Introduction
Recent studies have provided many new insights into the regulation and biology of inositide signaling thereby fueling interest in the cellular roles of water-soluble inositol polyphosphate (IP) pathways (Berridge, 1993; Hokin, 1985; Irvine and Schell, 2001; Kapeller and Cantley, 1994; Majerus, 1992; Mikoshiba, 1997; Nishizuka, 1986; Shears, 1998; York, 2006; York et al., 2001). The landmark studies demonstrating that cellular stimuli activate phosphoinositide specific phospholipase C (PLC) resulting in the production of intracellular messengers inositol 1,4,5-trisphosphate (IP3) and 1,2-diacylglycerol provided the rationale for understanding signaling pathways dependent on water soluble IP molecules (see reviews (Berridge, 1993; Nishizuka, 1986)). Over twenty such IP codes have been identified in cells, most of which are conserved through eukaryotes (see reviews (Irvine and Schell, 2001; Majerus, 1992; Shears, 1998). Progress in understanding the cellular roles of these putative regulatory molecules has come from the cloning and cellular manipulation of inositol polyphosphate kinase (IPKs) and inositol pyrophosphate synthase (IPS) gene products. IPK and IPS activities are required to produce many of these codes through conversion of IP3 to a variety of species including: inositol tetrakisphosphate (IP4), inositol pentakisphosphate (IP5), inositol hexakisphosphate (IP6) and inositol pyrophosphates (PP-IPs) (York, 2006).
This article will discuss the biochemistry and biology of IPK and IPS gene products and the processes influenced by these newly defined regulators of cellular communication pathways. Of interest, several labs have discovered that the IP and PP-IP products are required for proper regulation of nuclear processes and organism development. These new studies have invigorated the field and have been helpful in deciphering the inositide chemical codes.
Cloning of IPK and IPS gene products
The metabolic pathways for conversion of IP3 to IP4, IP5, IP6 and PP-IP molecules have been studied for decades (See Figure 1) (Irvine and Schell, 2001; Majerus, 1992; Shears, 1998). In the past ten years, the genes involved in their synthesis have been identified. Metabolomic studies in the budding yeast, Saccharomyces cerevisiae, identified two kinases, designated Ipk1 and Ipk2, that convert I(1,4,5)P3 to IP6 (Odom et al., 2000; Saiardi et al., 2000b; York et al., 1999). This pathway is dependent on yeast PI-specific phospholipase, Plc1, a dual-specificity IP3/IP4 6-/3-kinase (Ipk2) and an IP5 2-kinase (Ipk1). The cloning of Ipk2 and Ipk1 provided the gene products that accounted for activities initially identified by the Downes and Michell laboratories, who first reported a pathway of IP3 conversion to IP6 via 6-, 3- and 2-kinase activities, respectively (Estevez et al., 1994; Ongusaha et al., 1998; Ongusaha et al., 1997). Production of these molecules is lipid-dependent as deletion of Plc1 resulted in a failure to produce IP6, and other IPs, and over-expression of Plc1 induced IP6 synthesis over 20-fold (York et al., 1999). Metabolic analysis of ipk2 mutant yeast revealed a marked build-up of IP3 and a failure to produce IP4, IP5 and IP6 (Odom et al., 2000; Saiardi et al., 2000b; York et al., 1999). Analysis of ipk1 mutant cells showed an accumulation of IP5 and a failure to synthesize IP6 (York et al., 1999).
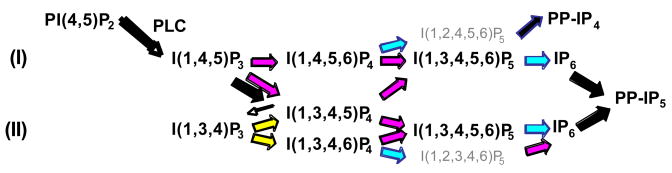
Fig. 1.
IPK and IPS metabolic pathways. Activation of phospholipase C triggers the conversion of phosphatidyinositol 4,5-bisphosphate, PI(4,5)P2, to the second messenger I(1,4,5)P3. Originally discovered in Saccharomyces cerevisiae, pathway I has been shown to exist in Arabidopsis thaliana, Drosophila melanogaster, and Rattus norvegicus. This pathway is initiated by the 6-/3-/5-kinase (Ipk2) that sequentially phosphorylates I(1,4,5)P3 to generate I(1,3,4,5,6)P5. Evidence supports the existence pathway II in Homo sapiens and Zea mays and occurs through a four step conversion of I(1,4,5)P3 to I(1,3,4,5,6)P5 through the activities of and IP3 3-kinase (IP3K), IP4 5-phosphatase, a I(1,3,4)P3 5-/6-kinase, 5-kinase of Ipk2 respectively. In either pathway, a 2-kinase (Ipk1) converts I(1,3,4,5,6)P5 to IP6 while also performing additional “branch” reactions. Inositol pyrophosphate synthase activities (IPS) generate high-energy pyrophosphate species PP-IP4 and PP-IP5.
In addition to the IPKs, budding yeast have two distinct inositol pyrophosphate synthase (IPS) activities that generate PP-IP4 and PP-IP5 branches from the core IP3 to IP6 pathway (see Figure 1) (Saiardi et al., 2000a; Saiardi et al., 1999; Seeds et al., 2005; York et al., 2005). These activities have been designated IP6 kinases (IP6K); however, since some utilize IP4 and IP5 as substrates it is suggested that IPS may be a more general nomenclature. The predominant IP6 kinase activity in yeast extracts capable of generating PP-IP5 was identified as Kcs1, a gene product originally found as a second-site suppressor of protein kinase C mutants (Huang and Symington, 1995; Saiardi et al., 1999). Kcs1 is related by sequence similarity to Ipk2 and IP3 3-kinases (see Figure 2). Additionally, Kcs1 has been found to utilize other IP substrates enabling the production of several high-energy pyrophosphates including PP-IP4, PP-IP5, PP2-IP3, and PP2-IP4 (Luo et al., 2002; Saiardi et al., 2000a, 2002; Seeds et al., 2005; York et al., 2005). Furthermore, Kcs1 appears to also function as an I(1,4,5)P3 3-kinase activity, or regulator thereof, in vivo (Seeds et al., 2005) that initiates a novel second minor metabolic pathway in yeast (Dubois et al., 2002; Seeds et al., 2005).
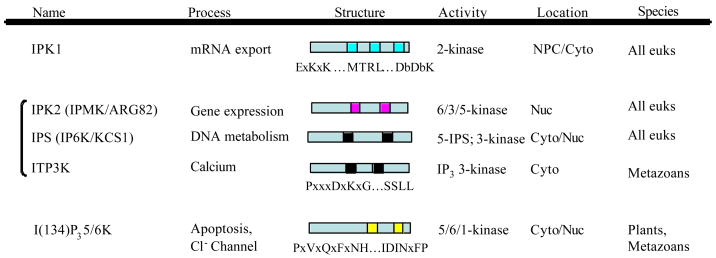
Fig. 2.
IPK family members. Sequence comparison of soluble inositol polyphosphate kinases indicate that there are at least three distinct classes, which do not show detectable similarity to each other or inositol lipid kinases. Ipk1 members act as selective 2-kinases and add a D-2 phosphate to IP4 and I(1,3,4,5,6)P5 substrates and harbor a ExKxK…MTRL…DbDbK motif. Ipk1 gene products have been found in all eukaryotes. The second class is comprised of related gene products that contain a signature PxxxDxKxG motif and include the 6-/3-/5-kinases (Ipk2/IPMK), IP3 3-kinases (IP3K), and inositol pyrophosphate synthases (IPS). Both Ipk2 and IPS gene products are found in all eukaryotes. IP3K is found in metazoans but not S. cerevisiae or Arabidopsis. The third class is the I(134)P35/6K/ITPK1 has a PxVxQxFxNH…IDINxFP motif and which originally was shown to designates inositol 1,3,4-trisphosphate 5/6-kinase, orthologs of which are present in plants and most metazoans (except Drosophila).
A second IPS activity, designated Ips1 (also referred to as Ids1 for inositol diphosphate synthase), was identified by metabolomic studies in yeast cells deficient for kcs1 and dpp1, an inositol pyrophosphate phosphatase (York et al., 2005). Ips1 is capable of producing several unique PP-IP4, PP-IP5 and PP2-IP4 species (Seeds et al., 2005; York et al., 2005); however its molecular identity has not yet been reported. Metabolic characterization of inositides in Dictyostelium have revealed multiple pyrophosphate species, although currently it is unclear if these are produced through the actions of Kcs1 and/or Ips1 orthologues (Albert et al., 1997; Laussmann et al., 1996, 1998, 2000, 1997).
The evolutionary conservation of the IPK and IPS gene products extends from yeast to man (Figure 2). Studies in plants, fruit flies, and mammals demonstrate that Ipk2, also known as inositol polyphosphate multi-kinase (IPMK), and Ipk1 are required for IP6 synthesis (Chang et al., 2002; Frederick, 2005; Fujii and York, 2005; Nalaskowski et al., 2002; Saiardi, 2001; Seeds et al., 2004; Stevenson-Paulik et al., 2005; Stevenson-Paulik et al., 2002; Verbsky et al., 2005a, 2005b, 2002; Xia et al., 2003). However, plants and mammals have been found to harbor alternate and/or redundant pathways for the synthesis of IP6 that involve two additional IPKs: 1) an I(1,4,5)P3 3-kinase and 2) an I(1,3,4)P3 5/6-kinase (Choi et al., 1990; Majerus, 1992; Shears, 1989, 1998; Shi et al., 2005, 2003; Takazawa et al., 1990; Wilson and Majerus, 1996, 1997). It has also been reported that some organisms produce IP6 through lipid-independent routes (Brearley and Hanke, 1996; Shi et al., 2005; Stephens and Irvine, 1990). These additional gene products do not appear to be present in the S. cerevisiae genome, and plants have a 5/6-K, but not an IP3K, gene product. Mice have three IP3K isoforms and single and double deletions alter I(1,3,4,5)P4 production but do not alter IP5 synthesis (Pouillon et al., 2003). In drosophila neither of the IP3K isoforms appear to be required to produce IP6 (Seeds et al., 2004). Disruption of 5/6-K decreased IP6 synthesis in maize (lpa mutants) and human cells (Shi et al., 2003; Verbsky and Majerus, 2005; Wilson and Majerus, 1997). Thus, even though plants and metazoans have may have more complex routes to synthesize inositides, it is clear that Ipk2/IPMK and Ipk1 appear to be required for IP5 and IP6 production in all eukaryotic species analyzed to date.
Nuclear localization of IPK and IPS proteins
A common theme in signaling biology relates to the spatial restriction of pathways to selective compartments, such as the nucleus. Nuclear specific inositol lipid pathways were among the first descriptions and have added to the complexities of inositide signaling – reviewed elsewhere (Cocco et al., 2002; D’Santos et al., 2000, 1998; Faenza et al., 2005; Manzoli et al., 2004; Martelli et al., 2004; Yagisawa, 2006). Of particular relevance to the IPK pathways presented in this article are the findings that some PLC isozymes undergo nucleo-cytoplasmic shuttling providing a direct mechanism for nuclear PIP2 hydrolysis and initiation of nuclear I(1,4,5)P3 and 1,2-diacylglycerol (DAG) signaling pathways (Faenza et al., 2005; Topham and Prescott, 2002; Yagisawa, 2006) (Jeremy Thorner, UC Berkeley, unpublished). The study of how nuclear kinases, lipases and phosphatases are locally activated – especially in the context of extracellular agonists – remains an active and exciting field. Are nuclear inositides, such as IP3, locally produced? This question has been difficult to answer as methods to spatially image IP molecules do not exist. Studies suggesting that IP3 receptors localize to the inner nuclear membrane and mediate nuclear calcium release provide indirect evidence that nuclear IP3 pathways are present (Gerasimenko and Gerasimenko, 2004). Interpretation of these types of studies is confounded by the rapid diffusion rates of IP molecules throughout the cell. Nonetheless, it has been suggested that rapid bursts of IP production within the nucleus may locally signal.
Subcellular localization studies of IPK and IPS proteins indicate that, in part, they function in the nucleus. Ipk2/IPMK has been shown to be primarily nuclear in a number of different cell types and species (Bercy et al., 1987; Fujii and York, 2005; Nalaskowski et al., 2002; Odom et al., 2000; Seeds et al., 2004; Xia et al., 2003). Yeast Ipk1 was shown to localize to the nuclear envelope (York et al., 1999). Kcs1 and mammalian IP6K have been found to be both nuclear and cytoplasmic (Luo et al., 2002; Saiardi et al., 2001).
Is the nuclear localization of the IPK and IPS proteins a requirement for production of IP and PP-IP molecules? The conservation of nuclear localization across species and their roles in regulating nuclear processes (see below section) indicates the answer to this question would be intuitively “yes”! However, based on cellular studies the answer is still unclear and there is evidence supporting both “no” and “yes” answers. Studies in which Ipk1 was artificially targeted to the plasma membrane still synthesize IP6 and appear to complement Gle1-mediated mRNA export (Miller et al., 2004). Heterologous expression of Arabidopsis thaliana, Drosophila melanogaster and Rattus norvegicus IPK2/IPMK gene products in ipk2 deficient yeast restore IP4/IP5 synthesis activity (Fujii and York, 2005; Seeds et al., 2005, 2004; Stevenson-Paulik et al., 2005, 2002; Xia et al., 2003). Studies in which yeast and rat GFP-Ipk2 were individually overexpressed in Rat-1 cells demonstrated that both resulted in a 4-fold elevation in cellular IP5 levels despite the fact they differentially localized to cytoplasmic and nuclear compartments, respectively (Fujii and York, 2005). These data indicate that localization does not appear to be required for metabolic function. In contrast, overexpression of a nuclear exclusive Plc1 that has a mutation in the nuclear export sequence, results in similar induction of IP4, IP5 and IP6 synthesis as compared to a wild-type Plc1 (J. Thorner and J. York laboratories, unpublished data). The Thorner lab has shown that Pik1 has both distinct essential nuclear and cytoplasmic functions of both nuclear and cytoplasmic pathways (Strahl et al., 2005); whereas studies of the Emr lab indicate that the PIP 5-kinase, Mss4p, appears to require cytoplasmic localization for its essential functions (Audhya and Emr, 2003). Thus there seems to be evidence supporting both arguments. We are cautious in interpreting these results as heterologous proteins were overexpressed and quantitative functional assays have not been adequately performed. Furthermore, the small size and rapid diffusion rates of soluble IPs make it difficult to reconcile exactly where the substrates for the IPK and IPS activities are generated. This emphasizes the need to generate tools capable of spatio-temporal imaging of these messengers in living cells.
Inositide signaling to the nucleus
Genetic and biochemical studies of a phospholipase C-dependent pathway in the budding yeast have provided compelling functional evidence for regulation of three distinct nuclear processes by higher IP and PP-IP molecules including: 1) transcriptional regulation/chromatin remodeling; 2) efficient mRNA export from the nucleus and 3) telomere length maintenance (Figure 3). The IPK and IPS gene products play a critical role in regulating these processes, although at this point the mechanisms as to how remain an active area of study.
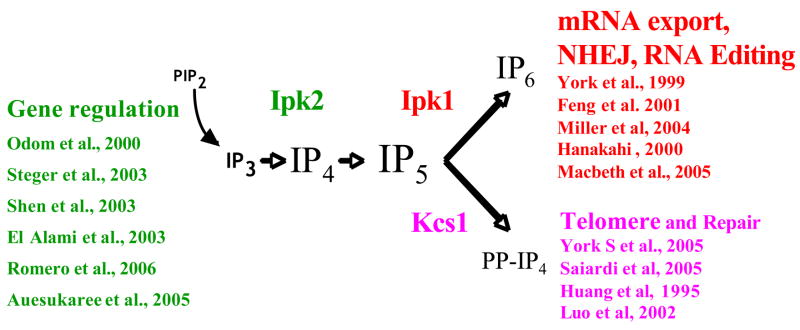
Fig. 3.
IPK signaling to the nucleus. Phospholipase C activation results in the cleavage of PI(4,5)P2 (PIP2) to generate I(1,4,5)P3 (IP3) Conversion of IP3 to IP4, IP5, IP6 and PP-IP4 occurs through the actions of Ipk2, Ipk1 and the inositol pyrophosphate synthase, Kcs1. Ipk2 and its IP4 and IP5 products have been shown to be required for proper regulation of gene expression/chromatin remodeling. Ipk1 and its IP6 product have been found to modulate mRNA export, non-homologous end joining (NHEJ), and RNA editing. Kcs1 and its PP-IP4 product have been shown to regulated telomere maintenance and DNA repair. Selected citations are listed for each process.
Gene expression
The regulation of gene expression by inositides has emerged from research performed within the past two decades. Among the initial links between inositol signaling and transcriptional control, were those made by the Henry lab through studies of the yeast INO1 gene product (Bailis et al., 1992; Carman and Henry, 1999; Hirsch and Henry, 1986; Lopes et al., 1991). They showed that regulation of Ino1 expression required cis-acting elements that link to sensing lipid and small molecules within the cell. Work of others has shown that in response to signals the physical location of the INO1 promoter with respect to the nuclear envelop is altered and that this movement changes the transcriptional rates (Brickner and Walter, 2004). Additionally, Crabtree and colleagues observed that PIP2 increases recruitment of the mammalian BAF chromatin remodeling complex to the nuclear matrix (Zhao et al., 1998). Subsequently, they also found that PIP2 binds directly to the BAF complex and increases its association with stable polymerized actin (Rando et al., 2003, 2002).
A direct link between gene expression and IP4/IP5 production has come from studies that found the yeast transcriptional regulator Arg82 is identical to the IP3 kinase – which prompted its renaming to inositol polyphosphate kinase 2, Ipk2 (Odom et al., 2000). As outlined above, yeast Ipk2/Arg82 functions as a 6-/3-kinase capable of converting IP3 to IP5 (Odom et al., 2000; Saiardi et al., 2000b). Ipk2/Arg82 was found to be conditionally essential for survival of yeast when grown on arginine or ornithine, but not glutamate, as the sole nitrogen source (Bechet et al., 1970; Dubois et al., 1987; Odom et al., 2000). Ipk2/Arg82 is one of four components (Mcm1, Arg80, Arg81, and Arg82) of the ArgR-Mcm1 complex required for arginine-specific transcriptional responses (Messenguy and Dubois, 2003). Ipk2/Arg82 also was found to directly associate with Mcm1, a MADS box DNA binding protein (El Bakkoury, 2000) and in response to arginine signals Ipk2/Arg82 is recruited to active regions of transcription as measured by chromatin immunoprecipitation (ChIP) assays (Yoon et al., 2004). Thus it appears that ArgR-Mcm1 controls the transcriptional activation of arginine catabolic gene products and repression of arginine anabolic pathway members, thereby allowing cells to reprogram nitrogen production under certain nutritional limitations. The role of Ipk2/Arg82 in transcriptional control extends beyond that of amino acid metabolism as subsequent studies have shown it is involved in controlling gene expression of other pathways including the chromatin remodeling within the PHO5 promoter (Steger et al., 2003) and of nitrogen responsive gene regulation pathways (El Alami et al., 2003).
Mechanistically, Ipk2/Arg82 may regulate gene expression through both kinase “dependent” and “independent” pathways. Prior to finding that Arg82 harbored IP3 kinase activity, it was shown that assembly of ArgR-Mcm1 protein complexes on “arginine box” site-specific DNA promoter elements in vitro requires Ipk2 protein (Dubois and Messenguy, 1994). Later it was shown that kinase activity is not required for assembly on DNA elements (Odom et al., 2000). Kinase activity of Ipk2/Arg82 and production of IPs through phospholipase C were found to be required for proper activation of ArgR-Mcm1 transcription complexes as determined through a phenotypic assay looking for cell growth on minimal medium supplemented with arginine or ornithine as the sole nitrogen source (Odom et al., 2000). Dubois and colleagues have reported that Ipk2 kinase activity and IP production were required for some, but not all, transcriptional regulation by Ipk2/Arg82 (Dubois et al., 2000; El Alami et al., 2003). It has been suggested that a poly aspartate region of Ipk2 (residues 286-301) accounts for a kinase-independent stabilization determinant for Mcm1 and/or ArgR-Mcm1 complexes (El Alami et al., 2003; El Bakkoury, 2000). Recent studies have shown that this model cannot fully account for regulation as it was shown that expression of Drosophila melanogaster or Arabidopsis Ipk2, both which lack the poly aspartate sequence, fully restore the growth of ipk2 null yeast in media containing arginine or ornithine as the sole nitrogen source (Odom, 2002; Seeds et al., 2005; Xia et al., 2003). Interestingly, the plant and fly Ipk2 sequences are less than 17% identical to yeast Ipk2 (over 300 mutations in 355 total residues), yet both retain dual-specific IP3/IP4 6-/3-kinase activity. Thus, the ability of the heterologous Ipk2 molecules to complement function provides a “genetic” add-back strategy. These data demonstrate that the kinase dependent function of Ipk2 is required for transcriptional control (as judged by a phenotypic assay) and that restoration of IP4/IP5 production is sufficient to bypass “kinase-independent” transcriptional roles for Ipk2.
A number of studies indicate a general role for Ipk2/Arg82 and its kinase activity in the transcriptional regulation of several pathways. For instance, Ipk2 is required for transcriptional responses other than through the ArgR-Mcm1 complex (Auesukaree et al., 2005; El Alami et al., 2003; Odom, 2002, 2000; Romero et al., 2006; Shen et al., 2003; Steger et al., 2003). It is also possible that some of the pleiotropic defects observed occur as a result of metabolic changes downstream of Ipk2, such as altered PP-IP4 and/or PP-IP5. Support for this hypothesis come from studies of kcs1 mutant yeast, which have altered gene expression profiles based on micro-array analysis (El Alami et al., 2003). The Snyder lab has also proposed that Ipk2/IPMK has “robust” PIP2 3-kinase activity and that this lipid kinase function is important for transcriptional control of some but not all genes (Resnick et al., 2005).
Possible mechanisms by which Ipk2 and its products, such as IP4 and IP5, regulate gene expression have been suggested by recent studies. Chromatin structures are precisely regulated to control access of nuclear machinery to DNA. De-repression of gene expression can occur when chromatin remodeling complexes mobilize nucleosomes from promoter regions and allow access of regulatory factors and general transcriptional machinery. The O’shea lab found that transcriptional regulation by IPs might occur through alteration of chromatin remodeling complexes. In their studies, Ipk2 was identified through a genetic screen looking for factors involved in the regulation of transcription at the PHO5 promoter (Steger et al., 2003). Pho5 expression was also repressed in strains containing plc1 or ipk2 mutations. Using ChIP methods, Ino80 and SWI/SNF chromatin remodeling complexes were identified as the mediators of nucleosome mobilization at the PHO5 promoter. Further, recruitment of these factors to promoters was impaired in strains that lacked IP4 and IP5 synthesis. Transcription factors were unable to access the promoter and transcription of the Pho5 was blocked. These results indicate that IP4 and IP5 may directly play a role in recruiting Ino80 and SWI/SNF complexes to specific promoters.
Shen and coworkers proposed a molecular basis for the action of the IPs on the chromatin remodeling complexes through the use of an in vitro nucleosome mobilization assay (Shen et al., 2003). Several classes of complexes are either stimulated or inhibited in their ability to mobilize nucleosomes. High concentrations of IP6 inhibited nucleosome mobilization by the NURF, INO80, and ISW2 complexes, potentially through inhibition of their ATPase activities. IP6 did not inhibit nucleosome mobilization by the SWI/SNF complex and it was shown that IP4 and IP5 were stimulatory. This study also demonstrated that transcription of the INO1 gene was significantly repressed in plc1 and ipk2 mutants and, partially repressed in ipk1 mutants. Together, these data indicate that IP4, IP5, and IP6 are required for proper transcriptional regulation of the Ino1 expression, possibly by directly targeting the chromatin remodeling complexes themselves.
Another possible nuclear receptor for IP or PIP products has come from a recently discovered family of PHD finger-containing proteins (Gozani et al., 2003). The PHD finger protein and tumor suppressor, inhibitor of growth protein-2 (ING2), was found to bind IP4-affinity resin and phosphoinositides, including PI5P (Gozani et al., 2003). Subsequent studies have furthered this work and have shown that nuclear PI5P alteration in cells lead to ING2 dependent changes in chromatin (Jones et al., 2006). Other links of inositide signaling to PHD proteins have come from studies of a Drosophila melanogaster PHD-containing protein, ASH2, which by two-hybrid analysis was found to associate with a putative PIP kinase called skittles (SKTL) (Cheng and Shearn, 2004). This may be the “tip of the iceberg” since PHD fingers are part of a large family of nuclear proteins (over 100 in metazoans and 16 in budding yeast) implicated in transcriptional regulation by interacting with or modifying chromatin, in part, through the regulation of histone acetylation (Feng et al., 2002).
This story has become even more exciting based on studies showing that PHD fingers bind to trimethyl-lysine residues of histones (H3K4me3) thereby playing a role in interpreting the histone-code hypothesis. Crystallographic studies defined a molecular basis for how PHD fingers of ING2 and NURF bind to H3K4me3 (Li et al., 2006; Pena et al., 2006; Shi et al., 2006; Wysocka et al., 2006). Yeast ING1 (Yng1p) also has been reported to bind to H3K4me3 (Martin et al., 2006). Mechanistically it is not clear whether or not IP/PIP and H3K4me3 binding to PHD fingers occur through the same pocket, overlapping or completely distinct sites. If the PHD fingers are bone fide receptors for both inositides and trimethyl-lysine residues on histone tails, then one can imagine a number of new scenarios regarding how inositide signaling may regulate the histone code and chromatin remodeling.
IP regulators of mRNA export
Another role for IP molecules in the regulation of nuclear function have emerged from the discovery that IP6 production is required for efficient mRNA export from the nucleus (York et al., 1999). A genetic screen was designed to gain insight into the function of GLE1, a factor thought to function as a mediator between splicing and nuclear export of poly(A)+ RNA. Plc1, Ipk2, and Ipk1 were identified as factors that are synthetically lethal with GLE1. Mutations in all three genes resulted in a common failure to both generate IP6 and export mRNA from the nucleus. Based on these data, IP6 was proposed as a mediator of mRNA export. Miller et al identified genes that were synthetic lethal with an ipk1 null strain including a subset of genes at the nuclear pore that had been previously tied to GLE1 function (Miller et al., 2004). Several of these nucleoporins reside on the cytoplasmic face of the nuclear pore and the author’s speculated that the IP6 site of action might reside there. Recent work by two different groups reveals that this hypothesis was correct as GLE1, a resident protein on the cytoplasmic face of the nuclear pore was found to interact directly with IP6 (Alcazar-Roman et al., 2006; Weirich et al., 2006). The GLE1/IP6 complex was found to recruit and activate the activity of the DExE/H-box ATPase Dbp5, a component that is required for mRNA export. When Ipk1 was expressed with a STE2 tag that localizes it to the plasma membrane it was able to rescue the growth defect of a gle1/ipk1 double mutant. Taken together these results suggest that that cytoplasmic production of IP6 is sufficient to stimulate mRNA export from the nucleus by binding GLE1 and activating the ATPase activity of DBP5.
Support for IP6 regulated mRNA export in mammalian cells comes from work of Majerus and coworkers (Feng et al., 2001). The Salmonella dublin virulence factor SopB is an inositol phosphatase that hydrolyzes a wide range of IP and PI species (Norris et al., 1998). When SopB was expressed in human cells, IP levels, including IP6, were greatly reduced and mRNA was found to accumulate in the nucleus (Feng et al., 2001).
IP regulators of DNA metabolism
Inositides have also been found to function as regulators of DNA metabolism through, homologous recombination, nonhomologous end joining (NHEJ), regulation of telomere length, and DNA repair. Yeast Kcs1 was originally cloned as a suppressor of a hyper-recombination phenotype conferred by a mutant pkc1 allele (Huang and Symington, 1995). The suppression was later attributed to PP-IP production through Kcs1 because a kinase dead mutant could not suppress the pkc1 allele (Luo et al., 2002). The mechanism through which the PP-IPs mediate recombination may be general because Kcs1 appeared to regulate all of the possible mechanisms tested. Another connection of IPs to DNA repair comes from identification of Ipk2 as a dosage suppressor of rad53 null and mec1 mutant yeast (Desany et al., 1998). Both Rad53 and Mec1 function to monitor genome integrity checkpoints suggesting a new link of IP production and these pathways.
A role for IP6 was proposed as a positive regulator of NHEJ in mammalian cells. The components that are required for NHEJ include the Ku70/80 heterodimer that binds to DNA ends and a DNA-dependent protein kinase catalytic subunit (DNA-PKcs) that shares significant homology to the PI3-kinase family. Ku recruits DNA-PK to the DNA and the break is repaired by a DNA ligase IV. An in vitro assay reveals that IP6 could activate the DNA-PKcs/Ku holoenzyme (Hanakahi et al., 2000). The IP6 interaction appeared to be mediated through its direct binding Ku (Hanakahi and West, 2002; Ma and Lieber, 2002). A possible link between cellular IP production and Ku was made using photobleaching experiments (Byrum et al., 2004). Cells with partially depleted IPs had correlative reduced mobility of GFP-Ku. The authors speculated IP6 induces a conformational change in Ku which caused dissociated from its binding partners and diffusion through the nucleus. Studies in yeast found that IP6 was unable to bind to yeast Ku and end joining is not defective in vivo when IP synthesis is disrupted (Hanakahi and West, 2002; York et al., 2005).
An additional role for inositides regulated DNA metabolism has been proposed in the modulation of telomere length in yeast (Saiardi et al., 2005; York et al., 2005). It was shown that plc1, ipk2, and kcs1 mutants have longer telomeres as compared with wild type cells, implicating PP-IP4 as a negative regulator of telomere length (York et al., 2005). Conversely, mutant cells with increased levels of PP-IP4 were shown to have shorter telomeres (York et al., 2005). Further, the author’s demonstrated that PP-IP4 production was epistatic with the PI 3-kinase related protein kinase Tel1 in mediating these effects of telomere length. Saiardi and coworkers also reported a role for PP-IPs in the regulation of telomere length through TEL1 (Saiardi et al., 2005). The mechanistic basis for inositol pyrophosphate regulation to telomere length remains an important area of study.
Roles for IPK proteins in organism development
Recent studies of IPK2 and IPK1 knockout animals and plants point to a more general role for IPs and PP-IPs in cellular signaling responses (Frederick, 2005; Sarmah et al., 2005; Stevenson-Paulik et al., 2005; Verbsky et al., 2005a). Deletion of Ipk2/IPMK in mice results in early embryonic lethality at E8.5 (Frederick, 2005). Ipk2/IPMK deficient mice exhibit multiple morphological defects, including: 1) abnormal folding of the neural tube, 2) failure of the allantois and chorion fusion, 3) failure of embryo turning, 4) abnormal elongation of the anterior-posterior axis, and 5) marked reduction/delay in somite formation (Frederick, 2005). Ipk2/IPMK expression increases significantly in neuronal specific tissues around day E8.5 and our data indicate that several tissue-patterning events require a functional Ipk2/IPMK protein. Of interest, our data indicate that calcium signaling is normal, and therefore the lethality and developmental defects are likely due to loss of IP5 and/or IP6 production.
Homozygous disruption of Ipk1 in mice through a gene-trap strategy developed by Bay Genomics (San Francisco, CA), which placed the gene trap in the first intron of IP5 2-kinase, resulted in early embryonic lethality and no homozygous embryos could be found even at day E8.5 (Verbsky et al., 2005a). Using the fact that heterozygous animals expressed b-galactosidase under control of the IP5 2-kinase promoter, it was shown that the enzyme was prominently expressed in embryonic neural tube, the myotome of the somites and in the yolk sac. In adult animals the enzyme was expressed in the hippocampus, the cortex, the Purkinje layer, cardiomyocytes, and the testes.
Disruption of Ipk2 and Ipk1 in Arabidopsis results in plants that lack the ability to generate phytate (IP6) in seeds, which has significant ramifications in the agricultural and signaling communities (Stevenson-Paulik et al., 2005). Phytate is a regulator of intracellular signaling, an anti-nutrient in animal feed, and a phosphate store in plant seeds. It was shown that AtIPK1 and AtIPK2β double mutant plants produced normal seed yield, strongly indicating that the disruption of ipk2 and ipk1 in cereal grains would represent a viable strategy for generating nutritionally improved crops. This study also provided indications that loss of AtIPK1 and IP6 production disrupted phosphate signaling and sensing (Stevenson-Paulik et al., 2005).
Furthermore, the work of the Wente laboratory has shown that reduction of zebrafish Ipk1 results in abnormal development (Sarmah et al., 2005). Strikingly, these mutants exhibited defects in heart left-right asymmetry and provide evidence that IP6 production is required for proper asymmetric calcium flux during left-right heart specification.
Summary
Our laboratory studies the biology and enzyme regulation of inositol signal transduction pathways, which are activated in response to a wide range of stimuli. As a six-carbon cyclitol, inositol and its numerous phosphorylated derivatives efficiently generate combinatorial ensembles of signaling molecules. Through the cloning and characterization of inositol polyphosphate kinases (IPK), novel roles for inositol tetrakisphosphate (IP4), inositol pentakisphosphate (IP5), and inositol hexakisphosphate (IP6) and inositol pyrophosphates (PP-IPs), have been identified. Studies have linked the IPKs and their inositide products to the regulation of nuclear processes including gene expression, chromatin remodeling, mRNA export, DNA repair and telomere maintenance. Analysis of IPK knockout animals has revealed a role for production of IPs in regulation of embryogenesis and organism development.
The discoveries of the IPK proteins and their connection to nuclear signaling have generated significant interest in the field. Furthermore, they have provided interesting clues into the evolution of inositide signaling pathways. Ipk2/IPMK and IPS/IP6K family members are conserved from yeast to man. In contrast, the IP3 3-kinase (ITPK) branch is observed in selected metazoans and not in plant or fungi. This may imply that Ipk2 and IPS activities evolved first among the group. The promiscuity of the Ipk2 protein further supports this notion and may provide the cell with a means to generate many IP species in a genetically economical fashion. Studies of yeast inositide signaling reveal that these simple eukaryotes do not have an IP3 receptor in their genome and do not utilize diacylglycerol to activate protein kinase C. Thus, it appears that the canonical “text book” aspects of inositide signaling pathways are not conserved throughout eukaryotic evolution. In light of the conservation of Ipk2/IPMK, Ipk1 and IPS/IP6K pathways from yeast to man it is interesting to speculate that a primordial role of phospholipase C-induced, IPK-dependent inositide signaling was to regulate nuclear processes. As calcium and PKC signaling evolved in metazoans, these may have greatly enhanced signaling capabilities. Recent studies demonstrating an essential role for IP5, IP6 and possibly PP-IP production in metazoan development highlight the importance of IPK signaling in cellular responses in metazoans. With these thoughts in mind, we eagerly await future studies aimed at further elucidating how these signaling codes participate in developmental processes and the control of gene expression, mRNA export, and DNA metabolism.
Acknowledgments
We wish to thank members of the lab, past and present, and numerous colleagues for helpful discussions. This work is supported by funds from the Howard Hughes Medical Institute, and from the National Institutes of Health grants HL-55672 and DK-070272
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Albert C, Safrany ST, Bembenek ME, Reddy KM, Reddy K, Falck J, Brocker M, Shears SB, Mayr GW. Biological variability in the structures of diphosphoinositol polyphosphates in Dictyostelium discoideum and mammalian cells. Biochem J. 1997;327 (Pt 2):553–60. doi: 10.1042/bj3270553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcazar-Roman AR, Tran EJ, Guo S, Wente SR. Inositol hexakisphosphate and Gle1 activate the DEAD-box protein Dbp5 for nuclear mRNA export. Nat Cell Biol. 2006;8:711–16. doi: 10.1038/ncb1427. [DOI] [PubMed] [Google Scholar]
- Audhya A, Emr SD. Regulation of PI4,5P2 synthesis by nuclear-cytoplasmic shuttling of the Mss4 lipid kinase. Embo J. 2003;22:4223–36. doi: 10.1093/emboj/cdg397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auesukaree C, Tochio H, Shirakawa M, Kaneko Y, Harashima S. Plc1p, Arg82p, and Kcs1p, enzymes involved in inositol pyrophosphate synthesis, are essential for phosphate regulation and polyphosphate accumulation in Saccharomyces cerevisiae. J Biol Chem. 2005;280:25127–133. doi: 10.1074/jbc.M414579200. [DOI] [PubMed] [Google Scholar]
- Bailis AM, Lopes JM, Kohlwein SD, Henry SA. Cis and trans regulatory elements required for regulation of the CHO1 gene of Saccharomyces cerevisiae. Nucleic Acids Res. 1992;20:1411–18. doi: 10.1093/nar/20.6.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechet J, Greenson M, Wiame JM. Mutations affecting the repressibility of arginine biosynthetic enzymes in Saccharomyces cerevisiae. European Journal of Biochemistry. 1970;12:31–39. doi: 10.1111/j.1432-1033.1970.tb00817.x. [DOI] [PubMed] [Google Scholar]
- Bercy J, Dubois E, Messenguy F. Regulation of arginine metabolism in Saccharomyces cerevisiae: expression of the three ARGR regulatory genes and cellular localization of their products. Gene. 1987;55:277–85. doi: 10.1016/0378-1119(87)90287-3. [DOI] [PubMed] [Google Scholar]
- Berridge M. Inositol triphosphate and calcium signaling. Nature. 1993;361:315–25. doi: 10.1038/361315a0. [DOI] [PubMed] [Google Scholar]
- Brearley CA, Hanke DE. Metabolic evidence for the order of addition of individual phosphate esters in the myo-inositol moiety of inositol hexakisphosphate in the duckweed Spirodela polyrhiza L. Biochem J. 1996;314 (Pt 1):227–33. doi: 10.1042/bj3140227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner JH, Walter P. Gene recruitment of the activated INO1 locus to the nuclear membrane. PLoS Biol. 2004;2:e342. doi: 10.1371/journal.pbio.0020342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrum J, Jordan S, Safrany ST, Rodgers W. Visualization of inositol phosphate-dependent mobility of Ku: depletion of the DNA-PK cofactor InsP6 inhibits Ku mobility. Nucleic Acids Res. 2004;32:2776–84. doi: 10.1093/nar/gkh592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carman GM, Henry SA. Phospholipid biosynthesis in the yeast Saccharomyces cerevisiae and interrelationship with other metabolic processes. Prog Lipid Res. 1999;38:361–99. doi: 10.1016/s0163-7827(99)00010-7. [DOI] [PubMed] [Google Scholar]
- Chang SC, Miller AL, Feng Y, Wente SR, Majerus PW. The human homolog of the rat inositol phosphate multikinase is an inositol 1,3,4,6-tetrakisphosphate 5-kinase. J Biol Chem. 2002;277:43836–843. doi: 10.1074/jbc.M206134200. Epub 42002 Sep 43839. [DOI] [PubMed] [Google Scholar]
- Cheng MK, Shearn A. The direct interaction between ASH2, a Drosophila trithorax group protein, and SKTL, a nuclear phosphatidylinositol 4-phosphate 5-kinase, implies a role for phosphatidylinositol 4,5-bisphosphate in maintaining transcriptionally active chromatin. Genetics. 2004;167:1213–23. doi: 10.1534/genetics.103.018721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi KY, Kim HK, Lee SY, Moon KH, Sim SS, Kim JW, Chung HK, Rhee SG. Molecular cloning and expression of a complementary DNA for inositol 1,4,5-trisphosphate 3-kinase. Science. 1990;248:64–66. doi: 10.1126/science.2157285. [DOI] [PubMed] [Google Scholar]
- Cocco L, Martelli AM, Vitale M, Falconi M, Barnabei O, Stewart Gilmour R, Manzoli FA. Inositides in the nucleus: regulation of nuclear PI-PLCbeta1. Adv Enzyme Regul. 2002;42:181–93. doi: 10.1016/s0065-2571(01)00030-9. [DOI] [PubMed] [Google Scholar]
- D’Santos C, Clarke JH, Roefs M, Halstead JR, Divecha N. Nuclear inositides. Eur J Histochem. 2000;44:51–60. [PubMed] [Google Scholar]
- D’Santos CS, Clarke JH, Divecha N. Phospholipid signaling in the nucleus: Een DAG uit het leven van de inositide signalering in de nucleus. Biochimica et Biophysica Acta. 1998;1436:201–32. doi: 10.1016/s0005-2760(98)00146-5. [DOI] [PubMed] [Google Scholar]
- Desany BA, Alcasabas AA, Bachant JB, Elledge SJ. Recovery from DNA replicational stress is the essential function of the S-phase checkpoint pathway. Genes Dev. 1998;12:2956–70. doi: 10.1101/gad.12.18.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois E, Bercy J, Messenguy F. Characterization of two genes, ARGRI and ARGRII required for specific regulation of arginine metabolism in yeast. Molecular and General Genetics. 1987;207:142–48. doi: 10.1007/BF00331501. [DOI] [PubMed] [Google Scholar]
- Dubois E, Dewaste V, Erneux C, Messenguy F. Inositol polyphosphate kinase activity of Arg82/ArgRIII is not required for the regulation of the arginine metabolism in yeast. FEBS Lett. 2000;486:300–04. doi: 10.1016/s0014-5793(00)02318-8. [DOI] [PubMed] [Google Scholar]
- Dubois E, Messenguy F. Pleiotropic function of ArgRIIIp (Arg82p), one of the regulators of arginine metabolism in Saccharomyces cerevisiae. Role in expression of cell-type-specific genes. Mol Gen Genet. 1994;243:315–24. doi: 10.1007/BF00301067. [DOI] [PubMed] [Google Scholar]
- Dubois E, Scherens B, Vierendeels F, Ho MM, Messenguy F, Shears SB. In Saccharomyces cerevisiae, the inositol polyphosphate kinase activity of Kcs1p is required for resistance to salt stress, cell wall integrity, and vacuolar morphogenesis. J Biol Chem. 2002;277:23755–63. doi: 10.1074/jbc.M202206200. [DOI] [PubMed] [Google Scholar]
- El Alami M, Messenguy F, Scherens B, Dubois E. Arg82p is a bifunctional protein whose inositol polyphosphate kinase activity is essential for nitrogen and PHO gene expression but not for Mcm1p chaperoning in yeast. Mol Microbiol. 2003;49:457–68. doi: 10.1046/j.1365-2958.2003.03562.x. [DOI] [PubMed] [Google Scholar]
- El Bakkoury M, Dubois E, Messenguy F. Mol Microbiol. 2000;35:15–31. doi: 10.1046/j.1365-2958.2000.01665.x. [DOI] [PubMed] [Google Scholar]; Recruitment of the yeast MADS-box proteins, ArgRI and Mcm1 by the pleiotropic factor ArgRIII is required for their stability. Molecular Microbiology. 2000;35:15–31. doi: 10.1046/j.1365-2958.2000.01665.x. [DOI] [PubMed] [Google Scholar]
- Estevez F, Pulford D, Stark MJ, Carter AN, Downes CP. Inositol trisphosphate metabolism in Saccharomyces cerevisiae: identification, purification and properties of inositol 1,4,5-trisphosphate 6-kinase. Biochem J. 1994;302 (Pt 3):709–16. doi: 10.1042/bj3020709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faenza I, Billi AM, Follo MY, Fiume R, Martelli AM, Cocco L, Manzoli L. Nuclear phospholipase C signaling through type 1 IGF receptor and its involvement in cell growth and differentiation. Anticancer Res. 2005;25:2039–41. [PubMed] [Google Scholar]
- Feng X, Hara Y, Riabowol K. Different HATS of the ING1 gene family. Trends Cell Biol. 2002;12:532–38. doi: 10.1016/s0962-8924(02)02391-7. [DOI] [PubMed] [Google Scholar]
- Feng Y, Wente SR, Majerus PW. Overexpression of the inositol phosphatase SopB in human 293 cells stimulates cellular chloride influx and inhibits nuclear mRNA export. Proc Natl Acad Sci U S A. 2001;98:875–79. doi: 10.1073/pnas.021558098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederick JP, Mattiske D, Wofford JA, Megosh L, Drake LY, Chiou ST, Hogan BL, York JD. An essential role for a nuclear inositol polyphosphate multikinase, Ipk2, in mouse embryogenesis and second messenger production. Proc Natl Acad Sci U S A. 2005;102(24):8454–9. doi: 10.1073/pnas.0503706102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii M, York JD. A role for rat inositol polyphosphate kinases rIPK2 and rIPK1 in inositol pentakisphosphate and inositol hexakisphosphate production in rat-1 cells. J Biol Chem. 2005;280:1156–64. doi: 10.1074/jbc.M412006200. [DOI] [PubMed] [Google Scholar]
- Gerasimenko O, Gerasimenko J. New aspects of nuclear calcium signalling. J Cell Sci. 2004;117:3087–94. doi: 10.1242/jcs.01295. [DOI] [PubMed] [Google Scholar]
- Gozani O, Karuman P, Jones DR, Ivanov D, Cha J, Lugovskoy AA, Baird CL, Zhu H, Field SJ, Lessnick SL, Villasenor J, Mehrotra B, Chen J, Rao VR, Brugge JS, Ferguson CG, Payrastre B, Myszka DG, Cantley LC, Wagner G, Divecha N, Prestwich GD. The PHD finger of the chromatin-associated protein ING2 functions as a nuclear phosphoinositide receptor. Cell. 2003;114:99–111. doi: 10.1016/s0092-8674(03)00480-x. [DOI] [PubMed] [Google Scholar]
- Hanakahi LA, Bartlet-Jones M, Chappell C, Pappin D, West SC. Binding of inositol phosphate to DNA-PK and stimulation of double- strand break repair. Cell. 2000;102:721–29. doi: 10.1016/s0092-8674(00)00061-1. [DOI] [PubMed] [Google Scholar]
- Hanakahi LA, West SC. Specific interaction of IP6 with human Ku70/80, the DNA-binding subunit of DNA-PK. Embo J. 2002;21:2038–44. doi: 10.1093/emboj/21.8.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch JP, Henry SA. Expression of the Saccharomyces cerevisiae inositol-1-phosphate synthase (INO1) gene is regulated by factors that affect phospholipid synthesis. Mol Cell Biol. 1986;6:3320–28. doi: 10.1128/mcb.6.10.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hokin LE. Receptors and phosphoinositide-generated second messengers. Annu Rev Biochem. 1985;54:205–35. doi: 10.1146/annurev.bi.54.070185.001225. [DOI] [PubMed] [Google Scholar]
- Huang KN, Symington LS. Suppressors of a Saccharomyces cerevisiae pkc1 mutation identify alleles of the phosphatase gene PTC1 and of a novel gene encoding a putative basic leucine zipper protein. Genetics. 1995;141:1275–85. doi: 10.1093/genetics/141.4.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvine RF, Schell MJ. Back in the water: the return of the inositol phosphates. Nat Rev Mol Cell Biol. 2001;2:327–38. doi: 10.1038/35073015. [DOI] [PubMed] [Google Scholar]
- Jones DR, Bultsma Y, Keune WJ, Halstead JR, Elouarrat D, Mohammed S, Heck AJ, D’Santos CS, Divecha N. Nuclear PtdIns5P as a transducer of stress signaling: an in vivo role for PIP4Kbeta. Mol Cell. 2006;23:685–95. doi: 10.1016/j.molcel.2006.07.014. [DOI] [PubMed] [Google Scholar]
- Kapeller R, Cantley LC. Phosphatidylinositol 3-Kinase. BioEssays. 1994;16:565–76. doi: 10.1002/bies.950160810. [DOI] [PubMed] [Google Scholar]
- Laussmann T, Eujen R, Weisshuhn CM, Thiel U, Vogel G. Structures of diphospho-myo-inositol pentakisphosphate and bisdiphospho-myo-inositol tetrakisphosphate from Dictyostelium resolved by NMR analysis. Biochem J. 1996;315 (Pt 3):715–20. doi: 10.1042/bj3150715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laussmann T, Hansen A, Reddy KM, Reddy KK, Falck JR, Vogel G. Diphospho-myo-inositol phosphates in Dictyostelium and Polysphondylium: identification of a new bisdiphospho-myo-inositol tetrakisphosphate. FEBS Lett. 1998;426:145–50. doi: 10.1016/s0014-5793(98)00329-9. [DOI] [PubMed] [Google Scholar]
- Laussmann T, Pikzack C, Thiel U, Mayr GW, Vogel G. Diphospho-myo-inositol phosphates during the life cycle of Dictyostelium and Polysphondylium. Eur J Biochem. 2000;267:2447–51. doi: 10.1046/j.1432-1327.2000.01264.x. [DOI] [PubMed] [Google Scholar]
- Laussmann T, Reddy KM, Reddy KK, Falck JR, Vogel G. Diphospho-myo-inositol phosphates from Dictyostelium identified as D-6-diphospho-myo-inositol pentakisphosphate and D-5,6-bisdiphospho-myo-inositol tetrakisphosphate. Biochem J. 1997;322 (Pt 1):31–33. doi: 10.1042/bj3220031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Ilin S, Wang W, Duncan EM, Wysocka J, Allis CD, Patel DJ. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature. 2006;442(7098):91–5. doi: 10.1038/nature04802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes JM, Hirsch JP, Chorgo PA, Schulze KL, Henry SA. Analysis of sequences in the INO1 promoter that are involved in its regulation by phospholipid precursors. Nucleic Acids Res. 1991;19:1687–93. doi: 10.1093/nar/19.7.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo HR, Saiardi A, Yu H, Nagata E, Ye K, Snyder SH. Inositol pyrophosphates are required for DNA hyperrecombination in protein kinase c1 mutant yeast. Biochemistry. 2002;41:2509–15. doi: 10.1021/bi0118153. [DOI] [PubMed] [Google Scholar]
- Ma Y, Lieber MR. Binding of inositol hexakisphosphate (IP6) to Ku but not to DNA-PKcs. J Biol Chem. 2002;277:10756–59. doi: 10.1074/jbc.C200030200. [DOI] [PubMed] [Google Scholar]
- Majerus PW. Inositol phosphate biochemistry. Annu Rev Biochem. 1992;61:225–50. doi: 10.1146/annurev.bi.61.070192.001301. [DOI] [PubMed] [Google Scholar]
- Manzoli L, Billi AM, Martell AM, Cocco L. Regulation of nuclear phospholipase C activity. Acta Biochim Pol. 2004;51:391–5. [PubMed] [Google Scholar]
- Martelli AM, Manzoli L, Cocco L. Nuclear inositides: facts and perspectives. Pharmacol Ther. 2004;101:47–64. doi: 10.1016/j.pharmthera.2003.10.003. [DOI] [PubMed] [Google Scholar]
- Martin DG, Baetz K, Shi X, Walter KL, Macdonald VE, Wlodarski MJ, Gozani O, Hieter P, Howe L. The yng1p plant homeodomain finger is a methyl-histone binding module that recognizes lysine 4-methylated histone h3. Mol Cell Biol. 2006;26:7871–79. doi: 10.1128/MCB.00573-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messenguy F, Dubois E. Role of MADS box proteins and their cofactors in combinatorial control of gene expression and cell development. Gene. 2003;316:1–21. doi: 10.1016/s0378-1119(03)00747-9. [DOI] [PubMed] [Google Scholar]
- Mikoshiba K. The InsP3 receptor and intracellular Ca2+ signaling. Curr Opin Neurobiol. 1997;7:339–45. doi: 10.1016/s0959-4388(97)80061-x. [DOI] [PubMed] [Google Scholar]
- Miller AL, Suntharalingam M, Johnson SL, Audhya A, Emr SD, Wente SR. Cytoplasmic inositol hexakisphosphate production is sufficient for mediating the Gle1-mRNA export pathway. J Biol Chem. 2004;279:51022–32. doi: 10.1074/jbc.M409394200. [DOI] [PubMed] [Google Scholar]
- Nalaskowski MM, Deschermeier C, Fanick W, Mayr GW. The human homologue of yeast ArgRIII protein is an inositol phosphate multikinase with predominantly nuclear localization. Biochem J. 2002;366:549–56. doi: 10.1042/BJ20020327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizuka Y. Studies and perspectives of protein kinase C. Science. 1986;233:305–12. doi: 10.1126/science.3014651. [DOI] [PubMed] [Google Scholar]
- Norris FA, Wilson MP, Wallis TS, Galyov EE, Majerus PW. SopB, a protein required for virulence of Salmonella dublin, is an inositol phosphate phosphatase. Proc Natl Acad Sci U S A. 1998;95:14057–59. doi: 10.1073/pnas.95.24.14057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odom AR. Doctoral Thesis. Duke University; 2002. Nuclear Functions of an Inositol Polyphosphate Kinase Pathway. [Google Scholar]
- Odom AR, Stahlberg A, Wente SR, York JD. A role for nuclear inositol 1,4,5-trisphosphate kinase in transcriptional control. Science. 2000;287:2026–29. doi: 10.1126/science.287.5460.2026. [DOI] [PubMed] [Google Scholar]
- Ongusaha PP, Hughes PJ, Davey J, Michell RH. Inositol hexakisphosphate in Schizosaccharomyces pombe: synthesis from Ins(1,4,5)P3 and osmotic regulation. Biochem J. 1998;335 (Pt 3):671–9. doi: 10.1042/bj3350671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ongusaha PP, Hughes PJ, Hirata M, Davey J, Michell RH. The inositol 1,4,5-trisphosphate 6-kinase of Schizosaccharomyces pombe. Biochem Soc Trans. 1997;25:105S. doi: 10.1042/bst025105s. [DOI] [PubMed] [Google Scholar]
- Pena PV, Davrazou F, Shi X, Walter KL, Verkhusha VV, Gozani O, Zhao R, Kutateladze TG. Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature. 2006;442(7098):100–3. doi: 10.1038/nature04814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouillon V, Hascakova-Bartova R, Pajak B, Adam E, Bex F, Dewaste V, Van Lint C, Leo O, Erneux C, Schurmans S. Inositol 1,3,4,5-tetrakisphosphate is essential for T lymphocyte development. Nat Immunol. 2003;4:1136–43. doi: 10.1038/ni980. [DOI] [PubMed] [Google Scholar]
- Rando OJ, Chi TH, Crabtree GR. Second messenger control of chromatin remodeling. Nat Struct Biol. 2003;10:81–83. doi: 10.1038/nsb0203-81. [DOI] [PubMed] [Google Scholar]
- Rando OJ, Zhao K, Janmey P, Crabtree GR. Phosphatidylinositol-dependent actin filament binding by the SWI/SNF-like BAF chromatin remodeling complex. Proc Natl Acad Sci U S A. 2002;99:2824–29. doi: 10.1073/pnas.032662899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnick AC, Snowman AM, Kang BN, Hurt KJ, Snyder SH, Saiardi A. Inositol polyphosphate multikinase is a nuclear PI3-kinase with transcriptional regulatory activity. Proc Natl Acad Sci U S A. 2005;102:12783–8. doi: 10.1073/pnas.0506184102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero C, Desai P, DeLillo N, Vancura A. Expression of FLR1 transporter requires phospholipase C and is repressed by Mediator. J Biol Chem. 2006;281:5677–85. doi: 10.1074/jbc.M506728200. [DOI] [PubMed] [Google Scholar]
- Saiardi A, Caffrey JJ, Snyder SH, Shears SB. The inositol hexakisphosphate kinase family. Catalytic flexibility and function in yeast vacuole biogenesis. J Biol Chem. 2000a;275:24686–92. doi: 10.1074/jbc.M002750200. [DOI] [PubMed] [Google Scholar]
- Saiardi A, Caffrey JJ, Snyder SH, Shears SB. Inositol polyphosphate multikinase (ArgRIII) determines nuclear mRNA export in Saccharomyces cerevisiae. FEBS Lett. 2000b;468:28–32. doi: 10.1016/s0014-5793(00)01194-7. [DOI] [PubMed] [Google Scholar]
- Saiardi A, Erdjument-Bromage H, Snowman AM, Tempst P, Snyder SH. Synthesis of diphosphoinositol pentakisphosphate by a newly identified family of higher inositol polyphosphate kinases. Curr Biol. 1999;9:1323–26. doi: 10.1016/s0960-9822(00)80055-x. [DOI] [PubMed] [Google Scholar]
- Saiardi A, Nagata E, Luo HR, Snowman AM, Snyder SH. Identification and characterization of a novel inositol hexakisphosphate kinase. J Biol Chem. 2001;276:39179–85. doi: 10.1074/jbc.M106842200. [DOI] [PubMed] [Google Scholar]
- Saiardi A, Nagata E, Luo HR, Sawa A, Luo X, Snowman AM, Snyder SH. Mammalian inositol polyphosphate multikinase synthesizes inositol 1,4,5-trisphosphate and an inositol pyrophosphate. Proc Natl Acad Sci U S A. 2001;98:2306–11. doi: 10.1073/pnas.041614598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiardi A, Resnick AC, Snowman AM, Wendland B, Snyder SH. Inositol pyrophosphates regulate cell death and telomere length through phosphoinositide 3-kinase-related protein kinases. Proc Natl Acad Sci U S A. 2005;102:1911–14. doi: 10.1073/pnas.0409322102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiardi A, Sciambi C, McCaffery JM, Wendland B, Snyder SH. Inositol pyrophosphates regulate endocytic trafficking. Proc Natl Acad Sci U S A. 2002;99:14206–11. doi: 10.1073/pnas.212527899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarmah B, Latimer AJ, Appel B, Wente SR. Inositol polyphosphates regulate zebrafish left-right asymmetry. Dev Cell. 2005;9:133–45. doi: 10.1016/j.devcel.2005.05.002. [DOI] [PubMed] [Google Scholar]
- Seeds AM, Bastidas RJ, York JD. Molecular definition of a novel inositol polyphosphate metabolic pathway initiated by inositol 1,4,5-trisphosphate 3-kinase activity in Saccharomyces cerevisiae. J Biol Chem. 2005;280(30):27654–61. doi: 10.1074/jbc.M505089200. [DOI] [PubMed] [Google Scholar]
- Seeds AM, Sandquist JC, Spana EP, York JD. A Molecular Basis for Inositol Polyphosphate Synthesis in Drosophila melanogaster. J Biol Chem. 2004;279:47222–32. doi: 10.1074/jbc.M408295200. [DOI] [PubMed] [Google Scholar]
- Shears SB. The pathway of myo-inositol 1,3,4-trisphosphate phosphorylation in liver. Identification of myo-inositol 1,3,4-trisphosphate 6-kinase, myo-inositol 1,3,4-trisphosphate 5-kinase, and myo-inositol 1,3,4,6-tetrakisphosphate 5-kinase. J Biol Chem. 1989;264:19879–86. [PubMed] [Google Scholar]
- Shears SB. The versatility of inositol phosphates as cellular signals. Biochim Biophys Acta. 1998;1436:49–67. doi: 10.1016/s0005-2760(98)00131-3. [DOI] [PubMed] [Google Scholar]
- Shen X, Xiao H, Ranallo R, Wu WH, Wu C. Modulation of ATP-dependent chromatin-remodeling complexes by inositol polyphosphates. Science. 2003;299:112–14. doi: 10.1126/science.1078068. [DOI] [PubMed] [Google Scholar]
- Shi J, Wang H, Hazebroek J, Ertl DS, Harp T. The maize low-phytic acid 3 encodes a myo-inositol kinase that plays a role in phytic acid biosynthesis in developing seeds. Plant J. 2005;42:708–19. doi: 10.1111/j.1365-313X.2005.02412.x. [DOI] [PubMed] [Google Scholar]
- Shi J, Wang H, Wu Y, Hazebroek J, Meeley RB, Ertl DS. The maize low-phytic acid mutant lpa2 is caused by mutation in an inositol phosphate kinase gene. Plant Physiol. 2003;131:507–15. doi: 10.1104/pp.014258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Hong T, Walter KL, Ewalt M, Michishita E, Hung T, Carney D, Pena P, Lan F, Kaadige MR, Lacoste N, Cayrou C, Davrazou F, Saha A, Cairns BR, Ayer DE, Kutateladze TG, Shi Y, Cote J, Chua KF, Gozani O. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature. 2006;442(7098):96–9. doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steger DJ, Haswell ES, Miller AL, Wente SR, O’Shea EK. Regulation of chromatin remodeling by inositol polyphosphates. Science. 2003;299:114–16. doi: 10.1126/science.1078062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens L, Irvine R. Stepwise phosphorylation of myo-inositol leading to myo-inositol hexakisphosphate in Dictyostelium. Nature. 1990;326:580–2. doi: 10.1038/346580a0. [DOI] [PubMed] [Google Scholar]
- Stevenson-Paulik J, Bastidas RJ, Chiou ST, Frye RA, York JD. Generation of phytate-free seeds in Arabidopsis through disruption of inositol polyphosphate kinases. Proc Natl Acad Sci U S A. 2005;102:12612–17. doi: 10.1073/pnas.0504172102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson-Paulik J, Odom AR, York JD. Molecular and biochemical characterization of two plant inositol polyphosphate 6-/3-/5-kinases. J Biol Chem. 2002;277:42711–18. doi: 10.1074/jbc.M209112200. [DOI] [PubMed] [Google Scholar]
- Strahl T, Hama H, DeWald DB, Thorner J. Yeast phosphatidylinositol 4-kinase, Pik1, has essential roles at the Golgi and in the nucleus. J Cell Biol. 2005;171:967–79. doi: 10.1083/jcb.200504104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takazawa K, Vandekerckhove J, Dumont JE, Erneux C. Cloning and expression in Escherichia coli of a rat brain cDNA encoding a Ca2+/calmodulin-sensitive inositol 1,4,5-trisphosphate 3-kinase. Biochem J. 1990;272:107–12. doi: 10.1042/bj2720107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topham MK, Prescott SM. Diacylglycerol kinases: regulation and signaling roles. Thromb Haemost. 2002;88:912–18. [PubMed] [Google Scholar]
- Verbsky J, Lavine K, Majerus PW. Disruption of the mouse inositol 1,3,4,5,6-pentakisphosphate 2-kinase gene, associated lethality, and tissue distribution of 2-kinase expression. Proc Natl Acad Sci U S A. 2005a;102:8448–53. doi: 10.1073/pnas.0503656102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbsky J, Majerus PW. Increased levels of inositol hexakisphosphate (InsP6) protect HEK293 cells from tumor necrosis factor (alpha)- and Fas-induced apoptosis. J Biol Chem. 2005;280:29263–8. doi: 10.1074/jbc.M503366200. [DOI] [PubMed] [Google Scholar]
- Verbsky JW, Chang SC, Wilson MP, Mochizuki Y, Majerus PW. The pathway for the production of inositol hexakisphosphate in human cells. J Biol Chem. 2005b;280:1911–20. doi: 10.1074/jbc.M411528200. [DOI] [PubMed] [Google Scholar]
- Verbsky JW, Wilson MP, Kisseleva MV, Majerus PW, Wente SR. The synthesis of inositol hexakisphosphate. Characterization of human inositol 1,3,4,5,6-pentakisphosphate 2-kinase. J Biol Chem. 2002;277:31857–62. doi: 10.1074/jbc.M205682200. [DOI] [PubMed] [Google Scholar]
- Weirich CS, Erzberger JP, Flick JS, Berger JM, Thorner J, Weis K. Activation of the DExD/H-box protein Dbp5 by the nuclear-pore protein Gle1 and its coactivator InsP6 is required for mRNA export. Nat Cell Biol. 2006;8:668–76. doi: 10.1038/ncb1424. [DOI] [PubMed] [Google Scholar]
- Wilson MP, Majerus PW. Isolation of inositol 1,3,4-trisphosphate 5/6-kinase, cDNA cloning and expression of the recombinant enzyme. J Biol Chem. 1996;271:11904–10. doi: 10.1074/jbc.271.20.11904. [DOI] [PubMed] [Google Scholar]
- Wilson MP, Majerus PW. Characterization of a cDNA encoding Arabidopsis thaliana inositol 1,3,4-trisphosphate 5/6-kinase. Biochem Biophys Res Commun. 1997;232:678–81. doi: 10.1006/bbrc.1997.6355. [DOI] [PubMed] [Google Scholar]
- Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, Kauer M, Tackett AJ, Chait BT, Badenhorst P, Wu C, Allis CD. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442(7098):86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- Xia HJ, Brearley C, Elge S, Kaplan B, Fromm H, Mueller-Roeber B. Arabidopsis inositol polyphosphate 6-/3-kinase is a nuclear protein that complements a yeast mutant lacking a functional ArgR-Mcm1 transcription complex. Plant Cell. 2003;15:449–63. doi: 10.1105/tpc.006676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagisawa H. Nucleocytoplasmic shuttling of phospholipase C-delta(1): A link to Ca(2+) J Cell Biochem. 2006;97:233–43. doi: 10.1002/jcb.20677. [DOI] [PubMed] [Google Scholar]
- Yoon S, Govind CK, Qiu H, Kim SJ, Dong J, Hinnebusch AG. Recruitment of the ArgR/Mcm1p repressor is stimulated by the activator Gcn4p: a self-checking activation mechanism. Proc Natl Acad Sci U S A. 2004;101:11713–18. doi: 10.1073/pnas.0404652101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- York JD. Regulation of nuclear processes by inositol polyphosphates. Biochim Biophys Acta. 2006;1761:552–9. doi: 10.1016/j.bbalip.2006.04.014. [DOI] [PubMed] [Google Scholar]
- York JD, Guo S, Odom AR, Spiegelberg BD, Stolz LE. An expanded view of inositol signaling. Adv Enzyme Regul. 2001;41:57–71. doi: 10.1016/s0065-2571(00)00025-x. [DOI] [PubMed] [Google Scholar]
- York JD, Odom AR, Murphy R, Ives EB, Wente SR. A phospholipase C-dependent inositol polyphosphate kinase pathway required for efficient messenger RNA export. Science. 1999;285:96–100. doi: 10.1126/science.285.5424.96. [DOI] [PubMed] [Google Scholar]
- York SJ, Armbruster BN, Greenwell P, Petes TD, York JD. Inositol diphosphate signaling regulates telomere length. J Biol Chem. 2005;280:4264–9. doi: 10.1074/jbc.M412070200. [DOI] [PubMed] [Google Scholar]
- Zhao K, Wang W, Rando OJ, Xue Y, Swiderek K, Kuo A, Crabtree GR. Rapid and phosphoinositol-dependent binding of the SWI/SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell. 1998;95:625–36. doi: 10.1016/s0092-8674(00)81633-5. [DOI] [PubMed] [Google Scholar]