LOA1, a yeast member of the glycerolipid acyltransferase family, encodes a novel lysophosphatidic acid acyltransferase associated with lipid droplets (LDs) and involved in triacylglycerol (TAG) accumulation. Loa1p, recruited during LD formation, preferentially directs oleic acid–containing phosphatidic acid species into the TAG biosynthetic pathway.
Abstract
For many years, lipid droplets (LDs) were considered to be an inert store of lipids. However, recent data showed that LDs are dynamic organelles playing an important role in storage and mobilization of neutral lipids. In this paper, we report the characterization of LOA1 (alias VPS66, alias YPR139c), a yeast member of the glycerolipid acyltransferase family. LOA1 mutants show abnormalities in LD morphology. As previously reported, cells lacking LOA1 contain more LDs. Conversely, we showed that overexpression results in fewer LDs. We then compared the lipidome of loa1Δ mutant and wild-type strains. Steady-state metabolic labeling of loa1Δ revealed a significant reduction in triacylglycerol content, while phospholipid (PL) composition remained unchanged. Interestingly, lipidomic analysis indicates that both PLs and glycerolipids are qualitatively affected by the mutation, suggesting that Loa1p is a lysophosphatidic acid acyltransferase (LPA AT) with a preference for oleoyl-CoA. This hypothesis was tested by in vitro assays using both membranes of Escherichia coli cells expressing LOA1 and purified proteins as enzyme sources. Our results from purification of subcellular compartments and proteomic studies show that Loa1p is associated with LD and active in this compartment. Loa1p is therefore a novel LPA AT and plays a role in LD formation.
INTRODUCTION
In mammals, lipid metabolism disorders have been implicated in several diseases. For instance, unlike adipocytes, nonadipose tissues have a limited capacity for storing excess fatty acids as neutral lipids. In obese patients, accumulation of excess lipid in these tissues as triacylglycerol (TAG) is associated with a number of medical complications, including type 2 diabetes, hypertension, and heart failure (Kopelman, 2000; Listenberger et al., 2003). A severe lipid storage disease, congenital generalized lipodystrophy 1 (CGL1), is characterized by an almost complete lack of adipose tissue and the development of insulin resistance (Garg and Agarwal, 2009). The CGL1 gene encodes a lysophosphatidic acid acyltransferase (LPA AT) that catalyzes the formation of phosphatidic acid (PA), a key intermediate in the biosynthesis of both phospholipids (PLs) and neutral glycerolipids. The sharp decrease in TAG accumulation observed in the CGL1 mutant indicates a link between PL synthesis and the control of adipogenesis. In all eukaryotic cells, due to their hydrophobicity, TAG and steryl esters (SEs) are organized in specialized structures forming the central core of cytosolic organelles, the lipid droplets (LDs). LDs are surrounded by a PL monolayer associated with a limited number of proteins (for reviews, see Murphy, 2001; Fujimoto et al., 2008; Olofsson et al., 2008). Neutral lipid biosynthesis is believed to occur in endoplasmic reticulum (ER) microdomains, where enzymes for neutral lipid synthesis are present. In most cells, the biosynthetic pathway for TAG begins with acylation of the precursor glycerol-3-phosphate with two acyl chains, which lead to PA. PA is then dephosphorylated to form diacylglycerol (DAG), which is subsequently converted into TAG by a third acylation step. Newly synthesized TAG is thought to accumulate between the two leaflets of the PL bilayer before budding from the ER membrane (for review, see Martin and Parton, 2006; see also Pol et al., 2005). For many years, LDs were considered to be inert storage for neutral lipids. However, recent data support the idea that LDs are highly dynamic organelles that play an important role in the biosynthesis and mobilization of neutral lipids. Under normal physiological conditions, cells are able to generate LDs in response to elevated fatty acid levels (Rosenberger et al., 2009) and to mobilize storage lipids during deprivation conditions, indicating a major role for LDs in maintaining lipid balance at the cellular level. Moreover, LDs participate in several cellular processes and interact with various other cellular compartments, including the ER and mitochondria (Goodman, 2008; Murphy et al., 2009; Stone et al., 2009; Jacquier et al., 2011).
At the cellular level, defects in TAG biosynthesis are related to dysfunctions in LD biogenesis and maintenance (Oelkers et al., 2002; Sandager et al., 2002). However, despite their central role in energy homeostasis, little is known about the cellular biology of LDs, such as the molecular mechanisms of their biogenesis, protein association, size and number control, or mobilization. In particular, the mechanisms that direct and control the flux of acyl chains either into membrane PLs or storage lipids remain largely unknown. Very recent findings indicate that the phosphorylation state of Pah1p, the yeast lipin that generates DAG, is a key regulatory mechanism for controlling PL and neutral glycerolipid homeostasis (Karanasios et al., 2010; Choi et al., 2011). Since glycerolipids are synthesized via pathways that are largely conserved throughout the eukaryotes, Saccharomyces cerevisiae is a practical model organism for understanding the regulatory aspects of eukaryotic lipid homeostasis. Using an extensive genomic database search, a set of putative acyltransferases belonging to the glycerolipid acyltransferase family has been identified. Among these genes, SLC1 encodes the sole acyl-CoA–dependent LPA AT characterized to date in yeast (Nagiec et al., 1993). TAZ1 and PSI1 were previously characterized by our group; TAZ1 encodes an acyl-CoA–independent lysophosphatidylcholine (LPC) acyltransferase involved in the remodeling of cardiolipin (Testet et al., 2005) and PSI1 is a lysophosphatidylinositol (LPI) acyltransferase that catalyzes the incorporation of stearate in the sn-1 position of neosynthesized phosphatidylinositol (PI; Le Guédard et al., 2009). Recently, high-throughput screening studies identified yeast mutant strains responsible for abnormalities in the morphology of LD (Szymanski et al., 2007; Fei et al., 2008). Among them, a mutant caused by the deletion of VPS66, alias YPR139c, was found to be affected in the number and size of LDs. The function of this gene is unknown. A phenotype in protein sorting to the vacuole was found to be associated with the deletion mutant (Bonangelino et al., 2002). VPS66 presents the distinctive features of the glycerolipid acyltransferase family. In this paper, we show that this gene encodes a novel LPA AT partly associated with and involved in LD biogenesis. Loa1p might be an enzyme preferentially channeling oleic acid into the TAG biosynthetic pathway.
RESULTS
VPS66/LOA1 belongs to the glycerolipid acyltransferase family
The gene previously named VPS66, alias YPR139c, was renamed LOA1 (lysophosphatidic acid: oleoyl-CoA acyltransferase 1) in the present study. The open reading frame (ORF) of Vps66p predicts a 300-amino acid protein of ∼33.8 kDa containing one or two transmembrane domains according to prediction programs. Specifically, five prediction models (TMHMM, TMPred, TopPred2, HMMTOP 2.0, and SOSUI) were used. The results show that four models out of five indicated that Loa1p contains two transmembrane helices (unpublished data). Only TMHMM predicted one transmembrane domain. In addition, according to all the prediction domain softwares, both the N-terminus and the C-terminus are cytosolic. The protein sequence bears motifs I and III, which are characteristic of plsC-domain–containing proteins and important for acyltransferase catalytic activity (Lewin et al., 1999). Supplemental Figure S1 shows a partial sequence of its amino acid sequence aligned with glycerolipid acyltransferases from Escherichia coli (plsC) and S. cerevisiae (Slc1p, Taz1p, Psi1p). Sequence analysis of Loa1p failed to predict a subcellular localization. The growth characteristic of the deletion mutant was determined in yeast–peptone–dextrose (YPD) and yeast–peptone–lactate (YPL) media and compared with that of the wild type. The mutant strain had a growth delay compared with the wild type. A generation time of 115 and 250 min for the mutant strain was found on YPD and YPL, respectively, as compared with 90 and 200 min for the wild type.
loa1Δ and overexpressed LOA1 strains display aberrant LD morphology
High-throughput screenings revealed that the absence of LOA1 results in abnormal LD morphology (Szymanski et al., 2007; Fei et al., 2008). loa1Δ mutant cells accumulated on average more LDs than wild-type cells in similar growth conditions (Fei et al., 2008). As shown in Figure 1A, we confirmed these observations. Cells were grown to stationary phase in YPD media; stained with Nile red, a lipophilic dye that specifically stains LDs; and monitored by fluorescence microscopy. The deletion strain loa1Δ displays an increase in the LD number (LDs per cell: 2.24 ± 0.11 in wild type vs. 5.21 ± 0.17 in loa1Δ; n = 30 cells examined; p < 0.001) and a trend toward a smaller size. In addition, the deletion mutant strain exhibited an increase in cell size compared with the wild-type strain, as reported in Jorgensen et al. (2002).
FIGURE 1:
Abnormal LDs in loa1Δ and overexpressed LOA1 strains. (A) Wild-type cells and loa1Δ mutant cells were grown in YPD medium until stationary phase. (B) Wild-type strain transformed with the empty expression vector or with the yeast expression vector pYES2/CT-LOA1 were grown on synthetic defined media. Nile red–stained cells were observed under a fluorescence microscope.
Next the wild-type strain was transformed with the yeast expression vector pYES2/CT-LOA1 or the empty vector in order to observe any changes in LD morphology. Cells were grown to stationary phase on synthetic defined media, stained with Nile red, and observed by fluorescence microscopy. The wild-type strain transformed with the empty vector displayed a slight LD accumulation (Figure 1B), with particles homogeneous in shape and size. These changes in morphology, which were observed when cells were grown in YPD or synthetic defined media, have been reported (Fei et al., 2008). In contrast, cells overexpressing LOA1 displayed a decrease in the number of LDs (Figure 1B; LDs per cell: 4.85 ± 0.19 in BY4742, pYES2/CT vs. 2.49 ± 0.11 in BY4742, pYES2/CT-LOA1; n = 30 cells examined, p < 0.001) and LDs seemed to be larger. This mutant overexpressing LOA1 completely reversed the phenotype observed with the deletion mutant strain loa1Δ with respect to size and number of LDs. In other words, these data clearly indicate that the function of Loa1p is essential for rescuing the morphology phenotype of the loa1Δ deletion mutant strain.
Knockout of LOA1 decreases the TAG cellular content
To gain insight into the metabolic function of Loa1p in the cell, we analyzed and compared the lipid composition of mutant and wild-type loa1Δ cells. For this purpose, the neutral and polar lipid compositions were assessed by steady-state labeling with [14C]acetate. In a first set of experiments, cells grown to logarithmic phase (∼0.5 OD600) were incubated with [14C]acetate and harvested again at the early stationary phase. Total lipids were then extracted and resolved in one-dimensional high-performance TLC (HPTLC) using successive polar and neutral solvent systems. The amount of [14C]acetate incorporated in neutral and polar lipids was similar in both strains (neutral lipids: 9.68 ± 0.61 nmol [14C]acetate in wild type vs. 9.51 ± 0.13 nmol in loa1Δ mutant; polar lipids: 7.29 ± 0.23 nmol in wild type vs. 8.52 ± 0.49 nmol in loa1Δ mutant). The quantification of the incorporation in the major lipid classes (Figure 2A) showed that the deletion of LOA1 led to a significant decrease in TAG incorporation (∼20%; 4.36 ± 0.32 nmol in wild type vs. 3.45 ± 0.08 nmol in loa1Δ mutant), accompanied by an increase in SE and PL. Because the main impact of the deletion was a decrease in TAG, we further examined the acetate incorporation of wild-type and deletion mutant cells during the stationary growth phase. During this phase, TAG synthesis predominated over PL synthesis. Wild-type cells and loa1Δ deletion mutant cells were grown on YPD medium until OD600 ∼7–8 and labeled with [14C]acetate for an additional 20 h. As can be seen from the inset (Figure 2B), quantification of total PL revealed a similar incorporation in both strains, whereas deletion of LOA1 resulted in a significant decrease in total neutral lipids compared with wild-type cells. The quantification of the major lipid classes is shown in Figure 2B. As observed in logarithmic growth phase labeling, the most striking change was that deletion of LOA1 resulted in a marked decrease (43%) in TAG labeling compared with the wild-type cells (7.84 ± 0.47 nmol label incorporated in wild-type cells vs. 4.45 ± 0.30 nmol in loa1Δ mutant). A slight decrease in label associated with SE and monoacylglycerol (MAG) plus DAG plus ergosterol (ERG) was also observed. The decrease in TAG content was also confirmed by gas chromatography (GC). Neutral lipids were purified from cells harvested at OD600 = 6 by HPTLC. The silica gel zone corresponding to the TAG was scraped from the plate and quantified by GC-flame ionization detector (GC-FID). In agreement with results obtained by acetate incorporation, the TAG content was strongly reduced in the mutant (0.89 ± 0.09 μg acyl chain/absorbance unit in wild type vs. 0.61 ± 0.06 μg acyl chain/absorbance unit in loa1Δ mutant; n = 5). This decrease (∼31%) observed when cells reached the end of the exponential phase was intermediate between that observed by steady-state labeling during the exponential phase (∼20%) and the stationary phase (∼43%). Moreover, there was a significant decrease in the percentage of 18:1 fatty acid in the mutant (30.10 ± 0.47% in wild-type cells compared with 21.53 ± 0.68% in the mutant). This decrease was compensated for by an increase in the percentage of 16:0 fatty acid (21.47 ± 0.31% compared with 32.97 ± 1.31%). Mitochondria from a mutant deleted for TAZ1 is devoid of acyl-CoA-independent LPC AT activity (Testet et al., 2005). As LOA1 (alias YPR139c) is localized in the immediate vicinity of TAZ1 (alias YPR140W), which expresses an enzyme involved in lipid metabolism, we checked whether putative changes in the lipidome observed in the loa1Δ mutant strain were due to a defect in the TAZ1 gene. We observed that the rate of phosphatidylcholine (PC) synthesis from labeled LPC in the absence of acyl-CoA was the same as that in mitochondria purified from loa1Δ mutant and wild-type cells, indicating that LOA1-deficient cells encode an active Taz1p (unpublished data).
FIGURE 2:
Effects of LOA1 on neutral lipid composition. (A) Logarithmic growth phase labeling: steady-state total lipid profiles of wild-type and loa1Δ mutant cells grown until logarithmic phase (OD600 0.5) and labeled with [14C]acetate for an additional 20 h. Lipids were extracted and separated by one-dimensional TLC using first a polar lipid solvent system (midplate) and then a neutral lipid system, as described in Materials and Methods. The results show the amount of [14C]acetate incorporated into individual neutral and polar lipid classes. Dark bar, wild-type cells; gray bar, loa1Δ mutant. Results are mean ± SD from one experiment performed in triplicate and are representative of two distinct experiments. (B) Stationary growth phase labeling: wild-type and loa1Δ mutant cells grown to end of logarithmic phase (OD600 ∼7–8) were labeled with [14C] acetate for 20 h. (B) Incorporation of label into individual lipid classes. Inset, incorporation of label into neutral and polar lipids. Dark bar, wild-type cells; gray bar loa1Δ mutant. Results are mean ± SD from one experiment performed in triplicate and are representative of two distinct experiments. NL, neutral lipids; LP, polar lipids.
Lipid profile of loa1Δ shows a specific defect in molecular species containing oleic acid
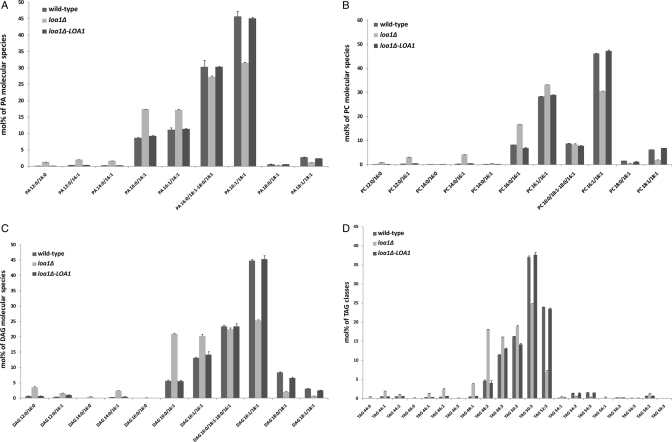
Tandem mass spectrometry enables lipid molecular species profiling (Brügger et al., 1997; Ejsing et al., 2009). In this study, a shotgun lipidomic approach was used to compare the lipidome of the deletion mutant strain loa1Δ with the lipidome of the wild-type strain, in order to detect specific perturbations and provide insight into the molecular function of the encoded protein. Lipid species were extracted and profiled as described previously (Ejsing et al., 2009). Lipid profiles were obtained with an LTQ-Orbitrap mass spectrometer using negative electrospray ionization for PA, PI, PE, and PS classes and positive electrospray ionization for PC, DAG, and TAG classes. The PL molecular species were determined by fragmentation experiments (multi-stage; MS) under operator control, given the location of the two fatty acyl chains on the glycerol backbone. In the present study, the PL 16:0/18:1 annotation indicated that the palmitic acyl chain is located at the sn-1 position of the glycerol backbone, whereas the oleic acyl chain is at the sn-2 position. Figure 3A shows the composition of the PA molecular species analyzed from wild type, deletion mutant, and deletion mutant overexpressing LOA1 strains. We observed that the PA molecular composition of the wild-type strain was close to that described in Ejsing et al. (2009), and in perfect agreement with the finding that in yeast, as in higher eukaryotes, unsaturated fatty acids are predominantly found at the sn-2 position (Wagner and Paltauf, 1994). Interestingly, the comparative analysis of the molecular species showed that the percentage of the total oleic acid–containing PA molecular species was significantly reduced in the deletion mutant: PA 16:1/18:1, PA 18:1/18:1, the isobaric combination PA 16:0/18:1-18:0/16:1, and to a small extent PA 18:0/18:1. This decrease in 18:1-containing molecular species was principally compensated for by an increase in 16:0/16:1 and 16:1/16:1 molecular species. The reduction in 18:1-containing molecular species was also observed in the other PL classes (PC, PI, PS, and PE; Figures 3B and S2, A–C) and in glycerolipids. Figure 3C shows that the molecular species DAG 16:1/18:1, DAG 18:0/18:1, and DAG 18:1/18:1, as well as the isobaric combination DAG 16:0/18:1-18:0/16:1, were significantly reduced in the mutant. The comparative TAG profiling shown in Figure 3D indicates a decrease in the 50:3, 52:3, 54:2, and 54:3 TAG species. Each TAG species decreasing in the mutant represents a combination of isobaric molecular species with theoretically at least one molecular species containing an oleic acyl chain, whatever its position on the glycerol backbone. This observation was confirmed by a neutral loss (NL) experiment based on the NL of oleic acid with NH4+ in positive ion mode (NL 299; Figure S3). This decrease in 18:1-containing TAG species was in agreement with the TAG analysis performed by GC. To verify that the absence of the gene was the basis for the observed phenotype, we profiled the deletion mutant overexpressing LOA1. As shown in Figures 3 and S2, the lipid phenotype associated with the deletion mutant was fully reverted to the wild-type lipidome, demonstrating that the low-oleic-content phenotype detected in the deletion strain was strictly linked to the absence of the gene.
FIGURE 3:
Lipid profiling of WT, loa1Δ deletion, and loa1Δ deletion overexpressing LOA1 strains. Cells were grown on synthetic defined media in the presence of 2% galactose and harvested at the early logarithmic phase. Lipids were extracted from yeast cells according to Ejsing et al. (2009). Species were profiled on a LTQ-Orbitrap mass spectrometer in negative mode: (A) PA; or in positive mode: (B) PC, (C) DAG, and (D) TAG. Lipid classes were identified and quantified by the software LipidXplorer (Herzog et al., 2011). Lipid molecular species were determined manually by fragmentation experiments. The abundance of lipid species is depicted in mol%. Error bars indicate ± SD (n = 3 independent analyses).
Overall, the lipidome of the deletion mutant showed a specific change in PL and glycerolipid molecular composition, confirming the involvement of Loa1p in lipid metabolism. These results suggested that this protein could be an acyltransferase responsible, at least in part, for the presence of oleic acid in PLs and glycerolipids. An easy way to explain this specific enrichment in lipid classes is to hypothesize that Loa1p is involved in the initial step in lipid biosynthesis, namely de novo PA synthesis, because PA is a key intermediate that serves as a general precursor for all PLs and glycerolipids, including DAG and TAG. In addition, the lipidomic approach used in this study revealed a defect in the incorporation of oleic acid at the sn-2 position of the PL glycerol backbones in mutant cells, suggesting that Loa1p is more likely an LPA AT than a glycerol-3-phosphate acyltransferase. To summarize, all these results (obtained by an in vivo approach at the molecular species level) clearly suggest that Loa1p is an acyltransferase involved in the acylation of LPA in the sn-2 position and that Loa1p has a substrate preference for oleic acid. Moreover, if these results are linked with those obtained in the acetate-labeling experiments, the qualitative results indicate that the defect in the incorporation of oleic acid in the deletion mutant was found in all polar and neutral glycerolipid classes, but the quantitative results show that only the TAG content was significantly reduced in the mutant, while the content of each class of PLs remained constant.
Loa1p displays an acyl-CoA:LPA AT activity
To determine whether Loa1p has an LPA AT activity, LOA1 was expressed in C41(DE3) E. coli strain. After induction with isopropylthio-β-galactoside, membranes were prepared and applied to LPA AT activity by using [14C]oleoyl-CoA and unlabeled 16:0 LPA. Figure 4 shows the synthesis of PA associated with membranes of bacteria transformed with the empty vector (synthesis of 10.6 ± 2.2 pmol of PA in our experimental conditions), corresponding to the endogenous LPA AT activity encoded by the E. coli plsC gene (Coleman, 1992). The LPA AT activity was doubled with the membranes of bacteria expressing LOA1 (synthesis of 22.7 ± 3.7 pmol of PA). These results suggest that Loa1p has an acyl-CoA–dependent LPA AT activity. In both conditions (empty vector and LOA1 expressed in E. coli), we observed the presence of labeled free fatty acid and PE in assays. The former activity suggests the presence of endogenous activities as acyl-CoA thioesterase. The latter activity may be due to an endogenous acyltransferase, because 1) PE was formed in E. coli membranes expressing or not expressing Loa1p (Figure 4); 2) following studies of acyltransferase assays as a function of time, a direct precursor–product relationship between PA and PE seemed to occur (Figure S4); and 3) this activity was absent in assays performed with partially purified Loa1p from yeast (see below). Overall, the synthesis of labeled PE suggests a transacylation reaction between the sn-2 position of neosynthesized PA and the sn-1 position of sn-2 acyl-LPE (Homma and Nojima, 1982; Harvat et al., 2005). This indicates that we should add the amounts of labeled PE and PA to determine the LPA AT activity.
FIGURE 4:

Acyltransferase activities associated with membranes of E. coli expressing LOA1. Membrane proteins were obtained from C41 (DE3) E. coli transformed with pET-15b (lane 1) and with pET-15b containing the coding sequence of LOA1 (lane 2). Lane 3: negative control without protein; lane 4: positive control with 1 μg yeast protein. LPA AT activities were determined using 0.5 nmol [14C]oleoyl-CoA, 1 nmol 16:0 LPA, and 10 μg membrane-bound proteins as described in Materials and Methods. After a 5-min incubation, lipids were extracted and analyzed by TLC using chloroform/methanol/1-propanol/methyl acetate/0.25% aqueous KCl (10:4:10:10:3.6) as solvent; this was followed by radioimaging. Results are from one experiment that is representative of three experiments performed with independent membrane preparations.
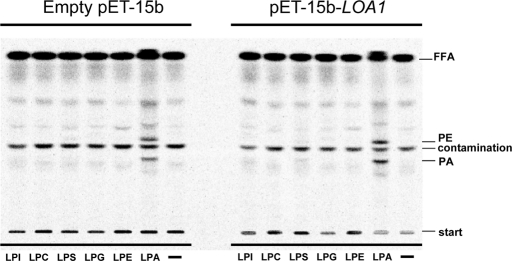
Next, using these E. coli membrane preparations, we measured the lysolipid acyltransferase activity of Loa1p by using [14C]oleyl-CoA as an acyl donor and various lysolipids as acyl acceptors. Figure 5 clearly shows that no detectable activity was found with LPC, lysophosphatidic ethanolamine (LPE), lysophosphatidyl glycerol (LPG), and LPI as substrates, indicating that, as expected, Loa1p is strictly specific for LPA. As a control, in the presence of LPA as substrate, assays confirm that LPA AT activity was doubled when membranes of bacteria expressing LOA1 were used(consider the sum of PE + PA, see above). In addition, various supplementary enzymatic activities were tested, including glycerol-3-phosphate acyltransferase, MAG AT, DAG AT, PA phosphatase, lipase, and transacylase activities; no other activities were detected (unpublished data). To confirm these results, the purification of Loa1p was undertaken. The wild-type strain was transformed with the yeast expression vector pYES2/CT-LOA1 (or the empty vector as a control) to produce a C-terminal V5-His–tagged fusion protein and to purify the protein by affinity chromatography with anti-V5 agarose affinity gel antibody. Overexpression of Loa1p was confirmed by Western blot analysis using an antibody against the V5 epitope. Unfortunately, the elution by competition with V5 peptide was not successful; the protein of interest remained attached to the beads regardless of the elution conditions used. Despite this, we decided to perform the LPA AT assay with the “anti-V5 beads” in which the recombinant Loa1p was immobilized. Proteomic analysis was done with this fraction in order to ensure that no other lysolipid acyltransferase was present in the fraction. All contaminant proteins were identified and their functions were characterized according to the Saccharomyces Genome Database (SGD). No lysolipid acyltransferase other than Loa1p was detected (Slc1p, Taz1p, Yor175cp, and Psi1p were absent). Thus, if any acyltransferase activity were to be detected in the assay, it would come from the protein of interest, Loa1p. As shown in Figure 6, several volumes of bead fractions were incubated in the presence of 16:0 LPA and [14C]oleoyl-CoA. PA synthesis increased proportionally with bead volume, indicating that Loa1p immobilized on the anti-V5 beads was able to form PA from LPA and oleoyl-CoA. The results obtained with an enzymatic assay by ectopic expression of the gene in E. coli or by expressing a tagged fusion gene in S. cerevisiae confirm the hypothesis formulated at the end of the lipidomic approach. To sum up, the results obtained by both in vivo and in vitro approaches indicate that LOA1 encodes an acyl-CoA–dependent LPA AT.
FIGURE 5:
Loa1p exhibits a strict specificity for LPA. Lysophospholipid acyltransferase activities were analyzed using LPI, LPC, LPS, LPG, LPE, and LPA as acyl acceptors (1 nmol). A control without exogenous lysophospholipid (−) was included. Other conditions were as described in Figure 4. Results are from one experiment representative of three experiments performed with independent membrane preparations.
FIGURE 6:
Enzymatic assay of recombinant protein Loa1-V5-6xHis immobilized onto anti-V5 agarose affinity gel antibody. The strain YSA01 (or YSA00 in control) was grown on synthetic defined media supplemented with galactose until the exponential phase. Purification of the recombinant protein was done with anti-V5 agarose affinity gel antibody. After incubation of the yeast lysate and the beads overnight at 4°C, the protein elution was performed with V5 peptide as indicated in Materials and Methods. LPA AT activities were determined using 0.5 nmol [14C]oleoyl-CoA, 1 nmol 16:0 LPA, and several volumes of the fraction of interest. After a 10-min incubation, lipids were extracted and analyzed by TLC using the polar system; this was followed by radioimaging.
Loa1p has a dual localization in LDs and ER
To gain further information on the role of LOA1, we determined its subcellular localization. A genomic-tagging construct expressed from its own promoter was engineered to append a hemagglutinin (HA) epitope to the C-terminus of Loa1p. Cells were grown in YPD media until exponential phase. In parallel, the genomic-tagging strain BY4742, ERG6-red fluorescent protein (RFP) was grown in the same conditions in order to serve as a control for LD isolation (Leber et al., 1998). After cell lysis, cell fractionation was performed using a sucrose cushion (Beaudoin et al., 2000). Protein fractions from the cell lysate and the top, middle, and bottom of the gradient were separated on SDS–AGE gel and analyzed by immunoblotting using antibodies specific for compartment markers (Figure 7). All of the markers were detected in the lysate and the bottom fraction, whereas they were absent from the middle fraction. As expected, the LD marker Erg6p was the only one found associated with the top fraction, indicating that the latter contains LD but is also devoid of the other compartments, including vacuole, plasma membrane, ER, and the trans-Golgi network/prevacuolar compartment. Therefore the presence of Loa1-HAp in this top fraction clearly indicated that this protein is associated with LD. Moreover, the presence of Erg6–RFPp in the bottom fraction could be attributed to an additional localization of this protein in a compartment forming the bottom fraction, which is consistent with the presence of this protein both in ER membranes and in LDs (Leber et al., 1998) or by the presence of LDs trapped with microsomal membranes. Since the abundance of the Erg6 protein in the top fraction was higher than that found in the bottom fraction and more Loa1p was observed in the bottom fraction compared with the top fraction, we deduced that Loa1p is also localized in at least one compartment distinct from the LDs, forming the bottom fraction. To further investigate the subcellular localization of Loa1p, we designed a genomic-tagging LOA1-mCherry strain that was expressed from its endogenous promoter at the LOA1 locus and compared its distribution with that of Erg1-GFPp, a protein marker with a dual localization in the ER and in LDs (Leber et al., 1998). The fatty acid content of the LOA1-mCherry strain was analyzed by GC and was compared with the fatty acid content of the wild-type and the loa1Δ deletion strains. The Loa1-mCherryp was able to restore the wild-type fatty acid composition, suggesting that the tagged protein is functional in vivo (unpublished data). Both Loa1-mCherryp and Erg1-GFPp showed a dynamic localization that changed according to the growth phase (Figure 8). In the early log phase, Loa1p exhibited a typical ER fluorescence pattern with enrichment in the perinuclear region and complete colocalization with Erg1-GFPp (Figure 8, top). When cells were grown to stationary phase, both Loa1p and Erg1p were mainly associated with spherical structures, likely LDs, although ER staining was still visible (Figure 8, bottom). To confirm that the spherical structures were LDs, fluorescence microscopy experiments were undertaken with cells expressing the genomic-tagging construct LOA1-DsRed1 from its own promoter. Cells harvested at the exponential phase were stained with BODIPY 493/503, a lipophilic dye specific for LDs (Guo et al., 2008). Micrographs shown in Figure 9 confirm that Loa1p colocalized with LD markers.
FIGURE 7:

Localization of Loa1-HAp in LD. Cells expressing the LOA1-HA fusion gene grown on YPD media until the exponential phase. The tagged gene obtained by homologous recombination was expressed from its own promoter. Cell lysate was loaded on top of a sucrose cushion and centrifuged at 100,000 × g for 90 min. Fractions were collected from the top (T), middle (M), and bottom (B). Proteins were separated by SDS–AGE 12.5% gel and immunoblotted with antisera for the compartment markers: VATPase for the vacuole, Pam1p for the plasma membrane, RFPp for the LDs, Dpm1p for the ER, and Pep12p for the trans-Golgi network/prevacuolar compartment.
FIGURE 8:
Loa1-mCherryp partitioned between ER and large spherical structures, likely LD. Cells expressing the LOA1-mCherry and ERG1-GFP fusion genes grown on YPD media until early log phase (top) or stationary phase (bottom) were examined under fluorescence microscopy as described in Materials and Methods. Merge indicates that fluorescence overlapped; DIC: differential interference contrast. Scale bars: 5 μm.
FIGURE 9:
Localization of Loa1-Ds red1p in LDs. Cells expressing the LOA1-Ds red1 fusion gene grown on YPD media until the exponential phase were stained with BODIPY 493/503 and examined under a fluorescence microscope as described in Materials and Methods. The fusion gene obtained by homologous recombination was expressed with its own promoter. Con A, concanavalin A-treated cell walls. BODIPY shows the LDs. The Ds red1 signal shows the localization of the fusion protein. Merge indicates that fluorescence overlapped.
In addition, highly purified LD fractions were prepared after three successive ultracentrifugation steps according to Connerth et al. (2009), and proteomic analysis was performed on the enriched LD fractions, which were treated with 4.5 M urea or left untreated. Following both of these treatments and further proteomic studies, results were analyzed by using a threshold corresponding to the lower value of the relative abundances of proteins previously shown to be present in LDs. The raw data are presented in Supplemental Table S1. Without urea pretreatment of purified LD, ∼327 proteins were detected. Using a threshold of 0.03%, we identified 213 putative LD proteins, including 42 proteins associated with LDs in previous studies (Athenstaedt et al., 1999) and annotated in the SGD database as LD–associated proteins. As expected, Loa1p was detected (Table 1), in accordance with a very recent LD proteomic analysis (Grillitsch et al., 2011).
TABLE 1:
Proteins identified from LD purification by a proteomic approach (strain YSA01) and previously annotated in the SGD database as LD-associated proteins.
| ORF | Gene | Function (hypothetical) | Metabolism | Presencea | Relative abundance (%) |
|---|---|---|---|---|---|
| ERG6 | YML008c | Δ(24)-Sterol C-methyltransferase | Ergosterol biosynthesis | + | 9.35 |
| PGK1 | YCR012w | Phosphoglycerate kinase | Glycolysis | − | 6.95 |
| FAA1 | YOR317c | Long-chain fatty acid-CoA ligase 1 | FA activation | + | 3.59 |
| AYR1 | YIL124w | NADPH-dependent 1-acyldihydroxyacetone | PL biosynthesis | + | 3.49 |
| PET10 | YKR046c | — | — | + | 2.85 |
| FAT1 | YBR041c | VLCFA transport protein | FA activation | + | 1.67 |
| TGL1 | YKL140w | Sterol esterase | TAG metabolism | + | 1.64 |
| TDH3 | YGR192c | GAPDH | Glycolysis | − | 1.63 |
| HFD1 | YMR110c | Putative fatty aldehyde dehydrogenase | Cellular process | + | 1.33 |
| TDH1 | YJL052w | GAPDH | Glycolysis | + | 1.06 |
| EHT1 | YBR177c | Medium-chain fatty acid ethyl ester synthase | FA metabolism | + | 0.98 |
| TGL4 | YKR089c | Lipase | TAG metabolism | + | 0.97 |
| YIM1 | YMR152w | Uncharacterized protein | — | + | 0.73 |
| PDI1 | YCL043c | Protein disulfide-isomerase | Cellular process | − | 0.72 |
| TGL3 | YMR313c | Lipase | TAG metabolism | + | 0.7 |
| ERG27 | YLR100w | 3-Keto-steroid reductase | Ergosterol biosynthesis | + | 0.6 |
| LOA1 | YPR139c | LPA AT | PL biosynthesis | + | 0.6 |
| SLC1 | YDL052c | LPA AT | PL biosynthesis | + | 0.42 |
| KES1 | YPL145c | Member of the oxysterol-binding protein family | Sterol transport | + | 0.39 |
| PDR16 | YNL231c | PI transfer protein | PL transport | + | 0.35 |
| TGL5 | YOR081c | Lipase | TAG metabolism | + | 0.35 |
| ERG7 | YHR072W | Lanosterol synthase | Ergosterol biosynthesis | + | 0.33 |
| YP147 | YPR147C | Uncharacterized protein | — | − | 0.26 |
| ACT1 | YFL039c | Actin | Cellular process | − | 0.25 |
| YJU3 | YKL094w | Serine hydrolase | Monoglyceride lipase | + | 0.25 |
| GST1 | YDR172w | Glutathione S-transferase | Cellular process | + | 0.24 |
| ERG1 | YGR175C | Squalene monooxygenase | Ergosterol biosynthesis | + | 0.23 |
| FAS1 | YKL182W | FA synthase subunit beta | FA metabolism | − | 0.2 |
| FAA4 | YMR246w | Long-chain fatty acid-CoA ligase | FA activation | + | 0.2 |
| NUS1 | YDL193w | Undecaprenyl pyrophosphate synthetase | Protein amino acid glycosylation | + | 0.13 |
| SAC1 | YKL212w | PIP phosphatase | PIP biosynthesis | + | 0.12 |
| YP091 | YPR091C | Uncharacterized PH domain–containing protein | — | + | 0.12 |
| — | YBR056W | Uncharacterized glycosyl hydrolase | — | − | 0.09 |
| — | YOR059C | — | — | + | 0.09 |
| NTE1 | YML059c | Lysophospholipase | PL metabolism | − | 0.09 |
| GPT2 | YKR067w | G-3-P acyltransferase 2 (GAT1) | PL biosynthesis | + | 0.09 |
| UBX2 | YML013w | Protein involved in ER-associated protein degradation | ER-associated protein catabolic process | + | 0.09 |
| DGA1 | YOR245c | Diacylglycerol O-acyltransferase 1 | DAG acyltransferase | + | 0.08 |
| — | YNR021W | ER membrane protein | — | − | 0.06 |
| CPR5 | YDR304c | Peptidyl-prolyl cis-trans isomerase D | Cellular process | − | 0.06 |
| PGC1 | YPL206C | Probable glycerophosphodiester phosphodiesterase | PL metabolism | + | 0.05 |
| CSR1 | YLR380w | PI transfer protein | PL transport | − | 0.03 |
Proteins identified in which the function has not been yet characterized were retained by default. FA, fatty acid; VLCFA, very long fatty acid; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; PIP, phosphatidylinositol phosphate; G-3-P, glycerol 3-phosphate.
a+ indicates the presence of the protein with or without urea treatment; − indicates the absence of protein after urea treatment.
Following urea pretreatment, ∼166 proteins were identified. Using a threshold of 0.07%, 92 putative LD proteins were retained, including 38 known to be associated with LDs. Only 31 of these 38 proteins were present in the previous list, and seven proteins were found not to be associated with LDs before urea treatment. As shown in Table 1, Loa1p was still detected, as well as acyltransferases already identified in this organelle, including Gat1p, Dga1p, and Slc1p (Sorger and Daum, 2003). By contrast, and as expected, the other glycerolipid acyltransferases previously localized in the ER, Gat2p, Lro1p, and Psi1p (Natter et al., 2005; Le Guédard et al., 2009; Jacquier et al., 2011) were not found associated with purified LDs.
As LDs have a distinctive feature, a hydrophobic core of neutral lipids surrounded by a monolayer of PL with embedded proteins, the question arose whether Loa1p, which is present in both the ER and the LD, is active in the latter compartment. We determined the LPA AT activities associated with purified LDs from the wild-type strain and loa1Δ deletion mutant. Figure 10 shows that LDs from wild-type and mutant cells were able to incorporate labeled oleic acid into LPA. Consistent with the localization of this protein in LDs, the activities decreased when LDs were purified from the loa1Δ deletion mutant. According to the proteomic analyses, this residual activity might correspond to the Slc1p enzyme. Since this activity represents one-half of the wild type, this result further indicates that both Slc1p and Loa1p enzymes have a similar specific activity in this compartment (∼0.5 nmol/min per mg PA synthesized in our experimental conditions).
FIGURE 10:
Purified LD catalyzed PA biosynthesis. Enzymatic activities were determined using 0.5 nmol [14C]oleoyl-CoA, 1 nmol 16:0 LPA, and LD proteins purified from wild-type and loa1Δ deletion strains. After a 10-min incubation, lipids were extracted and analyzed by TLC using the polar system; this was followed by radioimaging.
The above experiments, obtained with a lipidomic approach, suggest that Loa1p has a substrate preference for oleoyl-CoA. To confirm this selectivity, we determined the LPA AT activities associated with purified LDs from the slc1Δ deletion mutant using labeled palmitoyl-CoA or oleoyl-CoA as acyl donors. As shown in Figure 11, it appeared that the remaining LPA AT, that is, Loa1p, was able to incorporate oleoyl-CoA more efficiently than palmitoyl-CoA. Altogether, these in vitro assays clearly indicate that Loa1p is associated with LDs and confirm the preference for oleoyl-CoA.
FIGURE 11:
Loa1p is selective for oleoyl-CoA. LPA AT activities were determined by incubating 2 μg LD proteins purified from the slc1Δ deletion strain with 1 nmol 16:0 LPA and various amounts of [14C]oleoyl-CoA or [14C]palmitoyl-CoA. After a 10-min incubation time, lipids were extracted and analyzed by TLC using the polar system and monitored by radioimaging.
DISCUSSION
LPA AT catalyzes the second step of de novo TAG and PL synthesis. The lipid biosynthetic pathways have been well conserved through evolution. To date, all of the known eukaryotic and prokaryotic LPA ATs are homologous to the E. coli plsC and belong to the glycerolipid acyltransferase family characterized by the presence of four conserved domains (Lewin et al., 1999). In plants and mammals, the family comprises several isoforms, but the biological role of only a few isoforms has been characterized. In yeast, the sole LPA AT extensively studied is Slc1p, which has been described as the major LPA AT (Nagiec et al., 1993). In vitro data suggest that purified Slc1p displays not only LPA AT activity but also incorporates other lysolipids, including lysophosphatidylserine (LPS) and LPI (Benghezal et al., 2007). However, while cell-free lysates from slc1Δ overexpressing or not SLC1 incorporated low amounts of [14C]acyl-CoA into lipids in the presence of various lysolipids (other than LPA), a high incorporation of label into PA was observed in the presence of LPA (Zou et al., 1997). The authors suggest that SLC1 chiefly encodes a LPA AT. The deletion mutant slc1Δ is viable and exhibits significant residual activity, suggesting redundant LPA AT activities in yeast (Athenstaedt and Daum, 1997; Zou et al., 1997). Other enzymes presenting an LPA AT activity were recently identified in yeast. Investigations with another acyltransferase family, the membrane-bound O-acyltransferase family, revealed that the ORF YOR175c encodes an acyl-CoA lysolipid acyltransferase. In vitro microsomal assays determined a broad specificity as acceptor, including LPE, LPC, LPG, LPI, LPS, and LPA (Benghezal et al., 2007; Jain et al., 2007; Riekhof et al., 2007; Tamaki et al., 2007). Ghosh et al. (2008) reported that ICT1, a gene encoding for a soluble LPA AT, is expressed during organic solvent stress. More recently, Rajakumari and Daum (2010a) demonstrated that the yeast lipase Tgl5p, in addition to TAG hydrolysis, mediates the acyl-CoA–dependent acylation of LPA. These two enzymes, Ict1p and Tgl5p, share the HX4D acyltransferase motif. Interestingly, the Tgl4p lipase, reported by the same group to act also as an LPA AT, does not bear this motif (Rajakumari and Daum, 2010b). Using an extensive genomic database search, we previously identified several putative acyltransferases belonging to the glycerolipid acyltransferase family. In the present study, we focused on LOA1, a yeast gene that could be involved in LD morphology (Szymanski et al., 2007; Fei et al., 2008). The experimental approach chosen to characterize the enzymatic activity of the encoded protein included biochemicals and lipidomics based on mass spectrometry. Thus a global lipidomic approach to the loa1Δ deletion strain was very useful and determinant throughout, revealing that all the lipids (polar and neutral glycerolipids) containing oleic acid showed a significant decrease compared with the wild-type strain. This finding suggests that Loa1p could be an acyltransferase involved in the de novo synthesis of PA, the precursor of all membrane glycerophospholipids and storage lipid TAG in yeast. To discriminate between possible glycerol-3-phosphate acyltransferase and LPA AT activities, the oleic acid location on the PL glycerol backbone was investigated using tandem mass spectrometry. Fragmentation pathway analysis of each PL class indicated that oleic acid is mainly linked to the sn-2 position, a location in perfect agreement with previous studies (Wagner and Paltauf, 1994; Schneiter et al., 1999), thus evidencing an LPA AT activity. In total, this mass spectrometry approach suggests that Loa1p could be an LPA AT feeding both polar and neutral glycerides. Other experiments were then carried out to confirm this hypothesis. First, a significant increase in LPA AT activity associated with membrane extracts of E. coli expressing Loa1p was measured using [14C]oleoyl-CoA and LPA as substrates. Second, this enzyme displayed specificity for LPA, because no other acyltransferase activities were observed when it was tested against either glycerol-3-phosphate, DAG, or a variety of lysolipids. Third, a purified recombinant Loa1p immobilized on agarose beads demonstrated LPA AT activity. Finally, deletion of LOA1 reduced the LD LPA AT activity in highly purified LDs (see below for localization). Taken together, the data obtained by in vivo and in vitro approaches demonstrates that Loa1p acts primarily as an LPA AT.
Loa1p contains the four motifs of the glycerolipid acyltransferase family, except for motif I (HX4D). The histidine residue, which is highly conserved throughout this family, is replaced by a cysteine residue. Despite this characteristic, the protein was found to be active. It turns out that both residues are well-known for expressing affinity to metal ligands, and could determine a potential metal-binding site required for structure stabilization and acyltransferase activity. A lipidomic approach provides direct insight into the specificity of an acyltransferase for biological substrates. For instance, in our previous study (Le Guédard et al., 2009), the comparative lipidome analysis of psi1Δ and wild-type cells revealed highly specific changes in the molecular composition of PLs. Only the PI species containing stearic acid were dramatically decreased in yeast deleted for PSI1, indicating that Psi1p is responsible for the stearic acid–enrichment characteristic of PI in yeast. A similar approach was used to characterize the substrate preference for Slc1p. Disruption of the SLC1 gene led to moderate changes of the phospholipidome (Benghezal et al., 2007), apart from a decrease in oleic acid–containing species for a few PLs and a reduction in minor short chain–containing PI species (Guan and Wenk, 2006; Shui et al., 2010). In this paper, we show that the lipid phenotype observed in PA species for loa1Δ—a decreased content in molecular species containing oleic acid—was found in all polar and neutral lipids, suggesting that 1) oleoyl-CoA constitutes the preferential substrate for Loa1p (or the predominant substrate in the vicinity of Loa1p) and 2) PA generated by Loa1p is used in all lipid biosynthetic pathways. Biochemical assays, carried out with purified LDs from the slc1Δ deletion mutant, confirm the enzyme selectivity for oleoyl-CoA.
Establishing the precise subcellular localization of a protein represents a key step in the characterization of its function. According to the prediction tool used, one or two transmembrane domains are predicted for Loa1p. Moreover, an enzymatic assay performed on crude membrane extracts from E. coli expressing Loa1p revealed that the enzymatic activity is associated with membranes. However, large-scale analyses of protein localization based on the GFP fusion strategy localized Loa1p in the cytosol (Kumar et al., 2002). Owing to the lack of clear-cut results, the localization was undertaken using complementary approaches. Loa1p was localized to the ER and LDs when analyzed by subcellular fractionation, immunohistochemical analyses, and fluorescence microscopy. A proteomic approach on LDs associated with a biochemical activity assay confirmed the presence of Loa1p in this compartment (this study; Grillitsch et al., 2011). Both Slc1p and Loa1p localize to the same compartments in the cell, the ER and LDs. Moreover, the slc1Δ and loa1Δ deletion strains are viable, with a quite normal growth rate, indicating that they are able to supply all of the essential cell functions. However, despite this apparent redundancy, an altered LD morphology in the loa1Δ deletion mutant, which was not described for the slc1Δ mutant, indicates that both these LPA ATs are not functionally exchangeable in all respects. Indeed, PA and subsequent lipids generated by Loa1p, characterized by a specific fatty acid composition at the sn-2 position of the glycerol backbone, probably assume specific functions. A genome-wide RNA interference screen in Drosophila identified genes encoding enzymes of PL biosynthesis determinant for LD size and number (Guo et al., 2008), suggesting that the PL composition of the monolayer profoundly affects droplet morphology. For instance, Cct1 encoding a phosphocholine cytidylyltransferase, the enzyme that catalyzes the rate-limiting step in PC synthesis, localizes exclusively to the nucleus without treatment, whereas after treatment with oleate, that is, during LD formation, a significant portion of Cct1 localizes to the LD surface. This marked translocation of CCT enzyme to the vicinity of LD may provide building blocks for the PL monolayer of growing LD. Knockdown of Cct1 results in larger droplets, suggesting that PL monolayer supply is limiting compared with TAG biosynthesis during LD formation, and that the ratio of surface PL to core NL may regulate LD morphology. Similar interpretations could be drawn from our study. During the early logarithmic growth phase, Loa1p localizes exclusively to the ER, whereas in the stationary phase, that is, during LD formation, a major part of the Loa1p is associated with LDs. In loa1Δ deletion mutant cells, TAGs are limiting compared with PL monolayer supply and, in compensation, an increase in the number of smaller LDs leads to an increase in the ratio of PL monolayer to core neutral lipids. Conversely, in strains overexpressing LOA1, larger droplets allow a decrease in this ratio. In a very recent study, Fei et al. (2011) showed that an increase in the level of cellular PA is linked with fewer but bigger LDs, called “supersized” LDs. In perfect agreement, we show in this study that Loa1p is an LPA AT whose overexpression leads to an increase in PA level, and we observed a reduced number of cytoplasmic LDs and a trend to a larger diameter.
Oleic acid supplementation induces LD formation. In excess, or in cells defective for TAG biosynthesis, this fatty acid induces lipotoxicity, and it has been shown that TAG synthesis plays an important role in buffering excess toxic fatty acids (Petschnigg et al., 2009). In addition, the loa1Δ mutant was identified in a large-scale study as highly sensitive to unsaturated treatment (Lockshon et al., 2007). These findings strengthen the biological function proposed here, that is, Loa1p, which is recruited during LD formation, is an enzyme preferentially channeling oleic acid–containing PA species into the TAG biosynthetic pathway.
MATERIALS AND METHODS
Materials
TLC plates were HPTLC silica gel 60 F 254 10 cm × 10 cm or TLC silica gel 60 F 254 20 cm × 20 cm (Merck, Darmstadt, Germany). [1-14C]acetic acid, sodium salt and [oleoyl-1-14C]oleoyl-CoA were obtained from Perkin Elmer Life Sciences (Boston, MA). The 16:0 LPA (LIPID MAPS) was from Avanti Polar Lipids (Alabaster, AL). LPI, LPC, LPS, LPG, and LPE were purchased from Sigma-Aldrich (St. Louis, MO). For lipid quantification, mass spectrometry lipid standards (PA 17:0/17:0; PC 18:3/18:3; PE 17:0/17:0; PG 17:0/17:0; PS 17:0/17:0; DAG 17:0/17:0; TAG 17:1/17:1/17:1) were purchased from Avanti Polar Lipids–LIPID MAPS, except for PI 17:0/17:0, which was given by C. Thiele (LIMES, Bonn, Germany).
Yeast strains and growth conditions
The yeast strains used in this study are listed in Table S2. The strains used for lipid profiling were obtained from the EUROSCARF library. BY4742 is the wild-type strain; loa1Δ is the deletion mutant. In the rescue experiment, LOA1 gene was inserted into the pYES2 vector (Invitrogen, Carlsbad, CA) containing the GAL1-inducible promoter. The deletion strain loa1Δ was transformed with the plasmid construct (strain YSA04). For purification, proteomic experiments, and morphological studies, the ORF of LOA1 was inserted into the pYES2/CT vector (Invitrogen) containing the GAL1-inducible promoter. The plasmids constructed were transformed into BY4742 (strains YSA00 and YSA01). The cells were grown in a shaking incubator until early logarithmic phase at 30°C, in 250-ml Erlenmeyer flasks containing 50 ml of synthetic defined media composed of yeast nitrogen base without amino acids and with ammonium sulfate 0.67% (Difco, Detroit, MI), dropout mix minus uracil 0.2% (Sigma-Aldrich), KH2PO4 0.1% supplemented with raffinose 1% as a carbon source, and galactose 2% for induction. The pH was set at 5.5. The strains carrying LOA1-HA (strain YMS01), LOA1-DsRed1 (strain YMS02), and ERG6-RFP (strain YMS03) by genomic tagging were constructed as described in Janke et al. (2004) The strain carrying LOA1-mCherry and ERG1-GFP-HisMx (strain SL52) was constructed as described in Longtine et al. (1998). Cells were grown on YPD media (1% yeast extract, 1% peptone, 0.1% potassium phosphate, 0.12% ammonium sulfate, 2% glucose) as the carbon substrate. The pH was set at 5.5.
Lipidomic experiments
Yeast lipid extracts were prepared according to Ejsing et al. (2009) with slight modifications. Anionic glycerophospholipid species recovered in the 2:1 phase lipid extracts were detected by negative ion mode Fourier-transform mass spectrometry (FT MS) analysis by applying low m/z range scans (m/z 200–605) and high m/z range scans (m/z 505–1400) with target mass resolution of 100,000 (full-width at half-maximum [FWHM]) on a LTQ-Orbitrap mass spectrometer (Thermo Fisher Scientific, Lafayette, CO) equipped with the robotic nanoflow ion source TriVersa NanoMate (Advion Biosciences, Indianapolis, IN; Ejsing et al., 2006, 2009). The 2:1 phase lipid extracts were infused using methylamine (0.2 mM final in methanol). TAG, DAG, PC, and PE species recovered in the 15:1 phase lipid extracts were monitored by positive ion mode FT MS analysis (Ejsing et al., 2006; Schwudke et al., 2006, 2007) applying looped low m/z range scans (m/z 260–530) and high m/z range scans (m/z 500–1050) with target mass resolution of 100,000 (FWHM). The 15:1 phase lipid extracts were infused using ammonium acetate (7.5 mM final). Detected lipid species were identified and quantified using LipidXplorer (Herzog et al., 2011), as described in Graessler et al. (2009). The structures of detected lipid precursors were verified manually by MSn analysis. For NL experiments on TAG species, the experiments were performed on the AB SCIEX QTRAP 5500 system in direct infusion at 10 μl/min (AB Sciex, Foster City, CA).
Acyltransferase activity assays
Cloning, ectopic expression of the ORF LOA1 in E. coli, and isolation of E. coli membranes were done according to Testet et al. (2005). LPA AT reactions were conducted in 100 μl of assay mixtures (50 mM Tris-HCl, pH 8) containing 10 μg membrane proteins, 1 nmol 16:0 LPA, and 0.5 nmol [14C]oleoyl-CoA. After a 5-min incubation at 30°C, the reaction was stopped by adding 2 ml of chloroform/methanol (2:1, vol/vol) and 500 μl of water. The organic phase was isolated, and the aqueous phase was reextracted with 2 ml of chloroform. These combined lipid extracts were dried and redissolved in 50 μl of chloroform/methanol (2:1, vol/vol), and the lipids were separated by HPTLC, as described above. The radioactivity incorporated into PLs was analyzed using a STORM 860 PhosphorImager (GE Healthcare, Waukesha, WI) and quantified with ImageQuant TL software.
Purification
The yeast strains expressing the LOA1-V5-6xHis construct (YSA01) and the control (YSA00) were grown until exponential phase. Cells were then harvested, washed, and resuspended in phosphate-buffered saline (PBS)-NaCl buffer 0.3M supplemented with CHAPS 8 mM. Cells were disrupted by acid-washed beads (0.4–0.6 mm; Stratagene, Agilent, Santa Clara, CA) for 1 min (three times). After centrifugation, the soluble fraction of the lysate was recentrifuged 10 min at 4°C, 12,500 rpm. Two hundred microliters of suspension of the anti-V5 agarose conjugate (Sigma-Aldrich) was incubated with the clarified cell extract fraction overnight at 4°C. After incubation, the resin was washed several times with PBS buffer and incubated with 20 μg peptide V5 solution for 1 h. The fraction of interest was immediately used for acyltransferase assay. For Western blotting, the proteins were separated by SDS–AGE 12.5% and detected by Western blotting using anti-V5 mouse antibody (dilution 1:5000) and the goat anti-mouse immunoglobulin G linked to peroxidase. The immunoblots were photoactivated by a 40:1 mixture (reagents A and B) of ECL reagents plus (GE Healthcare). The membrane was covered with a transparent film and exposed to photographic AGFA Cronex 5 Medical X-Ray Film in a Kodak BioMax MS cassette.
Determination of steady-state neutral and polar lipid compositions by in vivo labeling
For logarithmic growth phase labeling, 5-ml samples of wild-type BY4742 strain and loa1Δ mutant cells grown at 30°C in YPD medium (0.5 OD600) were labeled with [14C]acetate (4 μCi/ml cell culture) for an additional 20 h for steady-state lipid labeling (about eight generations, according to Gaspar et al., 2006). Then 1.5-ml samples (OD600 9–10) were mixed with 100 μl of 100% acetic acid, cells were pelleted by centrifugation, and the supernatant was removed. To extract lipids from whole cells, 1 ml of chloroform/methanol (2:1, vol/vol) was added, and cell suspensions were vigorously shaken with glass beads (six times for 30 s with intermittent cooling on ice). One milliliter chloroform/methanol (2:1, vol/vol) and 500 μl 1% perchloric acid were added, and the cell suspensions containing beads were vigorously shaken for 30 s. After centrifugation, the organic phase was isolated, and the aqueous phase was reextracted with 2 ml of chloroform. These combined lipid extracts were dried and redissolved in 100 μl of chloroform/methanol (2:1, vol/vol), and the lipids were purified by TLC, using 20 cm × 20 cm TLC plates. PLs were separated using the solvent system chloroform/methanol/1-propanol/methyl acetate/0.25% aqueous KCl (10:4:10:10:3.6, vol/vol/vol/vol) until mid-height. The plates were then dried, and the neutral lipid classes were resolved using the second solvent system hexane/diethylether/acetic acid (90:15:2, vol/vol/vol). Lipid identity was based on the mobility of known standards, and the amounts of labeled lipids were analyzed by phosphorimaging. For stationary growth phase labeling, wild-type and loa1Δ mutant cells were grown on YPD medium until OD600 = 7–8. Cells were then labeled with [14C]acetate for 20 h in vivo labeling and lipid analyzed, as described above.
LD preparation, electrophoresis, and immunoblotting
For localization by cell fractionation and immunoblotting, LDs were prepared according to Beaudoin et al. (2000) from cells grown until the logarithmic phase, expressing the genomic construct LOA1-HA from its own promoter (strain YMS01) or, as a control, the genomic-tagging strain BY7442, ERG6-RFP (strain YMS03). Prior to cellular fractionation, cells were spheroblasted for 1 h at 30°C using spheroblasting buffer (20 mM KH2PO4, pH 7.5, 1.2 M sorbitol) plus Zymolyase 20T solution (5 mg/ml Zymolyase in 0.1 M KH2PO4, pH 7.5, 5 μl/ml β-mercaptoethanol). After flotation of LDs, equal amounts of top (LD fraction), middle, and bottom fractions were loaded for 12.5% SDS–AGE analysis followed by immunoblotting. Primary antibodies were used at the following dilutions: α-Pep12p (1:2000), α-VATPase (1:2000), α-RFP (1:10000), α-Dpm1p (1:500), α-Pma1 (1:5000), and α-HA (1:5000). Bound antibodies were developed using enhanced chemiluminescence Western blotting detection reagents (Amersham Pharmacia Biotech, Piscataway, NJ).
Localization of Loa1p by fluorescence microscopy
Cells expressing the genomic LOA1-mCherry and Erg1-GFP fusion genes (strain YSL52) were grown to early exponential phase or stationary phase. Photographs shown are both fluorescence and differential interference contrast images of the same cells. For microscopy with BODIPY staining, 1.5 OD units of cells expressing the genomic LOA1-DsRed1 (strain YMS02) from its own promoter were fixed and stained for LDs using 4% formaldehyde plus 0.5 μg/ml BODIPY 493/503 dye (Invitrogen) for 20 min. After being washed with distilled water, cells were incubated for 20 min in 20 μl PBS buffer supplemented with 0.1 mM CaCl2 and 1 mM MgCl2, with 20 μg concanavalin A, Alexa Fluor 647 (Invitrogen) conjugate to stain the cell wall. Cells were washed with PBS buffer-Ca-Mg and imaged directly after staining using an Olympus FV1000 confocal microscope.
Proteomic experiments
LDs were prepared according to (Connerth et al., 2009) from BY4742 cells overexpressing LOA1 (strain YSA01). After protein extraction, samples were loaded on a 10% one-dimensional SDS–AGE. Migration was stopped when samples entered the resolving gel. Bands were digested by trypsin, as indicated in Pocaly et al. (2008). Peptides were further analyzed on a Dionex U-3000 Ultimate nano LC system coupled to a nanospray LTQ-Orbitrap XL mass spectrometer (Thermo Finnigan, San Jose, CA). Ten microliters of peptide digests was separated on an analytical 75-μm inner diameter × 15-cm C18 PepMap column (LC Packing; Thermo Fisher Scientific, Lafayette, CO) with a 5–40% linear gradient of solvent B (H2O/ACN [20:80] supplemented with 0.1% formic acid) for 70 min. The separation flow rate was set at 200 nl/min. Data were acquired in a data-dependent mode, alternating an FT MS scan survey over the range m/z 300–1700 and five MS/MS scans in an exclusion dynamic mode. Peptides were identified with SEQUEST through the Bioworks 3.3.1 interface (Thermo Finnigan) against a subset of the UniProt SwissProt Saccharomyces cerevisiae database (downloaded 19 March 2010; 13,986 entries). The search parameters were as follows: mass accuracy of the monoisotopic peptide precursor and peptide fragments was set to 10 ppm and 0.5 atomic mass unit (amu), respectively. Only b- and y-ions were considered for mass calculation. Oxidation of methionines (+16 Da) was considered as a differential modification. Two missed trypsin cleavages were allowed. Trypsic peptides were validated using the following criteria: DeltaCN ≥ 0.1, Xcorr ≥ 1.5 (single charge), 2.0 (double charge), 2.5 (triple charge), 3.0 (quadruple charge), and peptide probability ≤ 0.001. Proteins were validated as soon as two different peptides were validated.
For LD morphology studies.
The BY4742 strain and the loa1Δ deletion strain were grown on YPD media until the stationary phase; the strains BY4742 transformed with the empty vector pYES2/CT (YSA00) or overexpressing LOA1 (YSA01) were grown on synthetic defined media until the stationary phase. Yeast cells (1.5 ml) were harvested and resuspended in 100 μl of PBS (pH 8). One microliter of Nile red dye (1 mg/ml in DMSO) was added, and the cell suspension was incubated for 20 min at room temperature. After two washes with PBS buffer, cells were resuspended in 20 μl of PBS and observed with an epifluorescence microscope (Zeiss Axioskop 2 plus, with a 100× oil objective and a GFP band-pass filter) directly after staining. The LD number estimation was performed using an unpaired t test (GraphPad software, Prism version 5.0; San Diego, CA).
LD preparation for acyltransferase activity assays.
LDs were prepared according to Connerth et al. (2009) from BY4742, loa1Δ, or slc1Δ strains. To show that LD catalyzed PA synthesis, LPA AT reactions were conducted in 100 μl of assay mixtures (50 mM Tris-HCl, pH 8) containing 1 nmol 16:0 LPA, 0.5 nmol [14C]oleoyl-CoA, and various amounts of LD proteins purified from the wild-type or the loa1Δ strains. For the selectivity study, LPA AT reactions were conducted using 2 μg LD proteins purified from the slc1Δ strain, 1nmol 16:0 LPA, and various amounts of [14C]oleoyl-CoA or [14C]palmitoyl-CoA. After a 10-min incubation, lipids were extracted and analyzed as described above.
Supplementary Material
Acknowledgments
This work was supported by the Ministère de l'Enseignement Supérieur et de la Recherche, the Center National de la Recherche Scientifique, the Université Bordeaux Segalen, and the Conseil Régional d'Aquitaine (PhD fellowships for S.A.). S.A.'s visit to A.S.'s laboratory was supported by an EMBO short-term fellowship (ASTF number 373–2008). The strain carrying Erg6-RFP genomically was a gift from P. Arvidson (Department of Chemistry and Chemical Biology, Harvard University, Cambridge, MA). Antibodies were a gift from Kai Simons' laboratory (MPI-CBG, Dresden, Germany). We thank J. M. Galan, R. Haguenauer, and S. Leon (Institut Jacques Monod, Paris, France) for providing the results shown in Figure 8. We thank M. Bonneu from the Plateforme Proteome (Université Bordeaux Segalen) for help with proteomics and F. Doignon (UMR5200) for help with the microscopy. We thank Julio Sampaio, Dominik Schwudke (NCBS, Bangalore), and Ronny Herzog (MPI-CBG, Dresden, Germany) for their input and help in the use of LipidXplorer software.
Abbreviations used:
- CGL1
congenital generalized lipodystrophy 1
- DAG
diacylglycerol
- ER
endoplasmic reticulum
- FT MS
Fourier-transform mass spectrometry
- FWHM
full-width at half-maximum
- GC
gas chromatography
- HA
hemagglutinin
- HPTLC
high-performance TLC
- LD
lipid droplet
- LPA
lysophosphatidic acid
- LPC
lysophosphatidylcholine
- LPE
lysophosphatidic ethanolamine
- LPG
lysophosphatidyl glycerol
- LPI
lysophosphatidylinositol
- LPS
lysophosphatidylserine
- MAG
monoacylglycerol
- NL
neutral loss
- ORF
open reading frame
- PA
phosphatidic acid
- PBS
phosphate-buffered saline
- PE
phosphatidic ethanolamine
- PI
phosphatidylinositol
- PL
phospholipid
- PS
phosphatidylserine
- RFP
red fluorescent protein
- SE
steryl ester
- SGD
Saccharomyces Genome Database
- TAG
triacylglycerol
- YPD
yeast–peptone–dextrose
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-07-0650) on November 16, 2011.
REFERENCES
- Athenstaedt K, Zweytick D, Jandrositz A, Kohlwein SD, Daum G. Identification and characterization of major lipid particle proteins of the yeast Saccharomyces cerevisiae. J Bacteriol. 1999;181:6441–6448. doi: 10.1128/jb.181.20.6441-6448.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athenstaedt K, Daum G. Biosynthesis of phosphatidic acid in lipid particles and endoplasmic reticulum of Saccharomyces cerevisiae. J Bacteriol. 1997;179:7611–7616. doi: 10.1128/jb.179.24.7611-7616.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudoin F, Wilkinson BM, Stirling CJ, Napier JA. In vivo targeting of a sunflower oil body protein in yeast secretory (sec) mutants. Plant J. 2000;23:159–70. doi: 10.1046/j.1365-313x.2000.00769.x. [DOI] [PubMed] [Google Scholar]
- Benghezal M, Roubaty C, Veepuri V, Knudsen J, Conzelmann A. SLC1 and SLC4 encode partially redundant acyl-coenzyme A 1-acylglycerol-3-phosphate O-acyltransferases of budding yeast. J Biol Chem. 2007;282:30845–30855. doi: 10.1074/jbc.M702719200. [DOI] [PubMed] [Google Scholar]
- Bonangelino CJ, Chavez EM, Bonifacino JS. Genomic screen for vacuolar protein sorting genes in Saccharomyces cerevisiae. Mol Biol Cell. 2002;13:2486–2501. doi: 10.1091/mbc.02-01-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brügger B, Erben G, Sandhoff R, Wieland FT, Lehmann WD. Quantitative analysis of biological membrane lipids at the low picomole level by nano-electrospray ionization tandem mass spectrometry. Proc Natl Acad Sci USA. 1997;94:2339–2344. doi: 10.1073/pnas.94.6.2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HS, Su WM, Morgan M, Han GS, Xu Z, Karanasios E, Siniossoglou S, Carman GM. Phosphorylation of phosphatidate phosphatase regulates its membrane association and physiological functions in Saccharomyces cerevisiae: identification of SER(602), THR(723), and SER(744) as the sites phosphorylated by CDC28 (CDK1)-encoded cyclin-dependent kinase. J Biol Chem. 2011;286:1486–1498. doi: 10.1074/jbc.M110.155598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman J. Characterization of the Escherichia coli gene for 1-acyl-sn-glycerol-3-phosphate acyltransferase (plsC) Mol Gen Genet. 1992;232:295–303. doi: 10.1007/BF00280009. [DOI] [PubMed] [Google Scholar]
- Connerth M, Grillitsch K, Köfeler H, Daum G. Analysis of lipid particles from yeast. Methods Mol Biol. 2009;579:359–374. doi: 10.1007/978-1-60761-322-0_18. [DOI] [PubMed] [Google Scholar]
- Ejsing CS, Duchoslav E, Sampaio J, Simons K, Bonner R, Thiele C, Ekroos K, Shevchenko A. Automated identification and quantification of glycerophospholipid molecular species by multiple precursor ion scanning. Anal Chem. 2006;78:6202–6214. doi: 10.1021/ac060545x. [DOI] [PubMed] [Google Scholar]
- Ejsing CS, Sampaio JL, Surendranath V, Duchoslav E, Ekroos K, Klemm RW, Simons K, Shevchenko A. Global analysis of the yeast lipidome by quantitative shotgun mass spectrometry. Proc Natl Acad Sci USA. 2009;106:2136–2141. doi: 10.1073/pnas.0811700106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei W, Shui G, Gaeta G, Du X, Kuerschner L, Li P, Brown AJ, Wenk MW, Parton RG, Yang H. Fld1p, a functional homologue of human seipin, regulates the size of lipid droplets in yeast. J Cell Biol. 2008;180:473–482. doi: 10.1083/jcb.200711136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei W, et al. A role for phosphatidic acid in the formation of “supersized” lipid droplets. PLoS Genet. 2011;7:e1002201. doi: 10.1371/journal.pgen.1002201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto T, Ohsaki Y, Cheng J, Suzuki M, Shinohara Y. Lipid droplets: a classic organelle with new outfits. Histochem Cell Biol. 2008;130:263–279. doi: 10.1007/s00418-008-0449-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg A, Agarwal AK. Lipodystrophies: disorders of adipose tissue biology. Biochim Biophys Acta. 2009;1791:507–513. doi: 10.1016/j.bbalip.2008.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar ML, Aregullin MA, Jesch SA, Henry SA. Inositol induces a profound alteration in the pattern and rate of synthesis and turnover of membrane lipids in Saccharomyces cerevisiae. J Biol Chem. 2006;28:22773–22785. doi: 10.1074/jbc.M603548200. [DOI] [PubMed] [Google Scholar]
- Ghosh AK, Ramakrishnan G, Rajasekharan R. YLR099C (ICT1) encodes a soluble Acyl-CoA-dependent lysophosphatidic acid acyltransferase responsible for enhanced phospholipid synthesis on organic solvent stress in Saccharomyces cerevisiae. J Biol Chem. 2008;283:9768–9775. doi: 10.1074/jbc.M708418200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman JM. The gregarious lipid droplet. J Biol Chem. 2008;283:28005–28009. doi: 10.1074/jbc.R800042200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan XL, Wenk MR. Mass spectrometry-based profiling of phospholipids and sphingolipids in extracts from Saccharomyces cerevisiae. Yeast. 2006;23:465–477. doi: 10.1002/yea.1362. [DOI] [PubMed] [Google Scholar]
- Graessler J, Schwudke D, Schwarz PE, Herzog R, Shevchenko A, Bornstein SR. Top-down lipidomics reveals ether lipid deficiency in blood plasma of hypertensive patients. PLoS One. 2009;4:e6261. doi: 10.1371/journal.pone.0006261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grillitsch K, Connerth M, Kofeler H, Arrey TN, Rietschel B, Wagner B, Karas M, Daum G. Lipid particles/droplets of the yeast Saccharomyces cerevisiae revisited: lipidome meets proteome. Biochim Biophys Acta. 2011;1811:1165–1176. doi: 10.1016/j.bbalip.2011.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Walther TC, Rao M, Stuurman N, Goshima G, Terayama K, Wong JS, Vale RD, Walter P, Farese RV. Functional genomic screen reveals genes involved in lipid-droplet formation and utilization. Nature. 2008;453:657–661. doi: 10.1038/nature06928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvat EM, Zhang YM, Tran CV, Zhang Z, Frank MW, Rock CO, Saier MH, Jr. Lysophospholipid flipping across the Escherichia coli inner membrane catalyzed by a transporter (LplT) belonging to the major facilitator superfamily. J Biol Chem. 2005;280:12028–12034. doi: 10.1074/jbc.M414368200. [DOI] [PubMed] [Google Scholar]
- Herzog R, Schwudke D, Schuhmann K, Sampaio JL, Bornstein SR, Schroeder M, Shevchenko A. A novel informatics concept for high-throughput shotgun lipidomics based on the molecular fragmentation query language. Genome Biol. 2011;19:R8. doi: 10.1186/gb-2011-12-1-r8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homma H, Nojima S. Transacylation between diacylphospholipids and 2-acyl lysophospholipids catalyzed by Escherichia coli extract. J Biochem. 1982;91:1093–1101. doi: 10.1093/oxfordjournals.jbchem.a133791. [DOI] [PubMed] [Google Scholar]
- Jacquier N, Choudhary V, Mari M, Toulmay A, Reggiori F, Schneiter R. Lipid droplets are functionally connected to the endoplasmic reticulum in Saccharomyces cerevisiae. J Cell Sci. 2011;124:2424–2437. doi: 10.1242/jcs.076836. [DOI] [PubMed] [Google Scholar]
- Jain S, Stanford N, Bhagwat N, Seiler B, Costanzo M, Boone C, Oelkers P. Identification of a novel lysophospholipid acyltransferase in Saccharomyces cerevisiae. J Biol Chem. 2007;282:30562–30569. doi: 10.1074/jbc.M706326200. [DOI] [PubMed] [Google Scholar]
- Janke C, et al. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21:947–962. doi: 10.1002/yea.1142. [DOI] [PubMed] [Google Scholar]
- Jorgensen P, Nishikawa JL, Breitkreutz BJ, Tyers M. Systematic identification of pathways that couple cell growth and division in yeast. Science. 2002;297:395–400. doi: 10.1126/science.1070850. [DOI] [PubMed] [Google Scholar]
- Karanasios E, Han GS, Xu Z, Carman GM, Siniossoglou S. A phosphorylation-regulated amphipathic helix controls the membrane translocation and function of the yeast phosphatidate phosphatase. Proc Natl Acad Sci USA. 2010;107:17539–17544. doi: 10.1073/pnas.1007974107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopelman PG. Obesity as a medical problem. Nature. 2000;404:635–643. doi: 10.1038/35007508. [DOI] [PubMed] [Google Scholar]
- Kumar A, et al. Subcellular localization of the yeast proteome. Genes Dev. 2002;16:707–719. doi: 10.1101/gad.970902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leber R, Landl K, Zinser E, Ahorn H, Spök A, Kohlwein SD, Turnowsky F, Daum G. Dual localization of squalene epoxidase, Erg1p, in yeast reflects a relationship between the endoplasmic reticulum and lipid particles. Mol Biol Cell. 1998;9:375–386. doi: 10.1091/mbc.9.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Guédard M,, et al. PSI1 is responsible for the stearic acid enrichment that is characteristic of phosphatidylinositol in yeast. FEBS J. 2009;276:6412–6424. doi: 10.1111/j.1742-4658.2009.07355.x. [DOI] [PubMed] [Google Scholar]
- Lewin TM, Wang P, Coleman RA. Analysis of amino acid motifs diagnostic for the sn-glycerol-3-phosphate acyltransferase reaction. Biochemistry. 1999;38:5764–5771. doi: 10.1021/bi982805d. [DOI] [PubMed] [Google Scholar]
- Listenberger L, Han X, Lewis SE, Cases S, Farese RV, Ory DS, Schaffer JE. Triglyceride accumulation protects against fatty acid-induced lipotoxicity. Proc Natl Acad Sci USA. 2003;100:3077–3082. doi: 10.1073/pnas.0630588100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockshon D, Surface LE, Kerr EO, Kaeberlein M, Kennedy BK. The sensitivity of yeast mutants to oleic acid implicates the peroxisome and other processes in membrane function. Genetics. 2007;175:77–91. doi: 10.1534/genetics.106.064428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Martin S, Parton RG. Lipid droplets: a unified view of a dynamic organelle. Nat Rev Mol Cell Biol. 2006;7:373–378. doi: 10.1038/nrm1912. [DOI] [PubMed] [Google Scholar]
- Murphy D. The biogenesis and functions of lipid bodies in animals, plants and microorganisms. Prog Lipid Res. 2001;40:325–438. doi: 10.1016/s0163-7827(01)00013-3. [DOI] [PubMed] [Google Scholar]
- Murphy S, Martin S, Parton RG. Lipid droplet-organelle interactions; sharing the fats. Biochim Biophys Acta. 2009;1791:441–447. doi: 10.1016/j.bbalip.2008.07.004. [DOI] [PubMed] [Google Scholar]
- Nagiec MM, Wells GB, Lester RL, Dickson RC. A suppressor gene that enables Saccharomyces cerevisiae to grow without making sphingolipids encodes a protein that resembles an Escherichia coli fatty acyltransferase. J Biol Chem. 1993;268:22156–22163. [PubMed] [Google Scholar]
- Natter K, Leitner P, Faschinger A, Wolinski H, McCraith S, Fields S, Kohlwein SD. The spatial organization of lipid synthesis in the yeast Saccharomyces cerevisiae derived from large scale green fluorescent protein tagging and high resolution microscopy. Mol Cell Proteomics. 2005;4:662–672. doi: 10.1074/mcp.M400123-MCP200. [DOI] [PubMed] [Google Scholar]
- Oelkers P, Cromley D, Padamsee M, Billheimer JT, Sturley SL. The DGA1 gene determines a second triglyceride synthetic pathway in yeast. J Biol Chem. 2002;277:8877–8881. doi: 10.1074/jbc.M111646200. [DOI] [PubMed] [Google Scholar]
- Olofsson SO, Boström P, Andersson L, Rutberg M, Levin M, Perman J, Borén J. Triglyceride containing lipid droplets and lipid droplet-associated proteins. Curr Opin Lipidol. 2008;19:441–447. doi: 10.1097/MOL.0b013e32830dd09b. [DOI] [PubMed] [Google Scholar]
- Petschnigg J, Wolinski H, Kolb D, Zellnig G, Kurat CF, Natter K, Kohlwein SD. Good fat, essential cellular requirements for triacylglycerol synthesis to maintain membrane homeostasis in yeast. J Biol Chem. 2009;284:30981–30993. doi: 10.1074/jbc.M109.024752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pocaly M, et al. Proteomic analysis of an imatinib-resistant K562 cell line highlights opposing roles of heat shock cognate 70 and heat shock 70 proteins in resistance. Proteomics. 2008;8:2394–2406. doi: 10.1002/pmic.200701035. [DOI] [PubMed] [Google Scholar]
- Pol A, Martin S, Fernández MA, Ingelmo-Torres M, Ferguson C, Enrich C, Parton RG. Cholesterol and fatty acids regulate dynamic caveolin trafficking through the Golgi complex and between the cell surface and lipid bodies. Mol Biol Cell. 2005;16:2091–2105. doi: 10.1091/mbc.E04-08-0737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajakumari S, Daum G. Janus-faced enzymes yeast Tgl3p and Tgl5p catalyze lipase and acyltransferase reactions. Mol Biol Cell. 2010a;21:501–510. doi: 10.1091/mbc.E09-09-0775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajakumari S, Daum G. Multiple functions as lipase, steryl ester hydrolase, phospholipase, and acyltransferase of Tgl4p from the yeast Saccharomyces cerevisiae. J Biol Chem. 2010b;285:15769–15776. doi: 10.1074/jbc.M109.076331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riekhof WR, Wu J, Jones JL, Voelker DR. Identification and characterization of the major lysophosphatidylethanolamine acyltransferase in Saccharomyces cerevisiae. J Biol Chem. 2007;282:28344–28352. doi: 10.1074/jbc.M705256200. [DOI] [PubMed] [Google Scholar]
- Rosenberger S, Connerth M, Zellnig G, Daum G. Phosphatidylethanolamine synthesized by three different pathways is supplied to peroxisomes of the yeast Saccharomyces cerevisiae. Biochim Biophys Acta. 2009;791:379–387. doi: 10.1016/j.bbalip.2009.01.015. [DOI] [PubMed] [Google Scholar]
- Sandager L, Gustavsson MH, Ståhl U, Dahlqvist A, Wiberg E, Banas A, Lenman M, Ronne H, Stymne S. Storage lipid synthesis is non-essential in yeast. J Biol Chem. 2002;277:6478–6482. doi: 10.1074/jbc.M109109200. [DOI] [PubMed] [Google Scholar]
- Schwudke D, Hannich JT, Surendranath V, Grimard V, Moehring T, Burton L, Kurzchalia T, Shevchenko A. Top-down lipidomic screens by multivariate analysis of high-resolution survey mass spectra. Anal Chem. 2007;79:4083–4093. doi: 10.1021/ac062455y. [DOI] [PubMed] [Google Scholar]
- Schneiter R, et al. Electrospray ionization tandem mass spectrometry (ESI-MS/MS) analysis of the lipid molecular species composition of yeast subcellular membranes reveals acyl chain-based sorting/remodeling of distinct molecular species en route to the plasma membrane. J Cell Biol. 1999;146:741–754. doi: 10.1083/jcb.146.4.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwudke D, Oegema J, Burton L, Entchev E, Hannich JT, Ejsing CS, Kurzchalia T, Shevchenko A. Lipid profiling by multiple precursor and neutral loss scanning driven by the data-dependent acquisition. Anal Chem. 2006;78:585–595. doi: 10.1021/ac051605m. [DOI] [PubMed] [Google Scholar]
- Shui G, Guan XL, Gopalakrishnan P, Xue Y, Goh JS, Yang H, Wenk MR. Characterization of substrate preference for Slc1p and Cst26p in Saccharomyces cerevisiae using lipidomic approaches and an LPAAT activity assay. PLoS One. 2010;5:e11956. doi: 10.1371/journal.pone.0011956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorger D, Daum G. Triacylglycerol biosynthesis in yeast. Appl Microbiol Biotechnol. 2003;61:289–299. doi: 10.1007/s00253-002-1212-4. [DOI] [PubMed] [Google Scholar]
- Stone SJ, Levin MC, Zhou P, Han J, Walther TC, Farese RV. The endoplasmic reticulum enzyme DGAT2 is found in mitochondria-associated membranes and has a mitochondrial targeting signal that promotes its association with mitochondria. J Biol Chem. 2009;284:5352–5361. doi: 10.1074/jbc.M805768200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanski KM, Binns D, Bartz R, Grishin NV, Li W-P, Agarwal AK, Garg A, Anderson RGW, Goodman JM. The lipodystrophy protein seipin is found at endoplasmic reticulum lipid droplet junctions and is important for droplet morphology. Proc Natl Acad Sci USA. 2007;104:20890–20895. doi: 10.1073/pnas.0704154104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaki H, Shimada A, Ito Y, Ohya M, Takase J, Miyashita M, Miyagawa H, Nozaki H, Nakayama R, Kumagai H. LPT1 encodes a membrane-bound O-acyltransferase involved in the acylation of lysophospholipids in the yeast Saccharomyces cerevisiae. J Biol Chem. 2007;282:34288–34298. doi: 10.1074/jbc.M704509200. [DOI] [PubMed] [Google Scholar]
- Testet E, Laroche-Traineau J, Noubhani A, Coulon D, Bunoust O, Camougrand N, Manon S, Lessire R, Bessoule J-J. Ypr140wp, “the yeast tafazzin”, displays a mitochondrial lysophosphatidylcholine (lyso-PC) acyltransferase activity related to triacylglycerol and mitochondrial lipid synthesis. Biochem J. 2005;387:617–626. doi: 10.1042/BJ20041491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner S, Paltauf F. Generation of glycerophospholipid molecular species in the yeast Saccharomyces cerevisiae. Fatty acid pattern of phospholipid classes and selective acyl turnover at sn-1 and sn-2 positions. Yeast. 1994;10:1429–1437. doi: 10.1002/yea.320101106. [DOI] [PubMed] [Google Scholar]
- Zou J, Katavic V, Giblin EM, Barton DL, MacKenzie SL, Keller WA, Hu X, Taylor DC. Modification of seed oil content and acyl composition in the Brassicaceae by expression of a yeast sn-2 acyltransferase gene. Plant Cell. 1997;9:909–923. doi: 10.1105/tpc.9.6.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.