Abstract
Anaplasma phagocytophilum is an obligate intracellular bacterium that invades neutrophils to cause the emerging infectious disease human granulocytic anaplasmosis. A. phagocytophilum undergoes a biphasic developmental cycle, transitioning between an infectious dense-cored cell (DC) and a noninfectious reticulate cell (RC). To gain insights into the organism's biology and pathogenesis during human myeloid cell infection, we conducted proteomic analyses on A. phagocytophilum organisms purified from HL-60 cells. A total of 324 proteins were unambiguously identified, thereby verifying 23.7% of the predicted A. phagocytophilum proteome. Fifty-three identified proteins had been previously annotated as hypothetical or conserved hypothetical. The second most abundant gene product, after the well-studied major surface protein 2 (P44), was the hitherto hypothetical protein APH_1235. APH_1235 homologs are found in other Anaplasma and Ehrlichia species but not in other bacteria. The aph_1235 RNA level is increased 70-fold in the DC form relative to that in the RC form. Transcriptional upregulation of and our ability to detect APH_1235 correlate with RC to DC transition, DC exit from host cells, and subsequent DC binding and entry during the next round of infection. Immunoelectron microscopy pronouncedly detects APH_1235 on DC organisms, while detection on RC bacteria minimally, at best, exceeds background. This work represents an extensive study of the A. phagocytophilum proteome, discerns the complement of proteins that is generated during survival within human myeloid cells, and identifies APH_1235 as the first known protein that is pronouncedly upregulated on the infectious DC form.
INTRODUCTION
Human granulocytic anaplasmosis (HGA) is an emerging and potentially deadly tick-borne disease (44, 53). HGA is an acute febrile illness, the clinical manifestations of which range from subclinical infection to severe disease, including death. Nonspecific symptoms include fever, chills, headache, malaise, and myalgia. Severe complications include prolonged fever, shock, leukopenia, thrombocytopenia, high levels of C-reactive protein and hepatic transaminases, pneumonitis, acute renal failure, and hemorrhages. Immunocompromised and elderly individuals are at greatest risk for fatal opportunistic infections that can be associated with HGA (53). The causative agent of HGA is Anaplasma phagocytophilum, an obligate intracellular bacterium that displays an unusual tropism for granulocytes in humans and several domestic and wild reservoir animal hosts. The annotated genome of the A. phagocytophilum HZ strain, a human isolate, is available (20). The bacterium can be cultivated in the promyelocytic cell line HL-60 (14), as well as other cell lines (15, 18, 37, 53), which enables it to be grown in large quantities and has permitted in vitro modeling of infection. Following cellular adhesion, A. phagocytophilum promotes its own uptake into a host cell-derived vacuole that it remodels into a protective safe haven in which it remains and replicates (44, 53).
A. phagocytophilum exhibits a biphasic developmental cycle in which it transitions between two morphologically distinct forms that execute different pathobiological roles (17, 36–38, 43, 45, 55, 58). Dense-cored cell (DC) organisms have electron-dense nucleoids, are spheroid, are ∼0.8 μm in diameter, and have outer membranes that are pronouncedly ruffled and often appear distended from the protoplast. Compared to the DC form, reticulate cell (RC) bacteria have electron-lucent nucleoids, are pleomorphic, are ∼0.7 by 1.0 μm, and have outer membranes that appear smooth to slightly ruffled and are tightly associated with the protoplast (55). The DC form is the adherent and infectious form, but it does not replicate. The RC stage replicates within the A. phagocytophilum-occupied vacuole (ApV) by binary fission, but it is nonadherent and noninfectious. We previously characterized A. phagocytophilum biphasic development over the course of HL-60 cell infection (55). DC organisms adhere to the host cell surface and facilitate their own uptake. Between 4 and 8 h, DC bacteria transition to the RC form, which replicates. By 24 h, the RC begin to transition to the DC form. The majority of RC organisms develop into DC bacteria between 28 and 32 h, with the remaining RC population transitioning to the DC form by 36 h. DC organisms either extrude from or lyse the host cell, after which the released bacteria initiate new waves of infection between 32 and 36 h. Studies designed to correlate differential expression of A. phagocytophilum factors involved in adhesion, invasion, and/or establishing infection by the DC form would benefit from identification of a DC-specific marker.
During intracellular replication, the pathogen subverts the microbial killing capacity of its host cell, remodels the ApV membrane with bacterial proteins, manipulates host cell membrane traffic to divert recycling endosome traffic to the ApV, alters host cell signaling, and delays host cell apoptosis (11, 12, 21–23, 44, 51). While the molecular bases for these strategies are beginning to be elucidated, a comprehensive profile of the proteins expressed by A. phagocytophilum during myeloid host cell infection would directly reveal proteins that are expressed during and are presumably critical for facilitating intracellular survival and altering host cell functions. Such work would also validate the genome annotation by confirming that predicted A. phagocytophilum genes encode bona fide proteins.
In this study, we developed a discontinuous gradient centrifugation method to purify A. phagocytophilum organisms. The isolated bacteria were subjected to multidimensional protein identification technology (MuDPIT) analysis using two-dimensional nanoAcquity ultraperformance liquid chromatography (2D-nanoLC) online with tandem mass spectrometry (MS-MS). Sequential analyses of NP-40- and urea-soluble bacterial fractions identified a total of 324 A. phagocytophilum proteins, including 53 hitherto hypothetical proteins. Taken together, these analyses assign 23.7% of the theoretical A. phagocytophilum proteome. The hitherto hypothetical protein APH_1235 is the second most abundant protein after the major surface protein 2 (P44) paralog, Msp2 (P44)-23. Because of its abundance and lack of characterization, we examined the differential transcriptional and protein expression profiles of APH_1235. APH_1235 is an Anaplasma and Ehrlichia species-specific protein, is considerably upregulated during late-stage A. phagocytophilum infection of HL-60 cells, and is pronouncedly expressed by DC organisms. This study offers insight into the A. phagocytophilum infection cycle and identifies APH_1235 as a marker for A. phagocytophilum organisms that are transitioning to the DC form as well as for the DC form itself.
MATERIALS AND METHODS
Cultivation of A. phagocytophilum.
The human promyelocytic cell line HL-60 (American Type Culture Collection [ATCC]; Manassas, VA; ATCC code CCL-240) and A. phagocytophilum (NCH-1 strain)-infected HL-60 were cultivated in Iscove's modified Dulbecco's medium supplemented with 10% fetal bovine serum (FBS) (IMDM-10; Invitrogen, Carlsbad, CA) at 37°C in a humidified incubator in the presence of 5% CO2.
Discontinuous density gradient purification of A. phagocytophilum.
A quantity of 1 × 109 A. phagocytophilum-infected (∼85%) HL-60 cells with high bacterial loads and uncompromised plasma membranes (unable to take up trypan blue) were pelleted at 500 × g for 10 min at 4°C. The supernatant containing naturally liberated host cell-free A. phagocytophilum organisms was centrifuged at 3,500 × g at 4°C for 15 min, after which the bacterial cell pellet was resuspended in 5 ml of chilled isotonic buffer (0.25 M sucrose,10 mM HEPES, 10 mM KCl, 1.5 mM MgCl2, pH 7.0) and saved on ice. The pelleted infected HL-60 cells were washed with ice-cold phosphate-buffered saline (PBS) and allowed to swell for 10 min on ice in hypotonic buffer (10 mM HEPES, 10 mM KCl, 1.5 mM MgCl2, pH 7.0) at a concentration of 5 × 107 cells ml−1. The swelled cells were homogenized by ≥20 strokes in a 7-ml Dounce homogenizer (Gerresheimer Kimble Chase LLC, Vineland, NJ) equipped with a tight-fitting pestle. Aliquots (7 ml) of swelled cells were processed, one at a time, and then all of them were combined and placed on ice. To return the liberated A. phagocytophilum organisms to an isosmotic state, an 0.33 volume of chilled hypertonic buffer (1 M sucrose, 10 mM HEPES, 10 mM KCl, 1.5 mM MgCl2, pH 7.0) was added.
The A. phagocytophilum cell pellet saved from the culture media was resuspended in isotonic buffer and Dounce homogenized for 10 strokes to break apart clumps of bacteria and host cellular debris. Both A. phagocytophilum homogenates were combined and distributed among several aliquots, the volumes of which corresponded to volumes that originally contained 1 × 108 infected HL-60 cells at the swelling step. Each homogenate aliquot was brought up to 45 ml with isotonic buffer. HL-60 cell nuclei and cellular debris were removed by centrifugation at 1,000 × g for 10 min. The supernatant was centrifuged at 4,500 × g for 15 min at 4°C to pellet A. phagocytophilum organisms. Bacterial cell pellets derived from 5 × 108 infected HL-60 cells were resuspended in 6 ml of chilled isotonic buffer and treated with DNase I (Roche Applied Science, Indianapolis, IN) at 250 μg ml−1 at 37°C for 30 min.
The DNase I-treated sample was overlaid on a discontinuous Optiprep (60% [wt/vol] iodixanol in water; Sigma-Aldrich, St. Louis, MO) gradient consisting of layers of 30% (wt/vol in isotonic buffer), 25%, 20%, 17.5%, 15%, and 12.5% Optiprep at volumes of 6 ml, 5 ml, 5 ml, 3.5 ml, 3.5 ml, and 3.5 ml, respectively. The sample was fractionated by discontinuous gradient centrifugation at 87,000 × g for 75 min at 4°C in a Sorvall WX Ultra series ultracentrifuge (Thermoscientific, Asheville, NC) using a Sorvall Surespin 630/36 swinging bucket rotor. Acceleration and deceleration rates were set to 8 and 1, respectively. The resulting band at the interface of the 25% and 30% layers, which contained A. phagocytophilum organisms, was aspirated and transferred in 1-ml aliquots. The purified bacterial samples were diluted 1:1 with SPG buffer (0.22 M sucrose, 3.7 mM KH2PO4, 7.0 mM K2HPO4, 5 mM l-glutamine, pH 7.0), inverted several times, and spun at 15,000 × g for 10 min at 4°C. A. phagocytophilum pellets were recombined in a final volume of 500 μl of SPG buffer per bacteria that originated from 5 × 108 infected HL-60 cells. Aliquots were removed and processed for transmission electron microscopy. The remaining purified A. phagocytophilum sample was stored at −80°C until use.
2D-nanoLC/tandem MS protein analysis of RIPA buffer-soluble A. phagocytophilum proteins.
Density gradient-purified A. phagocytophilum organisms derived from infected (6.0 × 108 cells ml−1) HL-60 cells were spun at 15,000 × g for 10 min. After the supernatant was removed, the resulting pellet was resuspended in 200 μl ice-cold radioimmunoprecipitation assay (RIPA) buffer (25 mM Tris-HCl [pH 7.6], 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 1 mM sodium orthovanadate, 1 mM sodium fluoride, 65 mM dithiothreitol [DTT] containing complete protease inhibitor cocktail [Roche Applied Science]) and incubated for 45 min on ice. The lysate was centrifuged at 10,000 × g for 10 min at 4°C. The insoluble pellet was retained and processed as described below. The protein concentration of the soluble fraction was determined by the Bradford assay (4).
Unless otherwise stated, all buffers were made with LC/MS grade solvents (Fisher Chemical, Fairlawn, NJ). A 50-μg portion of RIPA-soluble protein was purified using the OrgoSol DetergentOUT (Micro) detergent removal system (G-Biosciences, St. Louis, MO) according to the manufacturer's instructions, which resulted in the sample being reconstituted in a proprietary Tris-base buffer (pH 7.5). A DTT/EDTA stock solution was freshly added to achieve a final concentration of 100 mM and 5 mM, respectively. Reduction of disulfide bonds was performed at 90°C for 30 min. Mass spectrometry grade trypsin (Promega, Madison, WI) was added to a final 1:100 enzyme-protein ratio. The sample was vortexed, spun briefly, and incubated at 37°C overnight. The digested sample was dried within a speed vacuum and stored dry at −20°C.
The digest sample was reconstituted in 30 μl of 200 mM ammonium formate, confirmed at pH 10.0, and placed in the sample manager of a Waters (Milford, MA) 2D-nanoLC system. Peptide separation was accomplished via reversed-phase pH 10.0/reversed-phase pH 2.0 multidimensional separation. A 3-μl portion of the digest sample was injected onto the first-dimension column (BEH130 C18 medium; Waters), with the peptides step-eluted using ammonium formate (20 mM)-modified water and neat acetonitrile mobile phases, optimized to distribute the total peptide mass evenly into five fractions. After column injection, each fraction was mixed online with a 4-fold excess flow of 0.1% formic acid, allowing the majority of peptides to be recaptured onto a reversed-phase trap column (Symmetry C18 medium; Waters) at pH 2.0. Following trapping, peptides per first-dimension fraction were separated using a 75-μm diameter, 15-cm-long capillary reversed-phase column (BEH130 C18 medium; Waters) with gradient elution from a 7% to a 35% water-acetonitrile ratio (0.1% formic acid modified) in 60 min. Eluting peptides were transferred via online nano-scale electrospray through a 10-μm Pico-tip emitter (New Objective, Woburn, MA) into a Synapt HDMS orthogonal quadrupole/time of flight tandem mass spectrometer (Waters). Data were collected using a data-independent acquisition algorithm (13), whereby a mass spectrum of all eluting precursor ions was collected followed by collection of a mass spectrum of all product (i.e., fragmented) ions to all eluting precursor ions. The precursor and product ion acquisition scans were repeated every second. Every 30 s, the instrument acquired a mass spectrum of an external reference of glu1-fibrinopeptide to use for subsequent temporal mass accuracy correction. Analysis was performed on two technical replicates.
The resulting data were processed using Protein Lynx Global Surveyor (PLGS) software, v2.4 (Waters), as described in detail elsewhere (31). Briefly, charge-state and isotope reduction steps were performed for all precursor and product ion mass spectra, collapsing the data into a mono-isotopic, singly charged mass-to-charge value (MH+) per detected ion. MH+ values were mass corrected per the external reference data. Product ions were aligned via their retention time chromatographic profile with their precursor ion measures, which produced a fragment ion spectrum for each peptide ion analyzed. Processed data from the five 2D-nanoLC fractions per technical replicate were merged and superimposed. Data were then searched against an A. phagocytophilum-specific FASTA database (RefSeq, downloaded in February 2010) and its reversed sequences as a decoy database. Search parameters required a minimum precursor ion intensity of 500 counts, two or more peptide sequences per protein, and a minimum of seven matching fragment ions. Trypsin selectivity was specified, allowing for one missed cleavage event and variable methionine oxidation. Identified proteins were then filtered to an estimated 1% false-discovery rate based on the decoy-database method.
2D-nanoLC/tandem MS protein analysis of radioimmunoprecipitation assay (RIPA) buffer-insoluble A. phagocytophilum proteins.
The RIPA buffer-insoluble pellet was resuspended in 0.5% (vol/vol) SDS, 100 mM Tris-HCl, pH 7.0, that had been preheated to 95°C, and incubation at 95°C continued for 5 min. The sample was placed on ice for 5 min, after which it was mixed with 2.5 volumes of fresh urea lysis buffer (7 M urea, 2 M thiourea, 65 mM DTT). The mixture was mixed by end-over-end rotation at 4°C for 1 h. The sample was spun at 15,000 × g for 15 min at 4°C with no observed pellet discerned. The Bradford assay was used to determine the urea solubilized sample's protein concentration (4). The sample was diluted with urea lysis buffer to a final concentration of 0.5 μg/μl in 50 μl, after which it was submitted to the Virginia Commonwealth University (VCU) Department of Chemistry's Mass Spectrometry Core Facility. The sample was diluted to 250 μl with 100 mM ammonium bicarbonate to reduce the concentration of urea and detergents. The sample was reduced with 5 μl of freshly made 10 mM DTT in 0.1 M ammonium bicarbonate at room temperature for 30 min. Next, the sample was alkylated with 5 μl freshly made 50 mM iodoacetamide in 0.1 M ammonium bicarbonate at room temperature for 30 min. The sample was digested with 1 μg trypsin overnight twice and then quenched with 5% (vol/vol) glacial acetic acid. Half of the sample was desalted through a column self-packed with Oligo R3 packing material (Applied Biosystems, Foster City, CA). The final elution was dried down and then reconstituted with 20 μl 0.1% formic acid.
The LC-MS system consisted of a Thermo Electron LTQ-Orbitrap hybrid mass spectrometer system (Thermo Electron Corporation) with a nanospray ion source interfaced to a Waters NanoAcquity C18 reversed-phase capillary column. A 2-μg portion of the final solution was injected onto an SCX trap column placed in line with the C18 trap column on the NanoAcquity. Peptides that were not bound by the SCX column were retained on the C18 trap column, washed, and eluted to the C18 analytical column. Peptides were eluted from the analytical column into the mass spectrometer by an acetonitrile-0.1 M acetic acid gradient at a flow rate of 0.4 μl/min over 60 min. Subsequently, 5 μl of 1 mM ammonium acetate was injected onto the trap column to elute some of the less strongly bound peptides to the trap and analytical column, and a second 60-min gradient with mass spectrometric analysis was performed. This process was continued in a stepwise fashion with increasing concentrations of ammonium acetate, namely 2 mM, 5 mM 10 mM, 25 mM, 50 mM, 100 mM, and 1 M ammonium acetate, resulting in a total of nine fractions. The nanospray ion source was operated at 3.5 kV. The fractions were analyzed using data-dependent analysis acquiring full scan mass spectra to determine peptide molecular weights and product ion spectra of the four most intense precursor ions to determine amino acid sequence in sequential scans. This mode of analysis produces approximately 10,000 CAD spectra of ions ranging in abundance over several orders of magnitude, though not all CAD spectra are derived from peptides. The data were analyzed by database searching with Proteome Discoverer v.1.2 (Thermoscientific, West Palm Beach, FL) using the Sequest search algorithm against an Anaplasma phagocytophilum-specific FASTA database (RefSeq, downloaded in May 2011) and IPI Human databases (Refseq, downloaded in May 2011). Oxidation of methionine was allowed as a variable modification, while carbamidomethylation of the cysteine was used as a static modification. Trypsin selectivity was specified, allowing two missed cleavages. Search results from Proteome Discoverer were then imported into Scaffold (Proteome Software, Portland, OR) for ease of visualization. A minimum of two peptides was required for protein identification.
Electron microscopy.
A. phagocytophilum-infected HL-60 cells or aliquots of host cell-free A. phagocytophilum organisms released by syringe lysis or Dounce homogenization or following density gradient purification were analyzed by transmission electron microscopy as described previously (55). Immunoelectron microscopy using anti-APH_1235 serum or mouse anti-Msp2 (P44) monoclonal antibody (MAb) 20B4 (a gift from J. Stephen Dumler of Johns Hopkins University, Baltimore, MD) (42, 46) at a 1:10 dilution was performed in the VCU Department of Neurobiology and Anatomy Microscopy Core Facility as reported (23).
Analyses of APH_1235 expression over the course of infection.
Host cell-free A. phagocytophilum DC organisms from infected HL-60 cells were isolated by sonication, which destroys host cells and RC organisms but leaves DC bacteria intact, followed by differential centrifugation, and synchronous infections were established as described previously (55). Indirect immunofluorescence microscopic examination of aliquots recovered at 24 h postinfection confirmed that ≥60% of HL-60 cells contained morulae (intravacuolar colonies of A. phagocytophilum) and that the mean number of morulae per cell was 2.8 ± 0.6. The infection time course proceeded for 36 h at 37°C in a humidified atmosphere of 5% CO2. At the appropriate time points, aliquots were removed and processed for RNA isolation or indirect immunofluorescence microscopy. For RNA isolation, 5 × 106 infected HL-60 cells were stored in 4 ml RNAlater (Ambion) at 4°C until use to prevent RNA degradation. Total RNA was isolated using the RNaqueous kit (Applied Biosystems/Ambion). Genomic DNA was removed by treating total RNA with 150 units of DNase I (Invitrogen) for 1 h at 37°C. DNase I was subsequently removed from the reaction using DNase inactivating reagent (Applied Biosystems/Ambion). A 1-μg portion of RNA was reverse transcribed into cDNA using the iScript cDNA synthesis kit (Bio-Rad, Hercules, CA). Quantitative PCR assays were performed using the CFX96 real-time PCR detection system (Bio-Rad). cDNA samples from each time point were analyzed in triplicate using Ssofast SYBR green supermix (Bio-Rad), gene-specific primers [aph_1235-232F (5′-CGTGAAGAAATTTCTACAGCTGTGCC-3′) and aph_1235-405R1 (5′-CTAACCTTGGGTCGATGCCACAAT-3′) or Ap16S-527F (5′-TGTAGGCGGTTCGGTAAGTTAAAG-3′) and Ap16S-753R (5′-GCACTCATCGTTTACAGCGTG-3′)], and 1 μl of cDNA in a 20-μl reaction volume. The thermal cycling conditions used were 98°C for 2 min, followed by 39 cycles of 98°C for 5 s and 55°C for 5 s. Relative transcript levels for each target were normalized to the transcript levels of the A. phagocytophilum 16S rRNA gene using the 2−ΔΔCT (Livak) method (34).
Recombinant APH_1235 expression and antiserum production.
A. phagocytophilum aph_1235 was PCR amplified using primers aph_1235-001F-ENTR (5′-CACCATGAAAGGAAAGTCAGATTCTGAAATACGTACG-3′; the underlined segment corresponds to a Gateway Entry vector compatible sequence) and aph_1235-405R2 (5′-CTAACCTTGGGTCGATGCCACAATTTTC-3′) and Platinum Pfx DNA polymerase (Invitrogen). The amplicon was cloned into a pENTR/TEV/D-TOPO vector (Invitrogen) as described previously (22, 23) to yield p1235-ENTR. Plasmid inserts were sequenced by the Northwestern University Center for Genetic Medicine Genomics Core Facility to verify sequence integrity and cloning junctions. Recombination of the candidate gene insert downstream of and in frame with the gene encoding glutathione S-transferase (GST) was achieved using the pDEST-15 vector (Invitrogen) as described previously (23). The reaction was transformed into Escherichia coli BL21-AI One Shot cells (Invitrogen). Transformant cultures were grown as described previously (23), and GST-fusion protein expression was induced by the addition of l-arabinose at a final concentration of 0.2% at 25°C overnight. Induced bacterial suspensions were harvested by centrifugation at 4,500 × g for 10 min at 4°C, resuspended in ice-cold PBS containing complete protease inhibitors (Roche), and sonicated in an ice-water bath. Lysates were pelleted by centrifugation at 10,000 × g for 10 min at 4°C. Insoluble GST fusion proteins were solubilized in Sarkosyl (16, 52). To facilitate glutathione-Sepharose affinity purification, the supernatants were adjusted to 3% Triton X-100, 1 mM calcium chloride, and 30 mM 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS) and vortexed (52). The solubilized proteins were batch purified using glutathione-Sepharose beads (GE Healthcare, Piscataway, NJ). Eluted GST-tagged fusion protein fractions were collected, fractionated by SDS-PAGE, and visualized by Coomassie brilliant blue staining. Eluted fractions were pooled, dialyzed with PBS, and concentrated by centrifugation through an Amicon Ultra-4 centrifugal filter with a 30-kDa cutoff (Millipore, Bedford, MA). The Bradford assay and/or estimation against a bovine serum albumin standard on an SDS-polyacrylamide gel was used to quantify concentrated GST-fusion protein preparations (4). N-terminally His-tagged APH_1235 (His-1235) was generated by amplifying aph_1235 nucleotides 1 to 246 using primers aph_1235-001F-ENTR and aph_1235-246R (5′-CTAAGAAATTTCTTCACGCATTGATTTCTTAAGCTCACTG −3′; the underlined segment corresponds to an engineered stop codon), cloning the PCR product into pENTR/TEV/D-TOPO, and subcloning the insert sequence into pDEST-17 (Invitrogen). His-1235 expression was induced by the addition of l-arabinose followed by purification using His-bind resin (Novagen EMD Chemicals, Gibbstown, NJ). Murine polyclonal antiserum was generated against purified GST-1235 as described previously (22).
Western blotting.
HL-60 cells were synchronously infected with A. phagocytophilum DC organisms (55), after which the host cells were lysed by repeated passage through a 27-gauge needle at either 16 h or 28 h postinfection to obtain bacterial populations consisting exclusively of the RC or RC and DC stages, respectively, followed by solubilization in RIPA buffer (55). RIPA buffer lysates of A. phagocytophilum-infected HL-60 cells, A. phagocytophilum organisms, and density gradient-purified A. phagocytophilum organisms were fractionated by SDS-polyacrylamide gel electrophoresis (PAGE), transferred to nitrocellulose, and screened using MAb 20B4 (26) or anti-human actin MAb (Sigma, St. Louis, MO) as described previously (6). Immunoreactivity of anti-GST-1235 (here referred to as anti-APH_1235) serum against His-1235 and native A. phagocytophilum APH_1235, as well as lack of recognition of immunogens and A. phagocytophilum whole-cell lysates by preimmune serum, was confirmed.
Spinning disk confocal microscopy.
To monitor differential expression of APH_1235 over the course of infection, aliquots of HL-60 cells synchronously infected with A. phagocytophilum per time point were cytospun onto glass slides at 70 × g for 2 min followed by fixation and permeabilization in ice-cold methanol for 30 s, after which they were stored at −20°C until needed. Slides were screened using mouse anti-APH_1235 at a 1:200 dilution for 30 min followed by rabbit anti-Msp2 (P44) at a 1:500 dilution. Secondary antibody screening, initial blocking and washing steps after each antibody incubation, and mounting of slides were performed as described previously (5). Slides were examined and images were acquired by spinning disc confocal microscopy using a BX51 microscope (Olympus, Center Valley, PA) affixed with an Olympus disk spinning unit and an Orca-R2 charge-coupled-device (CCD) camera (Hammamatsu, Japan). Images were processed using the Slidebook 5.0 software package (Intelligent Imaging Innovations, Denver, CO). To determine the percentage of morulae (A. phagocytophilum-occupied vacuoles) that harbored bacteria that were positive for APH_1235, the number of APH_1235-positive morulae was divided by the total number of morulae (as denoted by the detection of intravacuolar Msp2 [P44]) and the result was multiplied by 100.
RESULTS
Density gradient purification of A. phagocytophilum.
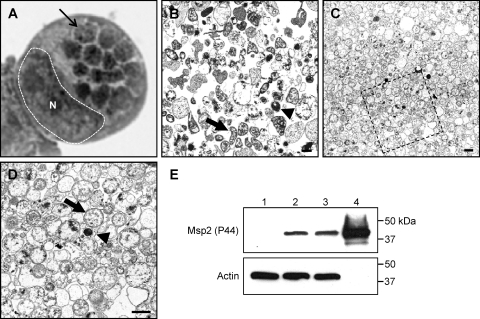
To maximize recovery of A. phagocytophilum proteins and minimize contamination with host cellular proteins, we purified Dounce homogenates of heavily infected (≥85%) HL-60 cells. Dounce homogenates from equal numbers of infected and uninfected control HL-60 cells were overlaid on a discontinuous Optiprep gradient. After ultracentrifugation, a large band found at the interface between the 25% and 30% layers present in the samples derived from infected but not those from uninfected host cells contained A. phagocytophilum organisms when visualized by transmission electron microscopy (Fig. 1 C and D). Bacterial yields were greatest when obtained from infected host cells that harbored many morulae each (Fig. 1A). The discontinuous gradient method yields a bacterial preparation that contains considerably less host cellular debris than A. phagocytophilum bacteria recovered by differential centrifugation following syringe lysis of infected HL-60 cells (Fig. 1B to D). The majority (∼85%) of purified A. phagocytophilum organisms were in the RC form. Western blot analysis of whole-cell lysates of A. phagocytophilum-infected HL-60 cells and A. phagocytophilum preparations obtained following syringe lysis or discontinuous gradient purification revealed enrichment for bacterial Msp2 (P44) and absence of host actin in the gradient-purified sample (Fig. 1E).
Fig. 1.
Dounce homogenization followed by discontinuous gradient centrifugation purifies A. phagocytophilum organisms while minimizing host cellular material contamination. (A) Light microscopic image of a Hema Fix-stained A. phagocytophilum-infected HL-60 cell that exhibits an infection load that is typical of the infected cells from which A. phagocytophilum organisms were purified for this study. A hatched line and an N demarcate the host cell nucleus, while a thin arrow denotes one of 14 morulae. (B) Transmission electron micrograph of A. phagocytophilum organisms and HL-60 cellular debris following syringe lysis and differential centrifugation to partially remove host cellular material. (C and D) Transmission electron micrograph of an A. phagocytophilum fraction obtained following centrifugation of a Dounce homogenate through a discontinuous Optiprep gradient. (D) Magnification of the region in panel C that is denoted by a hatched box. Thick arrows and arrowheads denote representative RC and DC organisms, respectively. Scale bars, 1 μm. (E) Western blot analyses demonstrating enrichment for A. phagocytophilum Msp2 (P44) by discontinuous gradient purification. Samples (10 μg) of whole-cell lysates of uninfected HL-60 cells (lane 1), A. phagocytophilum-infected HL-60 cells (lane 2), A. phagocytophilum organisms recovered following syringe lysis of infected HL-60 cells and differential centrifugation (lane 3), and A. phagocytophilum organisms recovered following Dounce homogenization and discontinuous density gradient centrifugation (lane 4) were resolved by SDS-PAGE, transferred to nitrocellulose, and screened with antibody against A. phagocytophilum Msp2 (P44) or human actin.
2D-nanoLC/tandem MS protein analysis.
Density gradient-pure A. phagocytophilum fractions were lysed in RIPA buffer, after which the soluble fraction was digested with trypsin and the insoluble fraction was saved. The digested soluble sample was separated by 2D-nanoLC and analyzed by data-independent tandem mass spectrometry. The resulting data were processed using PLGS software and searched against an A. phagocytophilum-specific database together with its reversed-sequence decoy database. Results were filtered to an estimated 1% false-discovery rate using matched sequences from the decoy database. The results included 87 A. phagocytophilum unique proteins with an average of 25 peptides per protein for a total of 2,160 unique peptides identified (see Table S1 in the supplemental material). Coverage of proteins ranged from 8.9 to 94.9%, with a greater than 95% confidence for each protein identification. Of the total amount of tryptic peptides detected (an estimated 26,903.2 fmol on-column), 51.1% was derived from Msp2 (P44) paralogs. Msp2 (P44) proteins constituted 13 of the 20 most abundant proteins, with Msp2 (P44)-23 being the most abundant of all detected proteins. Msp2 (P44)-23-derived peptides (4,141.3 fmol) represented 30.1% of all Msp2 (P44) peptides and 15.4% of total peptides. The uncharacterized protein, APH_1235, the peptides of which constituted 12.0% of all identified peptides, was the second most abundant protein.
To further our identification of the A. phagocytophilum proteome expressed during infection of myeloid host cells, we solubilized the RIPA buffer-insoluble pellet in urea lysis buffer and digested the solubilized sample with trypsin. We analyzed the resulting peptides by an orthogonal 2D-nanoLC method using ion-exchange instead of reversed-phase separation as the first dimension and an LTQ/Orbitrap-MS instrument that can provide enhanced identification of lesser abundant proteins, though with generally lower sequence coverage per protein. The resulting data were processed using the Sequest search algorithm against the annotated Anaplasma phagocytophilum proteome. Scaffolding was used to combine the nine SCX fractions into a single data set and was used to aid in visualization of this highly complex data set, as well as assigning a probability score to each protein identified, reflecting the likelihood that a given protein's assignment is correct. For the RIPA buffer-insoluble pellet, we identified 280 unique A. phagocytophilum proteins through 1,285 unique peptide assignments with a minimum of two peptides being assigned per protein (see Table S2 in the supplemental material). Protein coverage ranged from 1.8% to 65.9%. Forty-three of the proteins identified in the urea-soluble fraction were also identified in the RIPA buffer soluble fraction. Each of the 280 proteins that had two or more assigned peptides had a Sequest assigned probability score of 100%. Fifty-nine additional A. phagocytophilum proteins had single peptide hits and probability scores of 98 to 99% (data not shown).
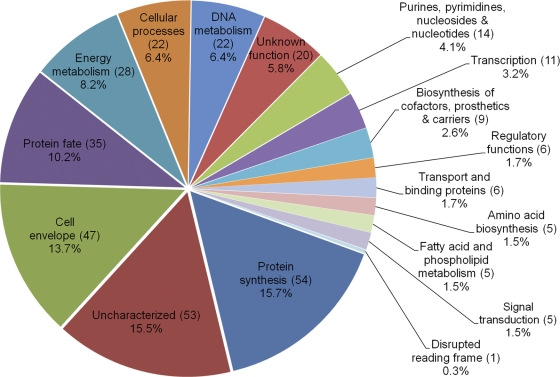
The two 2D-nanoLC-derived data sets identified a combined total of 324 proteins, which represents 23.7% of the annotated A. phagocytophilum proteome. A total of 28 different Msp2 (P44) paralogs were identified. The predicted functional roles to which these proteins are assigned are given in Tables S1 and S2 in the supplemental material, while the percentages of functional role categories represented by the identified proteins are presented in Fig. 2. Nineteen proteins are annotated as having more than one major role, which accounts for the total of 343 functional roles annotated in Fig. 2. Proteins categorized as being involved in protein synthesis, being localized to the cellular envelope, and determining protein fate, together with uncharacterized proteins, constitute 55.1% of detected proteins.
Fig. 2.
Graphic representation of A. phagocytophilum proteins expressed during HL-60 cell infection. The number of proteins identified for a given major role category was divided by the total number of proteins identified for all categories (343) times 100 to yield the percentage for each category. Uncharacterized refers to those proteins that were previously annotated as either hypothetical or conserved hypothetical by the J. Craig Venter Institute. The sum of all proteins per major role (343) is higher than the actual number of identified proteins (324) because 19 proteins are annotated as having more than one major role in the A. phagocytophilum proteome.
APH_1235 homologs in Anaplasma and Ehrlichia species.
Msp2 (P44)-23 and APH_1235 are the two most abundantly detected RIPA buffer-soluble proteins. Whereas the expression patterns and pathobiological roles of the A. phagocytophilum Msp2 (P44) paralogs and their homologs in other Anaplasma and Ehrlichia species have been extensively examined (2, 24, 27, 28, 32, 33, 35, 41, 42, 47–49, 61, 62), APH_1235 has yet to be studied. Therefore, we resolved to characterize APH_1235. APH_1235 is a 14.8-kDa protein that is 134 amino acids in length and has an isoelectric point of 5.3. A BLASTP (basic local alignment search tool for protein sequences; blast.ncbi.nlm.nih.gov/Blast.cgi) search revealed that it has homologs in Anaplasma and Ehrlichia species. A multiple sequence alignment using CLUSTAL W (54) determined that APH_1235 exhibits 28.9% identity, 31.1% strong similarity, and 17.8% weak similarity with its A. marginale and A. marginale subspecies centrale homologs and displays 16.9% identity, 19.1% strong similarity, and 14.0% weak similarity with its ehrlichial homologs (Fig. 3). With the exception of Microcoleus chthonoplastes, with which it exhibits 31.7% identity over just a 60-amino-acid stretch, APH_1235 hits no other protein in GenBank when it is the query sequence in a BLASTP search using default parameters.
Fig. 3.
Alignment of A. phagocytophilum APH_1235 with its homologs of other Anaplasma and Ehrlichia species. APH_1235 was aligned with its homologs from A. marginale, St. Maries strain (AM_1191), A. marginale subspecies centrale, Israel strain (ACIS_00207), Ehrlichia chaffeensis, Arkansas strain (ECH_0122), Ehrlichia canis, Jake strain (Ecaj_0073), and Ehrlichia ruminantium, Welgevonden strain (Erum_0730), using the CLUSTAL W algorithm (npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_clustalw.html) (54). Residues conserved among all six sequences are white with a black background. Residues conserved among four or five sequences are highlighted gray. Bold text denotes residues conserved only among Anaplasma species. Underlined residues are conserved only among Ehrlichia species. Among all six sequences: *, identical residues; colon(:), strongly similar residues; period (.), weakly similar residues.
Cloning and expression of recombinant APH_1235 and antibody production.
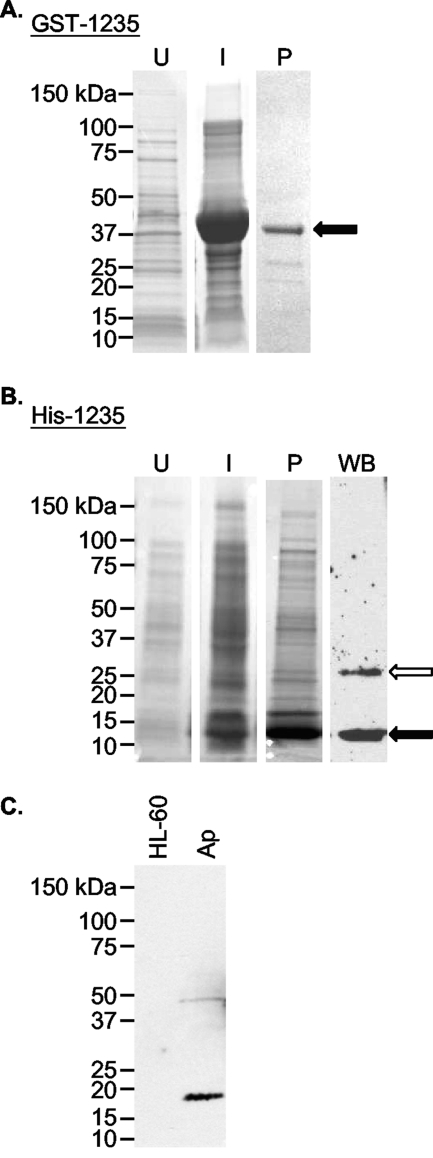
The aph_1235 coding region (14.8 kDa) was cloned and expressed in E. coli as an N-terminal GST-tagged fusion protein designated GST-1235 (Fig. 4 A). Nucleotides encoding amino acids 1 to 82 (9.1 kDa) were cloned and expressed as an N-terminally His-tagged fusion protein designated His-1235 (Fig. 4B). After glutathione-Sepharose and nickel affinity chromatography, purified GST-1235 and His-1235 appeared as 44.2- and 13.5-kDa bands, respectively, upon SDS-PAGE. GST-1235 was used to immunize mice. The resulting polyclonal antisera specifically bound to His-1235 and to what was presumably a His-1235 dimer (Fig. 4B). Anti-1235 recognized a protein that was slightly larger than the expected size for native APH_1235, but it did not bind any protein in an uninfected HL-60 cell lysate (Fig. 4C). The acidity of native APH_1235 likely accounts for its slower electrophoretic mobility (22, 23, 57). Preimmune mouse serum failed to recognize recombinant or native APH_1235 proteins (data not shown).
Fig. 4.
Generation of recombinant APH_1235 and antisera against APH_1235. (A and B) Whole-cell lysates of uninduced E. coli (lane U), E. coli induced (lane I) to express GST-1235 (A) or His-1235 (B), and GST-1235 (44.2 kDa) purified (lane P) by glutathione-Sepharose affinity chromatography (A) or His-1235 (13.5 kDa) purified by Ni2+ affinity chromatography (B) were separated by SDS-PAGE and stained with Coomassie blue. (B) Western blot (WB) analysis was performed using mouse anti-APH_1235 (raised against GST-1235) to detect His-1235 (lane WB). Black arrows denote GST-1235 (A) and His-1235 (B). The white arrow in panel B denotes what is presumably a His-1235 dimer. (C) Western blot analysis in which anti-APH_1235 was used to screen whole-cell lysates derived from uninfected HL-60 cells and A. phagocytophilum organisms (lane Ap).
aph_1235 transcription is pronouncedly upregulated between 28 and 36 h postinfection.
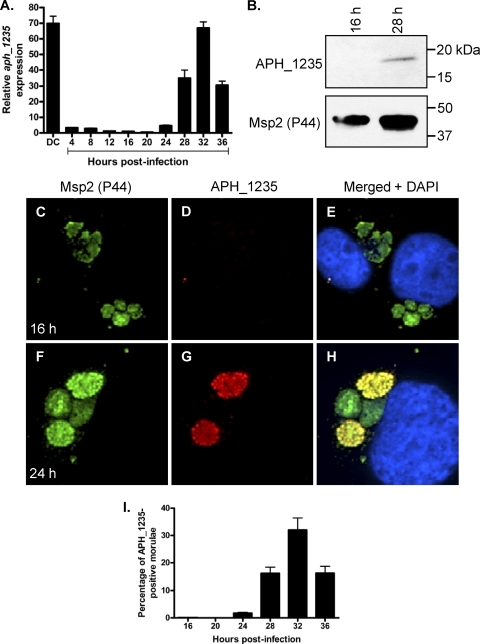
To determine the aph_1235 transcriptional expression profile over the course of A. phagocytophilum infection, we synchronously infected HL-60 cells with DC organisms and allowed the infection to proceed for 36 h, a time period that would allow for the bacteria to complete their biphasic developmental cycle and initiate a second round of infection (55). Quantitative reverse transcriptase PCR (qRT-PCR) with aph_1235-specific primers was performed on total RNA isolated from DC organisms used as the inoculum and from bacteria recovered at several postinfection time points. To determine the relative aph_1235 transcriptional pattern between RC and DC organisms, the gene expression results for each time point were calculated as the fold change in expression relative to expression at 16 h, a time point at which the A. phagocytophilum population consists exclusively of RC organisms (55). Low-level aph_1235 transcription was detected at 4 h, a time point by which the majority of A. phagocytophilum organisms entered HL-60 host cells (Fig. 5 A). The paucity of aph_1235 expression continued through 20 h, after which its expression exhibited a continual and pronounced increase that began at 24 h and continued until reaching a maximal 67.0 ± 3.9-fold increase at 32 h. The aph_1235 transcript level of DC organisms used as the inoculum was 69.8 ± 4.7-fold greater than those of RC organisms at 16 h.
Fig. 5.
APH_1235 is strongly expressed by DC organisms and between 28 and 36 h postinfection. HL-60 cells were synchronously infected with A. phagocytophilum. At the indicated postinfection time points, aliquots were removed and subjected to qRT-PCR (A), Western blot (B), and indirect immunofluorescent (C to I) analyses to monitor APH_1235 expression over the course of infection. (A) Total RNA was isolated from DC organisms used as the inoculum (DC) and from A. phagocytophilum-infected cells at several postinfection time points. qRT-PCR was performed using gene-specific primers. Relative aph_1235 transcript levels were normalized to A. phagocytophilum 16S rRNA gene transcript levels using the 2−ΔΔCT (Livak) method. To calculate the relative aph_1235 transcriptional pattern between RC and DC organisms, the gene expression results for each time point were calculated as the fold change in expression relative to expression at 16 h, a time point at which the A. phagocytophilum population consists exclusively of RC organisms. The results presented are the means and standard deviations of results for triplicate samples and are representative of two independent experiments that yielded similar results. (B) Anti-1235 serum recognized APH_1235 in a whole-cell lysate derived from A. phagocytophilum organisms obtained at 28 h but not 16 h. The blot was stripped and rescreened with antibody against Msp2 (P44). Results are representative of two experiments with similar results. (C to I) At 16, 20, 24, 28, 32, and 36 h postinfection, the cells were fixed and viewed by spinning disk confocal microscopy to determine immunoreactivity with antibodies against Msp2 (P44) (major bacterial surface protein; used to identify bacteria in panels C and F) and APH_1235 (D and G). Merged fluorescent images, including DAPI staining of host cell nuclei, are shown in panels E and H. (C to H) Representative images obtained at 16 (C to E) and 24 (F to H) h postinfection. (I) Percentages of morulae [based on the presence of Msp2 (P44)-positive A. phagocytophilum organisms] that are positive for APH_1235 staining of intravacuolar bacteria. The data are the means and standard deviations of results for two separate experiments. At least 869 Msp2 (P44)-positive ApVs were scored for APH_1235 for each time point.
APH_1235 is strongly expressed between 28 and 36 h postinfection.
We next assessed whether the aph_1235 transcriptional profile translates to late-stage APH_1235 protein expression. At 16 and 28 h postinfection, A. phagocytophilum populations consist exclusively of RC organisms or RC and DC bacteria, respectively (55). Western blot analysis of whole-cell lysates of A. phagocytophilum from infected HL-60 cells at 16 or 28 h detected APH_1235 at 28 h but not at 16 h (Fig. 5B). Indirect immunofluorescent microscopic examination of synchronously infected HL-60 cells revealed that 1.8 ± 0.4, 16.3 ± 3.0, 32.1 ± 6.2, and 16.4 ± 3.3 morulae contained APH_1235-positive organisms at 24, 28, 32, and 36 h, respectively (Fig. 5C to I). APH_1235 expression was undetectable at 16 and 20 h.
APH_1235 is expressed by A. phagocytophilum DC organisms.
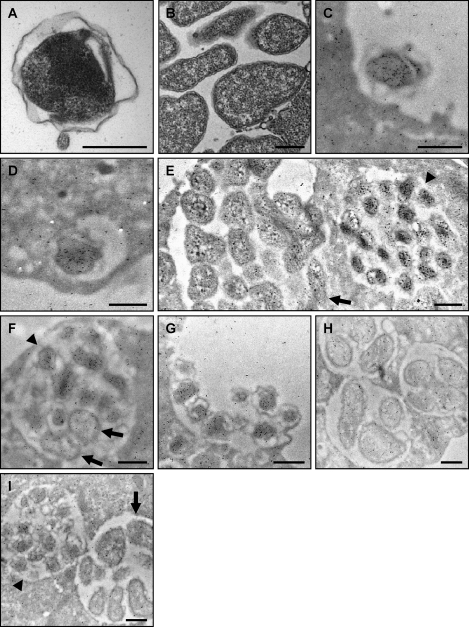
Because APH_1235 expression coincides with the period of A. phagocytophilum RC to DC transition, bacterial exit, and reinfection (55), we next employed immunoelectron microscopy to assess if DC and RC bacteria differentially express APH_1235. The fixation process that is requisite for immunoelectron microscopy partially compromises the clarities by which the characteristic distended and ruffled outer membrane of the A. phagocytophilum DC form and the more tightly associated outer membrane of the RC stage can be discerned (22). Accordingly, we have provided transmission electron micrographs (Fig. 6 A and B) that clearly present the distinctive differences between DC and RC outer membranes as a reference for viewing the immunoelectron micrographs (Fig. 6C to I). Considerable amounts of APH_1235 were detected on A. phagocytophilum DC organisms that were bound to the HL-60 cell surface (Fig. 6C), had internalized into a nascent host cell-derived vacuole (Fig. 6D), had transitioned to DC organisms postreplication (late-stage ApV; Fig. 6E and F), and were in the process of being released (Fig. 6G). The degree of anti-APH_1235-mediated immunogold labeling of DC organisms was more pronounced than the labeling observed for RC organisms (Fig. 6E, F, and H), which was comparable to the background labeling observed for the host cell cytoplasm and extracellular milieu (Fig. 6C to I). Figure 6F presents a late-stage morula in which A. phagocytophilum organisms are transitioning from the RC to DC form. Most of the bacteria have converted to the DC form, while two (denoted by arrows) are still in the RC form. Within this morula, the DC organisms prominently label for APH_1235, while labeling of the RC bacteria does not exceed background labeling of the adjacent host cell cytosol. Preimmune sera exhibited low-level background binding to DC and RC bacteria and to the host cell cytosol (Fig. 6I). Comparison of the immunoelectron images generated following screening with anti-APH_1235 (Fig. 6C to H) with those generated by screening with antibody against the confirmed surface protein, Msp2 (P44) (Fig. 7), provides no evidence of APH_1235 presentation on the A. phagocytophilum outer membrane.
Fig. 6.
Assessment of APH_1235 expression by immunoelectron microscopy. (A and B) Reference transmission electron micrographs of a DC (A) and multiple RC organisms (B) are provided because the fixation method that is requisite for immunoelectron microscopy compromises the clarity by which the distinctive DC and RC outer membranes can be discerned. (C to H) A. phagocytophilum-infected HL-60 cells were fixed and screened with anti-APH_1235 followed by goat anti-mouse IgG conjugated to 6-nm gold particles and examined by electron microscopy. Representative results of up to four experiments are shown. (C) A DC organism bound to the surface of a HL-60 cell. (D) At 40 min postinfection, a newly internalized DC organism is detected within a host cell-derived vacuole. (E) Two adjacent morulae. The morula on the left harbors RC organisms, while that on the right contains DC bacteria. (F) A morula harboring many DC organisms and two bacteria that are still in the RC form. (G) Remnant of an A. phagocytophilum-occupied vacuole that has ruptured at the host cell surface and from which DC organisms are being released. (H) Three morulae harboring RC bacteria at 18 h postinfection. (I) Two morulae harboring either DC (left) or RC (right) organisms screened with preimmune serum followed by goat anti-mouse IgG conjugated to 6-nm gold particles and examination by electron microscopy. (E, F, and I) Arrowheads denote individual DC bacteria or entire morulae consisting of DC organisms. Arrows denote single RC bacteria or entire morulae comprised of RC organisms. Scale bars, 0.5 μm.
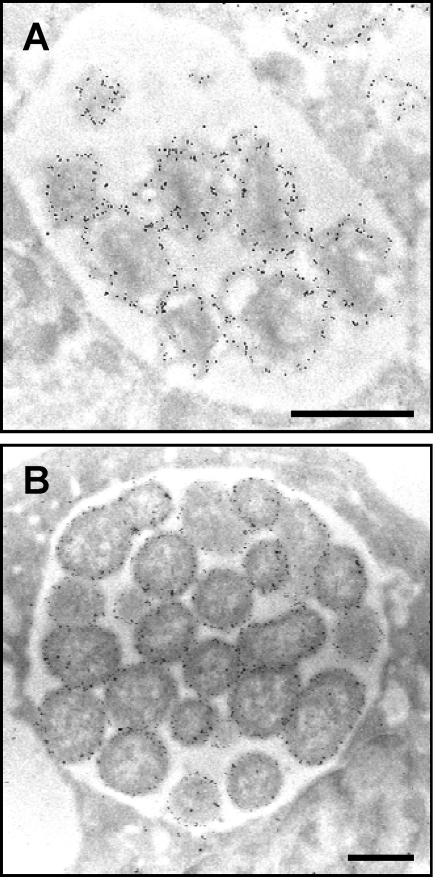
Fig. 7.
Msp2 (P44) is presented on the A. phagocytophilum outer membrane. A. phagocytophilum-infected HL-60 cells were fixed and screened with a Msp2 (P44)-specific MAb followed by goat anti-mouse IgG conjugated to 6-nm gold particles and examined by electron microscopy. Morulae harboring DC (A) and RC (B) organisms are presented. Scale bars, 0.5 μm.
DISCUSSION
Our analysis of the A. phagocytophilum proteome identifies proteins from all functional categories (as designated for the annotated A. phagocytophilum proteome by the J. Craig Venter Institute), thereby providing direct evidence for these processes and pathways with the exception of central intermediary metabolism and mobile and extrachromosomal element functions. A recent proteomic analysis of A. phagocytophilum in HL-60 cells detected peptides derived from all functional categories (31a). The data sets are derived from A. phagocytophilum organisms purified from asynchronously infected HL-60 cells. As isolated bacteria would have been at all stages (early, mid-, and late) of the infection cycle, the proteins identified presumably yield a profile of the A. phagocytophilum proteins expressed throughout replication in HL-60 cells. Msp2 (P44) paralogs constitute a major portion of the A. phagocytophilum myeloid host cell-specific proteome. Msp2 (P44)-23 is the most abundant RIPA buffer-soluble protein of the NCH-1 strain and is present in considerably higher abundance than other Msp2 (P44) paralogs. This observation is in agreement with our previous report that msp2 (p44)-23 represents 89.5% of all msp2 (p44) paralogs that are transcribed by NCH-1 during cultivation in HL-60 cells (56). The second most abundant RIPA buffer-soluble protein is APH_1235. Despite its small molecular mass, APH_1235 is so substantially expressed during A. phagocytophilum infection of HL-60 cells that it accounts for 12.0% of all RIPA buffer-soluble proteins. Based on its abundance, we hypothesize that APH_1235 plays an important role during infection of HL-60 cells and potentially other human myeloid primary cells or cell lines. APH_1235 homologs of other Anaplasma and Ehrlichia species may perform similar functions.
APH_1235 is the first identified A. phagocytophilum protein that is pronouncedly upregulated on the DC form. Its expression levels are highest at 32 h, a time point that coincides with RC to DC conversion and reinfection (55), and in the host cell-free DC organisms used to initiate infection. This indicates that the DC carries considerable amounts of carryover aph_1235 RNA. A similar phenomenon has been observed for late-stage chlamydial genes (1). aph_1235 transcript levels decline by 4 h, a time point by which the majority of bound A. phagocytophilum organisms enter into nascent host cell-derived vacuoles (3, 6, 25). APH_1235-positive morulae never exceeded 32.1% ± 6.2%, which suggests that the A. phagocytophilum DC stage is transient, at least during infection of HL-60 cells. APH_1235 and its homologs can be exploited as markers for the infectious DC forms of A. phagocytophilum and potentially for other pathogens of the Anaplasmataceae, respectively. A. phagocytophilum genes/proteins that exhibit expression profiles similar to APH_1235 would be worth investigating, as they may be critical for adhesion, invasion, and/or establishing infection.
In addition to expression of APH_1235, the expression of 52 other A. phagocytophilum proteins that were previously categorized as being hypothetical was confirmed. Many of these proteins are unique to Anaplasmataceae family members or to A. phagocytophilum itself and likely contribute to these organisms' pathogenic strategies and intracellular survival. Future studies of these unexplored proteins, such as when they are expressed during the course of infection, will open up new lines of investigation and will lead to a better understanding of A. phagocytophilum pathogenesis.
One-quarter of the A. phagocytophilum proteins that were detected are involved in protein synthesis and protein fate. This observation is in agreement with the RC, of which the majority of the isolated bacteria that served as input for proteomic analyses consisted, being the replicative and metabolically active form. All signal transduction proteins are expressed, which is consistent with recent studies that demonstrated the importance of two-component regulatory systems and signal transduction to A. phagocytophilum and Ehrlichia chaffeensis (9, 29, 30). Nearly one-third of the cellular envelope proteins were detected. These proteins are likely involved in mediating adhesion and invasion, providing a fitness advantage to A. phagocytophilum in HL-60 cells, facilitating antigenic variation in vivo, and maintaining cell wall structural integrity. Only 12.7% of proteins predicted to play roles in the biosynthesis of cofactors, prosthetic groups, and carriers were detected. An analysis of the A. phagocytophilum HL-60 cell-specific transcriptome revealed that the majority of genes encoding products involved in cofactor, prosthetic group, and carrier synthesis that were not detected in this study were not transcribed or were transcribed at very low levels (39), which verifies their expression profiles generated by our proteomic data. Only 1 of 55 proteins annotated as having a disrupted reading frame was detected, which hints that these truncated genes may represent genetic material that is not needed, at least not for survival in HL-60 cells, and may be in the process of being purged from the A. phagocytophilum genome. Alternatively, it cannot be ruled out that since these predicted truncated proteins are relatively small, if present in the digested sample, they would yield no to few tryptic peptides, which would prevent or limit their detection.
The lysis buffer used to solubilize A. phagocytophilum alters the complement of proteins that can be recovered. RIPA buffer does not solubilize the organism to nearly the extent of a urea/thiourea-based lysis buffer. Solubility is best reflected by the complete disappearance of the RIPA buffer-insoluble pellet in urea/thiourea lysis buffer, which contributed to the 3.2 times greater number of proteins identified from the urea/thiourea buffer lysate as the RIPA buffer lysate. Use of urea-based buffers requires a preseparation ahead of reversed-phase chromatography, accomplished here through a switch to an ion-exchange/reversed-phase 2D-nanoLC separation, which logistically required use of two different mass spectrometers. The former instrument utilizes a data-independent analysis method in comparison to a data-dependent method in the latter. A primary distinction between these two platforms was observed in our data—the former provides more detailed coverage of fewer identified proteins (2,160 peptides to 87 proteins), while the latter identifies a greater number of proteins but with fewer peptides per protein (1,285 peptides to 280 proteins). With that said, approximately half of those proteins identified from the RIPA extraction were also exclusively identified in that experiment, thereby indicating a prominent partitioning based on solubility and membrane association. The use of two protein extractions served as an additional dimension of separation, which extended our coverage of the A. phagocytophilum proteome.
Centrifugation through discontinuous Renografin (diatrizoate meglumine and diatrizoate sodium) gradients has been widely used to purify obligate intracellular bacterial species (8, 10, 13, 59). Since Renografin production has been discontinued, we developed a purification method using Optiprep that enables us to obtain a highly pure A. phagocytophilum fraction consisting primarily of RC organisms. Despite multiple attempts in which we varied the host cell lysis method, the lysis buffers used, centrifugation speeds, and/or the percentages of Optiprep used, we were unable to purify DC organisms as a distinct fraction. Their transiency is likely a contributing factor to our inability to purify a suitable abundance of DC organisms. Also, since DC organisms bind to P-selectin glycoprotein ligand-1 (PSGL-1) on the HL-60 cell surface (19), liberated DC bacteria may bind to PSGL-1 present in host cell membrane fractions following Dounce homogenization. If so, then the majority of DC bacteria would effectively be removed during the host cellular debris centrifugation step.
AnkA (APH_0740), AptA (APH_0233), Ats-1 (APH_0859), APH_0032, and APH_1387 are the known A. phagocytophilum secreted proteins (7, 22, 23, 40, 51). Of these, AptA, APH_1387, and APH_0032 localize to the ApV (22, 23, 51). AnkA, APH_1387, and Ats-1 are abundantly expressed throughout the course of infection (7, 23, 40) and, accordingly, were detected in the urea-solubilized and/or the RIPA buffer-solubilized bacterial homogenates. APH_0032, which is not expressed until late during the infection cycle (22), along with AptA, was not detected in either sample. It is likely that secreted proteins are efficiently exported during infection and therefore are not detectable in purified A. phagocytophilum organisms unless present in high abundances during their release. Similar results were reported for the Chlamydia trachomatis proteome, for which none of the bacterial encoded inclusion membrane proteins were detected (50). Thus, A. phagocytophilum-secreted proteins expressed during infection of HL-60 cells are underrepresented or were not recovered. Nonetheless, the identification of 23.7% of the predicted proteome of A. phagocytophilum yields extensive insight into the gene products that are important for this pathogen's survival in human myeloid cells.
Supplementary Material
ACKNOWLEDGMENTS
We thank J. Stephen Dumler of Johns Hopkins University for providing MAb 20B4.
This study was supported by NIH grants R01 AI072683, R01 AI072683-04S1, and R21 AI090170 (to J.A.C.).
Footnotes
Supplemental material for this article may be found at http://iai.asm.org/.
Published ahead of print on 15 August 2011.
REFERENCES
- 1. Abdelrahman Y. M., Belland R. J. 2005. The chlamydial developmental cycle. FEMS Microbiol. Rev. 29:949–959 [DOI] [PubMed] [Google Scholar]
- 2. Barbet A. F., et al. 2003. Expression of multiple outer membrane protein sequence variants from a single genomic locus of Anaplasma phagocytophilum. Infect. Immun. 71:1706–1718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Borjesson D. L., et al. 2005. Insights into pathogen immune evasion mechanisms: Anaplasma phagocytophilum fails to induce an apoptosis differentiation program in human neutrophils. J. Immunol. 174:6364–6372 [DOI] [PubMed] [Google Scholar]
- 4. Bradford M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254 [DOI] [PubMed] [Google Scholar]
- 5. Carlyon J. A. 2005. Laboratory maintenance of Anaplasma phagocytophilum, p. 3A.2.1–3A.2.30 In Coico R., Kowalik T. F., Quarles J. M., Stevenson B., Taylor R.(ed.), Current protocols in microbiology. J. Wiley and Sons, Hoboken, NJ: [DOI] [PubMed] [Google Scholar]
- 6. Carlyon J. A., Chan W. T., Galan J., Roos D., Fikrig E. 2002. Repression of rac2 mRNA expression by Anaplasma phagocytophila is essential to the inhibition of superoxide production and bacterial proliferation. J. Immunol. 169:7009–7018 [DOI] [PubMed] [Google Scholar]
- 7. Caturegli P., et al. 2000. ankA: an Ehrlichia phagocytophila group gene encoding a cytoplasmic protein antigen with ankyrin repeats. Infect. Immun. 68:5277–5283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen S. M., Dumler J. S., Feng H. M., Walker D. H. 1994. Identification of the antigenic constituents of Ehrlichia chaffeensis. Am. J. Trop. Med. Hyg. 50:52–58 [PubMed] [Google Scholar]
- 9. Cheng Z., Kumagai Y., Lin M., Zhang C., Rikihisa Y. 2006. Intra-leukocyte expression of two-component systems in Ehrlichia chaffeensis and Anaplasma phagocytophilum and effects of the histidine kinase inhibitor closantel. Cell. Microbiol. 8:1241–1252 [DOI] [PubMed] [Google Scholar]
- 10. Felsheim R. F., et al. 2006. Transformation of Anaplasma phagocytophilum. BMC Biotechnol. 6:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Garcia-Garcia J. C., Barat N. C., Trembley S. J., Dumler J. S. 2009. Epigenetic silencing of host cell defense genes enhances intracellular survival of the rickettsial pathogen Anaplasma phagocytophilum. PLoS Pathog. 5:e1000488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Garcia-Garcia J. C., Rennoll-Bankert K. E., Pelly S., Milstone A. M., Dumler J. S. 2009. Silencing of host cell CYBB gene expression by the nuclear effector AnkA of the intracellular pathogen Anaplasma phagocytophilum. Infect. Immun. 77:2385–2391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Geromanos S. J., et al. 2009. The detection, correlation, and comparison of peptide precursor and product ions from data independent LC-MS with data dependent LC-MS/MS. Proteomics 9:1683–1695 [DOI] [PubMed] [Google Scholar]
- 14. Goodman J. L., et al. 1996. Direct cultivation of the causative agent of human granulocytic ehrlichiosis. N. Engl. J. Med. 334:209–215 [DOI] [PubMed] [Google Scholar]
- 15. Granick J. L., Reneer D. V., Carlyon J. A., Borjesson D. L. 2008. Anaplasma phagocytophilum infects cells of the megakaryocytic lineage through sialylated ligands but fails to alter platelet production. J. Med. Microbiol. 57:416–423 [DOI] [PubMed] [Google Scholar]
- 16. Grieco F., Hay J. M., Hull R. 1992. An improved procedure for the purification of protein fused with glutathione S-transferase. Biotechniques 13:856–858 [PubMed] [Google Scholar]
- 17. Heimer R., Van Andel A., Wormser G. P., Wilson M. L. 1997. Propagation of granulocytic Ehrlichia spp. from human and equine sources in HL-60 cells induced to differentiate into functional granulocytes. J. Clin. Microbiol. 35:923–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Herron M. J., Ericson M. E., Kurtti T. J., Munderloh U. G. 2005. The interactions of Anaplasma phagocytophilum, endothelial cells, and human neutrophils. Ann. N. Y. Acad. Sci. 1063:374–382 [DOI] [PubMed] [Google Scholar]
- 19. Herron M. J., et al. 2000. Intracellular parasitism by the human granulocytic ehrlichiosis bacterium through the P-selectin ligand, PSGL-1. Science 288:1653–1656 [DOI] [PubMed] [Google Scholar]
- 20. Hotopp J. C., et al. 2006. Comparative genomics of emerging human ehrlichiosis agents. PLoS Genet. 2:e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Huang B., et al. 2010. The Anaplasma phagocytophilum-occupied vacuole selectively recruits Rab-GTPases that are predominantly associated with recycling endosomes. Cell. Microbiol. 12:1292–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Huang B., et al. 2010. Anaplasma phagocytophilum APH_0032 is expressed late during infection and localizes to the pathogen-occupied vacuolar membrane. Microb. Pathog. 49:273–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Huang B., et al. 2010. Anaplasma phagocytophilum APH_1387 is expressed throughout bacterial intracellular development and localizes to the pathogen-occupied vacuolar membrane. Infect. Immun. 78:1864–1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Huang H., Wang X., Kikuchi T., Kumagai Y., Rikihisa Y. 2007. Porin activity of Anaplasma phagocytophilum outer membrane fraction and purified P44. J. Bacteriol. 189:1998–2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. IJdo J. W., Mueller A. C. 2004. Neutrophil NADPH oxidase is reduced at the Anaplasma phagocytophilum phagosome. Infect. Immun. 72:5392–5401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. IJdo J. W., Wu C., Magnarelli L. A., Fikrig E. 1999. Serodiagnosis of human granulocytic ehrlichiosis by a recombinant HGE-44-based enzyme-linked immunosorbent assay. J. Clin. Microbiol. 37:3540–3544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. IJdo J. W., Wu C., Telford S. R., III, Fikrig E. 2002. Differential expression of the p44 gene family in the agent of human granulocytic ehrlichiosis. Infect. Immun. 70:5295–5298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jauron S. D., et al. 2001. Host cell-specific expression of a p44 epitope by the human granulocytic ehrlichiosis agent. J. Infect. Dis. 184:1445–1450 [DOI] [PubMed] [Google Scholar]
- 29. Kumagai Y., Cheng Z., Lin M., Rikihisa Y. 2006. Biochemical activities of three pairs of Ehrlichia chaffeensis two-component regulatory system proteins involved in inhibition of lysosomal fusion. Infect. Immun. 74:5014–5022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lai T. H., Kumagai Y., Hyodo M., Hayakawa Y., Rikihisa Y. 2009. The Anaplasma phagocytophilum PleC histidine kinase and PleD diguanylate cyclase two-component system and role of cyclic di-GMP in host cell infection. J. Bacteriol. 191:693–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li G. Z., et al. 2009. Database searching and accounting of multiplexed precursor and product ion spectra from the data independent analysis of simple and complex peptide mixtures. Proteomics 9:1696–1719 [DOI] [PubMed] [Google Scholar]
- 31a. Lin M., Kikuchi T., Brewer H. M., Norbeck A. D., Rikihisa Y. 2011. Gloabal proteomic analysis of two tick-borne emerging zoonotic agents: Anaplasma phagocytophilum and Ehrlichis chaffeensis. Front Microbiol. 2:article 24. doi: 10.3389/fmicb.2011.00024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lin Q., Rikihisa Y. 2005. Establishment of cloned Anaplasma phagocytophilum and analysis of p44 gene conversion within an infected horse and infected SCID mice. Infect. Immun. 73:5106–5114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lin Q., et al. 2002. Analysis of sequences and loci of p44 homologs expressed by Anaplasma phagocytophila in acutely infected patients. J. Clin. Microbiol. 40:2981–2988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Livak K. J., Schmittgen T. D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 35. Lohr C. V., et al. 2002. Expression of Anaplasma marginale major surface protein 2 operon-associated proteins during mammalian and arthropod infection. Infect. Immun. 70:6005–6012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Munderloh U. G., et al. 1999. Invasion and intracellular development of the human granulocytic ehrlichiosis agent in tick cell culture. J. Clin. Microbiol. 37:2518–2524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Munderloh U. G., et al. 2004. Infection of endothelial cells with Anaplasma marginale and A. phagocytophilum. Vet. Microbiol. 101:53–64 [DOI] [PubMed] [Google Scholar]
- 38. Munderloh U. G., et al. 1996. Isolation of the equine granulocytic ehrlichiosis agent, Ehrlichia equi, in tick cell culture. J. Clin. Microbiol. 34:664–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nelson C. M., et al. 2008. Whole genome transcription profiling of Anaplasma phagocytophilum in human and tick host cells by tiling array analysis. BMC Genomics 9:364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Niu H., Kozjak-Pavlovic V., Rudel T., Rikihisa Y. 2010. Anaplasma phagocytophilum Ats-1 is imported into host cell mitochondria and interferes with apoptosis induction. PLoS Pathog. 6:e1000774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Palmer G. H., Bankhead T., Lukehart S. A. 2009. ‘Nothing is permanent but change’—antigenic variation in persistent bacterial pathogens. Cell. Microbiol. 11:1697–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Park J., Choi K. S., Dumler J. S. 2003. Major surface protein 2 of Anaplasma phagocytophilum facilitates adherence to granulocytes. Infect. Immun. 71:4018–4025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Popov V. L., et al. 1998. Ultrastructural differentiation of the genogroups in the genus Ehrlichia. J. Med. Microbiol. 47:235–251 [DOI] [PubMed] [Google Scholar]
- 44. Rikihisa Y. 2010. Anaplasma phagocytophilum and Ehrlichia chaffeensis: subversive manipulators of host cells. Nat. Rev. Microbiol. 8:328–339 [DOI] [PubMed] [Google Scholar]
- 45. Rikihisa Y., et al. 1997. Ultrastructural and antigenic characterization of a granulocytic ehrlichiosis agent directly isolated and stably cultivated from a patient in New York state. J. Infect. Dis. 175:210–213 [DOI] [PubMed] [Google Scholar]
- 46. Scorpio D. G., Caspersen K., Ogata H., Park J., Dumler J. S. 2004. Restricted changes in major surface protein-2 (msp2) transcription after prolonged in vitro passage of Anaplasma phagocytophilum. BMC Microbiol. 4:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Shkap V., Molad T., Brayton K. A., Brown W. C., Palmer G. H. 2002. Expression of major surface protein 2 variants with conserved T-cell epitopes in Anaplasma centrale vaccinates. Infect. Immun. 70:642–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Singu V., Liu H., Cheng C., Ganta R. R. 2005. Ehrlichia chaffeensis expresses macrophage- and tick cell-specific 28-kilodalton outer membrane proteins. Infect. Immun. 73:79–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Singu V., et al. 2006. Unique macrophage and tick cell-specific protein expression from the p28/p30-outer membrane protein multigene locus in Ehrlichia chaffeensis and Ehrlichia canis. Cell. Microbiol. 8:1475–1487 [DOI] [PubMed] [Google Scholar]
- 50. Skipp P., Robinson J., O'Connor C. D., Clarke I. N. 2005. Shotgun proteomic analysis of Chlamydia trachomatis. Proteomics 5:1558–1573 [DOI] [PubMed] [Google Scholar]
- 51. Sukumaran B., et al. 2011. Anaplasma phagocytophilum AptA modulates Erk1/2 signalling. Cell. Microbiol. 13:47–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tao H., et al. 2010. Purifying natively folded proteins from inclusion bodies using sarkosyl, Triton X-100, and CHAPS. Biotechniques 48:61–64 [DOI] [PubMed] [Google Scholar]
- 53. Thomas R. J., Dumler J. S., Carlyon J. A. 2009. Current management of human granulocytic anaplasmosis, human monocytic ehrlichiosis and Ehrlichia ewingii ehrlichiosis. Expert Rev. Anti Infect. Ther. 7:709–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Thompson J. D., Higgins D. G., Gibson T. J. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Troese M. J., Carlyon J. A. 2009. Anaplasma phagocytophilum dense-cored organisms mediate cellular adherence through recognition of human P-selectin glycoprotein ligand 1. Infect. Immun. 77:4018–4027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Troese M. J., et al. 2009. Differential expression and glycosylation of Anaplasma phagocytophilum major surface protein 2 paralogs during cultivation in sialyl Lewis x-deficient host cells. Infect. Immun. 77:1746–1756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wakeel A., Zhang X., McBride J. W. 2010. Mass spectrometric analysis of Ehrlichia chaffeensis tandem repeat proteins reveals evidence of phosphorylation and absence of glycosylation. PLoS One 5:e9552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Webster P., IJdo J. W., Chicoine L. M., Fikrig E. 1998. The agent of Human Granulocytic Ehrlichiosis resides in an endosomal compartment. J. Clin. Invest. 101:1932–1941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yu C. S., Chen Y. C., Lu C. H., Hwang J. K. 2006. Prediction of protein subcellular localization. Proteins 64:643–651 [DOI] [PubMed] [Google Scholar]
- 60. Reference deleted.
- 61. Zhi N., Ohashi N., Rikihisa Y. 1999. Multiple p44 genes encoding major outer membrane proteins are expressed in the human granulocytic ehrlichiosis agent. J. Biol. Chem. 274:17828–17836 [DOI] [PubMed] [Google Scholar]
- 62. Zhi N., et al. 2002. Transcript heterogeneity of the p44 multigene family in a human granulocytic ehrlichiosis agent transmitted by ticks. Infect. Immun. 70:1175–1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.