Abstract
Cells of the innate immune system regulate immune responses through the production of antimicrobial peptides, chemokines, and cytokines, including human beta-defensins (hBDs) and CCL20. In this study, we examined the kinetics of primary human oral epithelial cell (HOEC) production of CCL20 and hBDs in response to Fusobacterium nucleatum, a commensal bacterium of the oral cavity, which we previously showed promotes HOEC induction of hBDs. HOECs secrete maximal levels of CCL20 at 6 h, following stimulation by F. nucleatum cell wall (FnCW). The kinetics of CCL20 release is distinct from that of hBD-2 and -3, which peaks after 24 h and 48 h of FnCW stimulation, respectively. FnCW-induced release of CCL20 by HOECs requires both transcriptional and translational activation. Release of CCL20 by HOECs is inhibited by brefeldin A, suggesting that it is secreted through a vesicle transport pathway. Other epithelium-derived agents that FnCW induces, such as hBD-2, hBD-3, tumor necrosis factor alpha (TNF-α) and interleukin-1β (IL-1β), are also able to release CCL20. By focusing on mitogen-activated protein kinases, we show that both extracellular signal-regulated kinase 1/2 and p38, but not JNK, are required for hBD-, TNF-α-, and IL-1β-induced secretion of CCL20 by HOECs. The ability of FnCW and its induced hBDs to produce proinflammatory cytokines and CCL20 suggests the broad role of F. nucleatum and human antimicrobial peptides in primary immune responses elicited by oral epithelium.
INTRODUCTION
Epithelial cells are the host's first line of defense against microbial invasion and pathogenesis (9, 12, 13, 18). At mucosal surfaces, including those of the oral epithelium, cells have evolved as part of the innate immune system to possess antimicrobial activity and to cross talk with the adaptive immune system. Cells of the innate immune system regulate immune responses through the production of chemokines and cytokines, including macrophage inflammatory protein 3α (MIP-3α), also known as CCL20. CCL20 is a 70-amino-acid chemokine that attracts immature dendritic cells and T cells via the chemokine receptor CCR6 (55), as has been reported for the epithelial cell-derived antimicrobial peptides (AMPs) human beta-defensin 1 (hBD-1) and -2 (60). We and others have reported on the ability of hBDs to cross talk with the adaptive immune system (21, 25, 52) and, most recently, we showed that overexpression of hBD-3 is linked to oral cancer (34, 35). CCL20 has also been linked to a variety of diseases, including cancer (39) and rheumatoid arthritis (54), and may have an important regulatory role in specific lymphocyte migration into inflamed periodontal tissues (30). By in situ hybridization, Abiko et al. (2) showed that CCL20 expression in oral squamous cell carcinoma (OSCC) is localized primarily at the epithelial pearls corresponding to the spinous layer. A recent study (6) also suggested that CCL20 may represent a potential immunohistochemical marker for prognostic prediction of OSCC. In periodontal diseased tissue, CCL20 has been shown to be distributed in the basal layer of gingival epithelial cells (30). The protein has direct antibacterial, antifungal, and antiviral activities, again comparable to hBDs (11, 37, 46, 61). Therefore, CCL20, along with hBDs, may play an important role in bridging the innate and adaptive immune responses.
The oral mucosa is in continuous contact with heterogeneous populations of diverse commensal and opportunistic microorganisms and yet, for the most part, it remains healthy and uninflamed. Recent research has highlighted the possibility that oral epithelium not only serves a barrier function as the first physical line of defense against colonizing pathogens but also orchestrates local immune responses (14, 26). Of the 700 species of oral bacteria estimated to colonize the oral cavity (1, 20, 48), a few have demonstrated the capacity to induce epithelial cell innate responses, while others have been more stealth-like (23, 40, 56). The ubiquitous oral bacterium Fusobacterium nucleatum and its cell wall extract (FnCW) induce expression of hBD-2 and -3 transcript in gingival epithelial cells (GECs) in vitro (15, 32, 33, 41). Recently, we identified an FnCW-associated peptide that induces hBD-2 in human oral epithelial cells (HOECs) (29).
FnCW has been shown to induce CCL20 transcript, and indeed induction of CCL20 by FnCW is highest (72.7-fold in 6 h) among all the cytokines/chemokines studied (62). In the present communication, we further characterize F. nucleatum-induced release of CCL20 by HOECs, the kinetics of beta-defensin release by HOECs upon stimulation with F. nucleatum, and their effects on CCL20 release by the host cells. The effects of proinflammatory cytokines on HOEC regulation of CCL20 are also investigated. Finally, we investigated the mitogen-activated protein kinase (MAPK) pathways regulating hBD- and cytokine-induced secretions of CCL20 by HOECs.
MATERIALS AND METHODS
Chemicals and reagents.
Synthetic hBD-2 and hBD-3 were purchased from Peptides International (Louisville, KY). Recombinant tumor necrosis factor alpha (TNF-α) and recombinant interleukin-1β (IL-1β) were purchased from Peprotech Inc. (Rocky Hill, NJ). All the MAPK inhibitors used in this study were purchased from Calbiochem (Gibbstown, NJ). Actinomycin D, brefeldin A (BFA), cycloheximide, and dimethyl sulfoxide (DMSO) were purchased from Sigma-Aldrich (St. Louis, MO).
Human primary oral epithelial cell cultures and stimulation.
Healthy human gingival tissue overlying impacted third molars was obtained from oral surgery clinics in the greater Cleveland area, in accordance with a Case Western Reserve University Institutional Review Board-approved protocol. Primary HOECs were isolated and grown in keratinocyte basal medium with supplements as previously described (10, 40, 41). To overcome interpersonal variability, epithelial cells from three donors were mixed at a ratio of 1:1:1 and grown to 80% confluence prior to challenge. Results were compared to HOECs grown to similar confluence from single donors. To avoid the effects of changes in epithelial cell physiology during growth in culture, each time point had a corresponding untreated, control group. Each experimental condition was conducted in triplicate.
Preparation of F. nucleatum cell wall fractions.
Cell wall from F. nucleatum was prepared as described previously (36). Briefly, F. nucleatum (ATCC 25586) was grown anaerobically in Columbia broth (BD Biosciences) overnight. Crude cell wall preparations were prepared by French pressure cell disruption of freshly harvested whole cells in phosphate-buffered saline (PBS; pH 7.2) at 15,000 lb/in.2. The cell walls were recovered after low-speed centrifugation (1,000 × g, 15 min), followed by high-speed centrifugation (138,000 × g, 30 min) of the supernatant.
Quantification of CCL20 in cell-free supernatant and in cell lysates.
CCL20 was quantified in culture supernatants and in cell lysates by sandwich enzyme-linked immunosorbent assay (ELISA) using antibody pairs from R&D Systems (Minneapolis, MN) following the vendor's instructions. CCL20 concentration was calculated from standard curves using recombinant human CCL20 (R&D Systems). The detection threshold was 250 pg/ml of CCL20.
Detection of hBD-2 and hBD-3 peptides in supernatant.
Human beta-defensins were measured in supernatants from FnCW-stimulated HOECs following our previously published protocol (27, 29).
RNA preparation and PCR.
Cells were lysed using TRIzol reagent (Invitrogen, Carlsbad, CA), and total RNA was isolated in accordance with the manufacturer's instructions. RNA concentration was measured by UV absorbance using the Nanodrop 1000 spectrophotometer (Thermo Fisher Scientific Inc.). Reverse transcription-PCR (RT-PCR) was performed with the SuperScript III one-step RT-PCR system (Invitrogen) according to the manufacturer's protocol, and the amplified product was analyzed on a 1.5% agarose gel. Real-time PCR was done using the Bio-Rad (Hercules, CA) MyIQ system with iScript cDNA synthesis kit and iQ SYBR green supermix according to the manufacturer's protocol. The results were analyzed to calculate the induction of CCL20, with human keratin 5 (HK5) as the internal control. The following primers were used to amplify CCL20 and HK-5: for CCL20, 5′-AATTTATTGTGGGCTTCACACG-3′ and 5′-ACCCAAGTCTGTTTTGGATTTG-3′; for HK5, 5′-GTCCTCTCCATGGACAACAAC-3′ and 5′-TGTCAATCTCGGCTCTCAGCC-3′.
PathScan MAPK multitarget sandwich ELISA.
Mixed-donor (N = 3) HOECs were treated by adding fresh medium containing the regulator for the desired time, rinsed with ice-cold phosphate-buffered saline (PBS), and lysed with ice-cold 1 lysis buffer (Cell Signaling, Danvers, MA) plus 1 mM phenylmethylsulfonyl fluoride (PMSF) for 10 min on ice. Cells were scraped off the plate, transferred to a microfuse tube, sonicated on ice, and clarified by centrifugation (13,000 rpm, 4°C, 15 min). The PathScan multitarget ELISA (targeting phospho-p38, phospho-ERK1/2, phospho-MEK1/2, phospho-SAPK/JNK, and MEK1) ELISA was then performed following the vendor's (Cell signaling, MA) instruction on the clarified supernatant.
Statistical analysis.
Statistical analyses were performed using either unpaired Student's t test or ANOVA test. The differences were considered significant when the probability value was less than 5% (P < 0.05).
RESULTS
F. nucleatum induces release of CCL20 by HOECs.
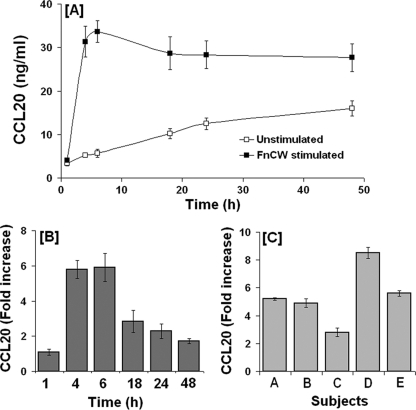
To determine baseline release of CCL20 by unstimulated HOECs, cell-free supernatants from epithelial cell cultures were collected at different time intervals and the CCL20 concentration was quantified by ELISA. Figure 1A demonstrates release of CCL20 protein by HOECs as early as 1 h after the addition of fresh medium that increased gradually with time. To examine the release of CCL20 protein upon stimulation with F. nucleatum, HOECs were treated with FnCW (10 μg/ml) for 1 to 48 h. Time course studies demonstrated a significant increase in CCL20 release at 4 h following F. nucleatum stimulation, with a plateauing of CCL20 release after 6 h of stimulation (Fig. 1A). We observed that the fold increase in secreted CCL20 from FnCW-treated cells, compared to untreated quiescent cells, reached its maximum by 6 h posttreatment (Fig. 1B, derived from A) and that interpersonal variability was demonstrated in HOECs from different donors toward release of CCL20 upon F. nucleatum challenge, i.e., from 2.8- to 8.5-fold above baseline (N = 5; variation coefficient [%CV], 37) (Fig. 1C).
Fig. 1.
Kinetics of FnCW-induced release of CCL20 by HOECs. (A) HOECs (from mixed donors; N = 3) were challenged with FnCW (10 μg/ml) for the indicated time periods, and the amount of CCL20 in cell-free supernatants was measured by ELISA. Results are the means ± standard deviations (SD) of three independent experiments. (B) Fold changes in CCL20 release by HOECs (mixed donors; N = 3) after challenge with FnCW (10 μg/ml) for the indicated time periods. Results are the means ± SD of three independent experiments. (C) Interpersonal variability in CCL20 release by HOECs from five different donors after challenge with FnCW (10 μg/ml) for 6 h. Results are the means ± SD of three independent ELISA measurements.
CCL20 release requires rapid de novo generation and is inhibited by brefeldin A.
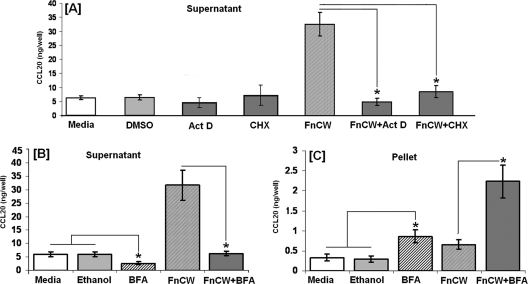
To determine if CCL20 is synthesized by HOECs in response to FnCW, near-confluent HOEC cultures were pretreated with actinomycin D (10 μg/ml) or cycloheximide (10 μg/ml) for 5 min. HOECs were then cultured for an additional 6 h with FnCW, as that was shown to be the optimal time for CCL20 release (Fig. 1A and B). Controls included HOECs with medium alone (or DMSO in medium), with FnCW alone, or with either actinomycin D or cycloheximide alone. After 6 h of exposure, medium was collected and analyzed for the presence of CCL20. As shown in Fig. 2A, FnCW was unable to induce the release of CCL20 by HOECs in the presence of actinomycin D or cycloheximide, indicating that the release of CCL20 by FnCW is transcriptionally and translationally regulated. To examine the effect of BFA, a protein release inhibitor, on FnCW-induced release of CCL20 by HOECs, near-confluent HOECs were treated with BFA (10 μg/ml) for 5 min followed by FnCW (10 μg/ml) for 6 h or with either agent alone for up to 6 h; supernatant and pellet samples were collected, and CCL20 levels were measured by ELISA. BFA inhibited CCL20 release from HOECs in the presence of FnCW stimulation (Fig. 2B). In contrast, BFA alone or in combination with FnCW caused an increase in intracellular CCL20 (Fig. 2C). Comparison of CCL20 levels in supernatants and in cell pellets indicated that the CCL20 is rapidly released upon synthesis.
Fig. 2.
(A) Effect of actinomycin D (Act D) and cycloheximide (CHX) on FnCW-induced release of CCL20 by HOECs. HOEC monolayers (mixed donors; N = 3) were pretreated with actinomycin D or cycloheximide for 5 min and then incubated with or without FnCW (10 μg/ml) for an additional 6 h. As a negative control, cells were incubated with medium or medium with vehicle control (DMSO) for actinomycin D. CCL20 levels in cell-free supernatants were measured by ELISA. (B and C) Effect of BFA on CCL20 release by HOECs. HOEC monolayers (mixed donors; N = 3) were pretreated with BFA (10 μg/ml) for 5 min, followed by incubation with or without FnCW (10 μg/ml) for an additional 6 h. As a negative control, cells were incubated with medium alone or medium with vehicle control (ethanol) for BFA. CCL20 levels in cell-free supernatants (B) and cell lysates (C) were measured by ELISA. Results are the means ± standard deviations of three independent experiments. *, P < 0.05. For comparisons of CCL20 content in supernatants and in cell pellets, the levels of CCL20 are expressed as ng/well instead of ng/ml. One ml of media per well was used in the six-well format.
Release of beta-defensins in HOECs by FnCW.
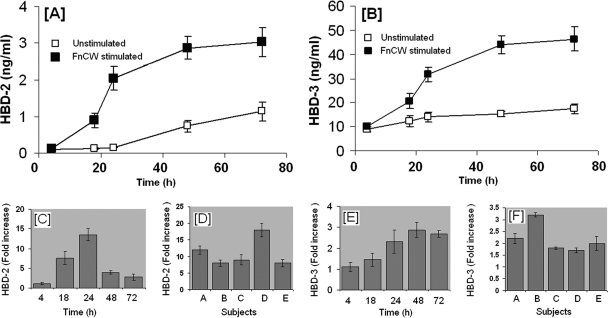
Cell-free supernatants from FnCW-treated (10 μg/ml) HOECs were collected at different time points (1 to 72 h) and hBD-2 and -3 peptide concentrations were quantified by ELISA. Figure 3A and B shows the kinetics of hBD-2 and -3 released from untreated versus FnCW-treated HOECs over the designated time periods, respectively. Compared to the untreated cells, hBD-2 increases of 1.1-, 7.5-, 13.5-, 3.9-, and 2.7-fold were observed at 4, 18, 24, 48, and 72 h, respectively (Fig. 3C). Increases for hBD-3 (Fig. 3E) were 1.1-, 1.4-, 2.3-, 2.9-, and 2.7-fold at the same respective time points as for hBD-2. Interpersonal variability was evident for hBD-2 and -3 release from HOECs induced by FnCW for 24 h (Fig. 3D and F) (N = 5 donors; CV, 38.5 and 27.5% for hBD-2 and hBD-3, respectively).
Fig. 3.
Kinetics of FnCW-induced release of hBD-2 and hBD-3 by HOECs. (A and B) HOEC monolayers (mixed donors; N = 3) were challenged with FnCW (10 μg/ml) for the indicated time periods, and the levels of hBD-2 and hBD-3 in cell-free supernatants were determined by ELISA. Results are the means ± standard deviations (SD) of three independent experiments. (C and E) Fold changes (calculated from panels A and B) in hBD-2 and -3 release by HOECs (mixed donors; N = 3) after FnCW challenge for the indicated time periods. Results are the means ± SD of three independent experiments. (D and F) Interpersonal variability in hBD-2 and -3 release by HOECs from five different donors after FnCW treatment for 24 h. Results are the means ± SD of three independent ELISA measurements.
In contrast to hBD-2 and -3, we observed no differences in the release of hBD-1 between FnCW-stimulated and unstimulated control cells (data not shown), supporting the constitutive nature of hBD-1 expression in HOECs. However, linear accumulation of hBD-1 in either stimulated or unstimulated culture medium over time was observed, with R2 values of 0.94 and 0.97, respectively.
Induction of CCL20 by beta-defensins (2 and 3).
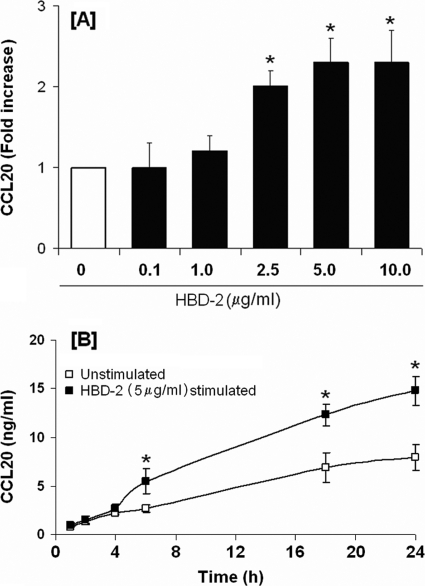
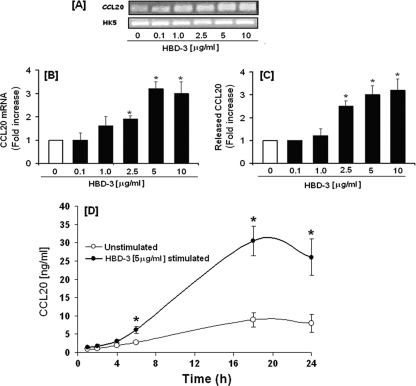
Since hBD-2 (2 μg/ml) has been shown to induce CCL20 transcription in primary oral epithelial cells (62), we looked for hBD-2-induced CCL20 release by HOECs. HOECs were incubated with synthetic hBD-2 (0.1 to 10 μg/ml) for 6 h. An hBD-2 dose-dependent release of CCL20 was evidenced, with the maximum release noted with 5 μg/ml hBD-2 (Fig. 4A). Moreover, CCL20 was seen to increase in the spent supernatant over time (1 to 24 h) when hBD-2 was added to HOECs at the optimal concentration of 5 μg/ml (Fig. 4B); a kinetic profile different from that seen with FnCW (Fig. 1A). To investigate the inductive capacity of hBD-3 to promote CCL20 expression and subsequent release from HOECs, we incubated near-confluent HOECs with increasing concentrations of hBD-3 (0.1 to 10 μg) for 6 h, followed by isolation of total RNA, amplification of CCL20 mRNA by PCR, and quantification of released CCL20 by ELISA. The increase in CCL20 mRNA expression was also determined by using real-time PCR. There was a direct correlation between the increase in CCL20 transcript expression (Fig. 5B) and protein release from HOECs with hBD-3 peptide concentration used over the 6-h period (Fig. 5C). CCL20 was seen to also increase in the spent supernatant over time (1 to 24 h) when hBD-3 was added to HOECs at the optimal concentration of 5 μg/ml, as seen with hBD-2, but different from that seen with FnCW (Fig. 1A). Interestingly, hBD-3 induced higher levels of released CCL20 than did hBD-2 (compare Fig. 4B and 5D), and there appeared to be an approximately 6-fold increase in CCL20 in the spent supernatant induced by hBD-3 at around 18 h of incubation (Fig. 5D).
Fig. 4.
hBD-2-induced release of CCL20 by HOECs. (A) HOEC monolayers (mixed donors; N = 3) were challenged with hBD-2 (0.1 to 10 μg/ml) for 6 h; released CCL20 in cell-free supernatants was determined by ELISA, followed by calculation of the fold increase above baseline. (B) HOEC monolayers (mixed donors; N = 3) were challenged with hBD-2 (5 μg/ml) for the indicated time periods, and the amount of released CCL20 in cell-free supernatants was determined by ELISA. Results are the means ± standard deviations of three independent experiments. *, P < 0.05 with respect to control (no treatment).
Fig. 5.
(A and B) hBD-3-induced upregulation of CCL20 transcript in HOECs. HOEC monolayers (mixed donors; N = 3) were treated with the indicated amounts of synthetic hBD-3 for 6 h. Total RNAs from both treated and untreated cells were isolated, the hBD-3 transcripts were amplified by RT-PCR (A), and the changes were quantified by real-time PCR (B). HK5 (human keratin 5) was used as a housekeeping gene. (C) hBD-3-induced release of CCL20 by HOECs. HOEC monolayers (mixed donors; N = 3) were treated with the indicated amounts of synthetic hBD-3 for 6 h, and the levels of CCL20 in the cell-free supernatants were determined by ELISA, followed by determination of the fold changes. (D) Kinetics of hBD-3-induced CCL20 release by HOECs. HOEC monolayers (mixed donors; N = 3) were treated with hBD-3 (5 μg/ml) for the indicated time periods, and release of CCL20 in cell-free supernatants for both treated and untreated cells was determined by ELISA. Results shown in panels B to D are the means ± standard deviations of three independent experiments. *, P < 0.05.
No lactate dehydrogenase activity was detected in the supernatants after stimulation with hBD-2 or hBD-3 (0.1 to 10 μg/ml) for 24 h (data not shown), confirming that cellular viability was not impaired by hBD-2 or -3 at the concentration range used and time periods studied.
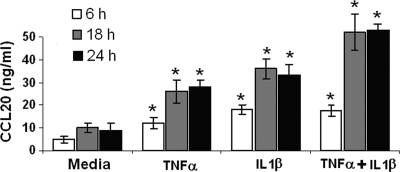
Cytokines (IL-1β and TNF-α) induce secretion of CCL20 by HOECs.
We and others have previously shown that FnCW induces TNF-α and IL-1β transcript expression in HOECs (41, 62), and these two cytokines have been shown to induce CCL20 release from several epithelial cell lines (8, 49, 51). Here, we looked to see if these cytokines could induce the release of CCL20 from HOECs. We incubated HOEC monolayers with 10 ng/ml of either IL-1β or TNF-α for three different time periods (6, 18, and 24 h), followed by ELISA analysis. We observed that both cytokines were capable of releasing increased amounts of CCL20 compared to the untreated control (Fig. 6). When cells were stimulated with a combination of TNF-α and IL-1β, an additive effect was observed in the release of CCL20. Since F. nucleatum- and hBD-induced CCL20 production is a relatively early event, it is unlikely that the production of intermediary cytokines are involved in F. nucleatum- or hBD-induced release of CCL20. We corroborated this by using antibodies against TNF-α and IL-1β in our assay system and saw that FnCW and hBDs, respectively, were not prevented from inducing CCL20 in HOECs (data not shown).
Fig. 6.
TNF-α- and IL-1β-induced release of CCL20 by HOECs. HOEC monolayers (mixed donors; N = 3) were separately treated with TNF-α (10 ng/ml), IL-1β (10 ng/ml), TNF-α plus IL-1β (5 ng/ml each), or medium alone for the indicated time periods, and the levels of released CCL20 in the spent supernatants were determined by ELISA. Results are the means ± standard deviations of three independent experiments. *, P < 0.05 with respect to control (no treatment).
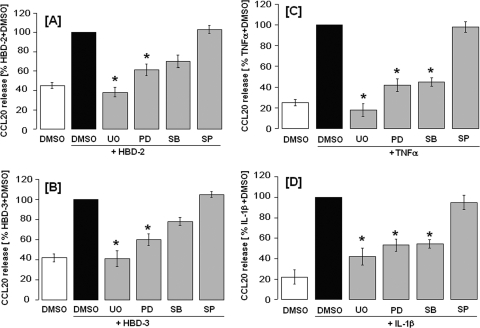
Effect of MAP kinase inhibitors on defensin- and cytokine-induced release of CCL20 by HOECs.
Different MAP kinases exist, and their impacts on CCL20 production in each cell line are different (31, 43, 51, 57). Hence, the signaling pathway involved in CCL20 production in HOECs needs further investigation. Since it was reported that beta-defensins may activate MAPK pathways (47), we used inhibitors of these pathways to see whether any are involved in hBD-induced release of CCL20. HOEC monolayers were pretreated for 1 h with different inhibitors before an 18-h incubation with stimulant. As shown in Fig. 7A and B, ERK1/2 activation inhibitors (UO126 [20 μM] or PD98059 [20 μM]) were effective in suppressing beta-defensin-induced (hBD-2 or hBD-3) release of CCL20 by HOECs. The specific p38 MAP kinase inhibitor SB203580 (20 μM) also had a modest inhibitory effect on defensin-induced CCL20 release. In contrast, the JNK-specific inhibitor SP600125 (20 μM) had no inhibitory effect on defensin-induced CCL20 release. Similar experiments were conducted to assess the importance of MAPK in TNF-α- and IL-1β-induced release of CCL20 by HOECs. We found that, like beta-defensins, these cytokines induced CCL20 via ERK1/2 and p38 but not via JNK (Fig. 7C and D). Interestingly, all four MAPK inhibitors showed little or no effect on FnCW-induced CCL20 release by HOECs (data not shown), suggesting that the signaling pathway(s) involved for FnCW-induced secretion of CCL20 is different from defensin- or cytokine-induced release of CCL20 by HOECs.
Fig. 7.
Inhibition of ERK, p38, and JNK in hBD-2-, hBD-3-, TNF-α-, and IL-1β-induced release of CCL20 by HOECs. HOEC monolayers (mixed donors; N = 3) were pretreated with inhibitors of ERK-specific (UO126 [20 μM] or PD98059 [20 μM]), p38-specific (SB203580 [20 μM]), or JNK-specific inhibitors (SP600125 [20 μM]), respectively, for 1 h, followed by treatment with hBD-2 (5 μg/ml) (A), hBD-3 (5 μg/ml) (B), TNF-α (10 μg/ml) (C), or IL-1β (10 μg/ml) (D) for an additional 18 h. DMSO was used as the negative control. CCL20 levels in the spent supernatants were measured by ELISA. Results are means ± standard deviations of three independent experiments. *, P < 0.05.
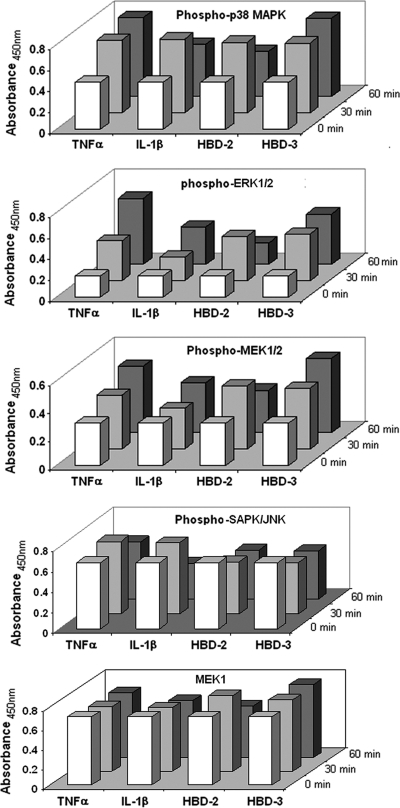
Since the inhibitor studies showed that beta-defensins (hBD-2 or hBD-3) and cytokines (TNF-α or IL-1β) induced release of CCL20 via ERK1/2 and p38 but not via JNK, we used the PathScan MAPK multitarget sandwich ELISA to compare the levels of phospho-ERK1/2 (Thr202/Tyr204), phospho-MEK1/2 (Ser217/221), phospho-p38 MAPK (Thr180/Tyr182), and phospho-SAPK/JNK (Thr183/Tyr185) in the cell lysates of HOECs after stimulating the cells with TNF-α, IL-1β, hBD-2, and hBD-3, respectively. As shown in Fig. 8, phosphorylation levels of ERK1/2, MEK1/2, and p38, but not of SAPK/JNK, were enhanced in HOECs after 30 or 60 min of challenge with the above-mentioned stimuli.
Fig. 8.
PathScan MAPK multitarget sandwich ELISA. HOEC monolayers (mixed donors; N = 3) were treated with either TNF-α (10 ng/ml), IL-1β (10 ng/ml), hBD-2 (5 μg/ml), or hBD-3 (5 μg/ml) for 0, 30, or 60 min, respectively. The cell lysates were analyzed for the indicated target using the PathScan MAPK multitarget sandwich ELISA kit (Cell Signaling, Danvers, MA) according to the vendor's instructions.
DISCUSSION
Commensal bacteria are usually regarded as beneficial to the host by displacing pathogens from a microbial niche or providing protection by continually stimulating epithelial surfaces to express and secret antimicrobial peptides (AMPs) at levels that kill opportunistic/pathogenic organisms (4). The release of antimicrobial chemokine CCL20 (51) requires stimulation for its release, and the discrete stimuli that lead to its upregulation may be cell specific (17, 31, 31, 57, 58). In these studies, we demonstrated the ability of primary oral epithelial cells to release CCL20 in response to the oral commensal bacterium F. nucleatum and other epithelium-derived agents that it also induces, i.e., hBD-2, hBD-3, TNF-α, and IL-1β. The kinetic studies of CCL20 release by HOECs suggested that this is an early event subsequent to challenge by any of the agents used. FnCW caused rapid release of CCL20 from HOECs, i.e., 6 h postchallenge there was a 6-fold-greater presence of CCL20 in the spent supernatant than control supernatant and twice the increase of a 24-h challenge (6-fold versus ∼3-fold). This is in agreement with a recent study that showed that F. nucleatum elicits rapid induction of the CCL20 gene (72.7-fold in 6 h compared to 21.7-fold in 24 h) (62). Lin et al. (43) also showed early release of CCL20 by human mast cells in response to Pseudomonas aeruginosa, which also peaked at 6 h. Adenylate and uridylate (A+U)-rich element (ARE)-mediated mRNA turnover is an important regulatory component of gene expression for innate and specific immunity (24). The transient and rapid upregulation of CCL20 gene expression by FnCW could be due to posttranscriptional regulation of mRNA stability via AREs (19, 53). Indeed, when we looked for AREs within the 3′-untranslated region of CCL20 mRNA, using the ARE website (http://rna.tbi.univie.ac.at/AREsite) (28), we found three ATTTA domains (see Fig. S1 in the supplemental material). Another rapidly induced gene by FnCW in HOECs is CXCL5 (40.5-fold in 6 h) (62), and by using the ARE site we found 11 ATTTA domains within its 3′-UTR (see Fig. S1). We could not find any AREs within the 3′-UTR of DEFB4 (hBD-2) or in DEFB103 (hBD-3) and, not surprisingly, these defensins were not released as rapidly as CCL20 by FnCW in HOECs.
In the present study, we found that actinomycin D and cycloheximide inhibited FnCW-induced release of CCL20 by HOECs, indicating that CCL20 release requires transcriptional and translational activation. BFA is a fungal metabolite able to rapidly and reversibly block intracellular vesicle transport from the endoplasmic reticulum to the Golgi apparatus and, therefore, inhibit protein secretion with minimal effect on protein synthesis (38). BFA blocks the release of newly synthesized proteins but has no effect on preformed granule-bound proteins (45). Although BFA has been widely used as an inhibitor of protein secretion, the information concerning its effect on CCL20 release has not been reported. Our study showed that BFA inhibited not only the FnCW-induced release of CCL20 by HOECs but also constitutive (unstimulated) release of CCL20. The increased levels of intracellular CCL20 in unstimulated and FnCW-stimulated cell lysates from BFA-pretreated HOECs further corroborate the inhibition of CCL20 release by BFA. Given the well-characterized properties of BFA, it is reasonable to conclude that CCL20 is secreted through the vesicle transport pathway.
Certain commensal bacteria are excellent inducers of beta-defensins (22, 41, 59), suggesting that the commensal bacterial community may act by priming innate immune readiness of the oral epithelium. We and others have shown that F. nucleatum can induce innate response elements, such as hBDs, in HOECs (29, 32, 41, 50). More recently, several immunoregulatory functions within the adaptive immune system have been attributed to hBDs, in addition to their antimicrobial capacity, i.e., their ability to cross talk with the adaptive immune system by acting as chemokines (34, 52) and by changing a cell's phenotype, either with activation, as seen with antigen-presenting cells (25), or antagonism, as seen with T cells (21). These findings conform to reports in the literature that innate responses usually precede, and are necessary for, the establishment of adaptive immunity (44). Upon stimulation by antimicrobial peptides, different immune and inflammatory cells have been shown to produce cytokines and/or chemokines. For example, the alpha-defensins human neutrophil peptide 1 and 3 stimulate the production of IL-1, -4, and -6, TNF-α, and gamma interferon in monocytes (5). Moreover, hBDs and LL-37 have been reported to enhance the generation of IL-18 in keratinocytes (47). In this study, we demonstrated that the inducible defensins hBD-2 and hBD-3 can induce the release of other AMPs/chemokines, i.e., CCL20, by HOECs. Therefore, the ability of hBDs to promote the release of the antimicrobial chemokine CCL20 suggests the broad role of human antimicrobial peptides in primary immune responses by oral epithelial cells. This regulatory loop could act in a complementary manner to provide added protection against microbial assault.
To gain initial insight into the cellular pathways through which defensins induce CCL20 release by HOECs, we focused on three major downstream MAPK cascades: ERK, p38, and JNK. In cells, these kinase cascades act as signal sorters for a variety of upstream signals before entering the nucleus. The p38, ERK, and JNK kinase cascades have been shown to be involved in a large variety of cellular activities (3, 7). Defensins have been shown to activate primary human keratinocytes through p38 and ERK1/2 (47). By focusing on these three MAPKs, we demonstrated that both ERK1/2 and p38, but not JNK, are required for hBD-induced CCL20 secretion by HOECs. The involvement of similar MAPKs was also observed in the release of CCL20 by TNF-α- or IL-1β-stimulated oral epithelial cells. The p38 MAPK pathway plays an important role in the posttranscriptional regulation of inflammatory genes and has been found to regulate both the translation and the stability of inflammatory mRNAs. The mRNAs regulated by p38 are known to have AREs present in their 3′-untranslated regions (16, 24) and thus, the presence of an ARE within the 3′-UTR of CCL20 supports the involvement of p38 in its induction by defensins and cytokines. Surprisingly, ERK1/2 and p38 inhibitors were found to have a negligible effect on FnCW-induced release of CCL20 by HOECs. Most recently, Yin and Chung (63) found a lack of involvement of p38 in F. nucleatum-induced CCL20 mRNA induction. At this stage, however, it is not clear why, in contrast to induction of CCL20 by defensins and cytokines, the induction of CCL20 by FnCW does not occur via p38, despite the fact that FnCW is capable of causing rapid and transient activation of p38 MAPK (42) in HOECs. Nevertheless, these observations further corroborate the fact that the induction of CCL20 by FnCW is not dependent on defensins (hBD-2 and -3), TNF-α, or IL-1β.
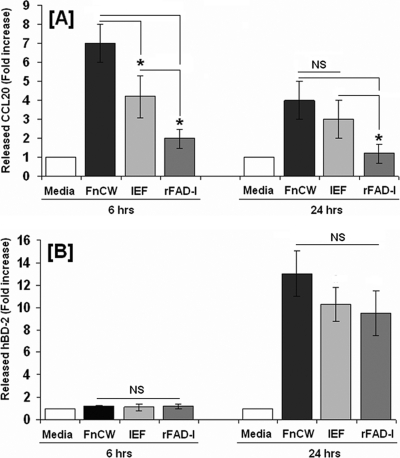
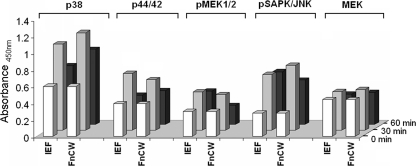
Kinetics of FnCW-induced CCL20 and β-defensin release are distinct, i.e., CCL20 release peaks at 4 to 6 h (Fig. 1B) by FnCW, while the peak time is 24 h for hBD-2 (Fig. 3C) and 48 h for hBD-3 (Fig. 3D). Thus, coordinated stimulation of CCL20 and beta-defensins by FnCW on oral epithelial cells might suggest a complementary activity in exerting the innate nature of F. nucleatum in host defense. Since FnCW is a complex mixture of proteins, it is tempting to speculate that different components within the FnCW fraction are responsible for the release of beta-defensins and CCL20. We have recently identified a Fusobacterium-associated beta-defensin inducer peptide (FAD-I) within FnCW that is responsible for the induction of hBD-2 in HOECs (29). When we tested FAD-I's ability to induce CCL20 release by HOECs, we found it to be negligible compared to FnCW (Fig. 9A), while its ability to induce hBD-2 release was comparable to that of FnCW (Fig. 9B). However, an isoelecric focusing (IEF) fraction of crude FnCW, from which FAD-I was further isolated (29), was capable of inducing CCL20 release by HOECs (Fig. 9A), indicating that the CCL20 inducing factor(s) is present in the IEF fraction. The results shown in Fig. 9A and B appear to suggest this. Although the IEF fraction was a semipurified fraction from crude FnCW, the phosphorylation patterns of different MAPKs elicited by the IEF fraction were similar to that elicited by crude FnCW (Fig. 10) and, not surprisingly, the effectiveness of the IEF fraction in inducing CCL20 or hBD-2 was close to that of FnCW. Isolation of the CCL20-inducing factor, in conjunction with FAD-I, may one day be used to bolster the innate defenses of vulnerable mucosae.
Fig. 9.
Comparison of FnCW, IEF fraction of FnCW-induced and recombinant FAD-I (rFAD-I)-induced CCL20 and hBD-2 release. HOECs (mixed donors; N = 3) were treated with medium alone, FnCW (10 μg/ml), IEF fraction (10 μg/ml), or rFAD-I (10 μg/ml) for the indicated time periods. The levels of CCL20 (A) and hBD-2 (B) in cell-free supernatants were measured by ELISA. The IEF fraction and rFAD-I were prepared as described by Gupta et al. (29). *, P < 0.05; NS, not significant.
Fig. 10.
PathScan MAPK multitarget sandwich ELISA results. HOEC monolayers (mixed donors; N = 3) were treated with either IEF (10 μg/ml) or FnCW (10 μg/ml) for 0, 30, and 60 min. The cell lysates were analyzed for the indicated target, using the PathScan MAPK multitarget sandwich ELISA kit (Cell Signaling, Danvers, MA) according to the vendor's instructions.
Supplementary Material
ACKNOWLEDGMENTS
We thank J. R. Blakemore, E. K. Schneider, W. S. Blood, and F. Faddoul for providing normal human oral tissue. We appreciate helpful discussions with Zhimin Feng, Ge Jin, and Jennifer Greene, Department of Biological Sciences.
This work was supported by National Institutes of Health grants R01DE016334 (A.W.) and RO1DE018276 (A.W.).
Footnotes
Supplemental material for this article may be found at http://iai.asm.org/.
Published ahead of print on 12 September 2011.
REFERENCES
- 1. Aas J. A., Paster B. J., Stokes L. N., Olsen I., Dewhirst F. E. 2005. Defining the normal bacterial flora of the oral cavity. J. Clin. Microbiol. 43:5721–5732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Abiko Y., et al. 2003. Expression of MIP-3α/CCL20, a macrophage inflammatory protein in oral squamous cell carcinoma. Arch. Oral Biol. 48:171–175 [DOI] [PubMed] [Google Scholar]
- 3. Ballif B. A., Blenis J. 2001. Molecular mechanisms mediating mammalian mitogen-activated protein kinase (MAPK) kinase (MEK)-MAPK cell survival signals. Cell Growth Differ. 12:397–408 [PubMed] [Google Scholar]
- 4. Boman H. G. 2000. Innate immunity and the normal microflora. Immunol. Rev. 173:5–16 [DOI] [PubMed] [Google Scholar]
- 5. Chaly Y. V., et al. 2000. Neutrophil-defensin human neutrophil peptide modulates cytokine production in human monocytes and adhesion molecule expression in endothelial cells. Eur. Cytokine Netw. 11:257–266 [PubMed] [Google Scholar]
- 6. Chang K. P., et al. 2011. Overexpression of macrophage inflammatory protein-3α in oral cavity squamous cell carcinoma is associated with nodal metastasis. Oral Oncol. 47:108–113 [DOI] [PubMed] [Google Scholar]
- 7. Chang L., Karin M. 2001. Mammalian MAP kinase signalling cascades. Nature 410:37–40 [DOI] [PubMed] [Google Scholar]
- 8. Choi S. C., et al. 2006. DA-9601, a standardized extract of Artemisia asiatica, blocks TNF-alpha-induced IL-8 and CCL20 production by inhibiting p38 kinase and NF-κB pathways in human gastric epithelial cells. World J. Gastroenterol. 12:4850–4858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Christ A. D., Blumberg R. S. 1997. The intestinal epithelial cell: immunological aspects. Springer Semin. Immunopathol. 18:449–461 [DOI] [PubMed] [Google Scholar]
- 10. Chung W. O., Dale B. A. 2004. Innate immune response of oral and foreskin keratinocytes: utilization of different signaling pathways by various bacterial species. 72:352–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cole A. M., et al. 2001. Cutting edge. IFN-inducible ELR-CXC chemokines display defensin-like antimicrobial activity. J. Immunol. 167:623–627 [DOI] [PubMed] [Google Scholar]
- 12. Cook D. N., et al. 2000. CCR6 mediates dendritic cell localization, lymphocyte homeostasis, and immune responses in mucosal tissue. Immunity 12:495–503 [DOI] [PubMed] [Google Scholar]
- 13. Crane-Godreau M. A., Wira C. R. 2004. Effect of Escherichia coli and Lactobacillus rhamnosus on macrophage inflammatory protein 3 alpha, tumor necrosis factor alpha, and transforming growth factor beta release by polarized rat uterine epithelial cells in culture. Infect. Immun. 72:1866–1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dale B. A. 2002. Periodontal epithelium: a newly recognized role in health and disease. Periodontology 2000 30:70–78 [DOI] [PubMed] [Google Scholar]
- 15. Dale B. A., et al. 2001. Localized antimicrobial peptide expression in human gingiva. J. Periodontal Res. 36:285–294 [DOI] [PubMed] [Google Scholar]
- 16. Dean J. L., Sully G., Clark A. R., Saklatvala J. 2004. The involvement of AU-rich element-binding proteins in p38 mitogen-activated protein kinase pathway-mediated mRNA stabilization. Cell Signal. 16:1113–1121 [DOI] [PubMed] [Google Scholar]
- 17. Dieu-Nosjean M. C., et al. 2000. Macrophage inflammatory protein 3α is expressed at inflamed epithelial surfaces and is the most potent chemokine known in attracting Langerhans cell precursors. J. Exp. Med. 192:705–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Eckmann L., Reed S. L., Smith J. R., Kagnoff M. F. 1995. Entamoeba histolytica trophozoites induce an inflammatory cytokine response by cultured human cells through the paracrine action of cytolytically released interleukin-1 alpha. J. Clin. Invest. 96:1269–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fan J., Heller N. M., Gorospe M., Atasoy U., Stellato C. 2005. The role of post-transcriptional regulation in chemokine gene expression in inflammation and allergy. Eur. Respir. J. 26:933–947 [DOI] [PubMed] [Google Scholar]
- 20. Faveri M., et al. 2008. Microbiological diversity of generalized aggressive periodontitis by 16S rRNA clonal analysis. Oral Microbiol. Immunol. 23:112–118 [DOI] [PubMed] [Google Scholar]
- 21. Feng Z., Dubyak G. R., Lederman M. M., Weinberg A. 2006. Cutting edge. Human beta defensin 3: a novel antagonist of the HIV-1 coreceptor CXCR4. J. Immunol. 177:782–786 [DOI] [PubMed] [Google Scholar]
- 22. Feng Z., Weinberg A. 2006. Role of bacteria in health and disease of periodontal tissues. Periodontology 2000 40:50–76 [DOI] [PubMed] [Google Scholar]
- 23. Feucht E. C., DeSanti C. L., Weinberg A. 2003. Selective induction of human beta-defensin mRNAs by Actinobacillus actinomycetemcomitans in primary and immortalized oral epithelial cells. Oral Microbiol. Immunol. 18:359–363 [DOI] [PubMed] [Google Scholar]
- 24. Frevel M. A., et al. 2003. p38 mitogen-activated protein kinase-dependent and -independent signaling of mRNA stability of AU-rich element-containing transcripts Mol. Cell. Biol. 23:425–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Funderburg N., et al. 2007. Human β-defensin-3 activates professional antigen-presenting cells via Toll-like receptors 1 and 2. Proc. Natl. Acad. Sci. U. S. A. 104:18631–18635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ganz T. 2003. Defensins: antimicrobial peptides of innate immunity. Nat. Rev. Immunol. 3:710–720 [DOI] [PubMed] [Google Scholar]
- 27. Ghosh S. K., et al. 2007. Quantification of human beta-defensin-2 and -3 in body fluids: application for studies of innate immunity. Clin. Chem. 53:757–765 [DOI] [PubMed] [Google Scholar]
- 28. Gruber A. R., Fallmann J., Kratochvill F., Kovarik P., Hofacker H. L. 2011. AREsite: a database for the comprehensive investigation of AU-rich elements. Nucleic Acids Res. 39(database issue):D66–D69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gupta S., et al. 2010. Fusobacterium nucleatum-associated beta-defensin inducer (FAD-I): identification, isolation, and functional evaluation. J. Biol. Chem. 285:36523–36531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hosokawa Y., et al. 2002. Macrophage inflammatory protein 3α-CC chemokine receptor 6 interactions play an important role in CD4+ T-cell accumulation in periodontal diseased tissue. Clin. Exp. Immunol. 128:548–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Izadpanah A., Dwinell M. B., Eckmann L., Varki N. M., Kagnoff M. F. 2001. Regulated MIP-3α/CCL20 production by human intestinal epithelium: mechanism for modulating mucosal immunity. Am. J. Physiol. Gastrointest. Liver Physiol. 280:G710–G719 [DOI] [PubMed] [Google Scholar]
- 32. Ji S., et al. 2009. Toll-like receptor 2 and NALP2 mediate induction of human beta-defensins by Fusobacterium nucleatum in gingival epithelial cells. Infect. Immun. 77:1044–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ji S., Kim Y., Min B.-M., Han S. H., Choi Y. 2007. Innate immune responses of gingival epithelial cells to nonperiodontopathic and periodontopathic bacteria. J. Periodont. Res. 42:503–510 [DOI] [PubMed] [Google Scholar]
- 34. Jin G., et al. 2010. An antimicrobial peptide regulates tumor-associated macrophage trafficking via the chemokine receptor CCR2, a model for tumorigenesis. PLoS One 5:e10993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kawsar H. I., et al. 2009. Overexpression of human beta-defensin-3 in oral dysplasia: potential role in macrophage trafficking. Oral Oncol. 45:696–702 [DOI] [PubMed] [Google Scholar]
- 36. Kennell W., Holt S. C. 1990. Comparative studies of the outer membranes of Bacteroides gingivalis, strains ATCC 33277, W50, W83, 381. Oral Microbiol. Immunol. 5:121–130 [DOI] [PubMed] [Google Scholar]
- 37. Kim B. E., et al. 2007. Macrophage inflammatory protein 3α deficiency in atopic dermatitis skin and role in innate immune response to vaccinia virus. J. Allergy Clin. Immunol. 119:457–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Klausner R. D., Donaldson J. G., Lippincott-Schwartz J. 1992. Brefeldin A: insights into the control of membrane traffic and organelle structure. J. Cell Biol. 116:1071–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kleeff J., et al. 1999. Detection and localization of Mip-3α/LARC/Exodus, a macrophage proinflammatory chemokine, and its CCR6 receptor in human pancreatic cancer. Int. J. Cancer 81:650–657 [DOI] [PubMed] [Google Scholar]
- 40. Krisanaprakornkit S., Weinberg A., Perez C. N., Dale B. A. 1998. Expression of the peptide antibiotic human beta-defensin 1 in cultured gingival epithelial cells and gingival tissue Infect. Immun. 66:4222–4228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Krisanaprakornkit S., et al. 2000. Inducible expression of human β-defensin 2 by Fusobacterium nucleatum in oral epithelial cells: multiple signaling pathways and role of commensal bacteria in innate immunity and the epithelial barrier. Infect. Immun. 68:2907–2915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Krisanaprakornkit S., Kimball J. R., Dale B. A. 2002. Regulation of human beta-defensin-2 in gingival epithelial cells: the involvement of mitogen-activated protein kinase pathways, but not the NF-κB transcription factor family. J. Immunol. 168:316–324 [DOI] [PubMed] [Google Scholar]
- 43. Lin T. J., et al. 2003. Selective early production of CCL20, or macrophage inflammatory protein 3α, by human mast cells in response to Pseudomonas aeruginosa. Infect. Immun. 71:365–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Medzhitov R., Janeway C. A., Jr 1997. Innate immunity: impact on the adaptive immune response. Curr. Opin. Immunol. 9:4–9 [DOI] [PubMed] [Google Scholar]
- 45. Miller S. G., Carnell L., Moore H. P. H. 1992. Post-Golgi membrane traffic: brefeldin A inhibits export from distal Golgi compartments to the cell surface but not recycling. J. Cell Biol. 118:267–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nakayama T., et al. 2006. Novel antiviral activity of chemokines. Virology 350:484–492 [DOI] [PubMed] [Google Scholar]
- 47. Niyonsaba F., Ushio H., Nagaoka I., Okumura K., Ogawa H. 2005. The human beta-defensins (-1, -2, -3, -4) and cathelicidin LL-37 induce IL-18 secretion through p38 and ERK MAPK activation in primary human keratinocytes. J. Immunol. 175:1776–1784 [DOI] [PubMed] [Google Scholar]
- 48. Paster B. J., Olsen I., Ass J. A., Dewhirst F. E. 2006. The breadth of bacterial diversity in the human periodontal pocket and other oral sites. Periodontology 2000 42:80–87 [DOI] [PubMed] [Google Scholar]
- 49. Pernet I., et al. 2003. Calcium triggers beta-defensin (hBD-2 and hBD-3) and chemokine macrophage inflammatory protein-3 alpha (MIP-3α/CCL20) expression in monolayers of activated human keratinocytes. Exp. Dermatol. 12:755–760 [DOI] [PubMed] [Google Scholar]
- 50. Peyret-Lacombe A., Brunel G., Watts M., Charveron M., Duplan H. 2009. TLR2 sensing of F. nucleatum and S. sanguinis distinctly triggered gingival innate response. Cytokine 46:201–210 [DOI] [PubMed] [Google Scholar]
- 51. Reibman J., Hsu Y., Chen L. C., Bleck B., Gordon T. 2003. Airway epithelial cells release MIP-3α/CCL20 in response to cytokines and ambient particulate matter. Am. J. Respir. Cell Mol. Biol. 28:648–654 [DOI] [PubMed] [Google Scholar]
- 52. Röhrl J., Yang D., Oppenheim J. J., Hehlgans T. 2010. Human beta-defensin 2 and 3 and their mouse orthologs induce chemotaxis through interaction with CCR2. J. Immunol. 184:6688–6694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rossi D. L., Vicari A. P., Franz-Bacon K., McClanahan T. K., Zlotnik A. 1997. Identification through bioinformatics of two new macrophage proinflammatory human chemokines: MIP-3α and MIP-3β. J. Immunol. 158:1033–1036 [PubMed] [Google Scholar]
- 54. Ruth J. H., et al. 2003. Role of macrophage inflammatory protein-3α and its ligand CCR6 in rheumatoid arthritis. Lab. Invest. 83:579–588 [DOI] [PubMed] [Google Scholar]
- 55. Schutyser E., Struyf S., Van Damme J. 2003. The CC chemokine CCL20 and its receptor CCR6. Cytokine Growth Factor Rev. 14:409–426 [DOI] [PubMed] [Google Scholar]
- 56. Socransky S. S., Haffajee A. D., Cugini M. A., Smith C., Kent R. L., Jr 1998. Microbial complexes in subgingival plaque. J. Clin. Periodontol. 25:134–144 [DOI] [PubMed] [Google Scholar]
- 57. Tanaka Y., et al. 1999. Selective expression of liver and activation-regulated chemokine (LARC) in intestinal epithelium in mice and humans. Eur. J. Immunol. 29:633–642 [DOI] [PubMed] [Google Scholar]
- 58. Tohyama M., Shirakara Y., Yamasaki K., Sayama K., Hashimoto K. 2001. Differentiated keratinocytes are responsible for TNF-α regulated production of macrophage inflammatory protein 3α/CCL20, a potent chemokine for Langerhans cells. J. Dermatol. Sci. 27:130–139 [DOI] [PubMed] [Google Scholar]
- 59. Vankeerberghen A., et al. 2005. Differential induction of human beta-defensin expression by periodontal commensals and pathogens in periodontal pocket epithelial cells. J. Periodontol. 76:1293–1303 [DOI] [PubMed] [Google Scholar]
- 60. Yang D., et al. 1999. Beta-defensins: linking innate and adaptive immunity through dendritic and T cell CCR6. Science 286:525–528 [DOI] [PubMed] [Google Scholar]
- 61. Yang D., et al. 2003. Many chemokines including CCL20/MIP-3α display antimicrobial activity. J. Leukoc. Biol. 74:448–455 [DOI] [PubMed] [Google Scholar]
- 62. Yin L., Dale B. A. 2007. Activation of protective responses in oral epithelial cells by Fusobacterium nucleatum and human beta-defensin-2. J. Med. Microbiol. 56:976–987 [DOI] [PubMed] [Google Scholar]
- 63. Yin L., Chung W. O. 2011. Epigenetic regulation of human β-defensin 2 and CC chemokine ligand 20 expression in gingival epithelial cells in response to oral bacteria. Mucosal Immunol. 4:409–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.