Abstract
Neisseria meningitidis NhhA (Neisseria hia/hsf homologue A) is an oligomeric outer membrane protein belonging to the family of trimeric autotransporter adhesins. NhhA mediates the interaction of N. meningitidis with human epithelial cells and components of the extracellular matrix. The recombinant protein is able to induce bactericidal antibodies and hence has also been considered a potential vaccine candidate. In this study, we analyzed the production of NhhA in a large panel of N. meningitidis strains belonging to different serogroups and clonal complexes. We found that trimeric NhhA was produced at different levels by the various strains tested. In some strains belonging to the clonal complex ST41/44, the protein is detectable only as a monomer. Sequencing of the nhhA gene and generation of complementing strains in different genetic backgrounds have proved that a single mutation (Gly to Asp) in the translocator domain affected both trimerization and surface localization of NhhA. In vitro infection assays showed that this mutation impairs meningococcal NhhA-mediated adhesion, suggesting that strains carrying the mutation may rely on different strategies or molecules to mediate interaction with host cells. Finally, we demonstrated that N. meningitidis ST41/44 strains producing the mutated form did not induce killing mediated by NhhA-specific bactericidal antibodies. Our data help to elucidate the secretion mechanisms of trimeric autotransporters and to understand the contribution of NhhA in the evolutionary process of host-Neisseria interactions. Also, they might have important implications for the evaluation of NhhA as a vaccine candidate.
INTRODUCTION
Neisseria meningitidis is a Gram-negative bacterium that specifically infects humans, causing meningitis and sepsis. Several surface-exposed proteins are produced by N. meningitidis in order to colonize and infect the human host; among them, adhesins are key factors that are required for initial colonization of the nasopharyngeal mucosa and subsequent attachment to the endothelium (31).
Neisseria hia/hsf homologue (NhhA) is a meningococcal outer membrane protein (OMP) similar to the Haemophilus influenzae Hia/Hsf proteins (20). NhhA was identified through a genome-based approach aimed at selecting new surface-exposed proteins able to induce protective immunity against the bacterium (21). The recombinant NhhA protein induces bactericidal antibodies and is recognized by sera of patients convalescing after meningococcal disease and healthy individuals, suggesting that it is produced in vivo during the development of invasive infection and potentially during asymptomatic carriage (15).
NhhA is a trimeric autotransporter adhesin that has a variety of functions in N. meningitidis pathogenesis, including mediation of bacterial attachment to heparan sulfate and laminin of the extracellular matrix and to human epithelial cells (24). In a murine model of meningococcal disease, NhhA was found to be essential for bacterial colonization of the nasopharyngeal mucosa, and it has also been shown to protect meningococci from phagocytosis and complement-mediated killing (28).
The trimeric autotransporter adhesin (TAA) family contains a continually increasing number of adhesins of Gram-negative bacteria (5, 12), such as Yersinia enterocolitica YadA (23); Moraxella catarrhalis UspA1 and UspA2 (4); H. influenzae Hia, Hsf, and HadA (6, 27, 29); N. meningitidis NadA (2); Bartonella henselae BadA (22); and Proteus mirabilis AipA and TaaP (1). TAAs have a head-stalk-anchor architecture (14) and are characterized by the ability to form highly stable trimers on the bacterial surface (6). The head is generally the primary mediator of attachment, the stalk functions as a spacer to project the head away from the bacterial cell surface, and the membrane anchor domain is homologous throughout TAAs and defines the family. Functional and structural studies conducted on YadA (10, 23), Hia (17, 18, 30), and NhhA (24) showed that members of the TAA family have a distinct mechanism of secretion compared with conventional monomeric autotransporters: three membrane anchor domains form a 12-stranded β-barrel pore, which mediates the translocation of the stalk and the head across the outer membrane.
Mutagenesis experiments performed to study the functionality of the translocator anchor domains of NhhA, Hia, and YadA (8, 18, 23, 24) revealed important domains and residues involved in the trimerization, translocation, and surface localization of TAAs. However, all the key residues identified so far in TAA translocator domains have been selected based on sequence homologies identified by in silico analysis and do not represent natural mutants.
Here, we investigated the expression of NhhA in a panel of N. meningitidis strains. Interestingly, in some N. meningitidis serogroup B strains, NhhA was detectable only in its monomeric and not its trimeric form, and we found that a single natural mutation of a glycine (Gly) to an aspartic acid (Asp) residue in the β-subdomain of the C-terminal translocator unit is responsible for this phenomenon. By genetic and functional studies, we demonstrated that this single-residue substitution affected trimerization, protein stability, and the surface localization and adhesive capabilities of NhhA and that it has strong implications in evaluating the role of NhhA as a vaccine antigen.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The N. meningitidis wild-type strains used in this study are listed in Table 1. The deletion mutants and recombinant strains generated in this study are listed in Table S1 in the supplemental material. N. meningitidis strains were cultivated on GC agar plates (Difco) with 5% CO2 at 37°C. For liquid cultures, bacteria grown overnight were used to inoculate GC broth medium, and then the bacteria were incubated as described above with shaking. When required, erythromycin, kanamycin, or chloramphenicol was used at final concentrations of 5, 100, and 5 μg/ml, respectively.
Table 1.
Differential production of NhhA among N. meningitidis serogroup B strains
| No. | Straina | Sgb | STc | Clonal complex | Pathogenicity | Yr of isolation | Countryd | NhhA productione | NhhA detection |
|---|---|---|---|---|---|---|---|---|---|
| 1 | MC58 | B | 74 | 32 | Invasive | 1985 | UK | ++ | Trimer |
| 2 | H44/76 | B | 32 | 32 | Invasive | 1976 | N | + | Trimer |
| 3 | CU385 | B | 33 | 32 | Invasive | 1980 | C | ++ | Trimer |
| 4 | BZ169 | B | 32 | 32 | Invasive | 1985 | NL | + | Trimer |
| 5 | N44/89 | B | 33 | 32 | Invasive | 1989 | BR | + | Trimer |
| 6 | 72/00 | B | 1346 | 32 | Invasive | 2000 | N | + | Trimer |
| 7 | 220173i | B | ND | 32 | Invasive | 1993 | I | + | Trimer |
| 8 | M1590 | B | 32 | 32 | Invasive | 1995 | US | + | Trimer |
| 9 | NGP165 | B | 11 | 11 | 1974 | N | + | Trimer | |
| 10 | M986 | B | 11 | 11 | Invasive | 1963 | US | +++ | Trimer |
| 11 | NZ98/254 | B | 42 | 41/44 | Invasive | 1998 | NZ | ± | Monomer |
| 12 | M1239 | B | 437 | 41/44 | Invasive | 1994 | US | + | Trimer |
| 13 | M1390 | B | 41 | 41/44 | Invasive | 1995 | US | + | Monomer |
| 14 | S90-307 | B | ND | 41/44 | Invasive | 1998 | S | ++++ | Trimer |
| 15 | NM008 | B | 41 | 41/44 | Invasive | 1995 | UK | + | Trimer |
| 16 | M01-0240149 | B | 41 | 41/44 | Invasive | 2001 | UK | + | Trimer |
| 17 | NZ05/33 | B | 42 | 41/44 | Invasive | 2005 | NZ | ± | Monomer |
| 18 | 67/00 | B | 1127 | 41/44 | Invasive | 2000 | N | ± | Monomer |
| 19 | ISS1026 | B | 44 | 41/44 | Invasive | 2000 | I | ± | Monomer |
| 20 | BZ198 | B | 41 | 41/44 | Invasive | 1986 | NL | ± | Monomer |
| 21 | M13203 | B | 44 | 41/44 | Invasive | 2005 | US | + | Trimer |
| 22 | M10994 | B | 44 | 41/44 | Invasive | 2003 | US | + | Trimer |
| 23 | C188 | B | 41 | 41/44 | Carrier | 2003 | I | ± | Monomer |
| 24 | M4287 | B | 44 | 41/44 | Invasive | 1996 | US | + | Trimer |
| 25 | OX99.30304 | B | 44 | 41/44 | Carrier | 1999 | UK | + | Trimer |
| 26 | M4105 | B | 154 | 41/44 | Invasive | 1996 | US | + | Trimer |
| 27 | M3279 | B | 136 | 41/44 | Invasive | 1997 | US | ++ | Trimer |
| 28 | M0579 | B | 43 | 41/44 | Invasive | 1993 | US | ++ | Trimer |
| 29 | ISS749 | B | 1127 | 41/44 | Invasive | 1996 | I | + | Monomer |
| 30 | M4407 | B | 6160 | 41/44 | Invasive | 1996 | US | +++ | Trimer |
| 31 | 1000 | B | 20 | 18 | Invasive | 1988 | CSI | +++ | Trimer/monomer |
| 32 | 2996 | B | 540 | 8 | Invasive | 1975 | UK | + | Trimer |
| 33 | BZ133 | B | 1 | 1 | Invasive | 1977 | NL | ++++ | Trimer |
| 34 | NGH15 | B | 43 | 41/44 | Carrier | 1988 | N | + | Trimer |
| 35 | BZ232 | B | 38 | 37 | Invasive | 1964 | NL | + | Trimer |
| 36 | 8047 | B | 11 | 11 | Invasive | 1978 | US | ++++ | Trimer |
| 37 | M01-0240101 | B | 1049 | 269 | Invasive | 2001 | UK | + | Trimer |
| 38 | M01-0240355 | B | 213 | 213 | Invasive | 2001 | UK | ++ | Trimer |
| 39 | M3369 | B | 1576 | Invasive | 1997 | US | + | Trimer | |
| N. meningitidis serogroup non-B strains | |||||||||
| 40 | M08-0240233 | A | 4789 | Sub.III | Invasive | 2008 | UK | + | Trimer |
| 41 | F6124 | A | 5 | Sub.III | 1988 | TCH | + | Trimer | |
| 42 | F8238 | A | 1989 | EAK | + | Trimer | |||
| 43 | M07-0240954 | C | 11 | ET37 | Invasive | 2007 | UK | − | Trimer |
| 44 | M07-0241093 | C | 11 | ET37 | Invasive | 2007 | UK | ++++ | Trimer |
| 45 | C11 | C | 345 | 1965 | C | ++ | Trimer/monomer | ||
| 46 | M2197 | C | 11 | 11 | US | − | No detectable level | ||
| 47 | M07-0240665 | C | 22 | 1158 | Invasive | 2007 | UK | ++ | Trimer |
| 48 | M07-0240845 | W | 22 | 1158 | Invasive | 2007 | UK | + | Trimer |
| 49 | 240070 | W | 22 | 184 | 2001 | UK | + | Trimer | |
| 50 | LNP17592 | W | 11 | 32 | 2000 | F | +++ | Trimer | |
| 51 | M07-0240625 | Y | 174/1466 | Invasive | 2007 | UK | +++ | Trimer | |
| 52 | M07-0240706 | Y | 174/1466 | Invasive | 2007 | UK | +++ | Trimer | |
| 53 | M07-0240771 | Y | 23 | Invasive | 2007 | UK | − | No detectable level | |
| 54 | 860800 | Y | 29/167 | 1986 | NL | + | Trimer |
All the strains tested contain the nhhA gene as assessed by PCR.
Sg, serogroup of N. meningitidis.
ST, sequence type. ND, not determined.
BR, Brazil; C, Cuba; CSI, Russia; EAK, Kenya; F, France; I, Italy; IS,: Iceland, N, Norway; NL, The Netherlands; NZ, New Zealand; TCH, Chad; S, Sweden; UK, United Kingdom; US, United States.
The level of NhhA production was evaluated by Western blotting using a polyclonal antiserun raised against recombinant NhhA-His. −, +, ++, +++, and ++++ represent an arbitrary estimation of NhhA production based on Western blot results. ±, the monomer was detected only after long exposure of the membrane.
Escherichia coli strain DH5α was cultured in Luria-Bertani (LB) agar or LB broth at 37°C, and when required, ampicillin or chloramphenicol was added up to final concentrations of 100 μg/ml and 20 μg/ml, respectively.
Cell fractionation and protein analysis.
N. meningitidis strains were grown in 6 ml of GC broth medium and incubated for approximately 2 h at 37°C and 5% CO2 with shaking until an optical density at 600 nm (OD600) of 0.45 to 0.50 was reached. For protein analysis under denaturing conditions, whole-cell lysates were prepared from pellets harvested, resuspended in phosphate-buffered saline (PBS), and then heat-cold shocked for bacterial lysis. Samples were mixed with sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer containing 2% SDS and heated for 10 min at 95°C prior to loading. The samples were separated by SDS-PAGE using the NuPAGE Gel System (Invitrogen) and transferred onto nitrocellulose membranes for Western blot analysis. The membranes were blocked overnight with PBS-10% milk powder at 4°C. For detection of NhhA, the primary antibodies used were polyclonal mouse antisera raised against the recombinant NhhA-His (diluted 1:2,000); the secondary antibody was goat anti-mouse serum conjugated to horseradish peroxidase (1:10,000; Dako). The primary and secondary antibodies were incubated at room temperature for 1 h and 30 min, respectively. Detection of bound antibodies was carried out with Super Signal Chemiluminescent Substrate (Pierce) following the manufacturer's instructions.
For analysis under seminative conditions, whole-cell lysates were prepared by alkaline lysis. The equivalent of ∼1 × 109 bacteria (based on the culture OD) were resuspended in 25 μl of buffer P1 and, after addition of 25 μl buffer P2 (QIAPrep spin minikit; Qiagen), treated with 10 μl buffer RDD and 2.5 μl of DNase (RNase-Free DNase Set; Qiagen). Protein samples analyzed under seminative conditions were resuspended in 10× native sample buffer (1 M Tris-HCl, pH 8.6, 0.025% bromophenol blue, 20% glycerol) and without β-mercaptoethanol. These samples were not heated and were kept at 4°C before loading. Ten microliters of each sample was loaded onto the gels. Electrophoresis was performed at 150 V at 4°C in Tris acetate SDS-PAGE. The gel was treated with 2% SDS for 10 min before transfer. For Western blot analysis, proteins were transferred from gels onto polyvinylidene difluoride (PVDF) membranes and fixed by air drying. The membranes were then treated for detection of NhhA as described above. Outer membrane protein preparations were recovered on the basis of Sarkosyl insolubility following the rapid procedure as previously described (3).
Isolation of bacterial RNA and quantitative real-time reverse transcriptase (qRT-PCR) analysis.
N. meningitidis serogroup B strains were grown on GC broth until mid-log phase and harvested by centrifugation at 5,000 × g for 10 min at 4°C. The pellets were processed for RNA isolation using the RNeasy Mini Kit (Qiagen) according to the manufacturer's protocol. DNA contamination was eliminated by on-column treatment and posttreatment with the RNase-Free DNase Set (Qiagen). Absence of bacterial DNA was confirmed by PCR with primers specific for the nhhA gene. The RNA concentration and integrity were determined by measuring the A260/A280 ratios and by electrophoretic analysis.
For analysis of in vitro nhhA transcripts derived from different N. meningitidis strains, two-step qRT-PCR analysis was performed. Primers nhhA-RTFwd and nhhA-RTRev for the nhhA gene were designed, and primer specificity was controlled following the denaturing protocol of the Mx3000P cycler PCR system software version 2.0 (Stratagene). cDNA synthesis was primed using 3 μg of total RNA, random-hexamer primers (Promega), and SuperScript II (Invitrogen) according to the manufacturer's instructions. Fluorescence PCR amplifications were performed using a 1-μl aliquot of each first-strand cDNA reaction mixture with Brilliant SYBR green QPCR Master mix (Stratagene) in a final volume of 25 μl on an Mx3000P cycler (Stratagene). qRT-PCRs were performed in triplicate. To normalize the data, 16S rRNA was used as an endogenous control of expression, as it is transcribed at constant levels. The specificity of all amplicons was confirmed by melting curve and gel analyses. Relative quantification was performed using the 2−ΔΔCT method, including an efficiency correction for the primers (Relative Expression Software Tool-REST). For analysis of degQ transcript, the reaction was performed as described above; the primers used for the reaction are listed in Table S2 in the supplemental material.
PCR and sequencing of the nhhA gene.
PCR amplification of the nhhA locus was performed using genomic DNA as a template. For preparation of genomic DNA, N. meningitidis strains were grown in liquid cultures until they reached an OD600 of 0.5, corresponding to the mid-logarithmic growth phase. Bacteria (approximately 2 × 109) were harvested for 10 min at 5,000 × g, and the pellets were used for DNA isolation with a DNeasy Blood and Tissue kit (Qiagen) according to the manufacturer's instructions. For PCR, the Expand High Fidelity PCR System (Roche) was used. Amplification reactions were carried out using the primers nhhA-Fwd and nhhA-Rev, mapping upstream and downstream of the nhhA locus, respectively. The PCR conditions were as follows: 5 cycles of denaturation at 94°C for 40 s, annealing at 54°C for 40 s, and extension at 72°C for 90 s; 30 cycles of denaturation at 94°C for 40 s, annealing at 56°C for 40 s, and extension at 72°C for 90 s; and a final cycle of elongation for 7 min. The PCR products were analyzed on 1% agarose gel electrophoresis, purified using the QIAquick PCR purification kit (Qiagen), and sequenced with an automated DNA analyzer (model 3730xl DNA analyzer; Applied Biosystems). The primers used for sequencing were as follows: forward, nhhA-Fwd, nhhA-A, nhhA-B, and nhhA-C, and reverse, nhhA-Rev (see Table S2 in the supplemental material). To sequence the degQ gene from strain M1390, we used the primers reported in Table S2 in the supplemental material and performed sequencing as for the nhhA gene.
In silico analysis and protein modeling.
The NhhA amino acid sequences of MC58 (GenBank NP_274028) and BZ198 (AAF42517) were retrieved from the NCBI Protein database (http://www.ncbi.nlm.nih.gov). The NhhA sequences of C188, ISS1026, NZ05/33, 67/00, M1390, ISS749, NZ98/254, M4287, OX99.30304, M1239, M10994, M13203, M01-024149, NM008, M4105, M3279, M0579, and M4407 were deduced from nucleotide sequences from the corresponding genomic DNA. Multiple-sequence alignment was performed with the predicted protein sequences, which were aligned using the ClustalW software included in the GCG package and shaded on the BoxShade server (http://www.ch.embnet.org/software/BOX_form.html). The computer model of the NhhA trimeric domain was obtained as previously described (24). Briefly, residues 504 to 592 of MC58 NhhA were threaded onto the X-ray coordinates of NalP (Protein Data Bank [PDB] code 1UYO). Gap filling and refinement of torsion angles were carried out with Deep View software (9). The trimeric coordinates were refined by energy minimization using the Gromos force field incorporated in the Deep View package. The stereochemical validity of both monomeric and trimeric final models was confirmed using PROCHECK (13).
Construction of plasmids and recombinant strains.
DNA manipulations and E. coli transformations were performed according to standard protocols. Restriction endonucleases were obtained from New England BioLabs and used according to the manufacturer's instructions.
To obtain deletion mutants for DegQ in strains MC58 and M1390, we constructed the plasmid pBS-UDdegQEryr (see Table S1 in the supplemental material). The flanking regions and the Eryr fragment were amplified by PCR using Platinum Taq DNA polymerase High Fidelity (Invitrogen) and synthetic primers with overhanging ends containing the appropriate restriction sites for cloning (see Table S2 in the supplemental material). The upstream, downstream, and Eryr fragments were cloned into the pBS-KS backbone by two-step cloning in E. coli. To obtain N. meningitidis recombinant strains expressing the wild-type and the single-residue mutated NhhA protein sequences, we constructed the strain-specific plasmids pILMC58-nhhA-G546 and pILM1390-nhhA-D549 and prepared their mutagenized derivatives pILMC58-nhhA-G546D (MC58, Gly546Asp) and pILM1390-nhhA-D549G (M1390, Asp549Gly), respectively. The plasmids contained the entire nhhA gene and were designed to allow integration into the bacterial chromosome. The constructs were prepared using the backbone of the plasmid pBluescript-II KS (pBS-KS) (Stratagene). Each strain-specific plasmid contained two flanking regions (upstream and downstream) to facilitate homologous recombination and a chloramphenicol resistance (Cmr) cassette for selection. The flanking regions and the Cmr cassette were amplified by PCR using Platinum Taq DNA polymerase High Fidelity (Invitrogen) and synthetic primers with overhanging ends containing the appropriate restriction sites for cloning. The upstream flanking region contains the entire nhhA gene and 133 bp upstream of the start codon site and was amplified using primers Up-nhhA-Fwd (XbaI) and Up-nhhA-Rev (PstI) with a PCR product of ∼1,900 bp (30 cycles at 94°C for 40 s, 60°C for 40 s, and 68°C for 2 min). The downstream flanking region corresponded to ∼850 bp downstream of the termination codon and was amplified using primers Dn-nhhA-Fwd (NsiI) and Dn-nhhA-Rev (XhoI) (30 cycles at 94°C for 40 s, 60°C for 40 s, and 68°C for 90 s). The flanking regions used to construct the strain-specific plasmids were amplified using the genomic DNA of strains MC58 and M1390 as a template. The chloramphenicol resistance cassette was amplified using primers Cm-Fwd (NsiI) and Cm-Rev (PstI) and plasmid pCompIND-Cmr (11) as a template (30 cycles at 94°C for 40 s, 60°C for 40 s, and 68°C for 1 min). All PCR fragments were purified and digested with the appropriate restriction enzymes. The upstream, downstream, and Cmr fragments were cloned into the pBS-KS backbone by two-step cloning in E. coli. The mutagenized plasmids with the single-residue substitution in the translocator domains, pILMC58-nhhA-G546D (MC58, Gly546Asp) and pILM1390-nhhA-D549G (M1390, Asp549Gly), were generated by site-directed mutagenesis. The reaction was carried out using the Gene Tailor site-directed mutagenesis kit (Invitrogen) and parental plasmids pILMC58-nhhA-G546 and pILM1390-nhhA-D549 as templates, according to the manufacturer's instructions. The mutagenesis reaction was performed with primers that aligned to the same region in the nhhA gene from MC58 and M1390 nucleotide sequences. For the plasmid pILMC58-nhhA-G546D, we used the primers Fwd-nhhA(D) and Rev-nhhA-mt. This reaction allows a single-nucleotide substitution of guanine to adenine, which is translated into replacement of glycine by aspartic acid in the MC58 NhhA protein sequence. For plasmid pILM1390-nhhA-D549G, the reaction used primers Fwd-nhhA(G) and Rev-nhhA-mt. This reaction permits a single-nucleotide substitution of adenine to guanine, which is translated into replacement of aspartic acid by glycine in the M1390 NhhA protein sequence. Once the plasmids were ready, naturally competent N. meningitidis strains were transformed as previously described with their strain-specific plasmids. Strain MC58 was transformed with pILMC58-nhhA-G546 and pILMC58-nhhA-G546D, and strains M1390 and NZ05/33 were transformed with pILM1390-nhhA-D549 and pILM1390-nhhA-D549G to maintain sequence homology. For the generation of deletion mutants in M1390 and NZ05/33, the nhhA gene was replaced by a kanamycin resistance (Kanr) cassette using plasmid pBS-UDnhhAKanr and controlled by PCR using previously described primers (24). As an alternative strategy for generation of deletion mutants in strains M1390 and NZ05/33, we prepared the plasmid pILM1390-nhhA-SC, in which a termination codon was introduced at nucleotide position 27 (AAT→TGA) by site-directed mutagenesis in the nhhA nucleotide sequence. For preparation of pILM1390-nhhA-SC, plasmid pILM1390-nhhA-D549 was used as a template, and the primers used were nhhA-SC-Fwd and nhhA-SC-Rev. All recombinant strains constructed using the strain-specific plasmid system were selected for chloramphenicol resistance resulting from a double-crossover event, and the correct insertion was confirmed by PCR using the primers nhhA-R-Fwd and nhhA-R-Rev. The expression of wild-type and mutant NhhA proteins and the correct nucleotide sequences for each strain were confirmed by Western blot analysis and sequencing. The various recombinant strains were also analyzed for growth kinetics in GC broth with respect to the isogenic wild-type strains. The genetic approach did not have any effect on the growth kinetics of N. meningitidis strains (see Fig. S1 in the supplemental material). The recombinant N. meningitidis serogroup B strains are listed in Table S1 in the supplemental material, and all the primers used in the PCRs described above are reported in Table S2 in the supplemental material.
Fluorescence-activated cell sorter (FACS) analysis of N. meningitidis.
N. meningitidis serogroup B wild-type and derivative strains expressing wild-type NhhA or mutated proteins were grown in GC broth medium to an OD600 of 0.5. The bacteria were pelleted, resuspended, and incubated with mouse polyclonal antibodies against NhhA-His for 1 h at 37°C. Following two washes in PBS, bacterial cells were incubated with fluorescein isothiocyanate (FITC)-conjugated rabbit anti-mouse IgG (Jackson ImmunoResearch Laboratories) for 30 min at room temperature. The bacterial cells were washed twice with PBS and fixed, and surface localization of NhhA was measured by flow cytometry on a FACSCalibur (Becton Dickinson). Membrane integrity controls were performed using polyclonal antibodies against the surface protein fHbp (16), a monoclonal antibody against the MenB capsule polysaccharide (SEAM12) as a positive control (6), and a preimmune serum as a negative control, and antibody was detected with FITC-conjugated rabbit anti-mouse IgG as a secondary antibody (Jackson ImmunoResearch Laboratories) (data not shown).
Immunogold electron microscopy.
For immunoelectron microscopy, bacterial cells were taken from GC agar plates and normalized to an OD600 of 0.8. One milliliter of the bacterial suspension was fixed with a filtered mixture of 4% paraformaldehyde (Invitrogen) and 0.02% glutaraldehyde (Invitrogen) in PBS, pH 7.2, for 1 h at room temperature. Whole-cell negative staining and immunogold labeling were performed as follows. Following fixation, cell droplets (10 μl) were placed on Formvar carbon-coated grids (Agar Scientific Ltd.) for 5 min. Excess fluid was wicked off, and blocking was accomplished in two stages using PBS containing 1% cold-water fish gelatin (Fluka) for 10 min. Excess aldehyde was quenched using 0.02 M glycine in PBS for 5 min. The grids with cells were inverted over anti-NhhA polyclonal antibodies diluted 1:250 in PBS-1% bovine serum albumin (BSA) for 1 h in a humidified chamber. The grids were rinsed five times for 1 min each time in PBS-1% BSA. Antigen was detected by incubation for 1 h with electron microscopy goat anti-mouse immunoglobulin G plus immunoglobulin M conjugated to 10-nm colloidal gold beads (British Biocell International) diluted 1:15 in PBS-1% BSA. Rinsing took place in PBS (four times for 1 min each time). The grids with cells were stabilized with 1% glutaraldehyde in PBS for 3 min, and finally, each sample was rinsed in distilled water five times for 1 min each time. The grids were finally treated with uranyl acetate and examined with a TEM GEOL 1200EX II transmission electron microscope.
Cell cultures.
Chang epithelial cells (a Wong-Kilbourne derivative, clone 1-5c-4; human conjunctiva; ATCC CCL-20.2) were maintained in Dulbecco's modified Eagle's medium (DMEM) (Gibco) supplemented with 25 mM HEPES, 15 mM l-glutamine, antibiotics (penicillin and streptavidin), and 10% (vol/vol) heat-inactivated fetal bovine serum (FBSi; Invitrogen). The cells were used between passages 5 and 15 and were grown at 37°C with 5% CO2.
Adhesion assays with Chang epithelial cells.
Adhesion experiments with N. meningitidis serogroup B wild-type and derivative strains with cultured Chang epithelial cells were performed as previously described (24). Briefly, 1.5 ×105 cells per well were seeded in 24-well tissue culture plates for 24 h in antibiotic-free DMEM plus 10% FBSi. Overnight cultures of bacteria were washed once and resuspended in infection medium (DMEM plus 1% FBSi) to a concentration of ∼2.25 × 107 bacteria ml−1 at a multiplicity of infection (MOI) of approximately 1:75. One-milliliter aliquots of each strain were added to monolayer cultures of Chang cells and incubated for 3 h at 37°C in 5% CO2. Nonadherent bacteria were removed by washing three times with DMEM plus 1% FBSi at 1-h intervals. Adherent bacteria were released by the addition of 1% Saponin (Sigma) and incubated at 37°C for 15 min. Adhesion capability was quantified by serial dilutions of the associated bacterial suspension plated on GC agar for CFU counting. At least three single clones were analyzed for each recombinant strain to confirm the adhesion phenotype.
Serum bactericidal assay (SBA).
Serum bactericidal activity against N. meningitidis serogroup B wild-type and derivative strains was evaluated as described previously (21), using pooled baby rabbit serum (CedarLane) as a complement source and mouse polyclonal antibodies raised against recombinant NhhA. Serum bactericidal titers were defined as the serum dilution resulting in a 50% decrease in CFU per ml after 60-min incubation of bacteria in the reaction mixture compared with control CFU per ml at time zero. Typically, bacteria incubated with the negative-control antibody in the presence of complement showed a 150 to 200% increase in CFU/ml during the 60 min of incubation. The monoclonal antibody SEAM12 was used as a positive control (7).
Nucleotide sequence accession numbers.
The nucleotide and protein sequences of the nhhA genes from N. meningitidis serogroup B strains have been entered into the GenBank database with accession numbers M01-0240149 (JF414785), M4287 (JF414786), OX99.30304 (JF414787), M1239 (JF414788), M4105 (JF414789), M3279 (JF414790), ISS749 (JF414791), M1390 (JF414792), 67/00 (JF414793), NZ05/33 (JF414794), M0579 (JF414795), M4407 (JF414796), ISS1026 (JF414797), NM008 (JF414798), M10994 (JF414799), M13203 (JF414800), and C188 (JF414801). The accession numbers for N. meningitidis non-B serogroup sequences are as follows: M08-0240233 (JN590750), F8238 (JN590751), M07-0240954 (JN590740), M07-0241093 (JN590742), C11 (JN590739), M2197 (JN590741), M07-0240665 (JN590749), M07-0240845 (JN590746), 240070 (JN590748), LNP17592 (JN590747), M07-0240625 (JN590743), M07-0240706 (JN590744), and M07-0240771 (JN590745).
RESULTS
Production of NhhA varies among different N. meningitidis serogroup B strains.
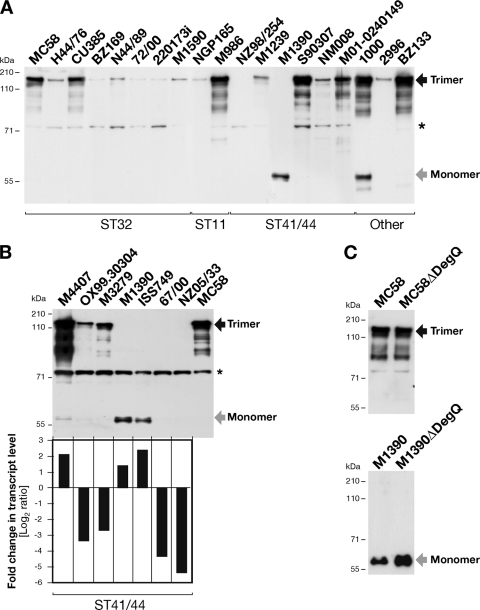
In a previous work, we demonstrated that NhhA is involved in the adhesion of encapsulated N. meningitidis to epithelial cells and extracellular matrix components (24). Some of the protein antigens being investigated as potential vaccine candidates for prevention of meningococcal disease exhibit variable production in different N. meningitidis strains (16, 19, 26). Here, we evaluated the production of the NhhA protein in different N. meningitidis serogroup B strains using polyclonal antibodies against recombinant NhhA that were previously shown to detect NhhA in N. meningitidis and E. coli (21, 24). A panel of 39 strains belonging to different clonal complexes (CCs) was analyzed for the presence of the nhhA gene and the expression level of the protein (Table 1). PCR analysis with specific primers for nhhA confirmed the presence of the gene in all the strains tested (data not shown). Whole-cell lysates prepared from mid-log-phase liquid cultures were analyzed by Western blotting under denaturing conditions. By this analysis, we identified a band with a molecular mass of ∼180 kDa corresponding to the trimeric form of NhhA. We observed that the production of trimeric NhhA varied considerably between the different strains tested (Fig. 1A and Table 1). Furthermore, we found two strains (M1390 and ISS749, belonging to the clonal complex ST41/44) in which only a band with an apparent molecular mass of ∼60 kDa was detected, likely corresponding, on the basis of its theoretical molecular mass, to the NhhA monomeric protein (Fig. 1A and B). In six other strains (NZ98/254, NZ05/33, 67/00, ISS1026, BZ198, and C188), all from the ST41/44 clonal complex, NhhA was detectable as a monomer only after long exposure of the membrane (Table 1 and data not shown). In the case of strain 1000, both trimeric and monomeric NhhA proteins were observed (Fig. 1A).
Fig. 1.
(A) Analysis of NhhA production in a panel of N. meningitidis serogroup B strains. Western blot analysis under denaturing conditions of whole-cell lysates showing the variation in protein production levels among different clinical isolates is presented. A subpanel of the strains tested is shown. The results for all the strains tested are reported in Table 1. (B) Comparison of NhhA production and nhhA transcript levels in a subset of ST41/44 N. meningitidis serogroup B strains. (Top) Western blot analysis of whole-cell lysates under denaturing conditions. Western blot analysis was performed using a polyclonal mouse serum raised against recombinant NhhA-His protein (N. meningitidis serogroup B strain 2996). (Bottom) Quantitative analysis of nhhA transcript levels analyzed by relative qRT-PCR. Both nhhA and the control gene were amplified from the same amount of total RNA, and the data are presented as fold change in nhhA mRNA after normalization to 16S rRNA expression levels, using MC58 as a reference strain, which is reported as a log2 value of 0. (C) Effect of DegQ on trimerization and detection of NhhA protein. Deletion mutants of DegQ in MC58 and M1390 were analyzed for the expression of NhhA in whole-cell lysates. NhhA trimer bands are observed at ∼180 kDa (black arrow) and monomer bands at ∼60 kDa (gray arrow). The asterisks mark a nonspecific cross-reactive band.
Finally, to have a broad picture of the NhhA expression pattern in N. meningitidis, the production of NhhA was additionally tested by Western blotting in a subpanel of 15 N. meningitidis strains belonging to other non-B serogroups. As shown for N. meningitidis serogroup B strains, expression of NhhA varies among these strains (Table 1; see Fig. S2 in the supplemental material). Also here, we found a strain, C11 (serogroup C), with dual monomer-trimer NhhA production.
To further investigate the variability of NhhA production, we performed additional analysis by relative qRT-PCR to measure the amount of nhhA gene transcript. We selected a subset of seven N. meningitidis serogroup B ST41/44 strains in which the NhhA protein was detected as a trimer or a monomer or was produced at very low levels. Total bacterial RNA and whole-cell lysates were prepared from cultures for each strain at the mid-log growth phase. Relative qRT-PCR data were obtained by normalizing the nhhA mRNA transcript to the 16S rRNA, and strain MC58 was used as a reference to compare nhhA transcripts and protein production levels. As shown in Fig. 1B, the amount of nhhA transcript correlated approximately with the production level of NhhA protein in all the strains tested. The strains M4407, M1390, and ISS749 had higher levels of nhhA transcript than strain MC58, and the RNA transcript levels were approximately 2-fold higher in strain M4407; this increase correlates with the amount of trimeric NhhA produced by the strain. In strains M1390 and ISS749, NhhA was detected as a monomer and a correlation was less evident. Strains OX99.30304, M3279, 67/00, and NZ05/33 had smaller amounts of nhhA transcript than MC58. For OX99.30304 and M3279, transcript levels of nhhA were approximately 3-fold lower than for MC58, which correlates with the smaller amount of trimeric NhhA protein produced by each strain. For strains 67/00 and NZ05/33, the transcript levels of nhhA were approximately 5-fold less than for MC58, and we were able to detect NhhA protein by Western blot analysis only after long-term exposure of the membrane (Fig. 1B and data not shown). Overall, these results show differential production of NhhA protein in the various strains tested. This analysis showed that among the different strains analyzed, few isolates from serogroup B belonging to ST41/44 produced NhhA protein detectable only as a monomer.
Although we found an association between the amounts of NhhA and RNA transcript levels, it is evident that strains expressing similar levels of nhhA transcript (e.g., M4407 and M1390) expressed different amounts of monomeric or trimeric NhhA (Fig. 1B). This might be caused by the activity of periplasmic proteases that target the monomeric NhhA protein. TAAs are target of different proteases present in the periplasm that control membrane homeostasis by degradation of unfolded monomers. Among these proteases, HtrA or DegP has been reported to have activity in degradation of unfolded YadA in E. coli (8). Based on this previous observation and on our results, we decided to explore if this family of proteins might have any effect on the production of NhhA. The genome sequence of N. meningitidis strain MC58 has only one HtrA homologue, which was previously described and referred to as DegQ (32). In order to evaluate a potential effect of DegQ on NhhA expression, we selected the strain M1390, in which the monomeric form of NhhA might be a candidate target of this protease. First, we sequenced the entire degQ gene in M1390 to confirm the absence of stop codons and tested its transcription by qRT-PCR (see Fig. S3 in the supplemental material). The degQ gene was transcribed more in M1390 than in MC58 (a 4-fold difference). To further investigate its role in NhhA expression, we constructed a deletion mutant of DegQ in strains M1390 and MC58. In MC58, the deletion of DegQ did not affect the expression and trimerization of the NhhA protein, as previously shown (32) (Fig. 1C). Interestingly, the deletion of DegQ in strain M1390 increased the detection level of monomeric NhhA (Fig. 1C). These results suggest that the monomeric form of NhhA might be a target of the DegQ protease.
A single-amino-acid substitution in the translocator domain correlates with detection of monomeric NhhA.
Since NhhA and all trimeric autotransporters form highly stable trimers even under denaturing conditions (10), our data suggest that the few ST41/44 strains in which only the NhhA monomer was detected, as well as strains 1000 and C11, showing the presence of trimeric and monomeric NhhA, might have variations in amino acid sequences that could affect protein trimerization and, consequently, NhhA-related functions. We first analyzed the amino acid sequences from 19 of the 21 ST41/44 strains that we considered in this study.
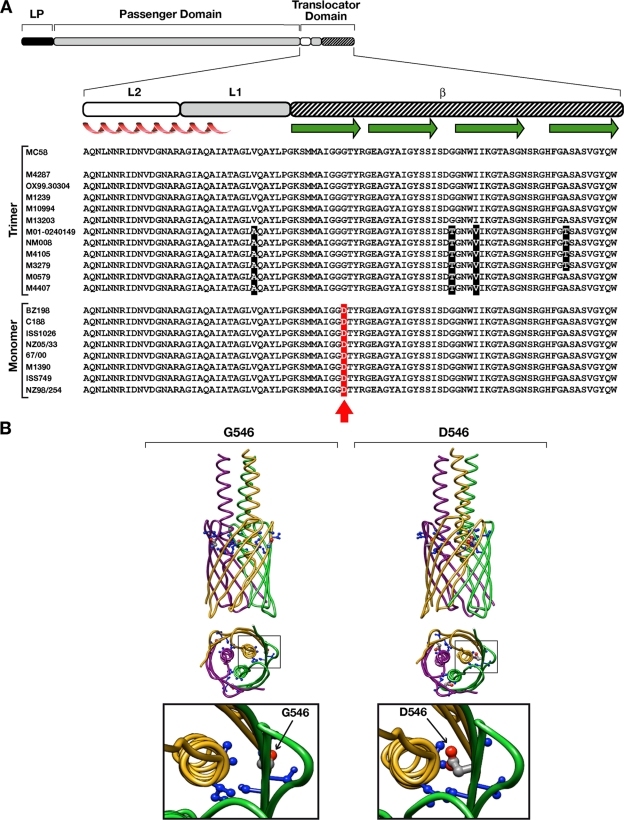
A comparison of NhhA predicted sequences showed that the majority of the diversity was largely confined to the passenger domain region (i.e., the first 200 amino acids [aa] of the mature protein). However, the capacity of the NhhA protein to trimerize and to be translocated on the bacterial surface resides in the C-terminal translocator domain (24). Multiple-sequence alignment of the translocator domain revealed a high degree of conservation between the different NhhA protein sequences (Fig. 2A), including strains 1000 and C11 (data not shown). Nonetheless, we noticed that strains where NhhA was detected only as a monomer shared a common single glycine-to-aspartic acid mutation (corresponding to Gly546 in the MC58 or Asp549 in the M1390 amino acid sequence) (Fig. 2A), establishing a correlation between a single mutation and monomer detection. Based on these observations, we hypothesized that this single mutated residue might be responsible for defective protein trimerization of NhhA.
Fig. 2.
Association between the Gly-to-Asp mutation in the translocator domain of NhhA and detection of monomeric protein. (A) Multiple-sequence alignment of the predicted protein sequences of the translocator domain of NhhA from ST41/44 N. meningitidis serogroup B strains. The predicted proteins included in the graph were obtained by sequencing the entire nhhA nucleotide sequences from the strains considered in the present study, M4287, OX99.30304, M1239, M10994, M13203, M01-0240149, NM008, M4105, M3279, M0579, M4407, C188, ISS1026, NZ05/33, 67/00, M1390, and ISS749; the additional NhhA sequences from strains NZ98/254, BZ198, and MC58 were reported previously (21). The MC58 NhhA amino acid sequence was included as a reference. The organization of the NhhA protein with the leader peptide (LP), passenger domain, and translocator domain region is shown. To map the localization of the single substitution, the subdivision of the translocator domain into L2, L1, and β regions was made according to previous reports (24). Predicted secondary structures are also indicated, with alpha helices in red and β-strands in green. The red arrow indicates the single mutation of a Gly residue to Asp. Shaded letters represent grouped nucleotide sequences that are not conserved among the different strains. (B) Modeling of the structure of the translocator domains from wild-type and mutated NhhA proteins. The prediction model was constructed using the membrane anchor crystal structure of the NhhA sequence from the MC58 wild-type strain. (Left) Model of trimeric NhhA harboring a glycine residue at position 546 (G546). (Right) Mutated trimeric NhhA with replacement of a glycine residue by aspartic acid (D546). The single-residue substitution confers an increase in the size of amino acid lateral chains that face into the internal cavity of the pore-forming β-barrel structure. Only side chains located within 5 Å from the residue in position 546 are explicitly represented as balls and sticks. In the model, each monomeric unit of the trimeric protein is represented by a different color.
The Gly-to-Asp substitution affects the trimerization of NhhA.
The hypothesis that the Gly-to-Asp mutation may affect protein trimerization was initially supported by the in silico modeling of the trimeric translocator domain of NhhA from strain MC58 (Fig. 2B). In the model, the presence of a Gly residue at position 546 (β-subdomain) produces a shorter amino acid side chain facing into the lumen of the pore formed by the β-barrel structure of the trimeric NhhA than the presence of an Asp residue at the same position (Asp546). This variation in the side chain could locally modify the protein by a different stearic hindrance, supporting the role of this mutation in affecting protein trimerization and/or trimer stability and, consequently, the localization of the autotransporter on the bacterial surface.
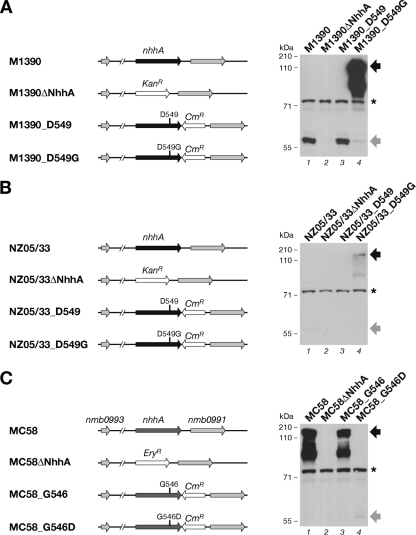
To evaluate whether the detection of NhhA as a monomer was caused strictly by this Gly-to-Asp mutation, we generated recombinant strains expressing the mutated proteins using strains from three different N. meningitidis genetic backgrounds: M1390, in which a large amount of monomeric NhhA was detected; NZ05/33, which produced small but detectable amounts of the monomer; and MC58, in which the protein was detected as a trimer. MC58 was considered our reference strain to exclude any effect on trimerization due to sequence variability in the N-terminal passenger domain. The NhhA sequence that contains Asp549 (M1390 and NZ05/33) was termed “natural mutated NhhA.”
We expressed the nhhA wild-type gene or the mutated genes, generated by site-directed mutagenesis, in the original locus under the control of their own promoter (Fig. 3). Western blot analysis of whole-cell lysates prepared from M1390 derivative strains showed that M1390_D549 produced amounts of monomeric NhhA comparable to those of the wild-type strain (Fig. 3A), indicating that the genetic strategy did not affect NhhA production. The single Asp-to-Gly amino acid substitution in strain M1390_D549G resulted in a shift from monomeric to trimeric form (Fig. 3A, lanes 3 and 4). Specific protein bands below the 180-kDa band were likely caused by degradation of the trimeric form.
Fig. 3.
The Gly-to-Asp substitution affects NhhA trimerization. Shown is the genetic organization of the nhhA loci of the wild-type, NhhA deletion mutant, and recombinant strains in the M1390 (A), NZ05/33 (B), and MC58 (C) genetic backgrounds. To generate deletion mutants, nhhA was replaced with an erythromycin resistance (Eryr) cassette in the MC58 genetic background or a kanamycin resistance (Kanr) cassette in the M1390 and NZ05/33 genetic backgrounds. In the recombinant strains, wild-type (G546 and D549) and mutated (G546D and D549G) alleles are indicated. NhhA expression is under the control of its own promoter, and downstream of the stop codon of the coding region, a chloramphenicol resistance (Cmr) cassette was inserted for selection. Western blot analysis of each group of strains is shown on the right. Whole-cell lysates were analyzed under denaturing conditions using a polyclonal mouse serum raised against recombinant NhhA-His protein. NhhA trimer bands are evidenced at ∼180 kDa (black arrows) and monomer bands at ∼60 kDa (gray arrows). The asterisks mark a nonspecific cross-reactive band.
Analysis of the whole-cell lysates revealed that in the NZ05/33 genetic background as well (the same NhhA variant as M1390), the Asp-to-Gly substitution (NZ05/33_D549G) resulted in detection of the trimeric protein (Fig. 3B, lanes 3 and 4). However, the amount of trimer detected in this case was considerably less than that of M1390_D549G, possibly caused by strain-specific gene expression levels. These data demonstrated a “gain of function” in trimerization of NhhA due to the replacement of Asp549 with Gly.
To further confirm that the effect of this mutation is crucial for NhhA trimerization and is not influenced by the sequence variability of the N-terminal passenger domain, we used the MC58 strain, where NhhA is naturally detected as a trimer. Western blot analysis of whole-cell lysates showed that replacement of Gly by Asp in the MC58 amino acid sequence (strain MC58_G546D) resulted in a shift of NhhA from a trimer to a monomer (Fig. 3C, lanes 3 and 4). This “loss of function” confirmed the pivotal role that this Gly plays in NhhA trimerization.
The Gly-to-Asp mutation affects surface localization of NhhA.
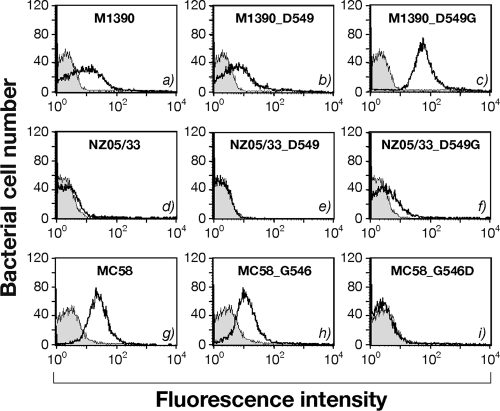
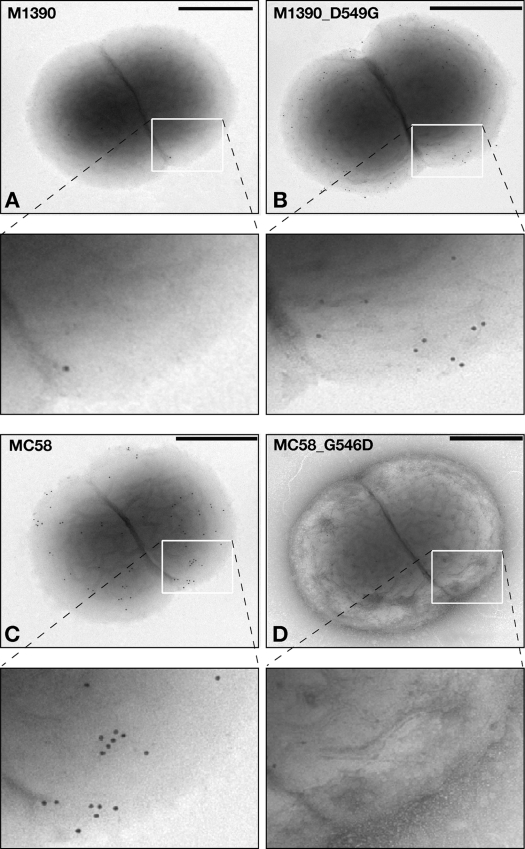
Since the mutation identified mapped in the β-subdomain region of the translocation unit (Fig. 2A) and affected trimerization, we expected that it might alter the surface localization of NhhA, which is mediated by the trimerization of the C-terminal translocator domain (5). To analyze protein localization on the bacterial surface, we performed whole-cell FACS analysis and immunogold electron microscopy on all the strains shown in Fig. 3 using polyclonal antibodies against NhhA.
Although trimerization was defective in the M1390 wild type and M1390_D549, FACS and immunogold analyses detected low levels of NhhA on the surfaces of these strains (Fig. 4a and b and 5A). Strain M1390_D549G, producing the trimeric form, showed increased staining on the bacterial surface (Fig. 4c and 5B). For strain NZ05/33, FACS analysis showed no NhhA on the meningococcal surface (Fig. 4d and e) for either the wild-type NZ05/33 or the NZ05/33_D549 strain; however, the production of NhhA with a single substitution in the recombinant strain NZ05/33_D549G, producing the trimer, resulted in little staining on the surfaces of the bacteria (Fig. 4f), as expected based on Western blot results (Fig. 3B, lane 4). Furthermore, MC58 and MC58_G546 showed a significant level of surface NhhA (Fig. 4g and h and 5C), while the Gly-to-Asp substitution in strain MC58_G546D abolished its detection on the bacterial surface (Fig. 4i and 5D). These data demonstrate that this single substitution contributes to NhhA localization.
Fig. 4.
Surface localization of NhhA in N. meningitidis strains. Shown is FACS analysis of NhhA on whole-cell bacteria. Deletion mutant strains in each genetic background were used as negative controls, and the corresponding (gray-shaded) histogram was superimposed on the results with the reported strains (solid line). The same polyclonal mouse serum anti-NhhA-His used for Western blot analysis was also used in FACS as a primary antibody.
Fig. 5.
Immunoelectron microscopy analysis for surface localization of NhhA on whole-cell bacteria. Shown are strains M1390 wild type (A), M1390_D549G (B), MC58 wild type (C), and MC58_G546D (D). Magnified images of the boxed areas are shown to allow better detection of the differential labeling on the surfaces of the bacteria. The same polyclonal mouse serum anti-NhhA-His used for Western blot and FACS analyses was also used in immunogold analysis as a primary antibody. Scale bars, 400 nm.
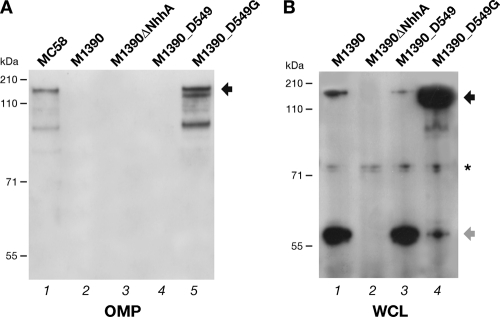
To confirm the defective export of mutated NhhA on the meningococcal surface, we examined if the NhhA monomeric form could reach the meningococcal outer membrane. As shown in Fig. 6A, Western blot analysis of an OMP preparation from strain M1390 showed that the NhhA protein was not detectable in the M1390 wild-type and M1390_D549 strains expressing the monomeric protein (Fig. 6A, lanes 2 and 4), while the trimer is present in the outer membrane of strain M1390_D549G, as in the MC58 wild-type strain, used as a positive control (Fig. 6A, lanes 1 and 5).
Fig. 6.
Gly-to-Asp substitution affects localization of NhhA in the outer membrane and trimer stability. (A) Western blot analysis of OMP preparations of the indicated M1390 and derivative strains subjected to denaturing conditions for subcellular detection of NhhA. Coomassie blue staining of the gel is shown in Fig. S4 in the supplemental material. Strain MC58 was used as a reference for trimeric localization of NhhA on OMPs. (B) Analysis of whole-cell lysates (WCL) from M1390 and its derivative strains prepared by alkaline lysis, subjected to seminative conditions, and analyzed by Western blotting. The Western blots were analyzed using serum anti-NhhA. The trimeric and monomeric forms are indicated by black and gray arrows, respectively. The asterisk marks a nonspecific cross-reactive band.
Finally, to confirm that the poor ability of monomeric NhhA to reach the bacterial surface may be ascribed to defective formation of the trimer or to its lower stability, we analyzed samples of whole-cell lysates prepared by alkaline lysis from M1390, M1390ΔNhhA, M1390_D549, and M1390_D549G by Western blotting under seminative conditions. As shown in Fig. 6B (lanes 1 and 3), we detected trimeric NhhA in strains M1390 and M1390_D549, together with the monomeric form, which suggests either that natural mutated NhhA can trimerize, although less efficiently, or that the trimer is less stable under denaturing conditions. We also observed in M1390_D549G an increase of the amount of trimeric NhhA (Fig. 6, lane 4).
Overall, these data demonstrate that the Gly-to-Asp mutation in the translocator domain of NhhA influences its trimer formation and/or stability and, consequently, its localization on the meningococcal surface.
NhhA trimerization is essential for killing mediated by NhhA-specific antibodies.
NhhA has been shown to be a protective antigen against meningococci because it is able to induce bactericidal antibodies in mice (21, 33) and is recognized by sera of patients convalescing after meningococcal disease (15).
To evaluate whether the Gly-to-Asp mutation could influence the recognition of NhhA-specific bactericidal antibodies, we tested a subpanel of N. meningitidis strains in a serum bactericidal assay using mouse polyclonal sera raised against recombinant NhhA. As shown in Table 2, when NhhA was surface expressed as a trimer, anti-NhhA antibodies activated complement-mediated killing of N. meningitidis strains MC58, M4407, 8047, and 2996. On the other hand, against the two N. meningitidis wild-type strains expressing the natural mutated monomer form of NhhA (M1390 and NZ05/33), titers were negative or very low (16 and <16, respectively). However, analysis of the recombinant strain M1390_D549G showed that the expression of a stable surface-localized trimer resulted in a positive bactericidal titer (1,024). In contrast, NZ05/33 recombinant strains showed negative titers even in the presence of a trimeric NhhA on the bacterial surface (NZ05/33_D549G), suggesting that the low level of production of NhhA trimer in this strain was not sufficient to efficiently mediate bacterial killing.
Table 2.
Bactericidal titers of NhhA-specific polyclonal antibodies
| Strain | Titera |
Remarks | |
|---|---|---|---|
| Anti-NhhA | Anti-SEAM 12b | ||
| Natural isolate | |||
| MC58 | 256 | 65,536 | Trimer |
| M4407 | 4,096 | 65,536 | Trimer |
| 2996 | 512 | 32,768 | Trimer |
| 8047 | 256 | 8,192 | Trimer |
| NZ05/33-derived recombinant strains | |||
| NZ05/33 | <16 | 16,384 | Monomer |
| NZ05/33ΔNhhA | <16 | 16,384 | Deletion mutant |
| NZ05/33_D549 | <16 | 16,384 | Monomer |
| NZ05/33_D549G | <16 | 8,192 | Trimer |
| M1390-derived recombinant strains | |||
| M1390 | 16 | 65,536 | Monomer |
| M1390ΔNhhA | <16 | 65,536 | Deletion mutant |
| M1390_D549 | 16 | 65,536 | Monomer |
| M1390_D549G | 1,024 | 65,536 | Trimer |
Bactericidal titers obtained using rabbit complement. The titers are expressed as the reciprocal of the serum dilution necessary to obtain >50% bacterial killing.
The SEAM12 monoclonal antibody against capsule polysaccharide was used as a positive control; a difference of a single dilution was not considered significant.
These data suggest that despite the sequence variations in the N-terminal region between the strains tested in this assay, polyclonal NhhA antibodies were able to induce bactericidal activity only against trimeric NhhA and that strains carrying a natural mutated monomeric NhhA (e.g., strains belonging to CC ST41/44) were not targets of NhhA-specific bactericidal antibodies, probably due to the small amount of protein localized at the bacterial surface.
A Gly-to-Asp single-residue substitution influences NhhA adhesive capabilities.
NhhA has been shown to promote adhesion of encapsulated meningococci to human epithelial cells (24). To evaluate the impact of the Gly-to-Asp mutation on the adhesive properties of NhhA, we tested in vitro the capability of wild-type MC58 and the derivative strains to adhere to monolayers of Chang epithelial cells. As shown in Fig. 7A, the production of the mutated NhhA (MC58_G546D) resulted in reduced bacterial attachment compared with the MC58 wild-type and MC58_G546 strains. The reduction in attachment of MC58_G546D was comparable to values obtained using the deletion mutant MC58ΔNhhA. Thus, in MC58, where NhhA contributes significantly to epithelial adhesion (24), the mutated NhhA protein strongly influences meningococcal attachment.
Fig. 7.
Effect of the Gly-to-Asp substitution on NhhA-mediated adherence. The adherence of N. meningitidis strains was calculated by counting the associated bacteria on cell monolayers. Chang epithelial cell monolayers were infected at an MOI of 1:75, and adherence was monitored after 3 h. (A) MC58 and derivatives strains. (B) M1390 and derivative strains. (C) NZ05/33 and derivative strains. Adherence is expressed as CFU per well, and the values represent the means and standard deviations of a representative experiment performed in triplicate. Each experiment was performed at least three times. For M1390 and NZ05/33, the data obtained for deletion mutants were confirmed using an additional strategy to generate an nhhA deletion mutant (see Materials and Methods).
As the natural mutated NhhA in M1390 is defectively localized on the bacterial surface, we studied whether the increased localization of trimeric NhhA in strain M1390_D549G could impact adhesion capability. As shown in Fig. 7B, the Asp-to-Gly single substitution in strain M1390_D549G resulted in a significant increase in bacterial attachment to epithelial cells compared to M1390 and M1390_D549, which express the mutated NhhA. Unexpectedly, the deletion of NhhA in M1390 increased the capacity for bacterial adhesion to epithelial cells compared with the wild-type strain (Fig. 7B).
Finally, in NZ05/33 strains, the replacement of the Asp residue (NZ05/33_D549G) and the deletion of NhhA did not significantly influence adherence to epithelial cells compared with the wild-type strain (Fig. 7C). This discrepancy between the data obtained with strains M1390 and NZ05/33 might be explained by the different levels of production of the mutated NhhA in the two strains, which is very high in M1390 but low in strain NZ05/33.
Overall, these results confirm that a single-residue mutation affects meningococcal NhhA-mediated adhesion and also suggest that NhhA function might have a strain-specific role, depending on its sequence variation and level of production.
DISCUSSION
In this study, we describe the importance of a natural single-residue mutation identified in the translocator domain of the NhhA autotransporter in impairing its trimerization and localization on the meningococcal surface. Our work ensued from the observation that in a subset of clinical isolates belonging to the clonal complex ST41/44 of N. meningitidis serogroup B strains, NhhA was detected only as a monomeric protein. This was considered an unusual and interesting finding because trimeric autotransporters are characterized by the ability to form stable trimeric structures, even under denaturing conditions (10). Trimerization of monomer subunits is a key event during the export of trimeric autotransporters. This step is mediated by the C-terminal translocator unit, whose trimerization is necessary to form a stable functional β-barrel pore through which the three monomer chains of the passenger domains cross the membranes and lean out of the bacterial surface (14). Sequence analysis of the translocator regions evidenced a single-residue mutation (glycine to aspartic acid substitution at position 549 in strain M1390) in the β-subdomains of all the strains expressing NhhA as a monomer. However, analysis under seminative conditions of total proteins from M1390, an ST41/44 isolate expressing a large amount of mutated NhhA (D549), showed that even this mutated protein was partially able to form trimers. Therefore, defective trimeric NhhA variants harboring the aspartic acid residue (D549) are susceptible to disruption under denaturing conditions, leading to detection of the monomer.
By FACS analysis and electron microscopy, we found a low level of surface localization of mutated monomeric NhhA in the M1390 wild-type strain. We suggest that on live bacteria, mutated NhhA (D549) is able to partially trimerize and is defective in localizing on the bacterial surface, most likely due to an incompletely folded or unstable β-barrel pore structure. Since we did not obtain the same results using strain NZ05/33, another ST41/44 isolate harboring mutated NhhA (D549) but expressing a small amount of the protein, we think that partial export of NhhA can be detected only when the NhhA protein is produced at a high level. Experiments in which the single Asp mutation was replaced with a Gly residue demonstrated an increase of trimerization and surface localization of NhhA in both the M1390 and NZ05/33 genetic backgrounds. These results are in accordance with data showing that the mutated protein is undetectable in the meningococcal outer membrane fraction of the M1390 wild-type strain, whereas the single substitution (D549G) permits the discovery of NhhA trimer in the outer membrane of the recombinant strain.
To confirm these data, we also used the opposite approach, replacing the Gly residue in strain MC58, where NhhA was detected as a stable trimer. We found that the replacement of the Gly residue in strain MC58 (G546D) abolished trimer formation and localization on the meningococcal surface, confirming an essential role of this mutation in the correct assembly, stability, and translocation of the trimeric protein. Furthermore, the expression of mutated NhhA (G546D) in strain MC58 resulted in the production of a very small amount of the mutated monomeric form. One explanation of this result could be the unstable nature of the mutated protein, which might be more efficiently degraded by periplasmic proteases, as reported for other trimeric autotransporters (8).
We explored the effect of the periplasmic protease DegQ on the NhhA protein in the MC58 and M1390 wild-type strains. We found that DegQ deletion did not influence the trimeric NhhA production in MC58. On the contrary, the effect of DegQ deletion in increasing monomeric NhhA in M1390 suggests that NhhA might be a target of DegQ protease for degradation of unfolded monomers in the periplasm of meningococci. Although this role in the control of protein folding could be exploited differently among N. meningitidis strains, we were able to prove that DegQ acts on monomeric NhhA protein. However, whether DegQ is also involved in the degradation of monomeric NhhA in other N. meningitidis strains remains to be demonstrated. These findings correlate with the lack of effect of DegQ in the assembly of trimeric NhhA and with the idea that chaperone/protease requirements may be species and autotransporter specific (32).
The translocator domain of trimeric autotransporters was subdivided into two structural regions: a β-domain (with four β-strands) and a linker, named L1 and L2 (a coiled-coil region). Previous works demonstrated the importance of the L1 β-subdomain of NhhA in trimer formation (18, 24) and the crucial role of the residues of the L1 β-loop in trimer stability (18). However, while these studies did not identify specific residues involved in trimer formation/stability, Grosskinsky et al. demonstrated that substitution of a single glycine residue located in the β-subdomain (β2 strand) of YadA (8), which was selected by in silico analysis because it is highly conserved among the translocation domains from different trimeric autotransporters, affected YadA trimer stability, surface localization, and virulence functions (8, 25).
The NhhA glycine residue that we characterized in this study is located in the β1 strand and is different from the glycine residue studied by Grosskinsky et al. Multiple-sequence alignment of the translocator domains of all the trimeric autotransporters described thus far suggests that this glycine residue is highly conserved, with few cases of replacement with serine or alanine residues (data not shown). These findings, together with modeling of the mutated translocator domain (Fig. 3B), suggest for the first time that in this particular position a long lateral chain of a charged amino acid may affect the formation and stability of the trimer and, consequently, the functionality of a trimeric autotransporter as a result of a natural mutation in a bacterial population.
We previously reported that NhhA promotes bacterial adhesion to host cells and extracellular matrix components (24). More recently, Sjölinder and coworkers reported the role of NhhA in colonization of the nasopharyngeal mucosa in a murine model of meningococcal disease (28).
The analysis of the adhesive capabilities of the different N. meningitidis strains used in this study suggests that this single Gly-to-Asp substitution has a direct influence on NhhA-mediated adhesion. In fact, we found that (i) only trimeric NhhA (G546 and D549G) has the capability to play a role in mediating meningococcal adherence to Chang epithelial cells (as shown in the case of MC58 wild-type and derivative M1390 strains expressing the trimeric protein); (ii) mutated NhhA (D549), when produced in large amounts (as shown in the case of the M1390 wild-type strain), seems to prejudice meningococcal adherence through a mechanism not yet understood, as shown by the result on the M1390 isogenic deletion mutant, where the loss of the protein seems to favor adhesion of the strain to epithelial cells (one can speculate that the presence of an incorrectly folded NhhA protein could interfere with the activities of multiple adhesins physiologically exported in the meningococcal outer membrane); and (iii) mutated NhhA (D549) does not play any functional role when produced at a low level (as shown in the case of strain NZ05/33). Further studies are needed to elucidate these findings.
The analysis conducted in this study using a panel of different N. meningitidis strains showed that the production of NhhA is variable among the strains. A similar observation was already reported by Peak and coworkers, but a mechanism was not proposed, except for two strains that showed a deletion at the 5′ end of the nhhA gene (20).
NhhA has been characterized as a potential protective antigen against meningococci (15, 21, 33). Despite the fact that the nhhA gene is present in all the strains of N. meningitidis tested so far (20, 21), we showed that the production of the protein is variable and that surface localization is affected by the D549 mutation, which, based on our analysis, seems overrepresented in strains belonging to the ST41/44 clonal complex of serogroup B strains. Our study demonstrates that N. meningitidis serogroup B strains producing natural mutated NhhA were not killed by NhhA-specific bactericidal antibodies, suggesting that effective surface exposure of trimeric NhhA is necessary in order to induce efficient complement-mediated killing. In the case of NZ05/33, the small amount of trimeric NhhA produced by the recombinant strain restored its trimeric form but was not sufficient to mediate killing, suggesting that the bactericidal activity is also dependent on the level of NhhA expression. These findings have important consequences for the evaluation of NhhA as a vaccine antigen against N. meningitidis serogroup B, particularly with respect to the need for broad strain coverage.
In conclusion, this study revealed that a single-amino-acid mutation in the translocator domain of NhhA affects trimerization, surface localization, and, consequently, NhhA-mediated functions. The finding that this mutation occurs in natural strains may help provide, through further studies, a better understanding of the contribution of NhhA to the pathogenesis of epidemiologically relevant N. meningitidis isolates, as well as provide new insights into the importance of a balanced distribution of amino acid charges in trimeric autotransporter assembly and stability.
Supplementary Material
ACKNOWLEDGMENTS
We thank Richard Moxon for very helpful discussions and Rino Rappuoli, Isabel Delany, and Kate Seib for critical reading of the manuscript. We thank Maurizio Comanducci and Stefania Bambini (Novartis Vaccines), Ray Borrow (HPA), Paola Mastrantonio (ISS), Carla Cellesi (University of Siena), Leonard Mayer (CDC), Dominique Caugant (NIPH), Simon Kroll (Imperial College), Diana Martin (ESR), Per Olcén (Orebro University Hospital), Martin Maiden (University of Oxford), Muhamed-Kheir Taha (Institut Pasteur), and Richard Moxon for providing Neisseria meningitidis strains. We thank Antonietta Maiorino and Lisa DeTora for manuscript editing and Giorgio Corsi for artwork.
H.E.-R. was the recipient of a Novartis fellowship from the Ph.D. program in Evolutionary Biology of the University of Siena.
Footnotes
Supplemental material for this article may be found at http://iai.asm.org/.
Published ahead of print on 15 August 2011.
REFERENCES
- 1. Alamuri P., et al. 2010. Adhesion, invasion, and agglutination mediated by two trimeric autotransporters in the human uropathogen Proteus mirabilis. Infect. Immun. 78:4882–4894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Capecchi B., et al. 2005. Neisseria meningitidis NadA is a new invasin which promotes bacterial adhesion to and penetration into human epithelial cells. Mol. Microbiol. 55:687–698 [DOI] [PubMed] [Google Scholar]
- 3. Carlone G. M., Thomas M. L., Rumschlag H. S., Sottnek F. O. 1986. Rapid microprocedure for isolating detergent-insoluble outer membrane proteins from Haemophilus species. J. Clin. Microbiol. 24:330–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cope L. D., et al. 1999. Characterization of the Moraxella catarrhalis uspA1 and uspA2 genes and their encoded products. J. Bacteriol. 181:4026–4034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cotter S. E., Surana N. K., St. Geme J. W., III 2005. Trimeric autotransporters: a distinct subfamily of autotransporter proteins. Trends Microbiol. 13:199–205 [DOI] [PubMed] [Google Scholar]
- 6. Cotter S. E., Yeo H. J., Juehne T., St. Geme J. W., III 2005. Architecture and adhesive activity of the Haemophilus influenzae Hsf adhesin. J. Bacteriol. 187:4656–4664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Granoff D. M., et al. 1998. Bactericidal monoclonal antibodies that define unique meningococcal B polysaccharide epitopes that do not cross-react with human polysialic acid. J. Immunol. 160:5028–5036 [PubMed] [Google Scholar]
- 8. Grosskinsky U., et al. 2007. A conserved glycine residue of trimeric autotransporter domains plays a key role in Yersinia adhesin A autotransport. J. Bacteriol. 189:9011–9019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Guex N., Peitsch M. C. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714–2723 [DOI] [PubMed] [Google Scholar]
- 10. Hoiczyk E., Roggenkamp A., Reichenbecher M., Lupas A., Heesemann J. 2000. Structure and sequence analysis of Yersinia YadA and Moraxella UspAs reveal a novel class of adhesins. EMBO J. 19:5989–5999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ieva R., et al. 2005. CrgA is an inducible LysR-type regulator of Neisseria meningitidis, acting both as a repressor and as an activator of gene transcription. J. Bacteriol. 187:3421–3430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kline K. A., Falker S., Dahlberg S., Normark S., Henriques-Normark B. 2009. Bacterial adhesins in host-microbe interactions. Cell Host Microbe 5:580–592 [DOI] [PubMed] [Google Scholar]
- 13. Laskowski R. A., Moss D. S., Thornton J. M. 1993. Main-chain bond lengths and bond angles in protein structures. J. Mol. Biol. 231:1049–1067 [DOI] [PubMed] [Google Scholar]
- 14. Linke D., Riess T., Autenrieth I. B., Lupas A., Kempf V. A. 2006. Trimeric autotransporter adhesins: variable structure, common function. Trends Microbiol. 14:264–270 [DOI] [PubMed] [Google Scholar]
- 15. Litt D. J., et al. 2004. Putative vaccine antigens from Neisseria meningitidis recognized by serum antibodies of young children convalescing after meningococcal disease. J. Infect. Dis. 190:1488–1497 [DOI] [PubMed] [Google Scholar]
- 16. Masignani V., et al. 2003. Vaccination against Neisseria meningitidis using three variants of the lipoprotein GNA1870. J. Exp. Med. 197:789–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Meng G., St. Geme J. W., III, Waksman G. 2008. Repetitive architecture of the Haemophilus influenzae Hia trimeric autotransporter. J. Mol. Biol. 384:824–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Meng G., Surana N. K., St. Geme J. W., III, Waksman G. 2006. Structure of the outer membrane translocator domain of the Haemophilus influenzae Hia trimeric autotransporter. EMBO J. 25:2297–2304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Moe G. R., Tan S., Granoff D. M. 1999. Differences in surface expression of NspA among Neisseria meningitidis group B strains. Infect. Immun. 67:5664–5675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Peak I. R., Srikhanta Y., Dieckelmann M., Moxon E. R., Jennings M. P. 2000. Identification and characterisation of a novel conserved outer membrane protein from Neisseria meningitidis. FEMS Immunol. Med. Microbiol. 28:329–334 [DOI] [PubMed] [Google Scholar]
- 21. Pizza M., et al. 2000. Identification of vaccine candidates against serogroup B meningococcus by whole-genome sequencing. Science 287:1816–1820 [DOI] [PubMed] [Google Scholar]
- 22. Riess T., et al. 2004. Bartonella adhesin a mediates a proangiogenic host cell response. J. Exp. Med. 200:1267–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Roggenkamp A., et al. 2003. Molecular analysis of transport and oligomerization of the Yersinia enterocolitica adhesin YadA. J. Bacteriol. 185:3735–3744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Scarselli M., et al. 2006. Neisseria meningitidis NhhA is a multifunctional trimeric autotransporter adhesin. Mol. Microbiol. 61:631–644 [DOI] [PubMed] [Google Scholar]
- 25. Schütz M., et al. 2010. Trimer stability of YadA is critical for virulence of Yersinia enterocolitica. Infect. Immun. 78:2677–2690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Serruto D., et al. 2010. Neisseria meningitidis GNA2132, a heparin-binding protein that induces protective immunity in humans. Proc. Natl. Acad. Sci. U. S. A. 107:3770–3775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Serruto D., et al. 2009. HadA is an atypical new multifunctional trimeric coiled-coil adhesin of Haemophilus influenzae biogroup aegyptius, which promotes entry into host cells. Cell Microbiol. 11:1044–1063 [DOI] [PubMed] [Google Scholar]
- 28. Sjölinder H., Eriksson J., Maudsdotter L., Aro H., Jonsson A. B. 2008. Meningococcal outer membrane protein NhhA is essential for colonization and disease by preventing phagocytosis and complement attack. Infect. Immun. 76:5412–5420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. St. Geme J. W., III, Cutter D. 2000. The Haemophilus influenzae Hia adhesin is an autotransporter protein that remains uncleaved at the C terminus and fully cell associated. J. Bacteriol. 182:6005–6013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Surana N. K., Cutter D., Barenkamp S. J., St. Geme J. W., III 2004. The Haemophilus influenzae Hia autotransporter contains an unusually short trimeric translocator domain. J. Biol. Chem. 279:14679–14685 [DOI] [PubMed] [Google Scholar]
- 31. Virji M. 2009. Pathogenic neisseriae: surface modulation, pathogenesis and infection control. Nat. Rev. Microbiol. 7:274–286 [DOI] [PubMed] [Google Scholar]
- 32. Volokhina E. B., et al. 2011. Role of the periplasmic chaperones Skp, SurA, and DegQ in outer membrane protein biogenesis in Neisseria meningitidis. J. Bacteriol. 193:1612–1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Weynants V. E., et al. 2007. Additive and synergistic bactericidal activity of antibodies directed against minor outer membrane proteins of Neisseria meningitidis. Infect. Immun. 75:5434–5442 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.