Abstract
For just over a decade, stochastic gene expression has been the focus of many experimental and theoretical studies. It is now widely accepted that noise in gene expression can be decomposed into extrinsic and intrinsic components, which have orthogonal contributions to the total noise. Intrinsic noise stems from the random occurrence of biochemical reactions and is inherent to gene expression. Extrinsic noise originates from fluctuations in the concentrations of regulatory components or random transitions in the cell’s state and is imposed to the gene of interest by the intra- and extra-cellular environment. The basic assumption has been that extrinsic noise acts as a pure input on the gene of interest, which exerts no feedback on the extrinsic noise source. Thus, multiple copies of a gene would be uniformly influenced by an extrinsic noise source. Here, we report that this assumption falls short when multiple genes share a common pool of a regulatory molecule. Due to the competitive utilization of the molecules existing in this pool, genes are no longer uniformly influenced by the extrinsic noise source. Rather, they exert negative regulation on each other and thus extrinsic noise cannot be determined by the currently established method.
Biology has been an immense source of problems demanding quantitative understanding. Efforts to take up this challenge have fueled the emergence of novel areas in mathematics and physics, including theories of random walks,1, 2 synchronization3 deterministic chaos,4, 5 biological networks,6, 7 and stochastic resonance.8 Without exception, each of these fields has been confronted with the issue of noise, defined as random and unpredictable fluctuations in the variables or parameters of the system being investigated. A remarkable example is stochastic gene expression, which has been the focus of increasing number of experimental and theoretical studies over the last two decades,9, 10, 11, 12, 13, 14, 15, 16 and is defined as the random variation of messenger RNA or protein levels in time in a single cell, or across a population of genetically identical cells at a given time. It is generally accepted that the sources of gene expression noise can be either extrinsic or intrinsic, which give rise to two orthogonal noise components, in the sense that their variances add up to the total variance.17, 18, 19Intrinsic noise originates from randomly occurring biochemical reactions during transcription and translation and is inherent to gene expression. Extrinsic noise can be attributed to fluctuations in the concentrations of regulatory components or random transitions in the cell’s state and is imposed to the gene of interest by the intra- and extra-cellular environment. The separation of total noise into these components assumes that extrinsic noise acts as a pure input on the gene of interest, which does not have any effect (feedback) on the extrinsic noise source. Thus, multiple copies of a gene exposed to a common extrinsic noise source should be identically influenced by their environment. Here, we report that this assumption cannot hold when multiple genes share a common pool of a regulatory molecule, because association with one of the target genes reduces the number of regulators available for other target genes. Therefore, genes are no longer uniformly influenced by the extrinsic noise source and exert indirect negative regulatory effect on each other. Consequently, extrinsic noise cannot be determined by the currently established method. We exemplify this conundrum by calculating the intrinsic and extrinsic noise in a simple theoretical system inspired from the original papers that described a method for separately measuring these noise components.17, 18 After some approximations, we obtain negative extrinsic noise values that have no physical meaning, both from analytically solving the Master equation and from performing stochastic simulations. These findings bring under question two key assumptions made when separating noise into intrinsic and extrinsic components: the “pure input” and the “independent genes” assumption. We conclude by discussing the implications of these results and proposing alternative strategies for defining noise components.
INTRODUCTION
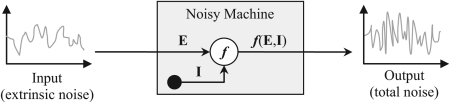
The basis for separating total gene expression noise into intrinsic and extrinsic components is the assumption that gene expression process is analogous to a noisy machine (Figure 1) that receives a noisy signal and performs a transformation of that signal while generating its own noise. Thus, the output of this machine will contain two noise components: the input (extrinsic) noise and the generated (intrinsic) noise. Analyzing the outputs of two identical and non-interacting such noisy machines exposed to the same input, one could safely assume that (i) the extrinsic components of these outputs would be identical since both machines receive the same input, (ii) the intrinsic components of the two outputs would be independent since the machines do not interact. Under these assumptions, the extrinsic variables can be defined as the noisy input variables to the noisy machines, while the intrinsic variables as the internal state variables of the noisy machines.
Figure 1.
A gene could be viewed as a noisy machine that accepts a random signal as input (extrinsic noise E) and transforms it through an inherently stochastic process (intrinsic noise, I), thereby generating a noisy output signal (f(E,I)). Consequently, the latter contains two orthogonal noise components: extrinsic, due to the input noise, and intrinsic due to the noise produced within the “machine.” In our discussion, the output is the protein expression level: f(E,I) = P(E,I).
In the case of gene expression, these two assumptions can be stated as follows. First, the “pure input assumption” states that the gene of interest cannot exert any feedback on the extrinsic noise source. Therefore, extrinsic noise acts as a pure input to the process of gene expression. This assumption allows for the partitioning into extrinsic and intrinsic variables. In particular, Swain et al.17 give examples of extrinsic variables being the state of the cell or the levels of regulatory components, and examples of intrinsic variables being the mRNA and protein contents for the gene of interest. Second, the “independent genes assumption” states that multiple identical copies of a gene do not influence one-another. This second assumption makes possible the calculation of the intrinsic and extrinsic noise components by simultaneously measuring the protein contents of two identical gene copies.17
Even though the decomposition of noise into extrinsic and intrinsic components has been a useful tool for analyzing stochasticity in gene expression, such an analysis is questionable unless both of the aforementioned assumptions hold true. In particular, this may not always be the case in biological systems: metabolic or signaling pathways, as well as synthetic gene networks often contain either explicit or implicit feedback loops.20, 21 The components of such feedback loops would contribute to their own extrinsic noise, which would invalidate the “pure input assumption”. Furthermore, autoregulation,22 namely, a gene exerting feedback to its own expression would also invalidate the “independent genes assumption.” Likewise, two identical copies of a gene that share a common pool of a low-abundance regulatory species23 could exert indirect negative regulatory effects on each other, due to the competitive utilization of the shared regulator pool. Such an effect is plausible in naturally occurring genetic networks, such as the lac operon in which the LacI repressor concentration is roughly 10 copies per cell.24
The aim of this paper is to examine a case involving a shared regulator pool and demonstrate that these two assumptions are violated; thus, even partitioning the overall noise into extrinsic and intrinsic components becomes problematic. We will apply mathematical modeling to a system similar to the one introduced to experimentally measure the intrinsic and extrinsic noise,18 which comprises of two reporter-gene variants under the influence of identical promoters repressed by LacI. We will show that the “pure input assumption” does not hold, because the state of each reporter gene’s promoter (repressed or not) affects the free repressor content. Likewise, the “independent genes assumption” fails, because if one promoter is in the repressed state, then this prevents one repressor molecule from repressing the other gene. Thus, the competitive utilization of a common repressor pool results in mutual indirect negative regulatory interactions, and subsequently in negatively correlated reporter protein contents. If one tries to calculate the extrinsic noise for this system, negative values with no physical meaning may be encountered.
The rest of the paper is organized as follows. We will first briefly overview the concepts of intrinsic and extrinsic noise. Second, we will propose a network of reactions that captures the salient features of the underlying biochemical interactions. Next, we will derive a stochastic model for the transitions of the two operators to the repressed and unrepressed states. We will then show that the “pure input assumption” and the “independent genes assumption” can only hold in the limit of an infinitely large repressor pool. Otherwise, meaningless negative extrinsic noise values may be encountered. Finally, we conclude by discussing when it is meaningful to consider partitioning the noise into extrinsic and intrinsic components and what alternatives may exist for properly defining them.
INTRINSIC AND EXTRINSIC NOISE: AN OVERVIEW
Much before the first gene expression measurements in single cells, the problem of information transfer through noisy nonlinear systems has confronted neuroscience for decades. Noise was originally considered a universal hindrance for information transfer in neurons,25 but this view had to be revised when noise-optimized weak signal detection and transfer were demonstrated experimentally26 and explained by theory27, 28 in neuronal systems. While externally added noise showed capable of optimizing signal transfer in biological neurons, a key question that remained open was whether noise internal to biological neurons, such as the noise originating from random ion channel dynamics29 could do the same. Theoretical work suggested that this may be possible,30, 31 but a rigorous experimental demonstration is still lacking. Posing this question treated neurons as noisy machines that could add their own intrinsic noise to the extrinsic noise already present in the incoming signal (Figure 1).
The issue of intrinsic and extrinsic noise arose again once gene expression noise became directly measurable in living cells. By using a chromosomally integrated fluorescent reporter, van Oudenaarden and colleagues studied the sources of random intrinsic gene expression variations in genetically identical Bacillus subtilis cells.32 Later that year, Elowitz and colleagues reported on the intrinsic and extrinsic components of gene expression noise in Escherichia coli using a cleverly designed double-reporter system18 that involved two fluorescent reporters located at equivalent chromosomal loci, following theoretical predictions by Swain et al.17 As sources of intrinsic noise, they propose random transcription and translation initiation events, as well as degradation of transcripts and proteins. Extrinsic noise, on the other hand originates from cell-wide fluctuations in the number of ribosomes, polymerases, regulators, cell cycle stage, etc. Since these original studies in bacteria, the double-reporter system has been successfully adapted to other organisms.33, 34, 35 Besides the double-reporter system, other methods proposed for estimating the intrinsic and extrinsic components of gene expression noise have relied on total noise measurements for various copy numbers of a reporter gene36, 37 or mutations altering biological parameters relevant for intrinsic noise only (such as the promoter sequence, and the rates of transcription and translation).32, 38, 39
Below we summarize the essence of the theory17 underlying the use of the double-reporter methodology18 to separate the intrinsic and extrinsic components of gene expression noise.
Let us consider a biological process for which the observable of interest is the expression level of a protein. Swain et al.17 propose that this level is a function P(E,I) of extrinsic and intrinsic variables E and I. As the variables E and I fluctuate, so does the observed (measured) expression level P(E,I), and the total variance of the observable will be
| (1) |
where ρ(E,I) is the joint probability density of E, I, and:
| (2) |
From Bayes’ theorem, the joint probability ρ(E,I) can be expressed as
| (3) |
Thus, from Eq. 1 by adding and subtracting a term and performing some algebraic manipulations they obtain
| (4) |
In order to calculate the extrinsic and intrinsic noise contributions just noted, Swain et al.17 suggest using two identical expression systems (noisy machines) for which both reporter genes are exposed to the same extrinsic variables while having individual intrinsic variables with identical probability densities
| (5) |
| (6) |
This allows the calculation of the coupling term that was added and subtracted
| (7) |
Let P(E1,I1) = c be the amount of cyan fluorescent protein (CFP) in the cell, and P(E2,I2) = y the amount of yellow fluorescent protein (YFP). These amounts can be inferred from the corresponding fluorescence values measured for individual cells by flow cytometry or fluorescence microscopy. Equations 5, 6 hold true, assuming that both reporter genes are under the influence of identical promoters and integrated into equivalent chromosomal loci. Consequently, according to Eq. 7, the coupling term is simply the cross-correlation between c and y measured in the same cell
| (8) |
where the angled brackets indicate averaging over the cell population. Furthermore, from the equivalence of the two reporter genes
| (9) |
and by substituting Eqs. 9 back to Eq. 4
| (10) |
In the above equation, the intrinsic variance can be further simplified as follows:
| (11) |
This procedure allows for the estimation of intrinsic and extrinsic noise components measured as the coefficient of variation (CV; standard deviation divided by the mean) based on the formula
| (12) |
Note that the first equality of Eq. 7 depends on the assumption that the expression systems are always exposed to identical external variables in Eq. 5. Is this always true or can there be exceptions? In the following, we will discuss a case when this assumption is not satisfied and show how this can give unphysical results when calculating the intrinsic and extrinsic noise components.
MATHEMATICAL MODELING AND STOCHASTIC SIMULATIONS
Competitive repressor utilization
Formulation of the model
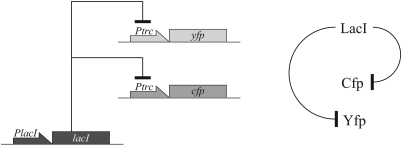
As an example for unequal extrinsic variables, we consider a two-promoter system regulated by the LacI repressor, similar to the system built to experimentally determine the intrinsic and extrinsic components of gene expression noise.18 The molecular mechanisms included in our model are depicted in Figure 2; species notation is shown in Table Table I., kinetic constants are shown in Table Table II., and the reaction network is summarized in Table Table III.. In constructing the reaction network, for simplicity we assume constitutive LacI production, single lacO operator sites per promoter,40 abundant RNA polymerases and ribosomes and 1st order degradation reactions. We ignore the effects of cell division and DNA duplication as we want to concentrate on the effects of competitive utilization of the LacI repressor pool. Thus, the stochastic model for this system is the chemical Master equation
| (13) |
which can be simulated with the Gillespie algorithm.41, 42
Figure 2.
Schematic representation of the interactions taken into account in the two promoter-reporter system. For species notation see Table Table I..
Table 1.
Symbols used for the species.
| Symbol | Species denoted |
|---|---|
| Lac | LacI repressor |
| OYfp | yfp lacO operator |
| OCfp | cfp lacO operator |
| RYfp | yfp m-RNA |
| RCfp | cfp m-RNA |
| Yfp | Yfp |
| Cfp | Cfp |
| ø | Generic source or sink |
Table 2.
Kinetic constants (deterministic and stochastic).
| Deterministic | Value | Units | Relation to stochastica |
|---|---|---|---|
| 0.23 | nM·min−1 | ||
| 15 | nM−1·min−1 | ||
| 50 | min−1 | ||
| 50 | min−1 | ||
| 50 | min−1 | ||
| 50 | min−1 | ||
| 50 | min−1 | ||
| 10−3 | min−1 |
Avogadro’s number: NA = 6.0221367·1014 nmol−1 and E. coli volume was taken VE. coli = 8·10−16 L
Table 3.
Reaction network and propensity functions.
| Reaction | Propensity functiona,b | |||
|---|---|---|---|---|
| (1) | α1 | = | ||
| (2) | α2 | = | ||
| (3) | α3 | = | ||
| (4) | α4 | = | ||
| (5) | α5 | = | ||
| (6) | α6 | = | ||
| (7) | α7 | = | ||
| (8) | α8 | = | ||
| (9) | α9 | = | ||
| (10) | α10 | = | ||
| (11) | α11 | = | ||
| (12) | α12 | = | ||
| (13) | α13 | = | ||
| (14) | α14 | = | ||
Variables without brackets denote number of molecules of the corresponding species.
All propensity functions have units of min−1.
The state vector containing numbers of molecules for each species is x = {Lac, OYfp, OYfpLac, OCfp, OCfpLac, RYfp, Yfp, RCfp, Cfp}, and we have n = 9 species participating in m = 14 reactions. The reactions’ propensity functions, αj(x) = cj·hj(x), j = 1, 2,…, m, are presented in Table Table III.. The vectors vj denote the change in the number of molecules for each species, e.g., for reaction (6), v6 = {−1, 0, 0, −1, 1, 0, 0, 0, 0}.
Let us now define the step operator as follows:43
| (14) |
A useful property of the step operator is the following:
| (15) |
Then the Master equation can be written as
| (16) |
where .
Next, we will apply a set of simplifying transformations to the Master equation, recasting it into a form that allows us to focus on the source of the problem defining extrinsic noise: the competition between Lac operator sites in the two promoters for LacI repressor molecules.
If we apply the summing operator to Eq. 16 using property 15 and taking into account boundary conditions such as
| (17) |
where is the marginal probability that allows us to focus on cfp and yfp promoter dynamics. We note that: (i) OYfp and OCfp are zero or unity, (ii) conservation of the operator sites holds
| (18) |
and (iii) the operator fluctuations between the free and the repressed state do not change the overall repressor content, defined as
| (19) |
We therefore only need three variables namely LacT, OYfp, and OCfp to refer to the marginal probability . Thus, we define a new probability as
| (20) |
and we recast the Master equation as follows:
| (21) |
If we now apply the summing operator to Eq. 21 we get an equation describing the probability of total repressor molecules
| (22) |
Fast-operator-fluctuations approximation
Next, based on the separation of time scales,44 we establish equations separately describing the probability of total repressor number and target promoter states. For this, let us assume fast operator fluctuations
| (23) |
This means that, due to fast repressor-operator binding, the probability of cfp and yfp operator states reaches steady state fast enough to be separable from the probability of having a certain level of total LacI repressor. Therefore, we introduce expansions for the time-dependent probabilities P0 describing the operator states conditioned on total LacI and P1 describing the distribution of total LacI repressor level
| (24) |
| (25) |
From the normalization conditions, it can be derived that
| (26) |
| (27) |
Also
| (28) |
and
| (29) |
Now let us go back to Eq. 21, in which we substitute expressions (28) and (23), and derive the expressions of order ε − 1 and 1
| (30) |
Note that and that we have no step operator in Eq. 30. Therefore, we can write
| (31) |
Furthermore, from Eq. 21
| (32) |
Equation 32 is easy to solve, thereby finding the probabilities for the states of both operators given LacT. Then, one can substitute these probabilities into the expression for F0 in the stationary form of Eqs. 32 finding the stationary probability distribution for LacT.
Solution of the Master equation for LacI and its target operators
In the following, we will obtain exact expressions for the probabilities of having either LacI-bound or LacI-free promoter states for both cfp and yfp at a predefined, constant LacI level (which is valid for the quasi steady-state approximation that we applied). These expressions will allow later the calculation of intrinsic and extrinsic noise in terms of these promoter states. Let us therefore solve equation 26 at stationary conditions given LacT fixed. We will set
| (33) |
Therefore, p10 will denote the probability of finding OYfp in the free state, and OCfp in the repressed state. Since we have two operators each of which can be found in two different states, Eq.31 is a 4 × 4 linear system
| (34) |
whose solution can be calculated (using the normalization condition p00 + p01 + p10 + p11 = 1)
| (35) |
Thus,
| (36) |
and Eq. 32 at stationary conditions can be written as
| (37) |
whose solution can be given as a recursion for q ≥ 1
| (38) |
Having calculated the stationary distribution for LacT we now know the approximate O(ɛ) stationary distribution for all three species (free operators and LacT) as well as the distribution for the operators only
| (39) |
| (40) |
Note that fixing the total LacI repressor concentration does not eliminate the repressor’s extrinsic noise contribution. The reason is that the target promoter dynamics depends on free LacI levels rather than total LacI. A LacI molecule bound to the cfp promoter is practically non-existent for the yfp promoter and vice versa, implying that free LacI will fluctuate even if total LacI is constant. Elimination of the LacI extrinsic noise contribution would result from assuming that the unbound (free) LacI remains constant.
Stochastic simulations
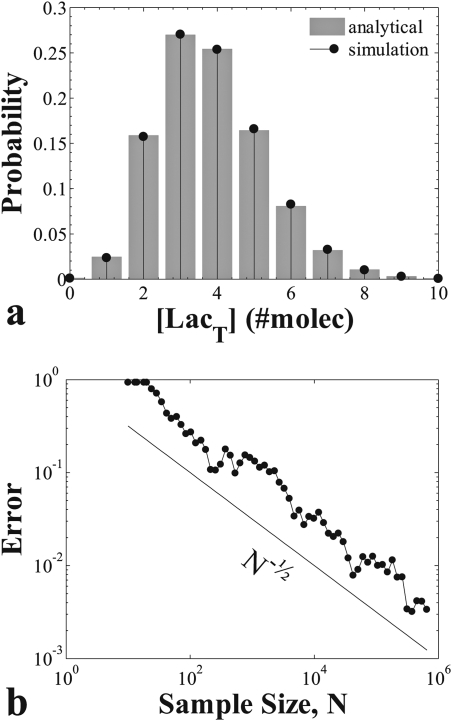
In order to compare the approximation with numerical results we performed simulations of the system using the Gillespie algorithm and the parameter values of Table Table II.. Equation 38 agrees with the distribution obtained from samples taken out of one long Gillespie simulation (compared in Figure 3a; for the Gillespie simulation sampling was done with constant Δt = 5·10−2). The error norm was calculated as
| (41) |
which is expected to scale with the inverse square root of the number of samples N taken from the Gillespie simulation. The simulation results are consistent with this expectation (Figure 3b).
Figure 3.
Comparison of the analytical O(ɛ) approximation for the stationary probability with simulation results. Panel (a): plot of Eq. 38 and of the stationary LacT probability estimate from samples obtained from Gillespie simulations. Panel (b): sampling error falls with the inverse square root of the sample size.
Furthermore, since the analytical calculations only refer to promoter states, we performed stochastic simulations to calculate levels of Cfp and Yfp proteins. In Figure 4a, a scatter plot of the Cfp versus Yfp concentration is shown, obtained from a stochastic simulation of the two promoter model (Eq. 16). For this simulation, we assume that only one LacI molecule exists, and constant total LacI content is assumed. Evidently, LacI can repress only one operator at a time, and thus the Cfp and Yfp protein contents are negatively correlated. This negative correlation results in a physically meaningless negative value for the extrinsic noise, namely ηext2 = -0.91. This situation is of course problematic, since a coefficient of variation should always be non-negative. The negative value obtained is a consequence of the definition of extrinsic noise (Eq. 12) not being applicable to this case. In panel 4b another simulation is shown with 4 LacI molecules. In this case even though there exist enough molecules to keep both promoters repressed at the same time, the extrinsic noise is still negative (ηext2 = −1.00). In the following, we will explain this behavior by focusing on the operators and performing analytical calculations for the operator extrinsic noise.
Figure 4.
(Color) Scatter plots of the Yfp versus Cfp concentration assuming constant repressor content (kLac = 0 nM/min, λLac = 0 min−1). Panel (a): total LacI content equal to 1 molecule; parameter values: kr = 10 (nM·min)−1, k−r = 1 min−1, km = 10 min−1, kp = 10 min−1, λm = 0.4 min−1, λp = 0.1 min−1. Panel (b): total LacI content equal to 4 molecules; parameter values: kr = 50 (nM·min)−1, k−r = 1 min−1, km = 10 min−1, kp = 1000 min−1, λm = 0.4 min−1, λp = 0.1 min−1. Colors indicate relative density of points on the Yfp-Cfp plane: warmer colors correspond to higher densities.
Calculation of extrinsic and intrinsic noise
Finally, we will analyze the extrinsic and intrinsic noise for the operators fluctuating between their free and bound states, using the results from the quasi steady-state approximation. This analysis will provide us with analytical expressions that will give insight into the protein noise since the free operators drive protein expression. Thus, the origin of the unphysical negative extrinsic noise values will be elucidated. Using the expressions for extrinsic and intrinsic noise that were also used by Elowitz et al.,18 we can calculate the operator noise components given LacT
| (42) |
| (43) |
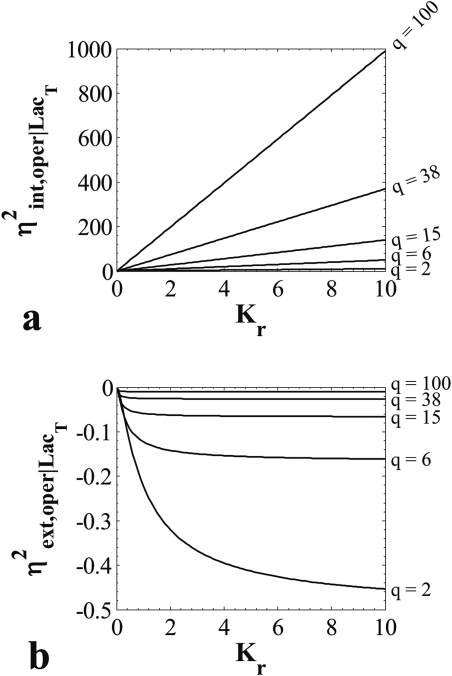
where we have used the symmetry p01 = p10, and p00, p01, p11 are given by Eqs. 35. Figure 5 presents plots of the intrinsic and extrinsic noise expressions given above. Immediately apparent is the fact that the calculated operator extrinsic noise is always negative, which has no physical meaning. This happens because of the negative correlations resulting from the competitive utilization of the common repressor pool. Furthermore, it is interesting to investigate the limiting behavior of the two noise components for high repressor contents (q → ∞) and for very strong repression (Kr → ∞)
| (44) |
Therefore, the negative correlations just mentioned become progressively negligible for abundant repressor contents. This happens because just a single molecule is required to repress an operator; thus, the change in repressor content brought about by that single molecule becomes negligible is the repressor is abundant. On the other hand, the negative correlations become larger and reach a limiting value for stronger repression. This result is also reasonable, since the tighter the repression, the more probable it is that a repressor molecule will be bound to one operator, thereby depleting the repressor pool.
Figure 5.
Plots of the intrinsic (panel a) and extrinsic (panel b) noise for given q = LacT concentration as calculated from Eqs. 42, 43.
The above expressions 42, 43 hold for a fixed value for the LacT content. In order to calculate the operator noise components for the case where Lac I is produced and degraded (assuming fast operator fluctuations), we have to use the probabilities for the operator states as given by Eq. 40
| (45) |
| (46) |
Numerical results indicate that the extrinsic noise as calculated from Eq. 46 is zero for an extended range of the parameter space even though there is clearly an extrinsic noise source due to the LacI fluctuations. Thus, the spurious negative extrinsic effect that we identified analytically for constant LacI content has a counterpart in the case where LacI is allowed to fluctuate.
Non-competitive repressor utilization
Finally, in order to show that indeed it is the competitive repressor utilization that results in the negative feedback between genes and the paradox of negative extrinsic noise values, we considered the case where genes do not alter the free repressor concentration. Thus, reactions 2, Table III. are substituted with the following:
| (47) |
| (48) |
where OYfpLac should now be thought of as the operator at an off-state rather than an operator–Lac I complex. Similar reactions can be written for the Cfp. Note that the reaction propensities do not change, but now the genes are not exerting any feedback on the free Lac I concentration. Thus, the Master equation for this system becomes (compare with Eq. 21)
| (49) |
Note that in this case the species OYfpLac merely stands for the Yfp operator at the off state, since free Lac I is not depleted after interacting with the free operator (similarly for the Cfp operator). Thus, now LacT is always equal to the free Lac I. If we now apply the summing operator to Eq. 49 we get (compare with Eq. 22)
| (50) |
which shows that, in this case, LacT follows Poisson distribution:
| (51) |
Furthermore, by applying the fast operator fluctuations assumption similarly as before we obtain the following linear system and its solution for the probabilities of the operator states (compare with Eq. 34 and the solution 35)
| (52) |
| (53) |
Now, the intrinsic and extrinsic noise values, given the total Lac I, are calculated to be
| (54) |
| (55) |
Evidently, the extrinsic noise is always zero in this case due to the fact that the free repressor concentration remains constant.
This analysis shows that the problem of negative correlations resulting from a common pool of a regulatory molecule could be circumvented by the following regulatory strategy. Assume that one could engineer a system in which the regulatory molecule, through a rapid interaction with the operator, induces a reversible change in the state of the latter. After this process, the operator is in the off-state in which it remains for some time, and the regulatory molecule is free to induce the same change to the other operator. Thus, the regulatory molecules are not occupied and the aforementioned negative correlations will no longer represent a problem. Note that the change from the on- to the off-state of the operator can involve a third species, provided that the latter exists at high copy numbers in the cell. If we denote this species by F, the pertinent mechanism could be
| (56) |
| (57) |
| (58) |
Similar reactions hold for Cfp. Since the second step is required to be fast, OYfpLac is a short-lived intermediate that exists in low concentrations, and thus competitive utilization of the Lac species is not an issue. Further, this hypothetic system requires that F exists in high copy numbers, thus, even though a common pool of F exists, the abundance of this species results in negligible competitive utilization when the OYfpLac complex is formed. A natural or engineered system that exhibits such a behavior could entail histone modifications of + 1 nucleosomes in eukaryotic promoters.45 More specifically, histone acetyltransferases or histone deacyltransferases could modify the accessibility of promoters to transcription factors as well as their transcriptional status, without having to occupy a binding site.
SUMMARY
It is widely accepted that noise in gene networks can be decomposed into extrinsic and intrinsic contributions. However, this is only valid under the premise that genes do not exert any feedback on their inputs; thus, they can neither modulate the extrinsic noise nor interact with each other through a common input.
Here, we have shown that this assumption falls short even in a simple case in which two genes share a common repressor pool. Starting from a simplified reaction network for the two promoter system invented by Elowitz et al.18 to quantify the contributions of extrinsic versus intrinsic noise, we focused on the operator states and calculated the corresponding noise values. We found that due to the competitive repressor utilization, the extrinsic noise value will always be negative if the total repressor is assumed constant. This unrealistic value is due to the negative correlations that result from the fact that a LacI molecule bound to the yfp operator, is unavailable for repressing the other operator (cfp). If production and depletion of repressor is accounted for (no assumption of constant repressor contents), these negative correlations result in an apparent underestimation of the extrinsic noise. Furthermore, similar conclusions can be reached by analyzing networks with activator species. Competitive utilization of these species by different genes will always result in gene interaction through the common pools and, consequently, in negative correlations in the expression levels of these genes.
Our analysis shows that these effects become less pronounced and finally disappear in the limit of abundant repressor contents (infinitely many molecules). Hence, the presumption that noise can be partitioned into extrinsic and intrinsic components is subject to limitations. In particular, the species’ concentrations that act as inputs to the genes of interest must be high enough so that depletion effects are negligible; otherwise, the calculated (or experimentally measured) extrinsic and intrinsic noise values may not be meaningful.
What may be a viable alternative to the intrinsic and extrinsic noise conundrum described above? One option is to rely on information theory to define intrinsic and extrinsic components of stochasticity. Specifically, with sufficient data points (as it is usually the case for flow cytometry data) the mutual information I(c,y) between the two reporters could be used to estimate extrinsic contributions to gene expression fluctuations. Then, one can calculate the normalized intrinsic and extrinsic entropy factors as follows:
| (59) |
| (60) |
where H denotes the Shannon entropy. These factors (Fint and Fext) will be always positive, will add up to 1, and may serve as robust descriptors of intrinsic and extrinsic contributions to gene expression noise.
While the drastic effects of competitive regulator utilization described above have not been observed in microbial systems so far,18, 35 further studies on different promoters or engineered systems may reveal the indeterminacy of extrinsic noise. This view is supported by the fact that the concentration of several transcription factors is very low within the cell, for instance, the copy number of the LacI repressor is roughly 10 in an E. coli cell.24 Moreover, these effects are likely to be more prominent in mammalian systems due to the complexity of gene regulation in higher eukaryotes, where complexes of many proteins (some of which may be in low abundance) must be assembled for transcription initiation. Indeed, scatter plots from two-color reporter assays in at least two recent studies suggest that the promoters driving the reporter genes may be competing for extrinsic regulators.33, 34 Downstream feedback loops may further amplify such competition effects.
Taken together, the above results and these recent studies indicate that gene expression may occasionally be reliant on limited intracellular resources. In such cases, negative regulatory effects stemming from competition for limited regulator pools alone may constitute mechanisms for gene regulation in addition to the biomolecular interactions between protein and DNA species. This emphasizes the integrated nature of all intracellular processes, and the fact that the expression of individual genes is embedded in a complex biochemical environment with implications that are still insufficiently understood.
ACKNOWLEDGMENTS
This work was supported by the NIH Director’s New Innovator Award Program [Grant No: 1DP2 OD006481-01] and by NSF grant BIO IOS (Integrative Organismal Systems) 1021675. The authors would also like to thank the anonymous reviewers for their constructive comments and suggestions.
References
- Berg H. C., Random Walks in Biology (Princeton University, Princeton, NJ, 1993). [Google Scholar]
- Einstein A., Ann. Phys. 17, 549 (1905). 10.1002/andp.v322:8 [DOI] [Google Scholar]
- Pikovsky A., Rosenblum M., and Kurths J., Synchronization: A Universal Concept in Nonlinear Sciences (Cambridge University Press, Cambridge, UK, 2003). [Google Scholar]
- Strogatz S. H., Nonlinear Dynamics And Chaos: With Applications To Physics, Biology, Chemistry, And Engineering (Westview Press, Cambridge, MA, USA, 2001). [Google Scholar]
- Anishchenko V. S., Astakhov V., Neiman A., Vadivasova T., and Schimansky-Geier L., Nonlinear Dynamics of Chaotic and Stochastic Systems: Tutorial and Modern Developments, 2nd ed. (Springer, New York, 2007). [Google Scholar]
- Barabási A.-L., Linked: How Everything Is Connected to Everything Else and What It Means (Plume, New York, NY, USA, 2003). [Google Scholar]
- Barabási A. L. and Oltvai Z. N., Nat. Rev. Genet. 5(2), 101 (2004). 10.1038/nrg1272 [DOI] [PubMed] [Google Scholar]
- Wiesenfeld K. and Moss F., Nature (London) 373(6509), 33 (1995). 10.1038/373033a0 [DOI] [PubMed] [Google Scholar]
- Balázsi G., van Oudenaarden A., and Collins J. J., Cell 144(6), 910 (2011). 10.1016/j.cell.2011.01.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eldar A. and Elowitz M. B., Nature (London) 467(7312), 167 (2010). 10.1038/nature09326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaern M., Elston T. C., Blake W. J., and Collins J. J., Nat. Rev. Genet. 6(6), 451 (2005). 10.1038/nrg1615 [DOI] [PubMed] [Google Scholar]
- Maheshri N. and O’Shea E. K., Annu. Rev. Biophys. Biomol. Struct. 36, 413 (2007). 10.1146/annurev.biophys.36.040306.132705 [DOI] [PubMed] [Google Scholar]
- Raj A. and van Oudenaarden A., Cell 135(2), 216 (2008). 10.1016/j.cell.2008.09.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao C.V., Wolf D. M., and Arkin A. P., Nature (London) 420(6912), 231 (2002). 10.1038/nature01258 [DOI] [PubMed] [Google Scholar]
- Murphy K. F., Balázsi G., and Collins J. J., Proc. Natl Acad. Sci. U.S.A. 104(31), 12726 (2007). 10.1073/pnas.0608451104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy K. F., Adams R. M., Wang X., Balázsi G., and Collins J. J., Nucleic. Acids Res. 38(8), 2712 (2010). 10.1093/nar/gkq091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain P. S., Elowitz M. B., and Siggia E. D., Proc. Natl Acad. Sci. U.S.A. 99(20), 12795 (2002). 10.1073/pnas.162041399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elowitz M. B., Levine A. J., Siggia E. D., and Swain P. S., Science 297(5584), 1183 (2002). 10.1126/science.1070919 [DOI] [PubMed] [Google Scholar]
- Thattai M. and van Oudenaarden A., Proc. Natl. Acad. Sci. U. S. A. 98(15), 8614 (2001). 10.1073/pnas.151588598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyson J. J. and Othmer H. G., Prog.Theor. Biol., 5, 1 (1978). [Google Scholar]
- Tan C., Marguet P., and You L. C.,. Nat. Chem. Biol., 5(11), 842 (2009). 10.1038/nchembio.218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacs F. J., Hasty J., Cantor C. R., and Collins J. J., Proc. Natl Acad. Sci. U.S.A. 100(13), 7714 (2003). 10.1073/pnas.1332628100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guptasarma P., BioEssays 17(11), 987 (1995). 10.1002/bies.v17:11 [DOI] [PubMed] [Google Scholar]
- Normanno D., Vanzi F., and Pavone F. S.,. Nucleic Acids Res. 36(8), 2505 (2008). 10.1093/nar/gkn071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bialek W., Rieke F., de Ruyter van Steveninck R. R., and Warland D., Science 252(5014), 1854 (1991). 10.1126/science.2063199 [DOI] [PubMed] [Google Scholar]
- Douglass J. K. et al. , Nature (London) 365(6444), 337 (1993). 10.1038/365337a0 [DOI] [PubMed] [Google Scholar]
- Gingl Z., Kiss L. B., and Moss F., Europhys. Lett. 29(3), 191 (1995). 10.1209/0295-5075/29/3/001 [DOI] [Google Scholar]
- Wiesenfeld K., Pierson D., Pantazelou E., Dames C., and Moss F., Phys. Rev. Lett. 72(14), 2125 (1994). 10.1103/PhysRevLett.72.2125 [DOI] [PubMed] [Google Scholar]
- Bezrukov S. M. and Vodyanoy I., Nature (London) 378(6555), 362 (1995). 10.1038/378362a0 [DOI] [PubMed] [Google Scholar]
- Gailey P. C., Neiman A., Collins J. J., and Moss F. M., Phys. Rev. Lett. 79(23), 4701 (1997). 10.1103/PhysRevLett.79.4701 [DOI] [Google Scholar]
- Collins J. J., Chow C. C., and Imhoff T. T., Nature (London) 376(6537), 236 (1995). 10.1038/376236a0 [DOI] [PubMed] [Google Scholar]
- Ozbudak E. M., Thattai M., Kurtser I., Grossman A. D., and van Oudenaarden A., Nat. Genet. 31(1), 69 (2002). 10.1038/ng869 [DOI] [PubMed] [Google Scholar]
- Neildez-Nguyen T. M., Parisot A., Vignal C., Rameau P., Stockholm D., Picot J., Allo V., Le Bec C., Laplace C., and Paldi A., Differentiation 76(1), 33 (2008). [DOI] [PubMed] [Google Scholar]
- Raj A., Peskin C. S., Tranchina D., Vargas D. Y., and Tyagi S., PLoS Biol. 4(10), e309 (2006). 10.1371/journal.pbio.0040309.sd001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raser J. M. and O’Shea E. K.,. Science 304(5678), 1811 (2004). 10.1126/science.1098641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becskei A., Kaufmann B. B., and van Oudenaarden A.,. Nat. Genet. 37(9), 937 (2005). 10.1038/ng1616 [DOI] [PubMed] [Google Scholar]
- Volfson D., Marciniak J., Blake W. J., Ostroff N., Tsimring L. S., and Hasty J., Nature (London) 439(7078), 861 (2006). 10.1038/nature04281 [DOI] [PubMed] [Google Scholar]
- Blake W. J., Balazsi G., Kohanski M. A., Isaacs F. J., Murphy K. F., Kuang Y., Cantor C. R., Walt D. R., and Collins J. J., Mol. Cell 24(6), 853 (2006). 10.1016/j.molcel.2006.11.003 [DOI] [PubMed] [Google Scholar]
- Blake W. J., Kærn M., Cantor C. R., and Collins J. J., Nature 422(6932), 633 (2003). 10.1038/nature01546 [DOI] [PubMed] [Google Scholar]
- Stamatakis M. and Mantzaris N. V., Biophys. J. 96(3), 887 (2009). 10.1016/j.bpj.2008.10.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie D. T., J. Comput. Phys. 22(4), 403 (1976). 10.1016/0021-9991(76)90041-3 [DOI] [Google Scholar]
- Gillespie D. T., J. Phys. Chem. 81(25), 2340 (1977). 10.1021/j100540a008 [DOI] [Google Scholar]
- van Kampen N. G., Stochastic Processes in Physics and Chemistry (North-Holland-Personal-Library, New York, Amsterdam, 1992). [Google Scholar]
- Rao C. V. and Arkin A. P., J. Chem. Phys. 118(11), 4999 (2003). 10.1063/1.1545446 [DOI] [Google Scholar]
- Jiang C. Z. and Pugh B. F., Nat. Rev. Genet. 10(3), 161 (2009). 10.1038/Nrg2522 [DOI] [PMC free article] [PubMed] [Google Scholar]