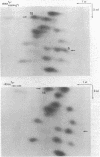
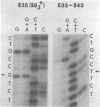
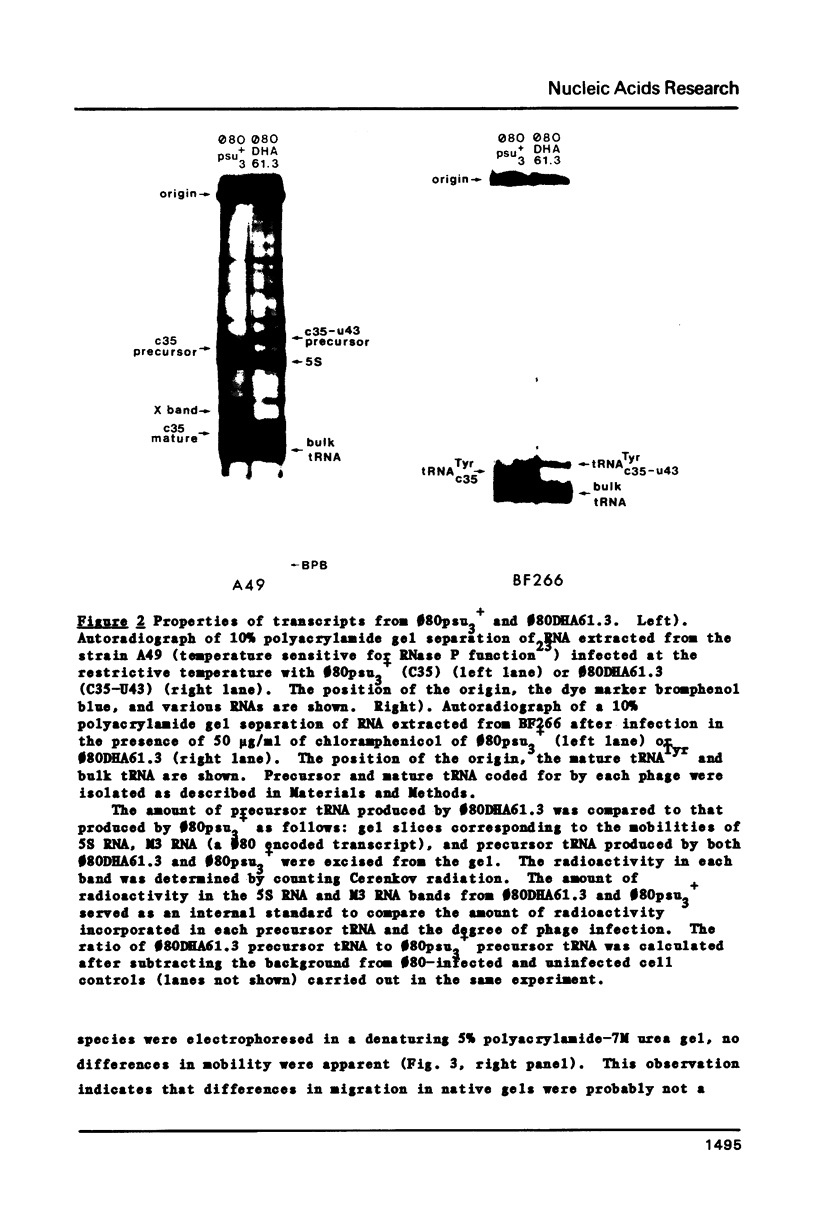
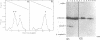Abstract
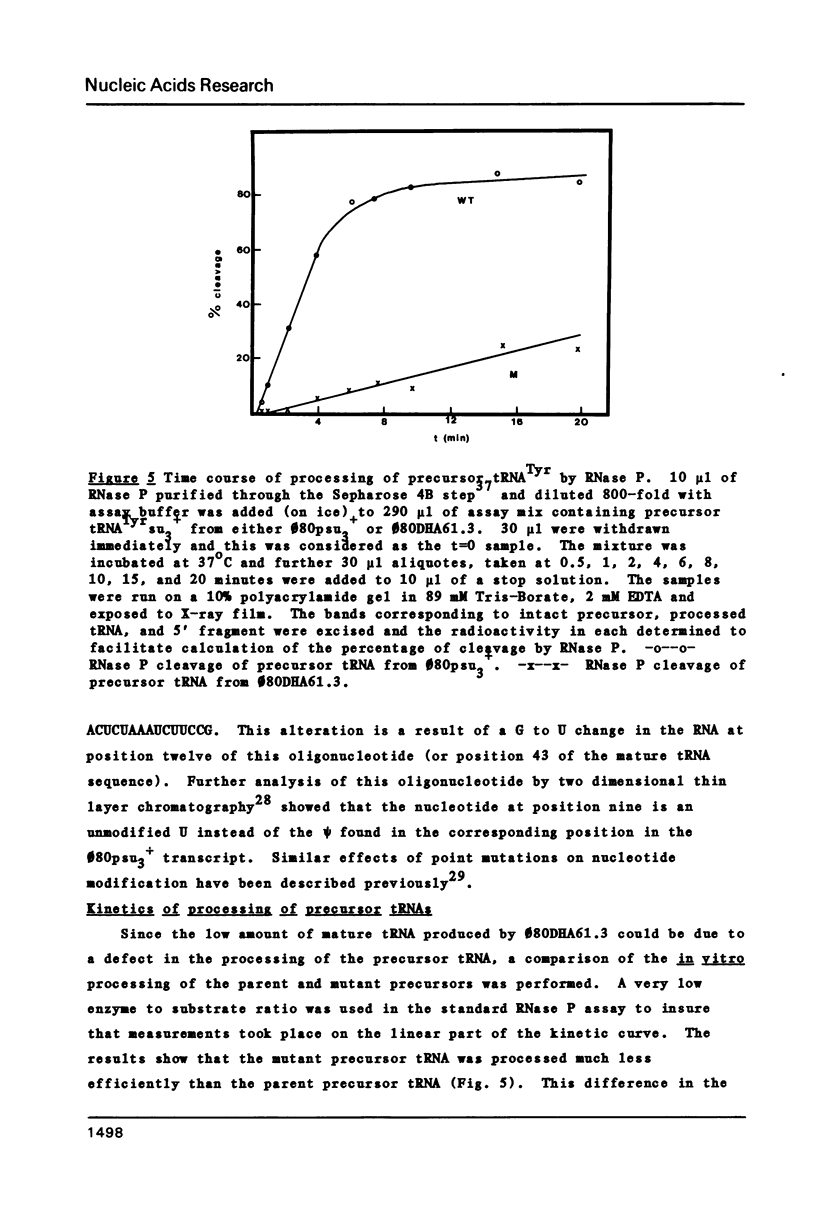
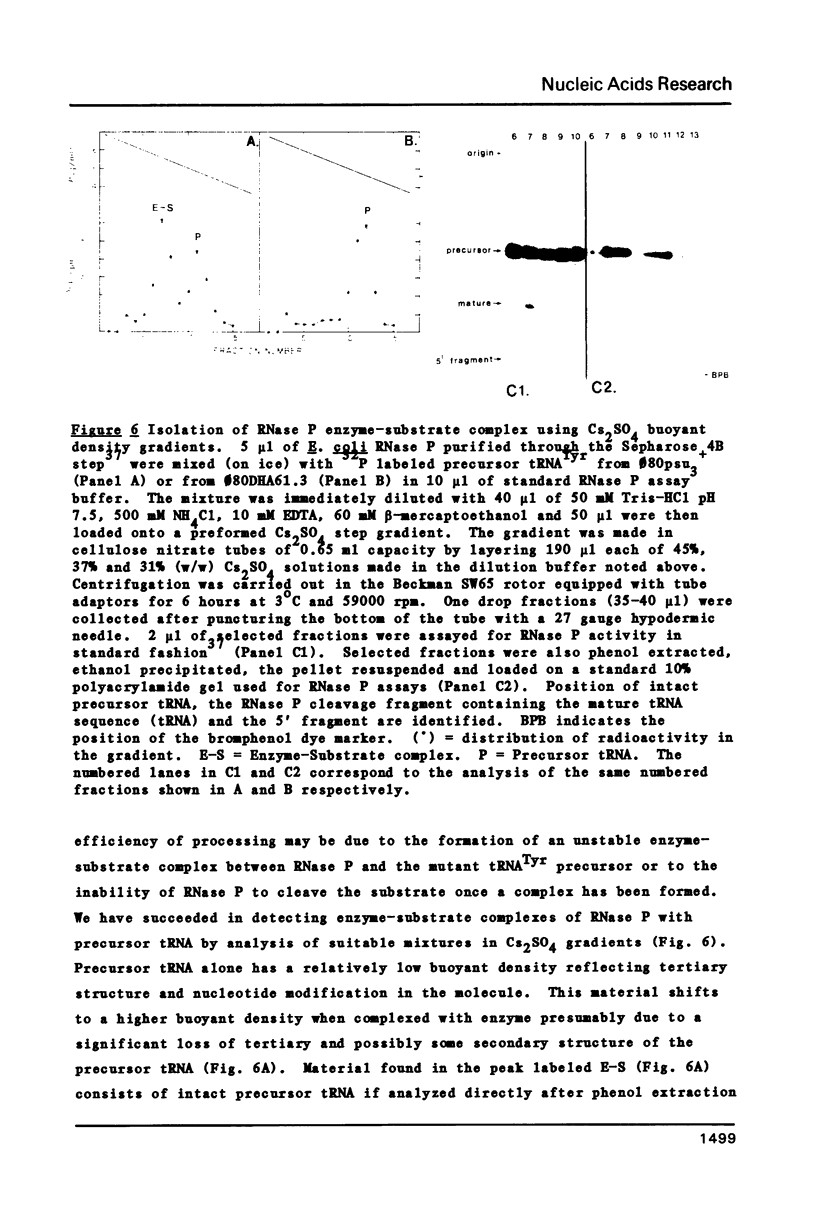
A novel mutation in the anticodon stem of E. coli tRNA1Tyrsu3+ (G43 to U43) has been characterized. The gene coding for the mutant tRNA, carried by phage phi 80DHA61.3 a derivative of phi 80psu3+su0, produces only 20% of mature suppressor tRNA as compared with phi 80psu3+. Both the mutant tRNA precursor and mature tRNA have an altered conformation. The precursor tRNA coded for by phi 80DHA61.3 is processed by RNase P more slowly than the su3+ precursor and does not form as stable an enzyme-substrate complex as does su3+ precursor. phi 80 DHA61.3 also contains a large deletion which begins in the spacer region between the su3+ gene and the su0- gene, extends through the su0- gene and includes most of the repeated region following the tRNA genes.
Full text
PDF











Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abelson J. N., Gefter M. L., Barnett L., Landy A., Russell R. L., Smith J. D. Mutant tyrosine transfer ribonucleic acids. J Mol Biol. 1970 Jan 14;47(1):15–28. doi: 10.1016/0022-2836(70)90398-0. [DOI] [PubMed] [Google Scholar]
- Altman S., Smith J. D. Tyrosine tRNA precursor molecule polynucleotide sequence. Nat New Biol. 1971 Sep 8;233(36):35–39. doi: 10.1038/newbio233035a0. [DOI] [PubMed] [Google Scholar]
- Altman S. Transfer RNA processing enzymes. Cell. 1981 Jan;23(1):3–4. doi: 10.1016/0092-8674(81)90262-2. [DOI] [PubMed] [Google Scholar]
- Daniel V. Biosynthesis of transfer RNA. CRC Crit Rev Biochem. 1981;9(4):253–292. doi: 10.3109/10409238109105436. [DOI] [PubMed] [Google Scholar]
- FREESE E., BAUTZ E., FREESE E. B. The chemical and mutagenic specificity of hydroxylamine. Proc Natl Acad Sci U S A. 1961 Jun 15;47:845–855. doi: 10.1073/pnas.47.6.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin R. M. Purification and properties of the replicative intermediate of the RNA bacteriophage R17. Proc Natl Acad Sci U S A. 1966 Jun;55(6):1504–1511. doi: 10.1073/pnas.55.6.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghysen A., Celis J. E. Joint transcription of two tRNA1Tyr genes from Escherichia coli. Nature. 1974 May 31;249(456):418–421. doi: 10.1038/249418a0. [DOI] [PubMed] [Google Scholar]
- Goodman H. M., Abelson J., Landy A., Brenner S., Smith J. D. Amber suppression: a nucleotide change in the anticodon of a tyrosine transfer RNA. Nature. 1968 Mar 16;217(5133):1019–1024. doi: 10.1038/2171019a0. [DOI] [PubMed] [Google Scholar]
- Koski R. A., Bothwell A. L., Altman S. Identification of a ribonuclease P-like activity from human KB cells. Cell. 1976 Sep;9(1):101–116. doi: 10.1016/0092-8674(76)90056-8. [DOI] [PubMed] [Google Scholar]
- Kudo I., Leineweber M., RajBhandary U. L. Site-specific mutagenesis on cloned DNAs: generation of a mutant of Escherichia coli tyrosine suppressor tRNA in which the sequence G-T-T-C corresponding to the universal G-T-pseudouracil-C sequence of tRNAs is changed to G-A-T-C. Proc Natl Acad Sci U S A. 1981 Aug;78(8):4753–4757. doi: 10.1073/pnas.78.8.4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Küpper H., Sekiya T., Rosenberg M., Egan J., Landy A. A rho-dependent termination site in the gene coding for tyrosine tRNA su3 of Escherichia coli. Nature. 1978 Mar 30;272(5652):423–428. doi: 10.1038/272423a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel M., Higa A. Calcium-dependent bacteriophage DNA infection. J Mol Biol. 1970 Oct 14;53(1):159–162. doi: 10.1016/0022-2836(70)90051-3. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McClain W. H., Seidman J. G. Genetic perturbations that reveal tertiary conformation of tRNA precursor molecules. Nature. 1975 Sep 11;257(5522):106–110. doi: 10.1038/257106a0. [DOI] [PubMed] [Google Scholar]
- McCorkle G. M., Altman S. Large deletion mutants of Escherichia coli tRNATyr1. J Mol Biol. 1982 Feb 25;155(2):83–103. doi: 10.1016/0022-2836(82)90438-7. [DOI] [PubMed] [Google Scholar]
- Meagher R. B., Shepherd R. J., Boyer H. W. The structure of cauliflower mosaic virus. I. A restriction endonuclease map of cauliflower mosaic virus DNA. Virology. 1977 Jul 15;80(2):362–375. doi: 10.1016/s0042-6822(77)80012-3. [DOI] [PubMed] [Google Scholar]
- Nishimura S., Harada F., Narushima U., Seno T. Purification of methionine-, valine-, phenylalanine- and tyrosine-specific tRNA from Escherichia coli. Biochim Biophys Acta. 1967 Jun 20;142(1):133–148. doi: 10.1016/0005-2787(67)90522-9. [DOI] [PubMed] [Google Scholar]
- Nishimura S. Minor components in transfer RNA: their characterization, location, and function. Prog Nucleic Acid Res Mol Biol. 1972;12:49–85. [PubMed] [Google Scholar]
- Platt T., Yanofsky C. An intercistronic region and ribosome-binding site in bacterial messenger RNA. Proc Natl Acad Sci U S A. 1975 Jun;72(6):2399–2403. doi: 10.1073/pnas.72.6.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed R. E., Baer M. F., Guerrier-Takada C., Donis-Keller H., Altman S. Nucleotide sequence of the gene encoding the RNA subunit (M1 RNA) of ribonuclease P from Escherichia coli. Cell. 1982 Sep;30(2):627–636. doi: 10.1016/0092-8674(82)90259-8. [DOI] [PubMed] [Google Scholar]
- Rossi J., Egan J., Hudson L., Landy A. The tyrT locus: termination and processing of a complex transcript. Cell. 1981 Nov;26(3 Pt 1):305–314. doi: 10.1016/0092-8674(81)90199-9. [DOI] [PubMed] [Google Scholar]
- Russell R. L., Abelson J. N., Landy A., Gefter M. L., Brenner S., Smith J. D. Duplicate genes for tyrosine transfer RNA in Escherichia coli. J Mol Biol. 1970 Jan 14;47(1):1–13. doi: 10.1016/0022-2836(70)90397-9. [DOI] [PubMed] [Google Scholar]
- Schedl P., Primakoff P., Roberts J. Processing of E. coli tRNA precursors. Brookhaven Symp Biol. 1975 Jul;(26):53–76. [PubMed] [Google Scholar]
- Smith H. O., Birnstiel M. L. A simple method for DNA restriction site mapping. Nucleic Acids Res. 1976 Sep;3(9):2387–2398. doi: 10.1093/nar/3.9.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J. D., Abelson J. N., Clark B. F., Goodman H. M., Brenner S. Studies on amber suppressor tRNA. Cold Spring Harb Symp Quant Biol. 1966;31:479–485. doi: 10.1101/sqb.1966.031.01.062. [DOI] [PubMed] [Google Scholar]
- Smith J. D., Barnett L., Brenner S., Russell R. L. More mutant tyrosine transfer ribonucleic acids. J Mol Biol. 1970 Nov 28;54(1):1–14. doi: 10.1016/0022-2836(70)90442-0. [DOI] [PubMed] [Google Scholar]
- Soberon X., Covarrubias L., Bolivar F. Construction and characterization of new cloning vehicles. IV. Deletion derivatives of pBR322 and pBR325. Gene. 1980 May;9(3-4):287–305. doi: 10.1016/0378-1119(90)90328-o. [DOI] [PubMed] [Google Scholar]
- TESSMAN I., ISHIWA H., KUMAR S. MUTAGENIC EFFECTS OF HYDROXYLAMINE IN VIVO. Science. 1965 Apr 23;148(3669):507–508. doi: 10.1126/science.148.3669.507. [DOI] [PubMed] [Google Scholar]
- Tessman I. Mutagenic treatment of double- and single-stranded DNA phages T4 ans S13 with hydroxylamine. Virology. 1968 Jun;35(2):330–333. doi: 10.1016/0042-6822(68)90275-4. [DOI] [PubMed] [Google Scholar]
- Topal M. D., Fresco J. R. Complementary base pairing and the origin of substitution mutations. Nature. 1976 Sep 23;263(5575):285–289. doi: 10.1038/263285a0. [DOI] [PubMed] [Google Scholar]