as follows
| (4) |
where Δt is the simulation time step, tb is the total time that the particle has been bound, and tref is a reference time (included to preserve unit consistency and is arbitrarily defined as 1 second). During each time step, a constant number of particles are added to the system based on the experimentally determined attachment rate (kACw), and detachment is sampled for each bound particle by randomly generating a number between 0 and 1 and comparing the value to PD. If the random number is less than PD the particle detached and tB is recorded, otherwise tB is updated by Δt and the process is repeated during the subsequent time step. Binding profiles are constructed by totaling the number of bound particles at each time step. Detachment experiments are simulated analogously, however particles are no longer added to the system.
Simulations were conducted using Matlab with a time step of 1 second, and for each condition three independent simulations were averaged and the standard deviation was calculated for data fitting purposes. Detachment parameters (•D0 and •) were then chosen to simultaneously fit binding and detachment experiment data.
Microparticle Shear Detachment Assays
Detachment assays were performed with 9.95 µm particles using the same straight channel parallel plate flow chamber described for nanoparticle binding experiments, and were analogous to previous studies.19, 30, 31 Particles were drawn into the flow chamber at 0.2 ml/min and flow was halted to allow particles to settle and bind. After a 30 min period to allow binding to reach equilibrium, unbound particles were removed by initiating flow at 0.1 ml/min and images were captured at two separate positions in the flow chamber using 10× magnification and an exposure time of 500 milliseconds to distinguish between bound and flowing particles. Flow rate was then increased incrementally every 2 minutes followed by a scan of the chamber, similar to Ham.32 At each flow rate monitored, the fraction particles remaining with respect to the initial 0.1 ml/min scan was determined and plotted versus the shear rate (calculated using equation 1). This enabled identification of the critical shear rate (•c), which is defined as the shear rate at which 50% of the particles remain bound.30, 31
Statistics
ELISA measurements, bound nanoparticle densities from flow chamber adhesion experiments, fraction particles remaining from microparticle shear detachment assays, and critical shear rates are given ± the standard error from at least 3 independent experiments. Curve fits to determine the attachment rate and attachment rate constant are given ± the standard error of the data. All other curve fits are given ± the 95% confidence interval.
RESULTS
Single-Chain Antibody Production and Characterization
The VIII scFv was cloned from a hybridoma expressing the VCAM-1 specific VIII-6G10 antibody and inserted into a pCT302 plasmid (Supplemental Fig. 1a) for yeast surface display. Flow cytometry confirmed that yeast produced full-length and functional VIII scFv based on staining with an anti-c-myc antibody and purified VCAM-1 protein, respectively (Supplemental Fig. 1b). Expression and fluorescein binding for the wild-type and mutant 4-4-20 scFvs has previously been reported.13, 14 For soluble production, the 4-4-20, 4M2.4, 4M5.3, and VIII scFv sequences were inserted into a pRS324-based plasmid (Fig. 2a) to induce secretion from yeast. Culture supernatants were purified using a His-tag affinity column and size exclusion chromatography and scFv protein was assessed by SDS-PAGE and anti-c-myc Western blot of (Figs. 1b and c). The scFv samples were highly pure and migrated at approximately 35 kD, as predicted. The scFv-mRFP constructs migrated at approximately 65 kD, close to the predicted value of 66 kD, and contained some residual BSA protein. Protein yields were determined by micro-BCA assay as 47, 48, 32, 457, 28, and 1060 µg for the 4-4-20 scFv, 4M2.4 scFv, 4M5.3 scFv, 4-4-20 scFv-mRFP, VIII scFv, and VIII scFv-mRFP samples, respectively. The higher values obtained for the mRFP fusions in part reflects the presence of BSA, but the Western blot does suggest that considerably more scFv protein is present as well. It is not known whether the higher yields obtained for the mRFP fusions are due to greater production rate from yeast or decreased losses during processing.
Nanoparticle Binding and Detachment Assays
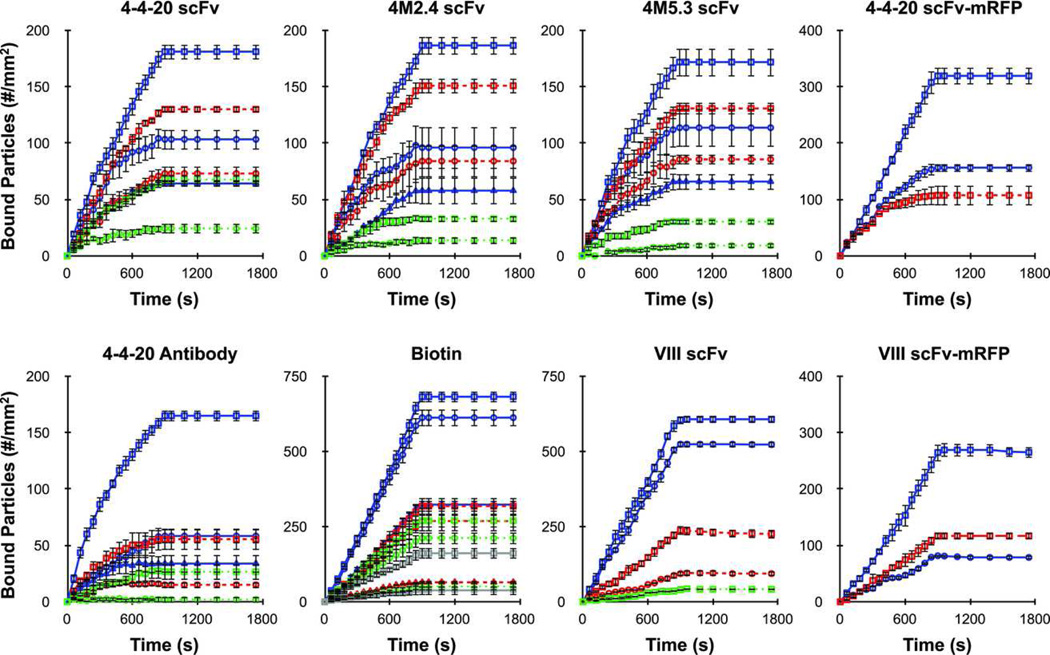
Nanoparticles were prepared by attaching biotinylated receptor protein to avidin-coated, 210 nm diameter spheres at various densities (nr). Counter-ligand substrates were prepared at various densities (nl) by adsorbing protein directly to glass coverslips (avidin, avidin-fluorescein) or by binding to adsorbed protein G (VCAM-1/Fc). Receptor and ligand densities employed were measured by ELISA, and are listed in Table 2. Nanoparticle binding and detachment experiments were performed in a parallel plate flow chamber at 100 s−1 shear rate, and the binding profiles are given in Figure 3. Included are separate plots for each receptor/ligand interaction investigated, and each trace represents a different particle receptor density and/or substrate ligand density condition. Background nanoparticle binding levels, as determined using control nanoparticles and substrate surfaces, were minimal and were subtracted from the data in Figure 3. Nanoparticle binding experiments were followed directly by detachment experiments using buffer only to monitor dissociation.
Figure 3.
Binding profiles for 210 nm particles obtained from parallel plate flow chamber adhesion assays the different receptor/ligand systems, as indicated. Adhesion assays were performed at 100 s−1 shear rate using a particle concentration of 5 × 106/ml for 15 minutes (binding period) and buffer flow for 15 minutes (detachment period). Background levels were determined using control particles and substrates, and were averaged and subtracted from the above profiles. Colors represent different receptor density conditions (solid blue - high, dashed red - medium, dotted green - low, solid gray - lowest; see Table 2) and shapes represent different ligand density conditions (square - high, circle - medium, triangle - low; see Table 2). Error bars represent the standard error of at least 3 independent adhesion experiments.
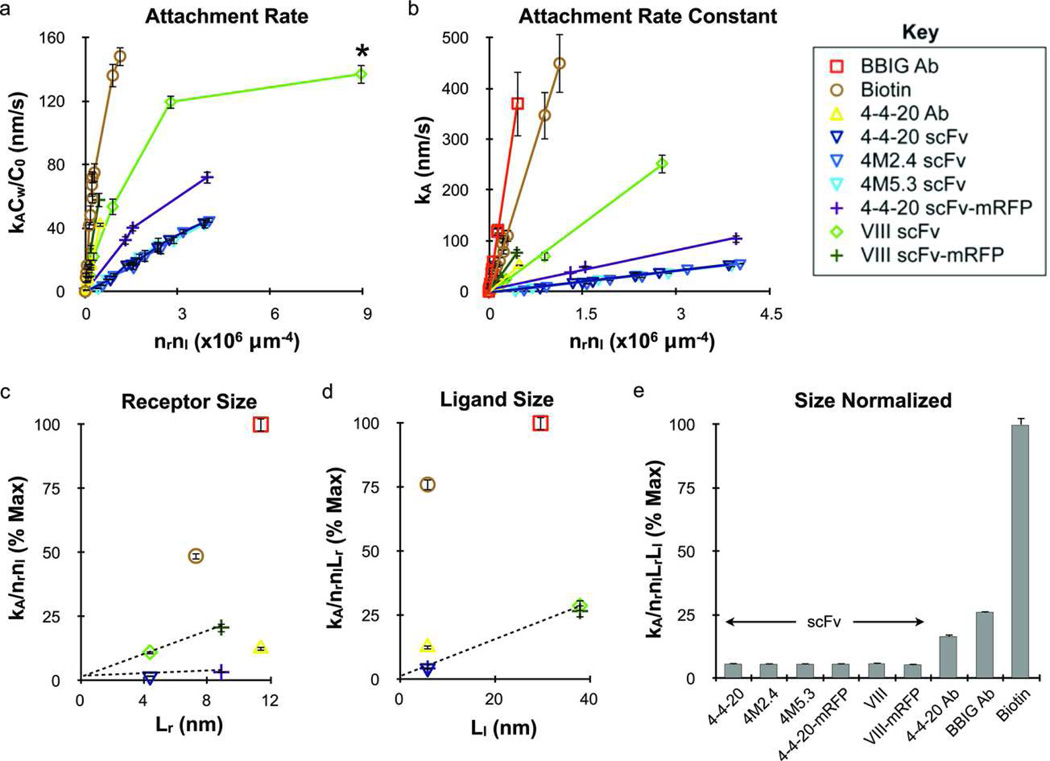
Evaluation of Nanoparticle Attachment
The net nanoparticle attachment rate (kACw) was determined for each experimental condition by linear least-squares fit of the attachment data in accordance with equation 3. These results are presented in Figure 4a as a function of the molecular densities (nrnl) to indicate attachment efficiency per adhesion molecule. Attachment rates (kACw/C0) appeared to reach a plateau above 100 nm/s, indicating that nanoparticle attachment was transport-limited above this point. The attachment rate constant (kA) was then calculated from the attachment rate using a transport-reaction model (equations S1–S3); the results are presented in Figure 4b. The kA values obtained from a similar study of 210 nm particles binding via monoclonal antibody (BBIG)/ICAM-1 interaction are also included in Figure 4b for comparison.2 Attachment efficiency was greatest for the BBIG antibody/ICAM-1 interaction, followed by avidin/biotin, VIII scFv-mRFP/VCAM-1, 4-4-20 antibody/fluorescein, VIII scFv/VCAM-1, 4-4-20 scFv-mRFP/fluorescein and finally by the 4-4-20 scFv/fluorescein (wild-type and mutants). The Identical attachment rates observed for the 4-4-20, 4M2.4 and 4M5.3 scFvs was unexpected given the substantial differences in bond kinetics, and suggest either that kinetic effects were already maximal in the regime investigated or were mitigated by another factor such as multivalency or insufficient bond mechanical strength. The small size of scFvs did enable attachment at much higher densities however (Table 2); thus nanoparticles coated with the maximum number of scFv molecules demonstrated comparable or superior binding rates than particles coated with the maximum coating of antibody (Figure S2).
Figure 4.
Nanoparticle attachment efficiency depends on adhesion molecule valency and size but not necessarily reaction kinetics. (a) Attachment rates for each receptor/ligand interaction were obtained from attachment profiles and normalized by inlet concentration (kACw/C0) plotted versus the molecular densities to indicate binding efficiency. The nanoparticle adhesion rate for the VIII scFv/VCAM-1 interaction approached saturation, and therefore the data point marked with a * was not used for subsequent analysis. Addition of the mRFP increased attachment rate for both scFv species, but modulating kinetics did not have an effect for the 4-4-20 scFv family of mutants. (b) Similar to part a but using the attachment rate constant (kA) determined using a transport-reaction model. The results from a previous study using a monoclonal antibody (clone BBIG) that is specific for ICAM-1 is included for comparison.2 (c) Attachment efficiency (slope kA/nrnl from part b) plotted versus Lr (Table 1) to illustrate the effect of receptor size, demonstrating that the addition of mRFP to the scFv constructs increased attachment efficiency in direct proportion to the increase in size. (d) Similar plotting of the receptor size-normalized attachment efficiency (slope kA/nrnlLr from part c) against ligand size (Ll, Table 1), converging all scFv data. (e) Complete normalization of the attachment rate by molecular valency and size effects (slope kA/nrnlLrLl from part d), further illustrating that identical attachment efficiencies were obtained for the scFv species. The antibody/antigen and avidin/biotin interactions had much higher attachment efficiencies after normalization that reflected their chemical kinetic values (Table 1). The different receptor/ligand species are defined in the legend key. Error bars for attachment rate represent the standard error of at least 3 independent adhesion experiments.
The addition of the mRFP extender significantly enhanced the nanoparticle attachment efficiency for both the 4-4-20 and VIII scFvs, clearly indicating a role for molecular size. To quantify this effect, we first normalized the attachment results by receptor size (Lr) as determined by measurements of published molecular structures (Table 1, Figure 1). As Figure 4c illustrates, the slope values for kA/nrnl from Figure 4b (listed in Table S1) scaled directly with Lr for both the 4-4-20 and VIII scFvs after addition of the mRFP extender, demonstrating that the rate of nanoparticle attachment is proportional to receptor size in the range tested. Similar normalization to ligand size (Ll, Table 1) effectively converged all scFvs to the same value (Figures 4d and e), suggesting that a linear relationship holds for ligand size as well. While the molecular size and valency-normalized attachment rate values (kA/nrnlLrLl) for all scFvs were similar, significant differences were observed for the other receptors that could be related to bond kinetics or other factors.
Microparticle Shear Detachment Assays to Assess Bond Adhesion Strength
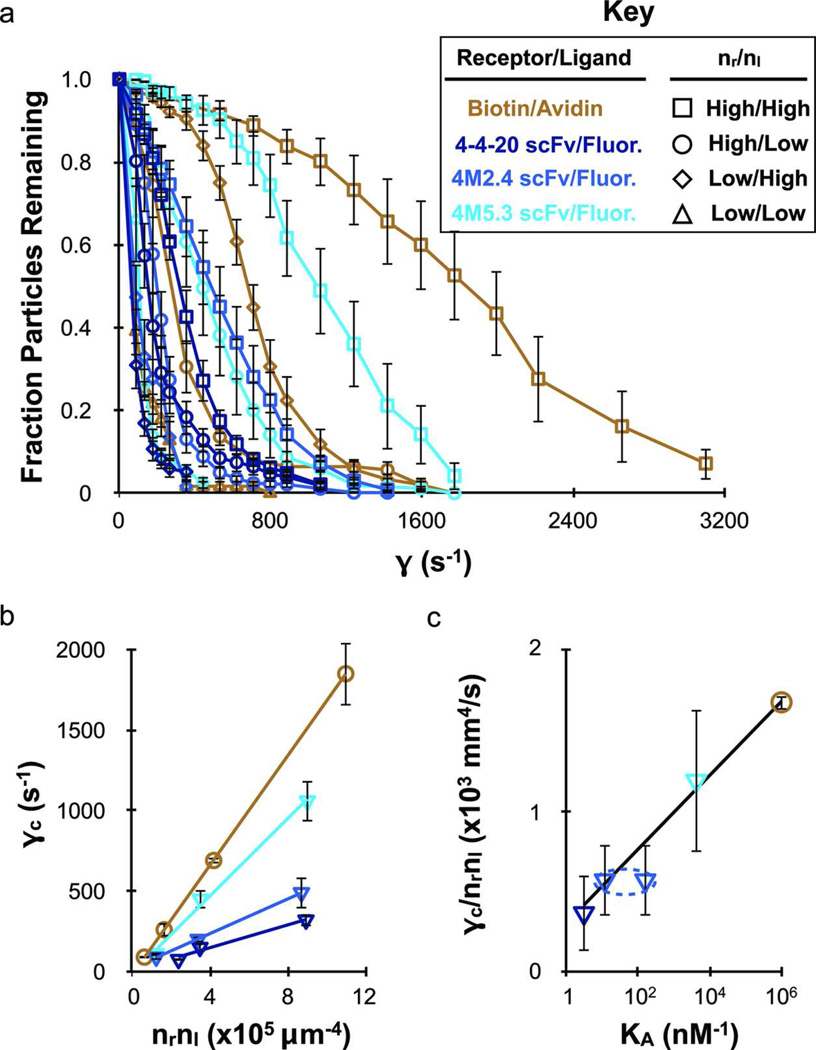
To investigate whether the differences in nanoparticle attachment efficiency observed for the scFvs and the other targeting molecules studied were related to bond mechanics, we tested the adhesion strength of the 4-4-20 scFv/fluorescein (wild-type and mutants) and avidin/biotin interactions using microbead shear detachment assays. Microbeads (9.95 µm diameter) were coated with varying densities of biotinylated BSA as well as the 4-4-20, 4M2.4, and 4M5.3 scFvs, and allowed to bind statically to substrates functionalized with varying densities of counter-ligand. The fraction of microbeads that remained bound as shear rate was increased is plotted in Figure 5a. At the maximum receptor and ligand densities, the beads bound via avidin/biotin demonstrated the greatest ability to withstand shear force, followed in turn by the 4M5.3, 4M2.4 and 4-4-20 scFv-coated beads bound to fluorescein. To quantify adhesion strength, the critical shear rate (•c) was determined based on the shear rate in Figure 5a at which 50% of the beads remained bound. Next, the values of •c were plotted versus the molecular densities (nrnl) to eliminate the influence of bond number (Figure 5b). The slopes from Figure 5b (•c/nrnl) were then plotted versus the bond affinity (KA) in Figure 5c, revealing a logarithmic dependence. Thus, in microbead detachment assays, the scFvs exhibit adhesion strengths that are comparable to avidin/biotin after appropriately accounting for the limits of chemistry.
Figure 5.
The wild-type and mutant 4-4-20 scFvs exhibit adhesion strengths that scale directly with chemical affinity in microparticle shear detachment assays. Receptor functionalized 9.95 µm diameter particles bound to equilibrium under static conditions and were then removed by shear flow. (a) Fraction of particles remaining bound at each shear rate investigated. Background binding was determined for control particle and substrates, and was subtracted from the specific adhesion data. (b) The critical shear rate (•c), determined from (a) as the shear rate at which 50% of the particles remained bound, plotted against the molecular densities to represent adhesion strength per available adhesion molecule. Adhesion strength was greatest for avidin/biotin and decreased in accordance with the different 4-4-20 scFv mutant generations back down to the wild-type. (c) The adhesion strength per bond (slope •c/nrnl from part b) are plotted versus the logarithm of the equilibrium affinity constant (KA, Table 1). Since kf is not known for the 4M2.4 scFv, representative data points were added using the kf values of the 4-4-20 and 4M5.3 scFvs to indicate the expected bounds. The linear fit depicted was performed without using the 4M2.4 data, but suggests that the actual kf value is similar to the 4M5.3 scFv. The different receptor/ligand species are defined in the legend key. Error bars represent the standard error of at least 3 independent detachment experiments for the fraction particles remaining and •c, and the 95% confidence interval from linear fits of the •c versus nrnl plots.
Evaluation of Nanoparticle Detachment Dynamics
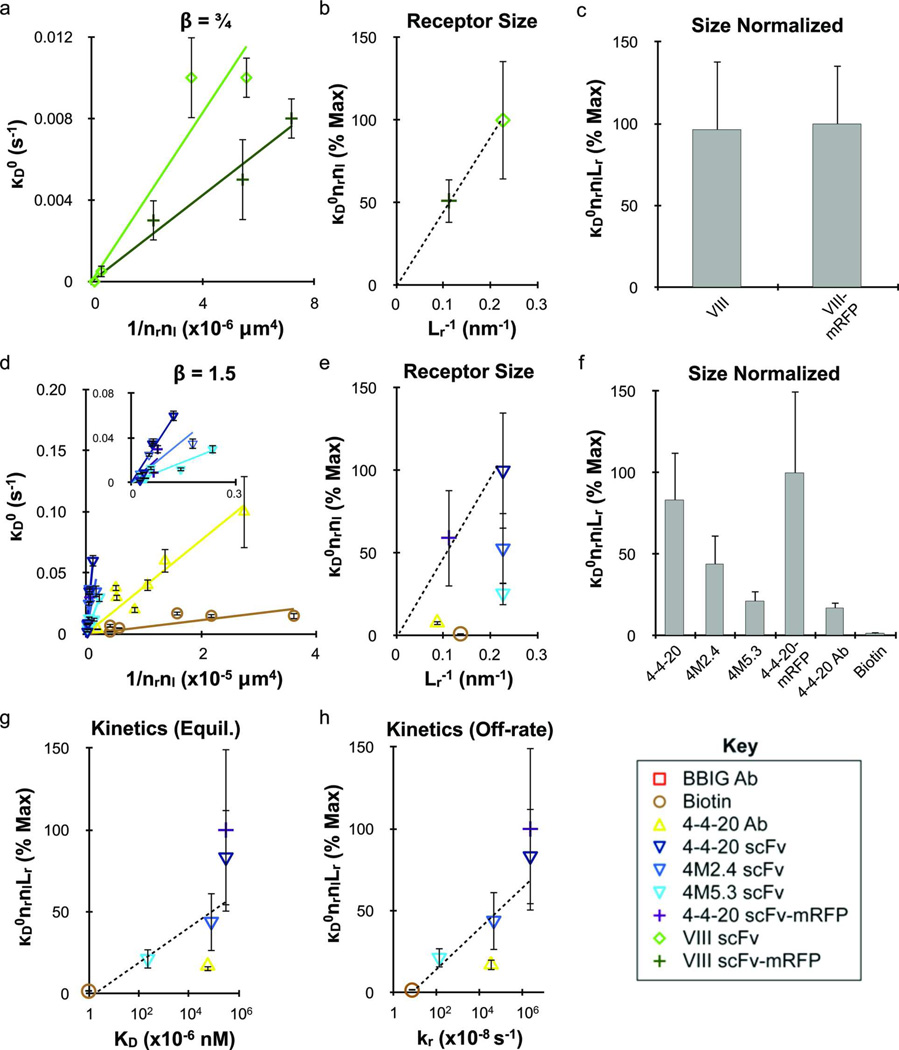
Experimental binding data was recreated using a Monte Carlo simulation to track bound particle histories and stochastically sample for detachment events. This enabled determination of detachment rate parameters, including the functional dependence of dissociation on time (•) and the magnitude of dissociation (•D0), as described in equation 4. Our findings revealed that all adhesion molecules fell into one of two categories with respect to their best-fit value of •. The VIII scFv and VIII scFv-mRFP cases were best characterized by • = ¾, as seen previously for nanoparticle adhesion mediated by an antibody/ICAM-1 interaction.2, 3 The remaining interactions, including the 4-4-20 scFv/fluorescein (wild-type and mutants, mRFP fusion), 4-4-20 Ab/fluorescein and avidin/biotin interactions, were characterized by infrequent nanoparticle dissociation during detachment experiments. Simulation fits could only conclusively establish that • was ≥ 1.5, and therefore • = 1.5 was used for these cases.
Best fit •D0 values are plotted in Figures 6a (• = ¾) and d (• = 1.5) against (nrnl)−1 to account for adhesion molecule density effects. Linear least-square fits of •D0 versus (nrnl)−1 are included in Figures 6a and d, and slope values from these fits (•D0nrnl) are listed in Table S1. To account for the role of receptor size, the slope values for •D0nrnl were plotted against 1/Lr (Figures 6b and e). Based on the linear relationships observed for the scFvs relative to their mRFP fusion proteins, detachment rate decreases directly with receptor size. Finally, since significant differences between the different binding interactions still remained after size normalization (Figure 6c and f), the slope values for •D0nrnlLr in Figure 6e were compared to the known kinetic parameters listed in Table 1. Reasonable correlations could be made to both KD (Figure 6g) and kr (Figure 6h), but appeared stronger for kr.
Figure 6.
Nanoparticle detachment probability depends on molecular valency, size, and reaction kinetics. Detachment results were determined using the stochastic simulation for functional time constant • values of (a–c) ¾ and (d–h) 1.5. (a and d) Detachment rate constant κD0 plotted against the molecular densities to normalize for valency effects. Addition of mRFP decreased detachment rate for both the 4-4-20 and VIII scFvs, which was accounted for in parts (b and e) by plotting the detachment efficiency (slope κD0nrnl from parts a and d) against receptor size (1/Lr, Table 1). (c and f) The receptor size-normalized detachment efficiencies (slope κD0nrnlLr from parts b and e) for each species. The results in part part f correlate with the measured kinetic rates (Table 1), and were therefore plotted against their respective (g) equilibrium affinity (KA) and (h) reverse reaction rate (kr) values. In both cases, reasonable fits were obtained with logarithmic dependence, however the relationship with kr appeared stronger. The kf value for the 4M5.3 scFv was used as an estimate for the 4M2.4 scFv based on the microparticle shear detachment assays. The effect of ligand size on κD0 could not be addressed because different • values were obtained for the two classes of ligands employed. The different receptor/ligand species are defined in the legend key. Error bars indicate the standard error from simulation fits for κD0 and 95% confidence intervals for linear regression analysis.
DISCUSSION
In this study we quantified nanoparticle adhesion mediated by different receptor/ligand systems in an attempt to understand the effects of bond properties such as molecular kinetics, mechanics, and size. To isolate specific bond properties, we employed a set of scFv proteins that were engineered to display different kinetic binding rates. Furthermore, we directly investigated the effect of receptor size by inserting mRFP into the scFv fusion construct to act as a linker that extends the binding domain further from the particle surface. We also compared two different scFvs that are specific for ligands with disparate sizes. Finally, monoclonal antibody/antigen and avidin/biotin interactions were also included in our analysis. A concise summary of our findings with respect to nanoparticle attachment probability are as follows: the attachment rate (1) increases proportionally with receptor and ligand valencies, (2) increases proportionally with receptor and ligand size, and (3) does not depend on kinetics for recombinant scFv proteins but does appear to be affected by kinetics for the antibody/antigen and avidin/biotin interactions. Our findings with respect to nanoparticle detachment probability are as follows: the detachment rate (1) transiently decreases with the time bound based on a power law relationship (β) that varies with ligand size but not receptor size, (2) decreases in magnitude in proportion to receptor and ligand valencies, (3) decreases in magnitude in proportion to receptor size, and (4) decreases in magnitude in proportion with the logarithm of the bond kinetics.
Our finding that molecular size exerts a strong influence over nanoparticle attachment was expected because bond length controls the length scale and effective contact area over and within which bonds can form.22, 23 Both of these factors should scale directly with bond length, and thus a linear relationship is reasonable. However, more work with engineered receptor and ligand sizing is needed to validate our finding. We also observed that molecular size can influence particle attachment through its control over the maximum coating density. For instance, scFvs could be incorporated at approximately 10-fold higher densities in comparison to antibodies (Table 2), which compensated for lower intrinsic binding efficiency and led to similar maximum attachment rates for the 4-4-20 scFv and full 4-4-20 antibody (Figure S2). Adding a linker to the 4-4-20 scFv fusion construct in the form of the mRFP protein did not affect loading capacity but did increase adhesion rate, and thus we were able to successfully engineer a recombinant targeting receptor that performed superior to the full antibody.
Following normalization by molecular sizes, all scFv species displayed identical nanoparticle attachment rates. This result was surprising for two reasons: (1) we employed two scFvs that bound to very different ligands (small molecule fluorescein versus large protein VCAM-1) and (2) the wild-type and mutant 4-4-20 scFvs displayed kinetic differences that spanned one order of magnitude in forward reaction rate (kf) and four orders of magnitude in reverse reaction rate (kr) (Table 1). This result firmly establishes that multivalent nanoparticle adhesion is not necessarily proportional to the receptor binding rates. One possible explanation could be that within the kinetic regime tested, nanoparticle attachment was so fast that transport limits dominated, and thus improving kinetics did not increase binding efficiency. There is evidence this is not the case however; transport limitations did not appear in Figure 4a until the attachment rate was much greater than that exhibited by the scFvs. An alternative explanation may be related to multivalency, in which the formation of multiple bonds masked kinetic effects. This explanation is likely insufficient on its own though, because we expect that the antibody/antigen and avidin/biotin interactions would support a similar extent of multivalent binding, but these interactions were able to attain significantly higher nanoparticle attachment efficiencies (Figure 4e) that appeared to reflect bond kinetics (Table 1). A secondary consideration may be bond mechanical strength, in which the relatively simple structure of scFvs (two polypeptide domains connected by a short peptide linker) prevented a single scFv tether from mediating nanoparticle capture under the applied shear force. Under this scenario scFv-targeted nanoparticles would need to encounter the substrate in an orientation such that multiple bonds form simultaneously. Increasing receptor number and size would increase the likelihood of achieving a required orientation, but changing kinetics would not provided that a minimum threshold is exceeded. This behavior is consistent with our data, and recent computational modeling of multivalent nanoparticle binding has indicated that distinct ligand arrangements can dictate adhesion under certain conditions.33 Future work will investigate scFv-mediated binding further using nanoparticles of different size and various flow rates to modulate hydrodynamic force. In addition, different receptor/ligand systems, including natural and recombinant proteins as well as synthetic molecules such as peptides and aptamers, will be studied and mechanical strength measurements will be made using techniques such as atomic force microscopy. Finally, computational simulation techniques such as Adhesive Dynamics will be needed to further elucidate the complex relationships between force, bond kinetics and mechanics, and ligand arrangements.34, 35
Shear detachment assays using micron-scale particles provided additional insight into scFv bond strength. We found that the 4-4-20 scFvs/fluorescein (wild-type and mutants) and avidin/biotin interactions all bound with adhesion strengths per bond that correlated directly with equilibrium affinity (Figure 5c), as expected based on previous work.18, 19 We believe that the disparity in results from these two assays, where bond kinetics did not affect nanoparticle attachment but did influence microbead detachment, is related to differences in context. Nanoparticle attachment must occur under dynamic conditions with initial capture mediated by the formation of a limited number of bonds. For microbead detachment, a large number of bonds are allowed to form under equilibrium conditions before force is applied. Thus we believe that the small, but not insignificant, shear force acting on the nanoparticles during capture is great enough to prevent single (or even a few) scFv bonds from successfully forming, resulting in adhesion that is independent of kinetics. Once bonds have formed and particle translation stops however, chemical kinetics dictates adhesion strength. We believe that we have uncovered a unique phenomenon that influences adhesion at the nano-scale.
Consistent with our previous findings,2, 3 multivalent nanoparticles bound transiently with a detachment probability that decreased with the time bound to the substrate. We previously established for the BBIG antibody/ICAM-1 interaction that nanoparticle dissociation rate decreased with time according to a power law (β = ¾) that was independent of adhesion molecule density, shear rate, or particle size. For the receptor/ligand systems used here, we found a similar power law for the VIII scFv (which binds to VCAM-1, of similar size to ICAM-1) but a substantially different power law (β = 1.5) for the other interactions. The source of these differences is unclear, but may be related to bond length (Table 1) or the manner in which the ligand substrates were prepared. For the cases in which β = ¾ (VIII scFv and BBIG antibody), the ligands were large proteins (VCAM-1 and ICAM-1) that were bound as chimeras with human IgG1 Fc to protein G. Conversely, for the systems in which β = 1.5, the smaller Neutravidin protein was adsorbed and used directly (biotin) or reacted with FITC-biotin to create fluorescein substrates. Studies employing more diverse molecular sizes and a consistent tethering strategy will be required to further elucidate these time-dependent dissociation effects.
We have previously shown that the detachment rate constant (κD0) for multivalent nanoparticles scales inversely with adhesion molecule density, and specifically with the relationship (nrnlφ)−1.2, 3 The factor φ is used to account for limitations in ligand accessibility due to geometric mismatch between the spherical particle and planar substrate. Here we find φ = 1 for interactions studies. This is in contrast to the results obtained using the BBIG antibody/ICAM-1 interaction, where φ was found to be ⅓. This difference in φ is unlikely to be related exclusively to ligand size or the manner in which the ligand was tethered, as ICAM-1 (BBIG antibody) and VCAM-1 (VIII scFv) have similar sizes and were attached using the method. Instead, it appears that the use of either a relatively smaller receptor (VIII scFv/VCAM-1), smaller ligand (4-4-20 antibody/fluorescein), or both (avidin/biotin, 4-4-20 scFv/fluorescein) reduced the difference in relative accessibility between receptors on the nanoparticle and ligand on the substrate within the contact zone. Rephrased, the contact zone appears more planar when smaller adhesion molecules were used. After normalization of κD0 by adhesion molecule densities and sizes, we uncovered a general scaling relationship with kinetics (Figure 6g and h). This included good correlation with the logarithm of the equilibrium dissociation constant (KD), although this effect could be driven by the reverse reaction rate (kr). Thus, bond kinetics and mechanics may dictate nanoparticle attachment, but nanoparticle dissociation is universally dictated by chemical affinity.
In summary, we have elucidated the contributions of receptor/ligand bond properties, specifically kinetics and size, on nanoparticle adhesion dynamics under fluid flow conditions. While the flow chamber model that we employed lacks many in vivo characteristics that can influence nanoparticle transport, such as the presence of erythrocytes or recirculation regimes caused by vessel branching and stenoses,36 and binding, such as a non-planar reaction surface and mobile ligands on cells, our results illustrate general guidelines that should hold in various contexts. For example, we demonstrate that the factors that increase encounter probability (molecular numbers and sizes) control nanoparticle attachment at all times, while the influence of bond kinetic rates depends on a combination of multivalency and bond mechanical strength. Specifically, kinetics did not affect nanoparticle adhesion mediated by scFv proteins, and thus binding was much less efficient on a per molecule basis than other interactions studied. The scFvs did however compensate for lower binding efficiency with higher loading capacity on the nanoparticle. Conversely, the factors that increase bond number (molecular numbers and sizes, bond kinetics) directly decrease the likelihood of nanoparticle dissociation. Finally, we have established that scFvs can function as effective targeting receptors for the delivery of therapeutic nanocarriers, displaying nanoparticle adhesion characteristics that are comparable to or even surpass full antibodies. These tunable recombinant proteins are thus viable alternatives to antibodies for nanoparticle targeting applications. However, more work is needed to address the shortcomings of scFvs for targeting nanoparticles, such as structural modifications or the addition of stabilizing elements that improve mechanical strength, so that their tunable kinetic properties can be fully exploited and binding efficiency maximized. These findings could also be applicable to other recombinant protein or synthetic construct (peptide, aptamer, affibody, avimer) targeting receptors.
Supplementary Material
ACKNOWLEDGMENT
The authors thank E.J. Johnston and A.W. Nields for technical assistance and helpful discussions. This work was supported by NIH grant GM081444 and NSF Penn MRSEC grant DMR05-20020.
Footnotes
Supporting Information Available: Supporting methods describing the verification of VIII scFv specificity for VCAM-1, biotinylation of the 4-4-20 antibody, characterization of nanoparticle and microbead surface molecule densities, characterization of flow chamber surface ligand densities, and description of transport-reaction model. Supporting Figures S1 and S2 showing yeast surface display of VIII scFv and specificity for VCAM-1 by flow cytommetry and maximal nanoparticle attachment rates for each receptor/ligand interaction studied. Supporting Table S1 listing attachment (kA/nrnl) and detachment (•, •, •D0nrnl) parameter values determined for each receptor/ligand system. This materials is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Davis ME, Chen ZG, Shin DM. Nat Rev Drug Discov. 2008;7:771–782. doi: 10.1038/nrd2614. [DOI] [PubMed] [Google Scholar]
- 2.Haun JB, Hammer DA. Langmuir. 2008;24:8821–8832. doi: 10.1021/la8005844. [DOI] [PubMed] [Google Scholar]
- 3.Haun JB, Robbins GP, Hammer DA. J Adhesion. 2010;86:131–159. [Google Scholar]
- 4.Ackerson CJ, Jadzinsky PD, Jensen GJ, Kornberg RD. J Am Chem Soc. 2006;128:2635–2640. doi: 10.1021/ja0555668. [DOI] [PubMed] [Google Scholar]
- 5.Zhou Y, Drummond DC, Zou H, Hayes ME, Adams GP, Kirpotin DB, Marks JD. J Mol Biol. 2007;371:934–947. doi: 10.1016/j.jmb.2007.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lo YS, Nam DH, So HM, Chang H, Kim JJ, Kim YH, Lee JO. ACS Nano. 2009;3:3649–3655. doi: 10.1021/nn900540a. [DOI] [PubMed] [Google Scholar]
- 7.Kirpotin DB, Drummond DC, Shao Y, Shalaby MR, Hong K, Nielsen UB, Marks JD, Benz CC, Park JW. Cancer Res. 2006;66:6732–6740. doi: 10.1158/0008-5472.CAN-05-4199. [DOI] [PubMed] [Google Scholar]
- 8.Qian X, Peng XH, Ansari DO, Yin-Goen Q, Chen GZ, Shin DM, Yang L, Young AN, Wang MD, Nie S. Nat Biotechnol. 2008;26:83–90. doi: 10.1038/nbt1377. [DOI] [PubMed] [Google Scholar]
- 9.Yang L, Mao H, Wang YA, Cao Z, Peng X, Wang X, Duan H, Ni C, Yuan Q, Adams G, Smith MQ, Wood WC, Gao X, Nie S. Small. 2009;5:235–243. doi: 10.1002/smll.200800714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang X, Peng X, Wang Y, Wang Y, Shin DM, El-Sayed MA, Nie S. ACS Nano. 2010;4:5887–5896. doi: 10.1021/nn102055s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Y, Zhu X, Zhang X, Liu B, Huang L. Mol Ther. 2010;18:1650–1656. doi: 10.1038/mt.2010.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoogenboom HR. Nat Biotechnol. 2005;23:1105–1116. doi: 10.1038/nbt1126. [DOI] [PubMed] [Google Scholar]
- 13.Boder ET, Wittrup KD. Nat Biotechnol. 1997;15:553–557. doi: 10.1038/nbt0697-553. [DOI] [PubMed] [Google Scholar]
- 14.Boder ET, Midelfort KS, Wittrup KD. Proc Natl Acad Sci U S A. 2000;97:10701–10705. doi: 10.1073/pnas.170297297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hammer DA, Lauffenburger DA. Biophys J. 1987;52:475–487. doi: 10.1016/S0006-3495(87)83236-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bell GI. Science. 1978;200:618–627. doi: 10.1126/science.347575. [DOI] [PubMed] [Google Scholar]
- 17.Evans EA. Biophys J. 1985;48:175–183. doi: 10.1016/S0006-3495(85)83770-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dembo M, Torney DC, Saxman K, Hammer D. Proc R Soc Lond B Biol Sci. 1988;234:55–83. doi: 10.1098/rspb.1988.0038. [DOI] [PubMed] [Google Scholar]
- 19.Kuo SC, Lauffenburger DA. Biophys J. 1993;65:2191–2200. doi: 10.1016/S0006-3495(93)81277-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuo SC, Hammer DA, Lauffenburger DA. Biophys J. 1997;73:517–531. doi: 10.1016/S0006-3495(97)78090-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang KC, Tees DF, Hammer DA. Proc Natl Acad Sci U S A. 2000;97:11262–11267. doi: 10.1073/pnas.200240897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jeppesen C, Wong JY, Kuhl TL, Israelachvili JN, Mullah N, Zalipsky S, Marques CM. Science. 2001;293:465–468. doi: 10.1126/science.293.5529.465. [DOI] [PubMed] [Google Scholar]
- 23.Huang J, Chen J, Chesla SE, Yago T, Mehta P, McEver RP, Zhu C, Long M. J Biol Chem. 2004;279:44915–44923. doi: 10.1074/jbc.M407039200. [DOI] [PubMed] [Google Scholar]
- 24.Green NM. Methods Enzymol. 1990;184:51–67. doi: 10.1016/0076-6879(90)84259-j. [DOI] [PubMed] [Google Scholar]
- 25.Barbas CF, Burton DR, Scott JM. Phage Display: A Laboratory Manual. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 26.Sikorski RS, Hieter P. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang D, Shusta EV. Biotechnol Prog. 2005;21:349–357. doi: 10.1021/bp0497482. [DOI] [PubMed] [Google Scholar]
- 28.Clements JM, Catlin GH, Price MJ, Edwards RM. Gene. 1991;106:267–271. doi: 10.1016/0378-1119(91)90209-t. [DOI] [PubMed] [Google Scholar]
- 29.Gietz RD, Schiestl RH, Willems AR, Woods RA. Yeast. 1995;11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Y, Eniola AO, Graves DJ, Hammer DA. Langmuir. 2003;19:6905–6911. [Google Scholar]
- 31.Pangu G, Johnston E, Petkov J, Parry N, Leach M, Hammer DA. Langmuir. 2007;23:10682–10693. doi: 10.1021/la700603u. [DOI] [PubMed] [Google Scholar]
- 32.Ham AS, Goetz DJ, Klibanov AL, Lawrence MB. Biotechnol Bioeng. 2007;96:596–607. doi: 10.1002/bit.21153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martinez-Veracoechea FJ, Frenkel D. Proc Natl Acad Sci U S A. 2011;108:10963–10968. doi: 10.1073/pnas.1105351108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hammer DA, Apte SM. Biophys J. 1992;63:35–57. doi: 10.1016/S0006-3495(92)81577-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trister AD, Hammer DA. Biophys J. 2008;95:40–53. doi: 10.1529/biophysj.107.118430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blackwell JE, Dagia NM, Dickerson JB, Berg EL, Goetz DJ. Ann Biomed Eng. 2001;29:523–533. doi: 10.1114/1.1376697. [DOI] [PubMed] [Google Scholar]
- 37.Kranz DM, Herron JN, Voss EWJ. J Biol Chem. 1982;257:6987–6995. [PubMed] [Google Scholar]
- 38.Piran U, Riordan WJ. J Immunol Methods. 1990;133:141–143. doi: 10.1016/0022-1759(90)90328-s. [DOI] [PubMed] [Google Scholar]
- 39.Eniola AO, Willcox PJ, Hammer DA. Biophys J. 2003;85:2720–2731. doi: 10.1016/s0006-3495(03)74695-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.