Abstract
Sub-Saharan Africa has consistently been shown to be the most genetically diverse region in the world. Despite the fact that a substantial portion of this variation is partitioned between groups practicing a variety of subsistence strategies and speaking diverse languages, there is currently no consensus on the genetic relationships of sub-Saharan African populations. San (a subgroup of KhoeSan) and many Pygmy groups maintain hunter-gatherer lifestyles and cluster together in autosomal-based analysis, whereas non-Pygmy Niger-Kordofanian speakers (non-Pygmy NKs) predominantly practice agriculture and show substantial genetic homogeneity despite their wide geographic range throughout sub-Saharan Africa. However, KhoeSan, who speak a set of relatively unique click-based languages, have long been thought to be an early branch of anatomically modern humans based on phylogenetic analysis. To formally test models of divergence among the ancestors of modern African populations, we resequenced a sample of San, Eastern, and Western Pygmies and non-Pygmy NKs individuals at 40 nongenic (∼2 kb) regions and then analyzed these data within an Approximate Bayesian Computation (ABC) framework. We find substantial support for a model of an early divergence of KhoeSan ancestors from a proto-Pygmy-non-Pygmy NKs group ∼110 thousand years ago over a model incorporating a proto-KhoeSan–Pygmy hunter-gatherer divergence from the ancestors of non-Pygmy NKs. The results of our analyses are consistent with previously identified signals of a strong bottleneck in Mbuti Pygmies and a relatively recent expansion of non-Pygmy NKs. We also develop a number of methodologies that utilize “pseudo-observed” data sets to optimize our ABC-based inference. This approach is likely to prove to be an invaluable tool for demographic inference using genome-wide resequencing data.
Keywords: sub-Saharan Africa, resequencing, Approximate Bayesian Computation, KhoeSan, Pygmy, demographic history
Introduction
Sub-Saharan Africa has consistently been shown to be the most genetically diverse region in the world, with high levels of both within- and between-group variability (Olerup et al. 1991; Vigilant et al. 1991; Seielstad et al. 1999; Jorde et al. 2000; Tishkoff et al. 2009). These observations have been cited as genetic support for the “out-of-Africa” model for the origins of anatomically modern humans (AMH)—a model that posits larger long-term effective population sizes (and population structure) for sub-Saharan African populations and one or more bottlenecks that resulted in a subset of African diversity being carried by non-Africans as their ancestors dispersed from Africa approximately 65 thousand years ago (kya) (Relethford and Jorde 1999; Mellars 2006). The continent of Africa is also linguistically and culturally diverse, consisting of four distinct language families and over 1,500 languages (Lewis 2009). Niger-Kordofanian, one of the major language families with almost 400 million speakers, predominates throughout much of sub-Saharan Africa. This is due, in part, to the recent expansion of a subgroup of Niger-Kordofanian—known as Bantu (languages that are now spoken by more than 200 million people)—from the Cameroon/Nigeria border region to eastern and southern Africa approximately 5 kya (Blench 2006). Interestingly, the distribution of genetic variation among Niger-Kordofanian speakers is relatively homogenous across most of sub-Saharan Africa (Tishkoff et al. 2009; Bryc et al. 2010; Alves et al. 2011). This isolation-by-distance pattern is consistent with recurrent gene flow within this language family and may have been strongly affected by the rapid spread of Bantu-speaking farmers and a male-biased dispersal process (Underhill et al. 2001; Cruciani et al. 2002; Wood et al. 2005).
Two groups of culturally and phenotypically distinct peoples, KhoeSan (located predominantly in southwest Africa) and Pygmies (found throughout central Africa), appear to possess a distribution of genetic variation that distinguishes them from neighboring Niger-Kordofanian speakers as well as from each other (Tishkoff et al. 2009). Bolstered by evidence of long-term archaeological continuity and the possible persistence of a hunter-gatherer lifestyle in the San subgroup (though we note that there is still considerable debate regarding the latter, see Barnard (2006)), some researchers suggest that Khoisan languages, with their unique click sounds, may be modern representatives of those first spoken by early AMH (Blench 2006; Mitchell 2010). Most speakers of Khoisan languages also display certain characteristics in their physical appearance, such as reduced height, lighter skin color, and the presence of epicanthic eye folds (Cavalli-Sforza et al. 1994) that distinguish them from other sub-Saharan Africans. Their particular morphology may have been adapted to suit the semiarid conditions of the savanna, deserts, and plains that they inhabit (Nurse et al. 1985; Morris 2003). Genetic studies further underscore the importance of KhoeSan with regard to human origins. Analysis of classical (Cavalli-Sforza et al. 1994), mitochondrial DNA (mtDNA) (Ingman and Gyllensten 2001; Salas et al. 2002; Gonder et al. 2007; Behar et al. 2008), and Y-chromosome (Hammer et al. 2001; Cruciani et al. 2002; Semino et al. 2002) markers have consistently identified basal lineages associated with the KhoeSan, and the recent whole-genome sequencing of a KhoeSan individual revealed a large number of private polymorphisms (Schuster et al. 2010).
Phylogenetic analysis suggests that African Pygmy groups, many of whom also practice mobile hunter-gatherer lifestyles, are also substantially diverged from other AMH populations. Pygmies have no known indigenous language of their own and appear to have adopted the languages of their neighbors (Blench 2006). As many Pygmy groups thus now speak Niger-Kordofanian languages, from this point forward, we distinguish between Pygmies and non-Pygmy Niger-Kordofanian speakers (abbreviated non-Pygmy NKs). The short stature phenotype shared by most Pygmies (mean adult heights of less than 150–160 cm) may have evolved in response to the harsh environment of the central African rain forests where most Pygmy groups live, with many different hypotheses proposed for the exact mechanism of adaptation (reduction in caloric intake, thermoregulation, increased mobility, and earlier reproductive age) (Blench 2006; Perry and Dominy 2009). A recent study suggests that differential admixture could account for a significant part of height variation among Pygmy groups (Becker et al. 2011).
Though populations historically recognized as African Pygmies exhibit considerable between group morphological and cultural variation (including significant linguistic and subsistence strategy differences) (Hewlett 1996; Richards 2006; Perry and Dominy 2009), they have traditionally been divided into the broad groupings of Western and, though somewhat less frequently, Eastern Pygmies. Western Pygmies, which include Biaka, Baka, Bakola, Bezan, Bakoya, and a number of Babongo groups, live mainly west of the Congo Basin. Eastern Pygmies consist of a diverse group that includes Mbuti, Asua, and Efe living in and around the Ituri rainforest and various distinct Twa groups that inhabit an area further south extending toward Lake Victoria. Genetic data have generally supported the validity of this Western/Eastern grouping, though it is possible that this may simply represent sample collection-based ascertainment bias as there are Pygmy groups living in intermediate locations that have not been sampled for genetic analysis (e.g., those living in the Lake Tumba region in the Democratic Republic of Congo) (Pagezy 1998). Although both groups possess relatively basal Y chromosome (Hammer et al. 2001; Knight et al. 2003) and mtDNA (Salas et al. 2002; Quintana-Murci et al. 2008) lineages, Eastern and Western Pygmies appear to be genetically differentiated at the autosomal level (Patin et al. 2009). An earlier investigation of mtDNA variation (Destro-Bisol, Coia, et al. 2004) and more recent simulation-based population divergence estimates suggest a Western–Eastern Pygmy split time of approximately 20 kya (Patin et al. 2009; Batini, Lopes, et al. 2011).
Understanding the evolutionary relationships among KhoeSan, Pygmies, and non-Pygmy NKs is clearly important for understanding African prehistory; however, no general consensus has been reached regarding the order and timing of population divergence events or the evolution of the various characters that define these groups. Conventional thinking has tended toward a model where KhoeSan initially diverged from the ancestors of all other AMH groups and remained relatively isolated. However, the KhoeSan demonstrate deep genetic connections with other click-speaking peoples in Tanzania (Henn et al. 2011), with proposed time to the most recent common ancestor (TMRCA) estimates ranging from 35 to 110 kya (Chen et al. 2000; Knight et al. 2003; Gonder et al. 2007; Tishkoff et al. 2007). In addition, a genetic link with contemporary Ethiopian populations has also been proposed (Cruciani et al. 2002; Salas et al. 2002; Semino et al. 2002). This, along with linguistic evidence, suggests that the KhoeSan territory once covered a much larger area, extending further northwest toward the Great Rift Valley (Cavalli-Sforza et al. 1994; Blench 2006; Scheinfeldt et al. 2010). Recent autosomal-based analyses show a tendency for KhoeSan and Pygmies to cluster together and away from other sub-Saharan Africans (Zhivotovsky et al. 2003; Tishkoff et al. 2009; Sikora et al. 2011), leading to the hypothesis that the ancestors of these two populations may have once formed a proto-KhoeSan–Pygmy hunter-gatherer group that was geographically widespread before being encroached upon by expanding agricultural populations.
To examine their autosomal genetic relationships, we generate autosomal resequencing data from 40 independent neutral 2 kb regions in 8 sub-Saharan African populations, including San, Western, and Eastern Pygmies and non-Pygmy NKs. We employ Approximate Bayesian Computation (ABC) (Beaumont et al. 2002; Bertorelle et al. 2010) to test the fit of a range of demographic models. Such exploration was previously difficult as the underlying likelihood function would be too complicated to evaluate theoretically. However, the use of ABC allows the likelihood function to be approximated by many simulations of the model being tested, a strategy that is particularly effective when examining complex population genetic models with many parameters (see, e.g., Fagundes et al. 2007; Ghirotto et al. 2010; Wegmann and Excoffier 2010). Our strategy is to use the ABC framework to 1) determine the most likely model of sub-Saharan African demographic prehistory and then 2) estimate demographic parameters from the most likely model. We test several of the above-mentioned models, including the early KhoeSan divergence model and a model where the lineage leading to KhoeSan and Pygmy groups diverged together from the ancestors of present day non-Pygmy NKs populations. Although the relationships of non-Pygmy NKs and Pygmies (both Eastern and Western) have been evaluated previously (Patin et al. 2009), this is the first study to incorporate resequencing data from a KhoeSan group (the San).
Materials and Methods
Sample Collection
This study primarily involves the examination of 119 individuals from 8 sub-Saharan African populations. The Mandenka (n = 16) and Biaka (n = 16) samples have been described previously (Wall et al. 2008) and, along with the Mbuti (n = 12) samples, come from publicly available cell lines administered by the Centre d'Etude du Polymorphisme Humain Human Genome Diversity Panel (Cann et al. 2002). The Luhya samples (n = 18) come from the National Human Genome Research Institute collection at the Coriell Institute for Medical Research. The Bakola Pygmies from Cameroon (n = 16), Ngoumba from Cameroon (n = 16), San from Namibia (n = 9), and Shona from Zimbabwe (n = 16) represent newly presented collections typed for this study. All sampling procedures were approved by the University of Arizona Human Subjects Committees. The San samples were obtained with either verbal or written consent with approval from the Committee for Research on Human Subjects, University of the Witwatersrand (protocol number M980553). The Bakola samples were collected by Gabriella Spedini and G.D.-B. with verbal informed consent and approval from the University of Rome “La Sapienza.” The individual identifiers for publicly available samples are provided in supplementary table 1, Supplementary Material online. Prior to this study, a superset of the sub-Saharan African samples were tested for relatedness using 18 autosomal microsatellites via the RELPAIR 2.0.1 (Epstein et al. 2000) software package. To check for consistency, the relatedness results of our Human Genome Diversity Project (HGDP) samples were checked against that of Rosenberg (2006) and based on these results, 119 unrelated samples were selected for resequencing. For ease of presenting the data in some circumstance, the following code names have be assigned to sample collections: Bakola Pygmies (BAK), Biaka Pygmies (BIA), Luyha (LUH), Mandenka (MAN), Mbuti Pygmies (MBI), Ngoumba (NGO), San (SAN), and Shona (SHN).
Resequencing
Previously, we identified a set of 61 autosomal regions of ∼20 kb in length that are far from genes and that lie within regions that experience moderately high rates of crossing over (Hammer et al. 2008, 2010; Wall et al. 2008), of which we sequenced ∼6 kb in each. This strategy minimized the possibility of sequencing regions that are linked to sites affected by natural selection. In this study, we sequenced ∼2 kb within each of 40 of these regions (supplementary table 2, Supplementary Material online) in all samples. Sequence data are freely available on request from the corresponding author.
Statistical and Population Genetic Analysis
Haplotype phasing was performed as described by Wall et al. (2008), though the additional African populations not described in that paper were phased independently. Pairwise FST (1 − Hw/Hb) for individual loci was calculated using in-house code. P values for significance of pairwise FST values were assessed by a permutation test. In brief, an FST P value was estimated for each population pair, for each locus, using 1,000 random permutations of the data. These P values were combined using the method of Fisher (Fisher 1925) allowing us to form a single P value for each population pair. A Bonferroni correction was then used to correct the P values for multiple hypothesis testing. Principal coordinates analysis (PCO) (Gower 1966) was performed on pairwise FST (averaged across all 40 loci) using the “R” statistical package (R Development Core Team 2007) via the “cmdscale” function found in the native “stats” package.
Resequencing data were recoded into haplotypes to resemble multiallelic microsatellite data following Patin et al. (2009). We then applied an unsupervised admixture model using STRUCTURE (Pritchard et al. 2000; Falush et al. 2003). We experimented with both “Independent” and “Correlated” allele frequency models with the latter visually giving a better fit of population structure. We applied a burn-in of 1,000,000 iterations, with 1,000,000 Markov chain Monte Carlo (MCMC) steps after burn-in. We ran these settings for K = [2 … 8] groups, with 30 independent runs for each K. Our settings for the admixture model were Infer Alpha (initial value 1.0) (the Dirichlet parameter for degree of admixture), use individual Alpha for each population, gamma prior for Alpha (A = 0.05, B = 0.001). Our settings for the Correlated allele frequency model were Infer lambda (uniform distributions, initial value 1.0), different FST for subpopulations, prior mean FST = 0.04, prior standard deviation (SD) FST = 0.05. Despite the large number of burn-in iterations, we visually observed some variation in predicted ancestry coefficients among runs. However, we also noted that runs with the highest likelihood values tended to show much more consistent convergence for a given K value. Therefore, we used CLUMPP (Jakobsson and Rosenberg 2007) to align cluster memberships across the ten runs with the highest likelihood values. We applied the LargeKGreedy algorithm using the G’ statistic and tested 1,000,000 random input orders of the ten runs.
We also performed the same STRUCTURE and CLUMPP analysis on microsatellite data taken from Tishkoff et al. (2009) with a subset of these data (158 individuals, 839 loci) used to match populations (but not necessarily individuals) within our own data set. Though sampling was independent between the two data sets, we were able to match the following populations: Bakola (n = 42), Biaka (n = 23), Luhya (n = 17), Mandenka (n = 22), Mbuti (n = 13), Ngoumba (n = 27), and San (n = 6). Because the Shona were not present in the Tishkoff et al. (2009) data set, we chose Bantu–South (n = 8) as a geographical and linguistic representative. Because of the much larger number of loci in this data set, we only applied a burn-in of 10,000 iterations, with 10,000 MCMC steps after burn-in.
Estimation of the number of heterozygotes in the HGDP panel (Cann et al. 2002) sub-Saharan Africans and Europeans was performed using PLINK routines (Purcell et al. 2007). An analysis of molecular variance was performed by using the computer program, Arlequin 3.5 (Excoffier and Lischer 2010). We randomly sampled only eight individuals (the lowest population size was 8 because one San individual failed sequencing for one locus) from each population so that sample size was equal when calculating summary statistics (SS) presented in table 3. FIS was also calculated in Arlequin.
Table 3.
SS across 40 Loci for All Sub-Saharan African Populations.
| Sb | Sa | Gene Diversitya | Theta Sa | Theta PIa | Tajima's Da | nb Haplotypesa | |
| NGO | 374 | 9 | 0.833 | 2.181 | 2.118 | −0.412 | 8 |
| LUH | 368 | 9 | 0.814 | 2.326 | 2.209 | −0.439 | 7 |
| SHN | 351 | 8.5 | 0.830 | 2.326 | 2.183 | −0.349 | 8 |
| MAN | 366 | 9 | 0.794 | 2.471 | 2.356 | −0.286 | 6.5 |
| BAK | 374 | 9 | 0.775 | 2.326 | 2.507 | −0.252 | 6.5 |
| BIA | 397 | 9 | 0.833 | 2.326 | 2.124 | −0.732 | 7 |
| MBI | 368 | 8 | 0.817 | 2.035 | 2.242 | −0.322 | 7 |
| SAN | 358 | 8.5 | 0.814 | 2.035 | 2.294 | −0.214 | 7 |
NOTE.—S = number of segregating sites, nb haplotypes = number of haplotypes.
Median across all 40 loci.
Total number across all 40 loci.
Approximate Bayesian Computation
ABC (Beaumont et al. 2002; Bertorelle et al. 2010) is a statistical framework that allows model testing and parameter estimation for models where the likelihood function does not need to be theoretically derived. Instead, posterior distributions can be approximated by simulation (usually hundreds of thousands or millions) of the model and the retention of a certain number of these simulations and associated parameter values closest to the observed data. This closeness is assessed by comparing the distribution of SS generated from the observed and simulated data, which in its raw form is often intractable for analysis purposes.
Mutation rates for each of the 40 loci were estimated based on the average divergence of human sub-Saharan African sequences (including additional individuals not presented in this study) from a chimpanzee outgroup (Nachman and Crowell 2000), assuming a divergence time for humans and chimpanzees of 6 My and 25 years per generation. We simulated 1 million data sets of 40 loci for each demographic model. Each of the 40 simulated loci was conditioned based on the estimated corresponding mutation rate and sequence length (supplementary table 2, Supplementary Material online). Simulations were performed using msABC (Pavlidis et al. 2011). Following Wegmann and Excoffier (2010), we utilized the mean and SD across all 40 loci for a number of SS (supplementary tables 3 and 4, Supplementary Material online). All prior ranges for model parameters are given in table 1. Effective population size, relative effective population size, and migration rate were set to a log10 scale, with values drawn from uniform distributions (Wegmann and Excoffier 2010).
Table 1.
Priors, Accuracy, and Posteriors Estimates for Model 1A.
| Parameter | Prior |
PQ Dist KS P value | HDPI CI fit |
Posterior Estimation |
||||||||||||||
| Maximum | Minimum | Distribution | HPDI 50 | HPDI 90 | HPDI 95 | HPDI99 | Mode | Median | HPDI 50 |
HPDI 90 |
HPDI 95 |
HPDI 99 |
||||||
| Lower | Upper | Lower | Upper | Lower | Upper | Lower | Upper | |||||||||||
| log(N1) | 3.0 | 5.0 | Uniform | 0.0372 | 0.51 | 0.91 | 0.95 | 0.99 | 4.23 | 4.24 | 4.07 | 4.39 | 3.87 | 4.64 | 3.81 | 4.72 | 3.69 | 4.84 |
| log(N2) | 3.0 | 5.0 | Uniform | 0.5456 | 0.48 | 0.90 | 0.96 | 0.99 | 4.27 | 4.30 | 4.05 | 4.54 | 3.79 | 4.86 | 3.69 | 4.94 | 3.55 | 5.00 |
| log(N3) | 3.0 | 5.0 | Uniform | 0.1600 | 0.50 | 0.90 | 0.95 | 0.98 | 3.69 | 3.70 | 3.46 | 3.91 | 3.18 | 4.21 | 3.10 | 4.31 | 3.00 | 4.47 |
| log(N4) | 3.0 | 5.0 | Uniform | 0.0840 | 0.52 | 0.90 | 0.95 | 0.99 | 4.31 | 4.31 | 4.17 | 4.45 | 3.95 | 4.68 | 3.89 | 4.74 | 3.77 | 4.86 |
| log(NT1)* | 3.0 | 5.0 | Uniform | 0.0000 | 0.47 | 0.87 | 0.93 | 0.98 | 4.33 | 4.19 | 3.97 | 4.70 | 3.42 | 4.98 | 3.28 | 5.00 | 3.12 | 5.00 |
| log(NT2) | 3.0 | 5.0 | Uniform | 0.0025 | 0.50 | 0.90 | 0.94 | 0.98 | 4.23 | 4.19 | 3.95 | 4.56 | 3.51 | 4.88 | 3.36 | 4.96 | 3.18 | 5.00 |
| log(Nanc) | 3.0 | 5.0 | Uniform | 0.6972 | 0.44 | 0.89 | 0.94 | 0.98 | 4.05 | 4.05 | 4.01 | 4.09 | 3.93 | 4.15 | 3.91 | 4.17 | 3.87 | 4.21 |
| T1_sc | 0.0 | 1.0 | Uniform | 0.0017 | 0.63 | 0.94 | 0.97 | 0.99 | 0.81 | 0.54 | 0.44 | 0.88 | 0.12 | 0.96 | 0.09 | 0.99 | 0.04 | 1.00 |
| T1* | 200 | T2 | (T1_sc × (Max − Min)) + Min | 0.0000 | 0.49 | 0.87 | 0.93 | 0.97 | 826 | 1286 | 356 | 1373 | 200 | 2703 | 200 | 3172 | 200 | 3720 |
| T2_sc | 0.0 | 0.1 | Uniform | 0.1323 | 0.59 | 0.93 | 0.97 | 0.99 | 0.19 | 0.36 | 0.07 | 0.41 | 0.00 | 0.77 | 0.00 | 0.86 | 0.00 | 0.95 |
| T2* | 400 | T3 | (T2_sc × (Max − Min))+Min | 0.0000 | 0.50 | 0.89 | 0.94 | 0.99 | 1551 | 1957 | 860 | 2241 | 400 | 3699 | 400 | 4236 | 400 | 5004 |
| T3 | 800 | 8000 | Uniform | 0.0755 | 0.55 | 0.92 | 0.97 | 0.99 | 4000 | 4431 | 3127 | 5164 | 2327 | 7127 | 2109 | 7491 | 1745 | 8000 |
| log(MNK-WPY) | −6.0 | −3.3 | Uniform | 0.1290 | 0.49 | 0.90 | 0.93 | 0.98 | −3.63 | −4.32 | −4.31 | −3.30 | −5.56 | −3.30 | −5.73 | −3.30 | −5.89 | −3.30 |
| log(MNK-EPY) | −6.0 | −3.3 | Uniform | 0.0612 | 0.51 | 0.89 | 0.94 | 0.98 | −3.74 | −4.54 | −4.72 | −3.57 | −5.65 | −3.38 | −5.75 | −3.33 | −5.89 | −3.30 |
| log(MNK-SAN) | −6.0 | −3.3 | Uniform | 0.0449 | 0.52 | 0.91 | 0.95 | 0.98 | −5.51 | −4.91 | −5.81 | −4.75 | −5.97 | −3.87 | −6.00 | −3.71 | −6.00 | −3.49 |
| log(MWPY-EPY) | −6.0 | −3.0 | Uniform | 0.1506 | 0.52 | 0.89 | 0.94 | 0.99 | −4.76 | −4.49 | −5.09 | −3.42 | −5.73 | −3.18 | −5.85 | −3.12 | −5.97 | −3.03 |
| log(MWPY/EPY-SAN) | −6.0 | −3.3 | Uniform | 0.0220 | 0.55 | 0.91 | 0.96 | 0.99 | −5.45 | −4.95 | −5.70 | −4.58 | −5.95 | −4.01 | −6.00 | −3.87 | −6.00 | −3.55 |
| log(N1anc/N1) | −1.0 | 0.0 | Uniform | |||||||||||||||
| log(N2anc/N2) | −1.0 | 0.0 | Uniform | |||||||||||||||
| log(N3anc/N3) | −1.0 | 0.0 | Uniform | |||||||||||||||
| log(N3anc/N4) | −1.0 | 0.0 | Uniform | |||||||||||||||
NOTE.—Parameter labels correspond to those given in figure 3. M = bidirectional migration. Parameters in italics and with asterisk show that parameter failed a KS test of uniformity of posterior quantiles (PQ) (P value less than 0.01 after Bonferonni Correction). Times are shown in generations.
ABC analysis was performed using ABCtoolbox (Wegmann et al. 2010), which implements a general linear model (GLM) regression adjustment (Leuenberger and Wegmann 2010) on retained simulations. In order to perform model selection, we use the marginal density for a particular model relative to the density for all models considered as an estimate of the posterior probability of that model. Power of inferring the correct model was estimated by generating pseudo-observed sets with known parameter values from each model and applying our ABC model choice pipeline. In order to estimate the probability that we chose the correct model, we extended the approach of Fagundes et al. (2007). In our methodology, we used multivariate kernel density estimation to condition this estimate on the posterior probabilities of all four models, rather than just one. We selected the set of SS (for two model comparisons) (supplementary figs. 1–4, Supplementary Material online) or family of SS (SSf) (for the four model comparison) (supplementary fig. 5, Supplementary Material online) and the number of retained simulations that maximized our power for model choice using a pseudo-observed approach that involved ranking SS or SSf by their model discriminatory power via a Kruskal–Wallis test. For parameter estimation from the best model, we transformed the full set of SS into partial least squares (PLS) components and used a pseudo-observed data set approach to choose the appropriate number of PLS components and number of retained simulations (supplementary figs. 6 and 7, Supplementary Material online) that gave parameter estimates that best fit the expected confidence interval (CI) behavior. ABCtoolbox generated a distribution of the posterior quantiles, and a Kolmogorov–Smirnov (KS) test was used to examine the uniformity of distributions (and thus reliability of parameter estimation) for individual parameters.
Principal component analysis (PCA) for comparing the multidimensional distribution of SS was performed using the “prcomp” function in R. A more in depth discussion of our ABC methodology, which involves a number of novel methods for optimizing performance, can be found in the supplementary information, Supplementary Material online.
Results and Discussion
Resequencing data from nongenic regions are likely to be particularly amenable for recovering demographic history via reconstruction of the underlying genealogy. High throughput SNP data, while available for many of the populations examined here, contain high levels of ascertainment bias (though attempts have been made to correct for this bias; Keinan et al. 2007; Wollstein et al. 2010). Examining HGDP 500K SNP microarray data, we find the highest heterozygosity in Europeans compared with sub-Saharan Africans (with particularly low values in Pygmies and San) (supplementary fig. 8, Supplementary Material online), counter to what we would expect from ascertainment bias-free data.
Given the advantages of resequencing data, we use results from our STRUCTURE and FST analyses, along with published inferences (see Introduction), to choose a set of plausible models portraying the demographic history of Pygmy (Western and Eastern), KhoeSan, and non-Pygmy NKs populations. We then use our resequencing data within an ABC framework to infer the best demographic model and estimate parameters from this model. In the supplementary information, Supplementary Material online, we describe the development of a number of methodologies that attempt to optimize the ABC performance.
Population Structure among San, Pygmies, and non-Pygmy NKs
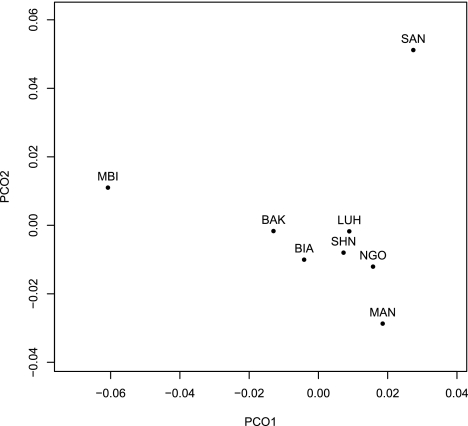
All pairwise FST values between non-Pygmy NKs speakers are nonsignificant (P > 0.01) except in the comparison between the Mandenka and Luhya, whereas all other FST values are significant. We visualized pairwise FST values (table 2) between 8 of our sub-Saharan African populations using the first two dimensions generated from PCO analysis (fig. 1). The Mbuti (along PCO 1) and San (along PCO 2) are clearly differentiated from each other and all other populations, whereas the Biaka and Bakola lie intermediate to the Mbuti and a cluster that contains all non-Pygmy NKs speakers. The non-Pygmy NKs cluster is orientated such that the three Bantu-speaking populations lie closest to the Western Pygmies, which would be expected given that previous studies have demonstrated considerable (and variable) gene flow between Western Pygmy populations and their non-Pygmy NKs neighbors (Coia et al. 2004; Destro-Bisol, Donati, et al. 2004; Quintana-Murci et al. 2008; Patin et al. 2009; Verdu et al. 2009), especially in comparison with Eastern Pygmies (Batini, Lopes, et al. 2011). FST values among the Biaka, Mandenka, and San are comparable to those observed in Wall et al. (2008) despite the use of a slightly different configuration of loci.
Table 2.
Pairwise FST Values between Sub-Saharan African Populations.
| FST*100 | NGO | LUH | SHN | MAN | BAK | BIA | MBI | SAN |
| NGO | * | P > 0.05 | P > 0.05 | P > 0.05 | P < 0.001 | P < 0.001 | P < 0.001 | P < 0.001 |
| LUH | 0.007 | * | P > 0.05 | P < 0.001 | P < 0.001 | P < 0.001 | P < 0.001 | P < 0.001 |
| SHN | 0.070 | 0.849 | * | 0.011 | P < 0.001 | P < 0.001 | P < 0.001 | P < 0.001 |
| MAN | 0.789 | 2.275 | 1.727 | * | P < 0.001 | P < 0.001 | P < 0.001 | P < 0.001 |
| BAK | 3.063 | 2.645 | 2.280 | 3.853 | * | 0.004 | P < 0.001 | P < 0.001 |
| BIA | 2.466 | 1.993 | 2.359 | 2.876 | 1.244 | * | P < 0.001 | P < 0.001 |
| MBI | 7.952 | 6.973 | 7.004 | 8.848 | 4.949 | 6.227 | * | P < 0.001 |
| SAN | 6.396 | 5.530 | 6.235 | 7.996 | 6.598 | 6.954 | 9.705 | * |
NOTE.—Pairwise FST*100 (lower diagonal) values and associated P values (upper diagonal) as assessed by permutation. Values with bold typeface are pairwise FST values that did not reach statistical significance.
FIG. 1.
PCO plot of pairwise FST values between sub-Saharan African populations.
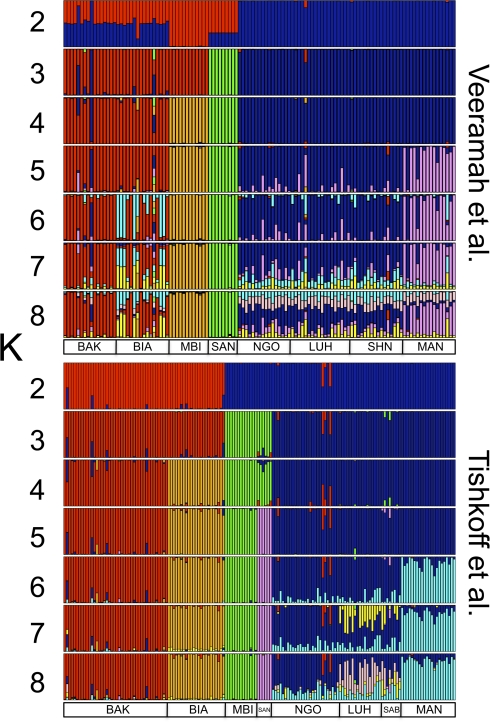
Next, we performed STRUCTURE analysis on our African data (fig. 2) as well as on a subset of the short tandem repeat data reported in Tishkoff et al. (2009). Note that the K = 2 results should be treated with great caution as different runs appeared to reach a number of different solutions (though all involved some distinction of San and Pygmies from non-Pygmy NKs). Therefore, CLUMPP had difficulty finding a clear optimum consensus ancestry solution.
FIG. 2.
Visualization of STRUCTURE and CLUMPP analysis of sub-Saharan African populations showing K = 2-8 for our data set and data from Tishkoff et al. (2009). Sample codes are as described in the method section.
When K = 5, both data sets clearly distinguish the Western Pygmies, Mbuti, San, and non-Pygmy NKs groups into clear clusters. However, although the San can easily be distinguished when K = 3 in our data, they remain grouped with Mbuti Pygmies until K reaches 5 in the Tishkoff et al. (2009) data set. In contrast, Tishkoff et al.’s (2009) data strongly separate the two Western Pygmy groups at K = 3. On the other hand, similar to Patin et al. (2009), we had difficulty obtaining separate Bakola and Biaka clusters, even when K = 6. Given that Western Pygmies are believed to have only differentiated very recently, perhaps within the last 3 kya (Patin et al. 2009; Verdu et al. 2009; Batini, Lopes, et al. 2011), and San divergence is almost certainly much more ancient, the different behaviors of these two data sets likely reflect their different sensitivity to the timing of these demographic processes. Because of its high mutation rate, the microsatellite data of Tishkoff et al. (2009) appears to allow good resolution of recent processes, while our sequence data may be more powerful for inferring ancient divergence.
As previously reported, we see the effects of recent admixture that apparently took place between non-Pygmy NKs and Western Pygmies but not between the former and the Mbuti (Patin et al. 2009). Despite the extremely large geographic distances that separate them, the non-Pygmy NKs populations are difficult to discriminate (the Bantu-speaking populations are completely indistinguishable using our sequence data), reflecting either high levels of geneflow or recent divergence. However, at higher K values, there appear to be some differences in the relative proportion of ancestry components between Bantu speakers and the Mandenka. Therefore, our results are highly consistent with the pairwise FST values reported above, despite the potential loss of information when conducting STRUCTURE analysis because of our strategy of treating haplotypes across entire loci as individual alleles to avoid the potentially detrimental effect of linkage disequilibrium between segregating sites within loci.
ABC Analysis
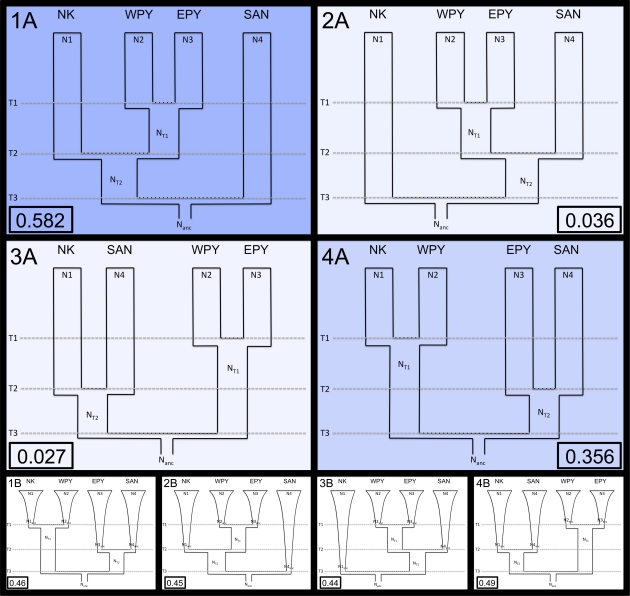
Based on these results and previous inferences in the literature, we identified four plausible demographic models (models 1–4) of Pygmy (Western and Eastern), KhoeSan, and non-Pygmy NKs population divergence (fig. 3). To minimize the possible confounding effects of population substructure, we removed the Mandenka sample and limited our non-Pygmy NKs to Bantu-speaking populations (Ngoumba, Shona, and Luhya). In this regard, we note that a previous study (Wegmann et al. 2009) indicated a relatively recent population divergence time (∼3.5 kya) between Mandenka and a more easterly non-Pygmy NKs population (the Yoruba from Nigeria), suggesting that the addition of Mandenka to our ABC analysis would not have a large effect on our model inference. Moreover, our Bantu-speaking populations demonstrate moderate median FIS (0.146 and −0.007 based on a haplotype frequency and pairwise difference model, respectively) and nonsignificant pairwise FST values suggesting low levels of population structure.
FIG. 3.
Demographic models tested. Estimated posterior probabilities are shown in each panel for each model within a box. NK = non-Pygmy NKs, WPY = Western Pygmies, EPY = Eastern Pygmies, and SAN = San populations, respectively.
As demonstrated by the STRUCTURE analysis above, our data likely lack power to accurately reflect recent processes. Therefore, the main focus of our ABC analysis was directed toward inferring ancient population divergence events. As a consequence, we followed the approach of Patin et al. (2009) and removed several individuals (eight Bakola and ten Biaka) from our Western Pygmy data set that showed evidence via STRUCTURE analysis of recent non-Pygmy NKs gene flow. To do this, we removed any individuals that did not demonstrate at least 95% Western Pygmy ancestry based on a STRUCTURE analysis that included additional sub-Saharan African populations (data not shown). Although this filtering approach may result in estimates of migration rates that are too low (which we have little power to evaluate with precision in any case), we believe it is a reasonable strategy to avoid the effects of very recent migration driven by the expansion of Bantu-speaking farmers ∼5 kya (Salas et al. 2002), which may obscure the signal of more ancient population divergence events (we do, however, examine the effect of removing these individuals later in the study).
Given that we took a relatively conservative approach and removed over half of our initial Western Pygmy data set, we chose to pool our two samples of Western Pygmies (which also has the advantage of reducing the complexity of the models considered). Although we previously found a significant FST between the Biaka and Bakola, and other studies have noted both genetic as well as cultural differences (Blench 2006; Patin et al. 2009; Verdu et al. 2009; Batini, Lopes, et al. 2011), the differentiation of Western Pygmy groups is likely very recent. As discussed above, we would not expect our data and consequent modeling to be particularly sensitive to such a relatively low level of genetic differentiation.
Finally, previous work has found evidence for asymmetric gene flow between non-Pygmy NKs and Pygmies (e.g., Verdu et al. 2009 and Batini, Lopes, et al. 2011, albeit in opposite directions depending on which genetic system was examined). Furthermore, Batini, Ferri, et al. (2011) have recently reported signatures of possible post Last Glacial Maximum male-mediated contacts between Pygmies and KhoeSan. Preliminary analysis of our data suggested that we had little power to detect asymmetric geneflow (which again is likely to be relatively recent). Therefore, we decided to exclude this parameter during our model choice testing phase of the analysis (although we briefly evaluate the effect of an asymmetric model of gene flow).
Our final data set for ABC analysis consisted of 100 non-Pygmy NKs (NK, consisting of Ngoumba, Luhya, and Shona), 28 Western Pygmies (WPY, consisting of Biaka and Bakola), 24 Eastern Pygmies (EPY, consisting of Mbuti), and 18 San (SAN, consisting of San) chromosomes.
Testing Models Incorporating Exponential Population Growth
All African populations in this study exhibit negative Tajima’s D values (table 3), consistent with studies inferring weak population growth in hunter-gatherer and agriculturalist populations (Voight et al. 2005; Cox et al. 2009). Thus, we initially used the ABC model choice framework to compare (for each of our four major models independently) a scenario with no growth (models 1A–4A) against a scenario with exponential growth in each population (models 1B–4B). We use a pseudo-observed data set approach to choose the set of SS (see Materials and Methods and Supplementary Material online) that maximizes the power of our ABC-based model choice methodology. For each of the four comparisons of the no growth (A) versus exponential growth (B) models, one SS (the best discriminatory summary statistic for all four models is in the FST family [supplementary table 5, Supplementary Material online]) was sufficient to give the best power (supplementary figs. 1–4, Supplementary Material online). However, in all cases, this increase in power was no more than 10% better than we would have obtained by chance (i.e., 60% power of obtaining the correct model compared with 50%).
Examining the data in detail reveals that Tajima’s D offers very little information (Pearson’s r < 0.02) about exponential growth on individual populations in our framework, as estimated by the log(N1anc/N1) parameter. It is instead highly correlated (r > 0.77–0.89) with the change in absolute population size from the oldest part of the entire African genealogy, Nanc, to the present population size, N1. This is consistent with Cox et al.’s (2009) inference that their negative Tajima’s D values may have been due in part to an expansion process that involved the common ancestor of some of the same sub-Saharan African populations examined here as well as with other studies that have previously found evidence of moderate ancient population growth in African populations (Voight et al. 2005; Atkinson et al. 2009; Gutenkunst et al. 2009; Gignoux et al. 2011) or those with recent African origins (Adams and Hudson 2004; Marth et al. 2004). Given the lack of power seen here, it is not surprising that the posterior probabilities for each pair of growth and nongrowth models (1A = 0.54, 2A = 0.55, 3A = 0.56, and 4A = 0.51) were almost identical. Therefore, we discontinued consideration of our growth models given the slightly higher posterior probability of the nongrowth models and the computational/statistical advantages of including fewer parameters.
Testing Different Models of Divergence
Next, we used ABC analysis to compare models 1A, 2A, 3A, and 4A. After again maximizing power by choosing the best set of SSf and the number of retained simulations via pseudo-observed sets (supplementary fig. 5 and table 6, Supplementary Material online) (see Materials and Methods and supplementary Material online), our power to correctly predict models 1A, 2A, 3A, and 4A was 0.68, 0.53, 0.57, and 0.72, respectively. These values are far higher than the expected 25% for each model if we had no discriminatory power, and this is confirmed via PCA of the multidimensional distribution of SS where each model explores a subtly different space at various PCA components (supplementary fig. 9, supplementary Material online). PCA also showed the observed data to lie within the major part of the simulated distribution for all major PCA components, demonstrating that simulations were exploring the appropriate space.
Model 1A has the highest posterior probability (0.582), substantially greater than that of the next most likely model, 4A (0.366). Models 2A and 3A have very low posterior probabilities (0.036 and 0.027, respectively). We extended the approach of Fagundes et al. (2007) to estimate the probability that model 1A is the correct model given the observed posterior probabilities of all four models using multivariate kernel density estimation. Although such estimates can be somewhat dependent on the applied bandwidth, our estimate for Pr (M1A = true | PM1A = 0.582, PM2A = 0.036, PM3A = 0.027, PM4A = 0.366) is relatively robust at 0.40–0.42 (for bandwidths between 0.05 and 0.2 and applying a product Gaussian kernel) and is always the most likely model. Estimates for the other three models, however, are more variable (Pr (M2A = true| … ) = 0.12–0.19, Pr (M3A = true| … ) = 0.11–0.19, Pr (M4A = true| … = 0.20–0.37)) (supplementary fig. 10a, Supplementary Material online).
However, it should be noted that the above estimates assume that all four models have the same prior probability. Model 4A invokes a disconnect between Eastern and Western Pygmies, yet two previous ABC-based analyses conducted by Patin et al. (2009) and Batini, Lopes, et al. (2011) with far larger collections of Pygmy populations than our own, have both strongly favored a common recent origin of these two Pygmy groups, as suggested by Model 1A. Therefore, while difficult to quantify, it could be argued that model 4A should have a lower prior probability than the other three models. Ignoring model 4A results in a posterior probability for model 1A of 0.903 with Pr (M1A = true| PM1A = 0.903, PM2A = 0.055, PM3A = 0.041) = 0.71–0.77 (for bandwidths 0.05–0.2) (supplementary fig. 10b, Supplementary Material online). Therefore, our data appear to support a scenario where the KhoeSan diverged first from the common ancestors of Pygmies and non-Pygmy NKs.
Parameter Estimation
Given our support for Model 1A, the distinguishing feature of which is an earlier KhoeSan divergence, we used ABC to estimate parameters of this model (tables 1 and 4). PCA of PLS components again shows the simulated data to be exploring a space that suitably surrounds the observed data (supplementary fig. 11, Supplementary Material online), there is generally good correspondence between raw retained and GLM regressed posterior distributions (supplementary fig. 12, Supplementary Material online) (which demonstrate substantial peakedness for the posteriors), the retained simulations are much closer to the observed data than the bulk of the other simulations (supplementary fig. 13, Supplementary Material online), and all parameters apart from three (log(NT1), T1, and T2) have relatively uniform posterior quantiles as assessed by a KS test (table 1). All the above demonstrate good performance of our ABC analysis and suggest our parameter estimates are relatively reliable. In addition, restricting our non-Pygmy NK population to one Bantu-speaking subpopulation, the Ngoumba, and rerunning our ABC parameter estimation did not alter the parameter estimates substantially (supplementary table 7, Supplementary Material online), suggesting that the effects of cryptic population structure within the Bantu-speaking populations are not a major concern.
Table 4.
Noteworthy Parameter Estimates from Model 1A.
| Median | HPDI 95 |
||
| Lower | Upper | ||
| Ne non-Pygmy NKs | 17,535 | 6,429 | 52,130 |
| Ne Western Pygmies | 19,898 | 4,864 | 86,960 |
| Ne Eastern Pygmies | 5,052 | 1,262 | 20,562 |
| Ne KhoeSan | 20,650 | 7,744 | 54,611 |
| Ne proto-Pygmies | 15,502 | 1,918 | 99,977 |
| Ne proto-non-Pygmy NKs-Pygmies | 15,505 | 2,311 | 91,100 |
| Ne ancestral African population | 11,150 | 8,113 | 14,847 |
| Time of Eastern/Western Pygmy split | 32,157 | 5,000 | 79,311 |
| Time of Pygmy divergence | 48,927 | 10,000 | 105,909 |
| Time of KhoeSan divergence | 110,781 | 52,727 | 187,273 |
NOTE.—Ne in individuals. Times translated into years using 25 years per generation.
Our estimate for the time of KhoeSan divergence is ∼110 kya (95% CI: 52–187 kya) (assuming 25 years per generation). Our estimate is, to best of our knowledge, the first direct measure of the population divergence time of Khoisan speakers from other sub-Saharan Africans. Though not directly comparable (see Edwards and Beerli (2000)), this divergence time agrees (at least with regard to being ancient) with previous TMRCA estimates from mtDNA (90–150 kya) (Behar et al. 2008), the Y-chromosome (70–154 kya) (Knight et al. 2003), and autosomal microsatellites (71–142 kya) (Zhivotovsky et al. 2003). As would be expected, the lower bound for our population divergence estimate is more recent by at least 20 ky than that of the previously described TMRCA estimates. We note that this lower range of ∼50 kya overlaps with the time when AMH are thought to have first left Africa, and thus, it is unclear how our inference contributes to the debate of ancestral population structure before the dispersal of AMH from Africa (Green et al. 2010).
Our estimates of divergence time between Pygmies and non-Pygmy NKs (48 kya, 95% CI: 10–105 kya) and between Eastern and Western Pygmies (32 kya, 95% CI: 5–79 kya) are comparable to previous estimates (Batini et al. 2007; Patin et al. 2009; Verdu et al. 2009; Wegmann et al. 2009; Batini, Lopes, et al. 2011). We note that these two estimates should be considered with some caution because of the particularly skewed posterior quantile distributions. However, the Pygmy–non-Pygmy NKs split time scaled by the divergence of the KhoeSan (T2_sc) suggests that there was a considerable time lag between the KhoeSan divergence and the common ancestors of the Pygmy and non-Pygmy NKs populations.
While previous estimates of Ne for sub-Saharan African populations vary widely depending on the methodology and type of marker used (Tenesa et al. 2007), our results are congruent in magnitude with many of these estimates. These estimates range from 7,500 to 17,500 (Tenesa et al. 2007; Wegmann et al. 2009; Henn et al. 2011). Interestingly, our estimates of Ne for the KhoeSan (20,650, 95% CI: 7,744–54,611), Western Pygmies (19,898, 95% CI: 4,864–86,960), and non-Pygmy NKs (17,535, 95% CI: 6,429–52,130) are similar. Given that the estimated present day census sizes of the KhoeSan and Pygmies (∼500,000) are only ∼0.25% that of Bantu speakers (Lewis 2009) and that Ne is often best modeled with a harmonic mean, our Ne estimates are consistent with the hypothesis of a very recent increase in population size for non-Pygmy NKs populations, a language family that is thought to have originated ∼10 kya (Blench 2006). Not unexpectedly, when we reran our ABC parameter estimation using only the Ngoumba samples to represent non-Pygmy NKs, the Ne estimate for this group was slightly lower (14,175), although the associated CI had almost complete overlap (95% CI: 4,230–52,129) with the combined Bantu-speaking non-Pygmy NKs group.
Particularly notable is the lower Ne for Mbuti Pygmies (5,052, 95% CI: 1,262–20,562), a finding that is consistent with studies suggesting a large bottleneck in this population (Patin et al. 2009; Wegmann et al. 2009; Batini, Lopes, et al. 2011; Henn et al. 2011). Interestingly, the demographic pattern expressed by our Ne estimates is in contrast to those found by Batini, Lopes, et al. (2011), which examined mtDNA resequencing data within a similar ABC framework. This study found a much larger Ne for the non-Pygmy NKs compared with those of Pygmy populations as well as a larger Eastern than Western Pygmy Ne estimate. This is consistent with different demographic histories for maternally versus paternally inherited systems (as noted by Batini, Lopes, et al. (2011) when comparing their results with those of Patin et al. (2009) and also observed in Pilkington et al. (2008)). However, this difference may also be a consequence of the previously discussed limitation of our resequencing data to infer recent demographic processes. In this regard, it is noteworthy that Verdu et al. (2009) also observed a non-Pygmy NKs Ne that was an order of magnitude greater than that of Western Pygmies using 28 autosomal microsatellites and a very large number of Western Pygmy population samples.
Also striking is the extremely peaked posterior for the ancestral African effective population size (Nanc), with an estimate of 11,150 (95% CI: 8,113–14,847). Interestingly, this estimate is highly consistent with the equivalent estimate of Patin et al. (2009) (11,402, 95% CI: 7,670–15,653), although they do not consider KhoeSan within their model. Comparing the point estimates of Nanc with populations of present day sub-Saharan Africans examined here (other than the Mbuti) suggests that ∼2-fold (and as high as 6-fold) growth has occurred since the earliest divergence in model 1A. These results thus support previous suggestions of a mild population expansion early in African prehistory (Voight et al. 2005; Cox et al. 2009; Gutenkunst et al. 2009).
To examine the effect of filtering admixed Western Pygmy individuals from the original data set, we conducted parameter estimation of model 1A that included the previously removed individuals. The addition of these admixed samples results in only a slight decrease in estimates of divergence times and highly overlapping CIs between the two estimates (supplementary table 8, Supplementary Material online). The divergence time estimate most affected by the addition of admixed samples is for the Western and Eastern Pygmy split (32 kya, 95% CI: 5–79 to 26 kya, 95% CI: 5–62 kya). Importantly, the divergence time estimate for the KhoeSan is largely unchanged (∼110 kya, 95% CI: 52–187 to ∼103 kya, 95% CI: 41–180 kya), and the T2_sc parameter still indicates a much earlier KhoeSan divergence time (0.36, 95% CI: 0.0–0.86 to 0.33 kya, 95% CI: 0.0–0.86 kya) before Pygmies split from non-Pygmy NKs. Most striking is that the median estimate of Ne for the Western Pygmies is noticeably larger (19,898–34,953), although the CIs are wide with considerable overlap between filtered and nonfiltered estimates (95% CI: 4,846–86,960 to 95% CI: 10,726–99,972). Other Ne estimates are almost completely unaltered, suggesting that the exclusion of admixed individuals has minimal impact on the overall topology of the demographic model.
One of the more interesting aspects of previous work examining Pygmy–non-Pygmy NKs demography has been the detection of asymmetric gene flow, with previous autosomal studies (Verdu et al. 2009) suggesting greater gene flow from non-Pygmy NKs, and mtDNA studies (Quintana-Murci et al. 2008; Batini, Lopes, et al. 2011) indicating more movement in the opposite direction. As discussed above, we do not believe that we have the power to observe this phenomenon with our sequence data, and thus, we initially only considered symmetric gene flow. Consistent with this expectation, our migration estimates are largely uninformative with very flat posterior distributions. However, given the observation of some gene flow into Western Pygmies via our STRUCTURE analysis, we further analyzed the data set including admixed individuals within an updated version of model 1A that incorporated asymmetric gene flow. Western Pygmy to non-Pygmy NKs gene flow was specified by a parameter, β, that scaled the initial non-Pygmy NKs to Western Pygmy migration rate from 0 to 1.0 (though Wegmann et al. (2009) did find evidence of asymmetric gene flow between non-Pygmy NKs and Eastern Pygmies, because of the lack of such a signal in our STRUCTURE analysis as well as previous work from Patin et al. (2009), we did not consider this possibility in our model). The inclusion of this β parameter has almost no effect on any of our other parameter estimates (supplementary table 9, Supplementary Material online), and the posterior estimate for β is very flat (95% CI: 0.05–0.96). Thus, although we are unable to provide any useful information with regard to asymmetric gene flow (and migration in general), our results are likely robust to the effects of recent elevated non-Pygmy NKs–Pygmy migration.
Conclusion
We demonstrate that autosomal resequencing data from multiple intergenic regions (i.e., polymorphism data that are relatively free from the effects of natural selection and ascertainment bias) support a demographic model that incorporates an early divergence of the lineage leading to the KhoeSan from a population that gave rise to both the ancestors of Pygmy and non-Pygmy NKs groups. This suggests perhaps a long period of independent evolution for the lineages leading to extant hunter gatherers and a longer period of shared history between Pygmy and non-Pygmy NKs groups than between either of these groups and KhoeSan. This has interesting implications for the evolution of physical and cultural characters that distinguish these populations. This work also helps to resolve the population divergence tree inferred by Zhivotovsky et al. (2003) and points to the power of using resequencing data over microsatellite data to infer more ancient demographic processes. We caution, however, that Khoisan languages are quite diverse despite the commonality of click consonants and that our small sample of San almost certainly does not represent the full spectrum of genetic diversity that exists among speakers of these languages. It will be interesting to investigate how other Khoisan-speaking groups and click speakers such as the Hadza and Sandawe from Tanzania, who exhibit ancient connections with southwestern KhoeSan, fit into this model of human evolution.
It should also be appreciated that KhoeSan and Pygmy populations, while clearly important for understanding African prehistory, represent only ∼0.2% of contemporary sub-Saharan Africans. More work is needed that explicitly models population relationships both within the Niger-Kordofanian family and between Niger-Kordofanian and other large language families in Africa. Not only will this help to elucidate details of the fine-scale relationships among agriculturalists and pastoralists, it will lead to a better understanding of past population structure in the region of the world with the most genetic diversity and further insights into human origins. Our resequencing data, while powerful for estimating divergence patterns, appears limited for estimating more recent processes (e.g., our posterior estimates for migration rate are relatively flat). A combined approach of using sequence data with microsatellites, as utilized by Wegmann et al. (2010) may improve our estimation, especially with regard to processes that occur near the tips of the genealogical tree, while whole-genome sequencing potentially offers even more power for all aspects of African demographic inference in the near future. ABC analysis should provide a particularly useful framework for simultaneously examining such complex demographic models and data.
Supplementary Material
Supplementary figures 1–13, tables 1–9, and supplementary information are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The authors thank all those who donated DNA samples. K.R.V. is funded by the National Institute of Health. D.W. is funded via a Scholar Program award to John Novembre. This research was supported by a HOMINID grant to M.F.H. Research was performed at the University of Arizona.
References
- Adams AM, Hudson RR. Maximum-likelihood estimation of demographic parameters using the frequency spectrum of unlinked single-nucleotide polymorphisms. Genetics. 2004;168:1699–1712. doi: 10.1534/genetics.104.030171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves I, Coelho M, Gignoux C, Damasceno A, Prista A, Rocha J. Genetic homogeneity across Bantu-speaking groups from Mozambique and Angola challenges early split scenarios between East and West Bantu populations. Hum Biol. 2011;83:13–38. doi: 10.3378/027.083.0102. [DOI] [PubMed] [Google Scholar]
- Atkinson QD, Gray RD, Drummond AJ. Bayesian coalescent inference of major human mitochondrial DNA haplogroup expansions in Africa. Proc R Soc B Biol Sci. 2009;276:367–373. doi: 10.1098/rspb.2008.0785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnard A. Kalahari revisionism, Vienna and the ‘indigenous peoples’ debate*. Soc Anthropol. 2006;14:1–16. [Google Scholar]
- Batini C, Coia V, Battaggia C, Rocha J, Pilkington MM, Spedini G, Comas D, Destro-Bisol G, Calafell F. Phylogeography of the human mitochondrial L1c haplogroup: genetic signatures of the prehistory of Central Africa. Mol Phylogenet Evol. 2007;43:635–644. doi: 10.1016/j.ympev.2006.09.014. [DOI] [PubMed] [Google Scholar]
- Batini C, Ferri G, Destro-Bisol G, et al. (17 co-authors) Signatures of the pre-agricultural peopling processes in sub-Saharan Africa as revealed by the phylogeography of early Y chromosome lineages. Mol Biol Evol. 2011;28:2603–2613. doi: 10.1093/molbev/msr089. [DOI] [PubMed] [Google Scholar]
- Batini C, Lopes J, Behar DM, Calafell F, Jorde LB, van der Veen L, Quintana-Murci L, Spedini G, Destro-Bisol G, Comas D. Insights into the demographic history of African Pygmies from complete mitochondrial genomes. Mol Biol Evol. 2011;28:1099–1110. doi: 10.1093/molbev/msq294. [DOI] [PubMed] [Google Scholar]
- Beaumont MA, Zhang W, Balding DJ. Approximate Bayesian computation in population genetics. Genetics. 2002;162:2025–2035. doi: 10.1093/genetics/162.4.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker NS, Verdu P, Froment A, Le BS, Pagezy H, Bahuchet S, Heyer E. Indirect evidence for the genetic determination of short stature in African Pygmies. Am J Phys Anthropol. 2011;145:390–401. doi: 10.1002/ajpa.21512. [DOI] [PubMed] [Google Scholar]
- Behar DM, Villems R, Soodyall H, et al. (35 co-authors) The dawn of human matrilineal diversity. Am J Hum Genet. 2008;82:1130–1140. doi: 10.1016/j.ajhg.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertorelle G, Benazzo A, Mona S. ABC as a flexible framework to estimate demography over space and time: some cons, many pros. Mol Ecol. 2010;19:2609–2625. doi: 10.1111/j.1365-294X.2010.04690.x. [DOI] [PubMed] [Google Scholar]
- Blench R. Archaeology, language, and the African Past. Lanham (MA): AltaMira Press; 2006. [Google Scholar]
- Bryc K, Auton A, Nelson MR, et al. (11 co-authors) Genome-wide patterns of population structure and admixture in West Africans and African Americans. Proc Natl Acad Sci U S A. 2010;107:786–791. doi: 10.1073/pnas.0909559107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cann HM, de Toma C, Cazes L, et al. (41 co-authors) A human genome diversity cell line panel. Science. 2002;296:261–262. doi: 10.1126/science.296.5566.261b. [DOI] [PubMed] [Google Scholar]
- Cavalli-Sforza LL, Menozzi P, Piazza A. The history and geography of human genes. Princeton (NJ): Princeton University Press; 1994. [Google Scholar]
- Chen YS, Olckers A, Schurr TG, Kogelnik AM, Huoponen K, Wallace DC. mtDNA variation in the South African Kung and Khwe-and their genetic relationships to other African populations. Am J Hum Genet. 2000;66:1362–1383. doi: 10.1086/302848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coia V, Caglia A, Arredi B, et al. (12 co-authors) Binary and microsatellite polymorphisms of the Y-chromosome in the Mbenzele pygmies from the Central African Republic. Am J Hum Biol. 2004;16:57–67. doi: 10.1002/ajhb.10236. [DOI] [PubMed] [Google Scholar]
- Cox MP, Morales DA, Woerner AE, Sozanski J, Wall JD, Hammer MF. Autosomal resequence data reveal Late Stone Age signals of population expansion in sub-Saharan African foraging and farming populations. PLoS One. 2009;4:e6366. doi: 10.1371/journal.pone.0006366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruciani F, Santolamazza P, Shen P, et al. (16 co-authors) A back migration from Asia to sub-Saharan Africa is supported by high-resolution analysis of human Y-chromosome haplotypes. Am J Hum Genet. 2002;70:1197–1214. doi: 10.1086/340257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Destro-Bisol G, Coia V, Boschi I, Verginelli F, Caglia A, Pascali V, Spedini G, Calafell F. The analysis of variation of mtDNA hypervariable region 1 suggests that Eastern and Western Pygmies diverged before the Bantu expansion. Am Nat. 2004;163:212–226. doi: 10.1086/381405. [DOI] [PubMed] [Google Scholar]
- Destro-Bisol G, Donati F, Coia V, Boschi I, Verginelli F, Caglia A, Tofanelli S, Spedini G, Capelli C. Variation of female and male lineages in sub-Saharan populations: the importance of sociocultural factors. Mol Biol Evol. 2004;21:1673–1682. doi: 10.1093/molbev/msh186. [DOI] [PubMed] [Google Scholar]
- Edwards SV, Beerli P. Perspective: gene divergence, population divergence, and the variance in coalescence time in phylogeographic studies. Evolution. 2000;54:1839–1854. doi: 10.1111/j.0014-3820.2000.tb01231.x. [DOI] [PubMed] [Google Scholar]
- Epstein MP, Duren WL, Boehnke M. Improved inference of relationship for pairs of individuals. Am J Hum Genet. 2000;67:1219–1231. doi: 10.1016/s0002-9297(07)62952-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Lischer HEL. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- Fagundes NJ, Ray N, Beaumont M, Neuenschwander S, Salzano FM, Bonatto SL, Excoffier L. Statistical evaluation of alternative models of human evolution. Proc Natl Acad Sci U S A. 2007;104:17614–17619. doi: 10.1073/pnas.0708280104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher R. Statistical methods for research workers. 13th ed. London: Oliver & Loyd; 1925. [Google Scholar]
- Ghirotto S, Mona S, Benazzo A, Paparazzo F, Caramelli D, Barbujani G. Inferring genealogical processes from patterns of Bronze-Age and modern DNA variation in Sardinia. Mol Biol Evol. 2010;27:875–886. doi: 10.1093/molbev/msp292. [DOI] [PubMed] [Google Scholar]
- Gignoux CR, Henn BM, Mountain JL. Rapid, global demographic expansions after the origins of agriculture. Proc Natl Acad Sci U S A. 2011;108:6044–6049. doi: 10.1073/pnas.0914274108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonder MK, Mortensen HM, Reed FA, de Sousa A, Tishkoff SA. Whole-mtDNA genome sequence analysis of ancient African lineages. Mol Biol Evol. 2007;24:757–768. doi: 10.1093/molbev/msl209. [DOI] [PubMed] [Google Scholar]
- Gower JC. Some distance properties of latent root and vector methods used in multivariate analysis. Biometrika. 1966;53:325–328. [Google Scholar]
- Green RE, Krause J, Briggs AW, et al. (56 co-authors) A draft sequence of the Neandertal genome. Science. 2010;328:710–722. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutenkunst RN, Hernandez RD, Williamson SH, Bustamante CD. Inferring the joint demographic history of multiple populations from multidimensional SNP frequency data. PLoS Genet. 2009;5:e1000695. doi: 10.1371/journal.pgen.1000695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer MF, Karafet TM, Redd AJ, Jarjanazi H, Santachiara-Benerecetti S, Soodyall H, Zegura SL. Hierarchical patterns of global human Y-chromosome diversity. Mol Biol Evol. 2001;18:1189–1203. doi: 10.1093/oxfordjournals.molbev.a003906. [DOI] [PubMed] [Google Scholar]
- Hammer MF, Mendez FL, Cox MP, Woerner AE, Wall JD. Sex-biased evolutionary forces shape genomic patterns of human diversity. PLoS Genet. 2008;4:e1000202. doi: 10.1371/journal.pgen.1000202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer MF, Woerner AE, Mendez FL, Watkins JC, Cox MP, Wall JD. The ratio of human X chromosome to autosome diversity is positively correlated with genetic distance from genes. Nat Genet. 2010;42:830–831. doi: 10.1038/ng.651. [DOI] [PubMed] [Google Scholar]
- Henn BM, Gignoux CR, Jobin M, et al. (19 co-authors) Hunter-gatherer genomic diversity suggests a southern African origin for modern humans. Proc Natl Acad Sci U S A. 2011;108:5154–5162. doi: 10.1073/pnas.1017511108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewlett BS. Cultural diversity among African Pygmies. In: Kent S, editor. Cultural diversity among twentieth-century foragers: an African perspective. Cambridge: Cambridge University Press; 1996. pp. 215–244. [Google Scholar]
- Ingman M, Gyllensten U. Analysis of the complete human mtDNA genome: methodology and inferences for human evolution. J Hered. 2001;92:454–461. doi: 10.1093/jhered/92.6.454. [DOI] [PubMed] [Google Scholar]
- Jakobsson M, Rosenberg NA. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics. 2007;23:1801–1806. doi: 10.1093/bioinformatics/btm233. [DOI] [PubMed] [Google Scholar]
- Jorde LB, Watkins WS, Bamshad MJ, Dixon ME, Ricker CE, Seielstad MT, Batzer MA. The distribution of human genetic diversity: a comparison of mitochondrial, autosomal, and Y-chromosome data. Am J Hum Genet. 2000;66:979–988. doi: 10.1086/302825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keinan A, Mullikin JC, Patterson N, Reich D. Measurement of the human allele frequency spectrum demonstrates greater genetic drift in East Asians than in Europeans. Nat Genet. 2007;39:1251–1255. doi: 10.1038/ng2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight A, Underhill PA, Mortensen HM, Zhivotovsky LA, Lin AA, Henn BM, Louis D, Ruhlen M, Mountain JL. African Y chromosome and mtDNA divergence provides insight into the history of click languages. Curr Biol. 2003;13:464–473. doi: 10.1016/s0960-9822(03)00130-1. [DOI] [PubMed] [Google Scholar]
- Leuenberger C, Wegmann D. Bayesian computation and model selection without likelihoods. Genetics. 2010;184:243–252. doi: 10.1534/genetics.109.109058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis MP, editor. Ethnologue: languages of the world. 16th ed. Dallas (TX): SIL International Online; 2009. [Google Scholar]
- Marth GT, Czabarka E, Murvai J, Sherry ST. The allele frequency spectrum in genome-wide human variation data reveals signals of differential demographic history in three large world populations. Genetics. 2004;166:351–372. doi: 10.1534/genetics.166.1.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellars P. Why did modern human populations disperse from Africa ca. 60,000 years ago? A new model. Proc Natl Acad Sci U S A. 2006;103:9381–9386. doi: 10.1073/pnas.0510792103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P. Genetics and southern African prehistory: an archaeological view. J Anthropol Sci. 2010;88:73–92. [PubMed] [Google Scholar]
- Morris AG. The Myth of the East African ‘Bushmen’. S Afr Archaeol Bull. 2003;58:85–90. [Google Scholar]
- Nachman MW, Crowell SL. Estimate of the mutation rate per nucleotide in humans. Genetics. 2000;156:297–304. doi: 10.1093/genetics/156.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurse GT, Weiner JS, Jenkins T. The peoples of southern Africa and their affinities. Oxford: Oxford University Press; 1985. [Google Scholar]
- Olerup O, Troye-Blomberg M, Schreuder GM, Riley EM. HLA-DR and -DQ gene polymorphism in West Africans is twice as extensive as in north European Caucasians: evolutionary implications. Proc Natl Acad Sci U S A. 1991;88:8480–8484. doi: 10.1073/pnas.88.19.8480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagezy H. Coping with uncertainty in food supply among the Oto and the Twa living in the equatorial flooded forest near Lake Tumba, Zaire. In: Garine ID, Harrison GA, editors. Coping with uncertainty in food supply. Oxford: Clarendon Press; 1998. pp. 175–209. [Google Scholar]
- Patin E, Laval G, Barreiro LB, et al. (15 co-authors) Inferring the demographic history of African farmers and pygmy hunter-gatherers using a multilocus resequencing data set. PLoS Genet. 2009;5:e1000448. doi: 10.1371/journal.pgen.1000448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlidis P, Laurent S, Stephan W. msABC: a modification of Hudson's ms to facilitate multi-locus ABC analysis. Mol Ecol Resour. 2011;10:723–727. doi: 10.1111/j.1755-0998.2010.02832.x. [DOI] [PubMed] [Google Scholar]
- Perry GH, Dominy NJ. Evolution of the human pygmy phenotype. Trends Ecol Evol. 2009;24:218–225. doi: 10.1016/j.tree.2008.11.008. [DOI] [PubMed] [Google Scholar]
- Pilkington MM, Wilder JA, Mendez FL, et al. (13 co-authors) Contrasting signatures of population growth for mitochondrial DNA and Y chromosomes among human populations in Africa. Mol Biol Evol. 2008;25:517–525. doi: 10.1093/molbev/msm279. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, et al. (11 co-authors) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana-Murci L, Quach H, Harmant C, et al. (23 co-authors) Maternal traces of deep common ancestry and asymmetric gene flow between Pygmy hunter-gatherers and Bantu-speaking farmers. Proc Natl Acad Sci U S A. 2008;105:1596–1601. doi: 10.1073/pnas.0711467105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. R: a language and environment for statistical computing. Vienna (Austria): R Foundation for Statistical Computing; 2007. [Google Scholar]
- Relethford JH, Jorde LB. Genetic evidence for larger African population size during recent human evolution. Am J Phys Anthropol. 1999;108:251–260. doi: 10.1002/(SICI)1096-8644(199903)108:3<251::AID-AJPA1>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Richards GD. Genetic, physiologic and ecogeographic factors contributing to variation in Homo sapiens: homo floresiensis reconsidered. J Evol Biol. 2006;19:1744–1767. doi: 10.1111/j.1420-9101.2006.01179.x. [DOI] [PubMed] [Google Scholar]
- Rosenberg NA. Standardized subsets of the HGDP-CEPH Human Genome Diversity Cell Line Panel, accounting for atypical and duplicated samples and pairs of close relatives. Ann Hum Genet. 2006;70:841–847. doi: 10.1111/j.1469-1809.2006.00285.x. [DOI] [PubMed] [Google Scholar]
- Salas A, Richards M, De la FT, Lareu MV, Sobrino B, Sanchez-Diz P, Macaulay V, Carracedo A. The making of the African mtDNA landscape. Am J Hum Genet. 2002;71:1082–1111. doi: 10.1086/344348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheinfeldt LB, Soi S, Tishkoff SA. Colloquium paper: working toward a synthesis of archaeological, linguistic, and genetic data for inferring African population history. Proc Natl Acad Sci U S A. 2010;107(Suppl 2):8931–8938. doi: 10.1073/pnas.1002563107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster SC, Miller W, Ratan A, et al. (48 co-authors) Complete Khoisan and Bantu genomes from southern Africa. Nature. 2010;463:943–947. doi: 10.1038/nature08795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seielstad M, Bekele E, Ibrahim M, Toure A, Traore M. A view of modern human origins from Y chromosome microsatellite variation. Genome Res. 1999;9:558–567. [PMC free article] [PubMed] [Google Scholar]
- Semino O, Santachiara-Benerecetti AS, Falaschi F, Cavalli-Sforza LL, Underhill PA. Ethiopians and Khoisan share the deepest clades of the human Y-chromosome phylogeny. Am J Hum Genet. 2002;70:265–268. doi: 10.1086/338306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikora M, Laayouni H, Calafell F, Comas D, Bertranpetit J. A genomic analysis identifies a novel component in the genetic structure of sub-Saharan African populations. Eur J Hum Genet. 2011;19:84–88. doi: 10.1038/ejhg.2010.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenesa A, Navarro P, Hayes BJ, Duffy DL, Clarke GM, Goddard ME, Visscher PM. Recent human effective population size estimated from linkage disequilibrium. Genome Res. 2007;17:520–526. doi: 10.1101/gr.6023607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff SA, Gonder MK, Henn BM, et al. (12 co-authors) History of click-speaking populations of Africa inferred from mtDNA and Y chromosome genetic variation. Mol Biol Evol. 2007;24:2180–2195. doi: 10.1093/molbev/msm155. [DOI] [PubMed] [Google Scholar]
- Tishkoff SA, Reed FA, Friedlaender FR, et al. (25 co-authors) The genetic structure and history of Africans and African Americans. Science. 2009;324:1035–1044. doi: 10.1126/science.1172257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underhill PA, Passarino G, Lin AA, Shen P, Mirazon LM, Foley RA, Oefner PJ, Cavalli-Sforza LL. The phylogeography of Y chromosome binary haplotypes and the origins of modern human populations. Ann Hum Genet. 2001;65:43–62. doi: 10.1046/j.1469-1809.2001.6510043.x. [DOI] [PubMed] [Google Scholar]
- Verdu P, Austerlitz F, Estoup A, et al. (14 co-authors) Origins and genetic diversity of pygmy hunter-gatherers from Western Central Africa. Curr Biol. 2009;19:312–318. doi: 10.1016/j.cub.2008.12.049. [DOI] [PubMed] [Google Scholar]
- Vigilant L, Stoneking M, Harpending H, Hawkes K, Wilson AC. African populations and the evolution of human mitochondrial DNA. Science. 1991;253:1503–1507. doi: 10.1126/science.1840702. [DOI] [PubMed] [Google Scholar]
- Voight BF, Adams AM, Frisse LA, Qian Y, Hudson RR, Di RA. Interrogating multiple aspects of variation in a full resequencing data set to infer human population size changes. Proc Natl Acad Sci U S A. 2005;102:18508–18513. doi: 10.1073/pnas.0507325102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall JD, Cox MP, Mendez FL, Woerner A, Severson T, Hammer MF. A novel DNA sequence database for analyzing human demographic history. Genome Res. 2008;18:1354–1361. doi: 10.1101/gr.075630.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegmann D, Excoffier L. Bayesian inference of the demographic history of chimpanzees. Mol Biol Evol. 2010;27:1425–1435. doi: 10.1093/molbev/msq028. [DOI] [PubMed] [Google Scholar]
- Wegmann D, Leuenberger C, Excoffier L. Efficient approximate Bayesian computation coupled with Markov chain Monte Carlo without likelihood. Genetics. 2009;182:1207–1218. doi: 10.1534/genetics.109.102509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegmann D, Leuenberger C, Neuenschwander S, Excoffier L. ABCtoolbox: a versatile toolkit for approximate Bayesian computations. BMC Bioinformatics. 2010;11:116. doi: 10.1186/1471-2105-11-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollstein A, Lao O, Becker C, Brauer S, Trent RJ, Nurnberg P, Stoneking M, Kayser M. Demographic history of Oceania inferred from genome-wide data. Curr Biol. 2010;20:1983–1992. doi: 10.1016/j.cub.2010.10.040. [DOI] [PubMed] [Google Scholar]
- Wood ET, Stover DA, Ehret C, et al. (11 co-authors) Contrasting patterns of Y chromosome and mtDNA variation in Africa: evidence for sex-biased demographic processes. Eur J Hum Genet. 2005;13:867–876. doi: 10.1038/sj.ejhg.5201408. [DOI] [PubMed] [Google Scholar]
- Zhivotovsky LA, Rosenberg NA, Feldman MW. Features of evolution and expansion of modern humans, inferred from genomewide microsatellite markers. Am J Hum Genet. 2003;72:1171–1186. doi: 10.1086/375120. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.