Abstract
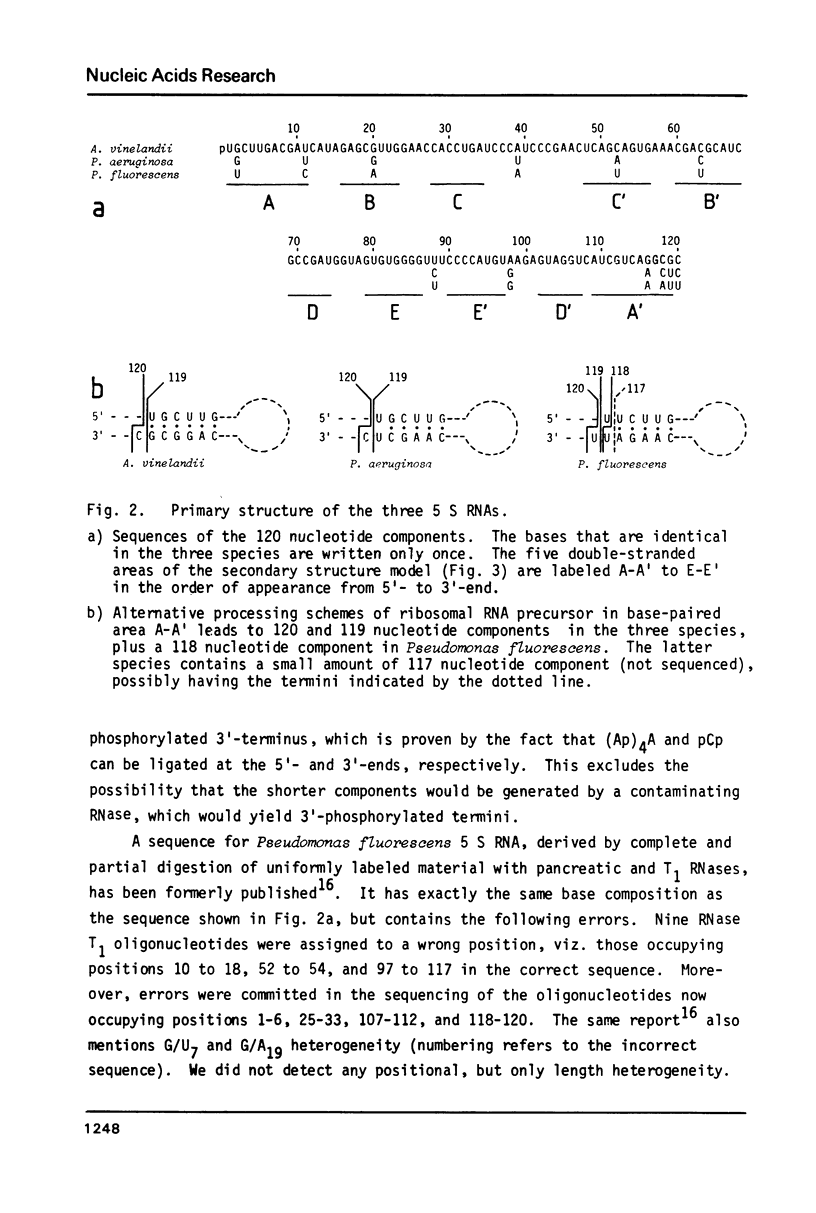
Recently published alignments of available 5 S rRNA sequences have shown that a rigid base pairing pattern, pointing to the existence of a universal five-helix secondary structure for all 5 S RNAs, can be superimposed on such alignments. For a few species, the alignment and the base pairing pattern show distortions with respect to the large majority of sequences. Their 5 S RNAs may form exceptional secondary structures, or there may just be errors in the published sequences. We have examined such a case, Pseudomonas fluorescens, and found the sequence to be in error. The corrected sequence, as well as those of the related species Azotobacter vinelandii and Pseudomonas aeruginosa, fit perfectly in the 5 S RNA sequence alignment and in the five-helix secondary structure model. There exists comparative evidence for the frequent presence of non-standard base pairs at several points of the 5 S RNA secondary structure.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Böhm S., Fabian H., Welfle H. Universal structural features of prokaryotic and eukaryotic ribosomal 5S RNA derived from comparative analysis of their sequences. Acta Biol Med Ger. 1982;41(1):1–16. [PubMed] [Google Scholar]
- Crick F. H. Codon--anticodon pairing: the wobble hypothesis. J Mol Biol. 1966 Aug;19(2):548–555. doi: 10.1016/s0022-2836(66)80022-0. [DOI] [PubMed] [Google Scholar]
- De Wachter R., Chen M. W., Vandenberghe A. Conservation of secondary structure in 5 S ribosomal RNA: a uniform model for eukaryotic, eubacterial, archaebacterial and organelle sequences is energetically favourable. Biochimie. 1982 May;64(5):311–329. doi: 10.1016/s0300-9084(82)80436-7. [DOI] [PubMed] [Google Scholar]
- Delihas N., Andersen J. Generalized structures of the 5S ribosomal RNAs. Nucleic Acids Res. 1982 Nov 25;10(22):7323–7344. doi: 10.1093/nar/10.22.7323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douthwaite S., Garrett R. A. Secondary structure of prokaryotic 5S ribosomal ribonucleic acids: a study with ribonucleases. Biochemistry. 1981 Dec 8;20(25):7301–7307. doi: 10.1021/bi00528a039. [DOI] [PubMed] [Google Scholar]
- DuBuy B., Weissman S. M. Nucleotide sequence of Pseudomonas fluorescens 5 S ribonucleic acid. J Biol Chem. 1971 Feb 10;246(3):747–761. [PubMed] [Google Scholar]
- Erdmann V. A., Huysmans E., Vandenberghe A., De Wachter R. Collection of published 5S and 5.8S ribosomal RNA sequences. Nucleic Acids Res. 1983 Jan 11;11(1):r105–r133. [PMC free article] [PubMed] [Google Scholar]
- FRESCO J. R., ALBERTS B. M., DOTY P. Some molecular details of the secondary structure of ribonucleic acid. Nature. 1960 Oct 8;188:98–101. doi: 10.1038/188098a0. [DOI] [PubMed] [Google Scholar]
- Fang B. L., De Baere R., Vandenberghe A., De Wachter R. Sequences of three molluscan 5 S ribosomal RNAs confirm the validity of a dynamic secondary structure model. Nucleic Acids Res. 1982 Aug 11;10(15):4679–4685. doi: 10.1093/nar/10.15.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox G. E., Stackebrandt E., Hespell R. B., Gibson J., Maniloff J., Dyer T. A., Wolfe R. S., Balch W. E., Tanner R. S., Magrum L. J. The phylogeny of prokaryotes. Science. 1980 Jul 25;209(4455):457–463. doi: 10.1126/science.6771870. [DOI] [PubMed] [Google Scholar]
- Fox G. E., Woese C. R. 5S RNA secondary structure. Nature. 1975 Aug 7;256(5517):505–507. doi: 10.1038/256505a0. [DOI] [PubMed] [Google Scholar]
- Garrett R. A., Olesen S. O. Structure of eukaryotic 5S ribonucleic acid: a study of Saccharomyces cerevisiae 5S ribonucleic acid with ribonucleases. Biochemistry. 1982 Sep 14;21(19):4823–4830. doi: 10.1021/bi00262a047. [DOI] [PubMed] [Google Scholar]
- Hancock J., Wagner R. A structural model of 5S RNA from E. coli based on intramolecular crosslinking evidence. Nucleic Acids Res. 1982 Feb 25;10(4):1257–1269. doi: 10.1093/nar/10.4.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori H., Osawa S. Evolutionary change in 5S RNA secondary structure and a phylogenic tree of 54 5S RNA species. Proc Natl Acad Sci U S A. 1979 Jan;76(1):381–385. doi: 10.1073/pnas.76.1.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori H., Osawa S., Iwabuchi M. The nucleotide sequence of 5S rRNA from a cellular slime mold Dictyostelium discoideum. Nucleic Acids Res. 1980 Dec 11;8(23):5535–5539. doi: 10.1093/nar/8.23.5535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan B. R., Galling G., Jourdan R. Sequence and conformation of 5 S RNA from Chlorella cytoplasmic ribosomes: comparison with other 5 S RNA molecules. J Mol Biol. 1974 Aug 5;87(2):205–225. doi: 10.1016/0022-2836(74)90144-2. [DOI] [PubMed] [Google Scholar]
- Kim S. H., Suddath F. L., Quigley G. J., McPherson A., Sussman J. L., Wang A. H., Seeman N. C., Rich A. Three-dimensional tertiary structure of yeast phenylalanine transfer RNA. Science. 1974 Aug 2;185(4149):435–440. doi: 10.1126/science.185.4149.435. [DOI] [PubMed] [Google Scholar]
- Kurland C. G. The requirements for specific sRNA binding by ribosomes. J Mol Biol. 1966 Jun;18(1):90–108. doi: 10.1016/s0022-2836(66)80079-7. [DOI] [PubMed] [Google Scholar]
- Küntzel H., Heidrich M., Piechulla B. Phylogenetic tree derived from bacterial, cytosol and organelle 5S rRNA sequences. Nucleic Acids Res. 1981 Mar 25;9(6):1451–1461. doi: 10.1093/nar/9.6.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luehrsen K. R., Fox G. E. Secondary structure of eukaryotic cytoplasmic 5S ribosomal RNA. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2150–2154. doi: 10.1073/pnas.78.4.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazar R. N., Matheson A. T. Nucleotide sequence of Thermus aquaticus ribosomal 5 S ribonucleic acid. Sequence homologies in thermophilic organisms. J Biol Chem. 1977 Jun 25;252(12):4256–4261. [PubMed] [Google Scholar]
- Ninio J. Prediction of pairing schemes in RNA molecules-loop contributions and energy of wobble and non-wobble pairs. Biochimie. 1979;61(10):1133–1150. doi: 10.1016/s0300-9084(80)80227-6. [DOI] [PubMed] [Google Scholar]
- Nishikawa K., Takemura S. Nucleotide sequence of 5 S RNA from Torulopsis utilis. FEBS Lett. 1974 Mar 15;40(1):106–109. doi: 10.1016/0014-5793(74)80904-x. [DOI] [PubMed] [Google Scholar]
- Peattie D. A. Direct chemical method for sequencing RNA. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1760–1764. doi: 10.1073/pnas.76.4.1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieler T., Erdmann V. A. Three-dimensional structural model of eubacterial 5S RNA that has functional implications. Proc Natl Acad Sci U S A. 1982 Aug;79(15):4599–4603. doi: 10.1073/pnas.79.15.4599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogin S. J., Sogin M. L., Woese C. R. Phylogenetic measurement in procaryotes by primary structural characterization. J Mol Evol. 1971;1(1):173–184. [PubMed] [Google Scholar]
- Sweeney W. V. Proton magnetic resonance studies of Azotobacter vinelandii ferredoxin I. Evidence for a difference in coordination of the 3Fe centers in azotobacter vinelandii ferredoxin I and desulfovibrio gigas ferredoxin II. J Biol Chem. 1981 Dec 10;256(23):12222–12227. [PubMed] [Google Scholar]
- Topal M. D., Fresco J. R. Complementary base pairing and the origin of substitution mutations. Nature. 1976 Sep 23;263(5575):285–289. doi: 10.1038/263285a0. [DOI] [PubMed] [Google Scholar]
- Traub W., Sussman J. L. Adenine-guanine base pairing ribosomal RNA. Nucleic Acids Res. 1982 Apr 24;10(8):2701–2708. doi: 10.1093/nar/10.8.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigne R., Jordan B. R. Partial enzyme digestion studies on Escherichia coli, Pseudomonas, Chlorella, Drosophila, HeLa and yeast 5S RNAs support a general class of 5S RNA models. J Mol Evol. 1977 Sep 20;10(1):77–86. doi: 10.1007/BF01796136. [DOI] [PubMed] [Google Scholar]