Abstract
The human microbiome plays a key role in a wide range of host-related processes and has a profound effect on human health. Comparative analyses of the human microbiome have revealed substantial variation in species and gene composition associated with a variety of disease states but may fall short of providing a comprehensive understanding of the impact of this variation on the community and on the host. Here, we introduce a metagenomic systems biology computational framework, integrating metagenomic data with an in silico systems-level analysis of metabolic networks. Focusing on the gut microbiome, we analyze fecal metagenomic data from 124 unrelated individuals, as well as six monozygotic twin pairs and their mothers, and generate community-level metabolic networks of the microbiome. Placing variations in gene abundance in the context of these networks, we identify both gene-level and network-level topological differences associated with obesity and inflammatory bowel disease (IBD). We show that genes associated with either of these host states tend to be located at the periphery of the metabolic network and are enriched for topologically derived metabolic “inputs.” These findings may indicate that lean and obese microbiomes differ primarily in their interface with the host and in the way they interact with host metabolism. We further demonstrate that obese microbiomes are less modular, a hallmark of adaptation to low-diversity environments. We additionally link these topological variations to community species composition. The system-level approach presented here lays the foundation for a unique framework for studying the human microbiome, its organization, and its impact on human health.
We humans are mostly microbes. Microbial communities populate numerous sites in the human anatomy and harbor over 100 trillion microbial cells (1). This complex ensemble of microorganisms, collectively known as the human microbiome, plays an essential role in our development, immunity, and nutrition, and has a tremendous impact on our health (2). Among the various body habitats, the most densely colonized is the distal gut. The normal gut flora alone consists of hundreds of bacterial species, collectively encoding an enormous gene set that is 150-fold larger than the set of human genes (3). The gut microbiome plays a key role in many essential processes, including vitamin and amino acid biosynthesis, dietary energy harvest, and immune development (4). Transferring a donor microbiota into a recipient can induce various donor phenotypes [including increased adiposity (5) and metabolic syndrome (6)] or prompt the recovery of a sick recipient (7), suggesting a promising avenue for clinical application via directed manipulation of the microbiome. Characterizing the capacity of the human microbiome, its interaction with the host, and its contribution to various disease states therefore has the potential to provide deep insight into both normal human physiology and human disease, and calls for a predictive systems-level understanding of community function and structure.
Addressing this challenge, worldwide research initiatives (3, 4) have recently started to map the human microbiome, providing insight into previously uncharted species and genes. Specifically, sequencing 16S ribosomal RNA allows researchers to determine the relative abundance of different taxonomic groups in a microbiome (8, 9). Such surveys have revealed, for example, marked associations between the species composition of the gut microbiome and a variety of host phenotypes (10–12). Species profiles, however, cannot be easily translated into function, because it is not clear how variation in the composition of species in the microbiome affects the metabolic activity of the community and, consequently, the host. In contrast, metagenomic shotgun sequencing of community DNA and a gene-centric comparative approach (8, 13, 14) may capture functional differences in the metabolic potential of the community. Yet, comparative metagenomic analysis of the gut microbiome frequently reveals high functional uniformity across samples and often identifies only a small set of genes or pathways that appear to be associated with certain host states (10, 15). Furthermore, such enriched sets offer preliminary insights into relevant functional differences but may not provide a comprehensive systems-level understanding of the variation and its potential effect on the host–microbiome supraorganism (16, 17).
Here, we introduce a unique framework for studying the human microbiome, integrating metagenomic data with a systems-level network analysis. This metagenomic systems biology approach goes beyond traditional comparative analysis, placing shotgun metagenomic data in the context of community-level metabolic networks. Comparing the topological properties of the enzymes in these networks with their abundances in different metagenomic samples and examining systems-level topological features of microbiomes associated with different host states allow us to obtain insight into variation in metabolic capacity. This approach extends the metagenomic gene-centric view by taking into account not only the set of genes present in a microbiome but also the complex web of interactions among these genes and by treating the microbiome as a single “independent” biological system (18).
Computational systems biology methods and complex network analyses have been applied widely to study microorganisms, and a variety of approaches have been developed to create genome-scale metabolic networks of various microbial species (19–21). In this study, we focus on simple connectivity-centered networks that are computationally derived from homology-based large-scale metabolic databases (22) coupled with a topological analysis. These networks form a simplification of the actual underlying metabolic pathways and may be relatively inaccurate and noisy. However, topology-based analysis of such networks has proved powerful for studying the characteristics of single-species metabolic networks and their impact on various functional and evolutionary properties, including scaling (23), metabolic functionality and regulation (24, 25), modularity (26, 27), essentiality and mutant viability (28), genetic and environmental robustness (29), adaptation (30, 31), and species interaction (32). To date, however, topological analysis has not been used to examine community-level metabolic networks and to study metagenome-scale metabolism.
Results
Datasets.
Illumina-derived shotgun metagenomic data from 124 unrelated Danish and Spanish individuals were analyzed (3). Of the 124 individuals, 82 were labeled as lean/overweight [body mass index (BMI) < 30] and 42 were labeled as obese (BMI ≥ 30). Additionally, 25 were diagnosed with inflammatory bowel disease (IBD) relative to 99 healthy individuals (SI Appendix, Table S1). Patients who had IBD were all of Spanish descent, and Spanish individuals were mostly labeled as lean. An additional dataset, comprising 454 FLX-derived data from six obese and lean monozygotic twin pairs and their mothers (10), was analyzed as well. When applicable, we applied our analysis to this second independent dataset to confirm the validity of our results (SI Appendix). A detailed description of each dataset is provided in Materials and Methods.
Obtaining Community-Level Metabolic Networks.
To construct a community-level metabolic network of the gut microbiome, metagenomic sequence reads were annotated using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database to identify enzymatic genes (Materials and Methods). In total, 1,610 enzymes were identified and annotated with a metabolic reaction. Overall, relative enzyme abundance across the 124 samples was highly concordant (average pair-wise correlation coefficient: R = 0.94, Spearman correlation test), in accordance with previous studies revealing intersample similarity in gene content (10). The annotation data from all samples were pooled, and a network was created in which nodes represented enzymes and enzymes catalyzing successive reactions were connected by directed edges. We excluded enzymes that were not part of the largest connected component of the network, resulting in a total of 1,570 enzymes (Materials and Methods).
Identifying Enzymes Associated with a Given Host State.
We compared the abundance of enzymatic genes across various samples to identify enzymes associated with a given host state (e.g., obesity, IBD). Specifically, we used an odds ratio (OR) test to measure the fold change in the abundance of an enzyme in samples taken from hosts with the given state compared with its abundance in other healthy samples (Materials and Methods; further details on the metric choice are provided in SI Appendix). The differential abundance score of each enzyme, defined as abs[log2(OR)], provides a measure of the extent to which an enzyme's abundance differs in samples from a given host state, relative to healthy samples. Enzymes with a differential abundance score higher than 1 (i.e., enzymes that are either 2-fold enriched or 2-fold depleted) are defined as being associated with the given host state.
To verify that the results reported below are not dependent on the specific choice of enrichment metric used, we further examined several alternative methods for identifying host state-associated enzymes (including significance analysis, presence/absence overrepresentation test, rank-based difference test, and distribution divergence analysis; more details are provided in SI Appendix). These enrichment metrics yielded qualitatively similar results (SI Appendix and SI Appendix, Table S3). Similarly, to confirm that our findings do not stem from potential noise in the read count data, we used a shuffling analysis to identify enzymes that are “consistently” enriched or depleted across samples (SI Appendix). Using this more stringent criterion for enzymes associated with a given host state did not qualitatively change the results below (SI Appendix and SI Appendix, Table S3).
An overrepresentation analysis (SI Appendix) showed that enzymes enriched in obese or IBD microbiomes are more frequently involved in membrane transport [P < 0.035 (obese), P < 0.006 (IBD); SI Appendix, Table S4]. These results are consistent with previous analysis of enriched functions in the smaller dataset of lean and obese twins (10). In contrast, enzymes that are depleted in obese microbiomes are more frequently involved in cofactors and vitamin metabolism (P < 0.03), nucleotide metabolism (P < 0.002), and transcription (P < 2.52 × 10−12), among other processes (SI Appendix, Table S4).
Linking Host State-Associated Enzymes to Centrality.
Using the community-level network outlined above, we examined whether enzymes that are associated with a specific host state exhibit unique topological features. We first focused on a topologically derived centrality measure termed betweenness centrality (25). This measure calculates the proportion of shortest paths in a complex network that pass through a given node, as a proxy for the node's location in relation to all other nodes (SI Appendix, Fig. S2B). High centrality values are typically associated with nodes located in the core of the network, whereas low centrality values indicate a more peripheral location.
We found that an enzyme's differential abundance score in obese samples is negatively correlated with its centrality in the network (R = −0.17, P < 1.3 × 10−12, Spearman correlation test). Partitioning the set of enzymes in the network into those that are associated with obesity (as defined above) and all other enzymes, we similarly found that centrality scores of obesity-associated enzymes are significantly lower (P < 8.9 × 10−6, Wilcoxon rank-sum test; Fig. 1A). As further validation, we note that decreased centrality is not associated with equivalent sets of randomly selected enzymes (P < 8 × 10−4). Significantly lower centrality scores can also be observed when examining obesity-enriched and obesity-depleted enzymes separately (P < 0.03 and P < 7.4 × 10−6, respectively, Wilcoxon rank-sum test; Fig. 1A), suggesting that obesity is characterized by both gain and loss of peripheral enzymes. Similarly, partitioning the enzymes in the network into three centrality-based tiers (Materials and Methods), we find a significant overrepresentation of obesity-associated enzymes in the peripheral tier of the network: 29.1% of the enzymes in this tier are associated with obesity compared with only 19.4% and 18.6% of the enzymes in the intermediate and central tiers, respectively (Fig. 1B). Using the more stringent criterion defined above for identifying enzymes that are consistently associated with obesity yields a similar trend: 13.6%, 10.4%, and 9.8% of the enzymes in the periphery, intermediate, and central tiers, respectively, are consistently associated with obesity (SI Appendix). This negative association between obesity-associated differential abundance and centrality was confirmed in the analysis of the smaller twin-mother trios dataset (R = −0.15, P < 9.7 × 10−8; additional results are presented in SI Appendix).
Fig. 1.
(A) Mean and SE of the centrality scores of obesity-associated enzymes vs. all other enzymes in the network. Obesity-associated enzymes are further divided into enzymes that are enriched or depleted in obese microbiomes. (B) Proportion of enzymes that are associated with obesity (main plot) and IBD (Inset) within three equally populated centrality-based network tiers. Each concentrical pie chart depicts the percent of enzymes within a specific centrality tier that are classified as enriched or depleted. Enzymes associated with obesity or IBD are found in significantly higher proportions in the peripheral tier (P < 5.6 × 10−6 [obesity], P < 4.8 × 10−5 [IBD]; Hypergeometric enrichment test). This result still holds considering alternative or stricter criteria for association with the host state (SI Appendix).
Interestingly, a similar pattern is observed in enzymes associated with IBD. An enzyme's differential abundance score in IBD is negatively correlated with its centrality (R = −0.15, P < 1.9 × 10−9, Spearman correlation test), and the centrality scores of IBD-associated enzymes are significantly lower than the centrality scores of enzymes not associated with IBD (P < 9.5 × 10−6, Wilcoxon rank-sum test; P < 0.003 and P < 0.0002 for IBD-enriched and IBD-depleted enzymes, respectively). Similarly, IBD-associated enzymes are significantly overrepresented in the peripheral tier of the network: 30.1% of the enzymes in this tier are associated with IBD compared with only 22.8% and 19.0% of the enzymes in the intermediate and central tiers, respectively (Fig. 1B, Inset). A similar trend is observed when considering only consistently associated enzymes (8.8%, 5.4%, and 6.1%, respectively).
We confirmed that the above patterns, linking host state-associated enzymes to centrality, are robust to several alternative network construction methods [e.g., using the SEED annotation framework (33) rather than KEGG] and are not affected by using different threshold values to filter out low count reads and potential noise (SI Appendix and SI Appendix, Table S3). To validate that the above results are not the outcome of population substructure, we repeated the analysis for obesity-associated differential abundance using only the Danish individuals and the analysis for IBD-associated differential abundance using only the Spanish individuals. Using these subpopulation samples, we still observed a significant correlation between centrality and differential abundance (SI Appendix, Table S3). We further confirmed that this correlation between differential abundance and centrality is not solely a product of the overrepresentation of transport enzymes (which are likely to be found at the periphery of the network) in obese microbiomes (SI Appendix and SI Appendix, Table S3).
Large-scale metabolic data (e.g., KEGG) are often based on automated, comparison-based, genome annotation (22), and are therefore bound to be incomplete and imprecise (34). Such inaccurate metabolic annotations may markedly affect various complex network properties and can potentially have a dramatic impact on our results. However, using a sensitivity analysis to examine the effect of missing or erroneous annotation data (SI Appendix and SI Appendix, Figs. S4 and S5), we verified that the calculated centrality scores and the pertaining results reported above are fairly robust to such inaccuracies in the raw metabolic annotations.
Linking Host State-Associated Enzymes to Additional Topological Features.
We next examined a number of additional topological measures for each enzyme in the network, including in-degree, out-degree, neighborhood connectivity, and clustering coefficient (Materials and Methods). In contrast to centrality, these measures are more local in nature, taking into account only the immediate neighborhood of each enzyme, and hence capture a different aspect of network topology. The seed set of the network was also identified using a previously published seed detection method (31), and it consisted of 126 enzymes. The seed detection method applies a graph theory-based algorithm to analyze the topology of a given network and identify the minimal set of topological “input” nodes sufficient to activate all other nodes in the network (more details are provided in SI Appendix). The seed sets of metabolite-based networks of a large array of microbial species were shown to be a successful proxy for the biochemical environments of these species and to provide insights into their ecology (31, 32, 35).
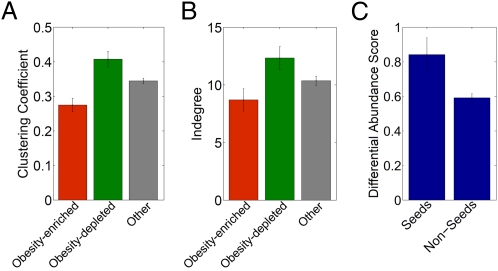
Although both enriched and depleted enzymes exhibit low centrality as described above, we found that enriched enzymes differ dramatically from depleted enzymes in respect to such local topological features. Specifically, enzymes enriched in obese microbiomes have a significantly lower clustering coefficient (P < 7.8 × 10−4, Wilcoxon rank-sum test) and lower in-degree (P < 0.004) compared with enzymes that are not associated with obesity (Fig. 2 A and B). In contrast, enzymes depleted in obese microbiomes have a significantly higher clustering coefficient (P < 0.006, Wilcoxon rank-sum test) and higher in-degree (P < 0.02) compared with nonassociated enzymes. IBD-associated enzymes follow similar trends but are not statistically significant because of smaller sample size (SI Appendix, Fig. S6). We additionally found that enzymes identified as network seeds have significantly higher differential abundance scores (P < 4.8 × 10−6, Wilcoxon rank-sum test) compared with non-seeds and that such network seeds are overrepresented among obesity- and IBD-associated enzymes [P < 2.7 × 10−4 (obesity) and P < 2.6 × 10−3 (IBD); more details are provided in SI Appendix].
Fig. 2.
Mean and SE of the clustering coefficient (A) and in-degree (B) of enriched (red; n = 170), depleted (green; n = 180), and other (gray; n = 1213) enzymes in obese microbiomes. Clustering coefficient is defined as the ratio between the total number of edges connecting a node's neighbors and the potential number of edges that could exist between them. In-degree denotes the number of edges terminating at a node. (C) Mean and SE of the differential abundance scores of seeds vs. non-seed enzymes.
Such distinct topological properties may additionally be used as potentially informative attributes and to highlight biomarkers for involvement in obesity and IBD. Specifically, we examined enzymes enriched in obese or IBD microbiomes, and within these sets, we focused on enzymes that also exhibit the topological features identified above (low centrality, low in-degree, and low clustering coefficient; SI Appendix, Table S2). We find that a large fraction of these enzymes within both the obesity-enriched and the IBD-enriched enzymes are involved in either the phosphotransferase system (PTS; 28.6% and 20.6% among obesity- and IBD-enriched enzymes, respectively) or the nitrate reductase pathway (17.1% and 17.6% among obesity- and IBD-enriched enzymes, respectively). Notably, the PTS is a Eubacteria-specific strategy for transporting sugar into the cell, and it has been specifically associated with members of the Firmicutes phylum (36). Use of this transport system has been implicated in regulation of carbohydrate uptake (37) and was found to be up-regulated following a switch to a high-fat/high-sugar “Western” diet in mice (38). Recently a PTS enzyme (FrvX) was found to be a biomarker for IBD (39). Similarly, nitrate reductase is a critical component in the conversion of nitrate into nitrite and nitric oxide, and it is not synthesized by human DNA. Elevated levels of nitric oxide have been associated with both IBD (40) and obesity-induced insulin resistance (41), as well as other serious carcinogenic and inflammatory effects (42). In this set, we additionally find enzymes for xenobiotic metabolism, most notably those for the metabolism of choline and p-cresol, which have been linked to various host diseases and metabolic phenotypes (SI Appendix).
Linking Topological Variation to Community Species Composition.
Shotgun metagenomic data and community-level models provide a functional view of community metabolism. Ultimately, however, differences in community gene content reflect differences in species composition. Understanding the link between variation in community-level topological properties and community composition can provide valuable insight into the mechanism by which community activity changes as a result of compositional shifts. Because a full decomposition of shotgun metagenomic data into species-specific data is not yet feasible, we studied the distribution of genes of interest across a large array of reference genomes. Specifically, examining the genomes of 326 fully sequenced, prevalent, gut-dwelling microbial species (Materials and Methods), we found that enzymes associated with either obese or IBD microbiomes tend to be present in fewer genomes than nonassociated enzymes [P < 10−54 (obesity), P < 10−56 (IBD), Wilcoxon rank-sum test; SI Appendix, Fig. S7]. Obesity-associated enzymes were also present in fewer genomes than randomly selected sets of enzymes (P < 10−4; SI Appendix). Moreover, the centrality of enzymes in the community-level metabolic network is correlated with the number of reference genomes in which these enzymes occur (R = 0.23, P < 10−17, Spearman correlation test; SI Appendix, Fig. S8). A universal association between centrality and prevalence has also been demonstrated recently for a smaller set of species that were not associated with the human microbiome (43). These findings suggest that the variation in community-level metabolism associated with obesity and IBD may be induced by an increase or decrease in the abundance of a relatively small subset of species.
Linking Host State to Network-Level Topological Properties.
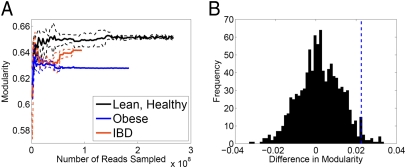
Finally, we examined whether host state-associated differences also translate into differences in network-level topological features. Sequence reads derived from lean-healthy, obese-healthy, and lean-IBD samples were pooled separately and used to construct state-specific metabolic networks. Calculating various network-level topological features for each of these networks, we found that the variation associated with host state goes beyond a limited set of enriched or depleted enzymes and also induces global differences in network topology. Specifically, obese microbiomes were found to induce a less modular metabolic network than lean microbiomes. Interestingly, reduced modularity in the metabolic networks of single species has recently been associated with lower variation in the environment (Discussion). A rarefaction analysis was performed to confirm that all networks derived from each of the three sample groups reached a stable topology within the available coverage (Fig. 3A). An extensive shuffling-based analysis (Fig. 3B, SI Appendix, and SI Appendix, Figs. S9–S11) demonstrated that the difference in the level of modularity between obese and lean microbiomes is statistically significant (P < 0.027) and is not expected at random from multiple individual realizations of networks with similar topological properties.
Fig. 3.
Modularity of host state-specific metabolic networks. (A) Rarefaction analysis of the modularity of pooled lean-healthy, obese-healthy, and IBD-lean microbiomes. The plot depicts the mean (solid lines) and SD (dotted lines) of five rounds of rarefaction analysis, obtained by calculating the modularity of networks derived from progressively smaller randomly selected sets of reads. (B) Difference between the modularity of the obese-specific and lean-healthy–specific network is plotted (dashed blue line) against a null distribution of differences obtained via random grouping of samples (more details are provided in SI Appendix). The observed difference in modularity is significantly greater than the expected difference according to this null distribution.
Discussion
Taken together, the topological features that were found to vary with obesity and IBD suggest a characteristic mode of deviation from a normal microbiome organization that may be associated with a disease state. This suggests that in addition to, or potentially as a consequence of, alterations in the abundance of individual genes or functional classes, disease may be associated with higher order modes of deviation in the microbiome. Clearly, such associations alone cannot directly implicate a mechanism for disease; both obesity and IBD are poorly understood diseases and embody extremely complex phenotypes. Accordingly, the system-level observations reported in this study can have multiple alternative interpretations and stem from mechanisms that are yet unknown. These observations, however, allow us to posit intriguing hypotheses for further study.
Specifically, we find that enzymes typifying various host states tend to have low centrality and are found mostly in the periphery of the network. As the topology of the network reflects metabolic interdependencies between enzymes (rather than physical location in the gut), the periphery of the network represents metabolic steps that are relatively remote (as measured by their distance along various metabolic pathways) from the core of the network and that are closer functionally to the microbiome environment (44). The most peripheral enzymes, for example, represent either the microbiome's first metabolic steps (i.e., enzymes that rely on substrates that are not produced by any other enzyme in the microbiome) or end points (enzymes that produce metabolites that are not utilized by other microbiome enzymes). Such enzymes are likely to directly use or produce metabolites that characterize the gut environment, forming an interface between microbial and human metabolism. Our results therefore suggest that much of the enzyme-level variation associated with obesity or IBD relates to changes in the way the microbiome interacts with the gut environment rather than variation in core metabolic processes. This variation corresponds to both gain and loss of certain peripheral metabolic enzymes, as suggested by the reduced centrality of both enriched and depleted enzymes. This is also supported by the reported link between differentially abundant enzymes and seed enzymes. Obesity-enriched enzymes, however, specifically possess further topological properties characteristic of network input points (low in-degree and low clustering coefficient). While several mechanisms that link the microbiota to obesity have been reported, this finding may suggest that obese microbiomes are capable of using a diverse repertoire of energy sources, accounting for their increased capacity for energy extraction from the diet (5). Interestingly, it has also been shown that functionally peripheral enzymes (those involved in nutrient uptake and first metabolic steps) are more likely to be horizontally transferred (44) and are gained and lost more frequently during the evolution of individual microbial organisms (31). This similarity between the adaptive variation that occurs in single species across an evolutionary time scale and community-level variation across samples further supports our treatment of the community as a comprehensive biological system.
Our topology-based system approach has also suggested candidate biomarkers involved in obesity and IBD. In addition to PTSs used for the import of dietary carbohydrates, both obesity and IBD were significantly associated with genes for the production of NO2 and the metabolism of choline and p-cresol. The unexpectedly high overlap between these disease-associated gene sets (SI Appendix, Table S2) may be indicative of some common underlying triggers of disease or, alternatively, a conserved response of the gut microbiome to disease. Follow-up studies using gnotobiotic mouse models colonized by microbial isolates with the ability to perform these key functions, “humanized” mouse models colonized with samples taken from paired healthy and diseased human donors, and human intervention studies will be critical to determine which aspects of the gut microbiome may contribute to disease and the precise mechanisms that link this complex microbial metabolic network to host physiology.
Our results further demonstrate that the variation associated with obesity and IBD induces a reduced network-wide modularity. Recent studies of metabolic network topologies across the bacterial tree of life revealed marked variation in network modularity and identified several genetic and environmental determinants affecting metabolic modularity (27, 45). Specifically, these studies demonstrated that reduced metabolic modularity in single-species networks is associated with organisms inhabiting less variable environments. Our analysis, however, presents a unique characterization of community-level modularity and demonstrates consistent differences that are associated with the host state. It is intriguing to extrapolate findings from single-species analyses and to hypothesize that reduced community-level modularity in obese microbiomes may be associated with decreased variability in the gut environment or with the lack of temporal regularities (46). Furthermore, this reduced modularity may be construed as a functional manifestation of the reported decrease in species diversity that has been observed in obese individuals (10).
In silico models of microbial communities are currently still scarce (19) and mostly focus on simulated communities comprising a handful of species and on pair-wise interactions among community members (21, 47–49). Here, in contrast, we take an integrative approach, treating the microbiome as a single supraorganismal system (17) and examining the metabolic network of the community as a whole (50). Moreover, this study focuses on the topology of this metagenome-based network and on the relationship between its topology and the host state. As with any attempt to represent a dynamic and stochastic set of biological processes via a simple model, our analysis is subject to various assumptions and simplifications. Our framework ignores boundaries between species and compartmentalization of various metabolic processes (information on analyzing communities as supraorganisms is provided in SI Appendix). Additionally, topological analysis of connectivity-based and static networks explicitly ignores several features of metabolic reactions, such as metabolic rates and dynamic regulation. Furthermore, our analysis considers metabolism alone and does not account for other processes that may be involved (e.g., immune response). Such simple models, however, are extremely useful for studying systems for which data are still limited and our ability to construct more involved models is hindered. Here, for example, they facilitate the integration of multiple modes of microbiome characterization and support analysis using the rich set of tools developed for systems biology and complex network analysis. As our understanding of the human microbiome improves, better models can be constructed, potentially using the collective effort of the research community (51, 52). Experimental validation of model components and parameters is crucial for a successful and accurate reconstruction. Moving forward, microbiome-wide models can further integrate transcriptional and metabolomics-based data. Such manually curated models may ultimately provide a predictive framework, similar to the one available for individual species, for targeted community manipulation and for informing clinical interventions.
In essence, this study represents an important step in the development of a “metagenomic systems biology” approach. Such an approach can potentially advance metagenomic research in the same way systems biology advanced genomics, appreciating not only the parts list of a system but the complex interactions among parts and the impact of these interactions on function and dynamics. Future work will also include identifying specific sets of enzymes responsible for systems-level patterns, characterizing the implications of various topological variations, and linking this variation to changes in species composition. Clearly, our understanding of the complexity of the gut microbiome is still lacking, and much work still remains to be done before exact mechanisms are identified. Future clinical applications may focus on specific functions rather than on system-level properties of the microbiome. Yet, this systems biology approach provides a complementary viewpoint to comparative and functional metagenomics in gaining valuable intuition concerning the function of the microbiome as a system and in identifying potential biomarkers for further validation.
Materials and Methods
Datasets.
Metagenomic data were obtained from two studies of the human gut microbiome. The first study (3) examined 576.7 gigabases of Illumina-derived sequences from 124 European individuals labeled with BMI (kg/m2) and IBD data. The second study (10) examined 454 FLX-derived sequences from six twin-mother trios from the Missouri Adolescent Female Twin Study binned according to BMI. All sequence data were mapped to KEGG orthologous groups (KOs) using BLASTX (additional details are provided in SI Appendix).
Enzyme Enrichment.
To identify enzymes (KOs) that are associated with obesity, the abundance of each enzyme in the set of samples obtained from obese individuals was compared with its abundance in lean/overweight individuals. To prevent the confounding effects of overlapping host states, samples labeled with IBD were excluded from this analysis. For each enzyme, k, an OR was calculated according to OR(k) = [∑s = obese Ask/∑s = obese (∑i≠k Asi)]/[∑s = lean Ask/∑s = lean (∑i≠k Asi)] where Ask denotes the abundance of enzyme k in sample s, obese denotes the set of obese samples, and lean denotes the set of lean/overweight samples (SI Appendix, Fig. S3). More details on this choice of enrichment metric are provided in SI Appendix. The differential abundance score was defined as the absolute value of the fold change in OR, abs[log2(OR)]. Obesity-associated enzymes were those with a differential abundance score >1. Obesity-associated enzymes were further classified as obesity-enriched (OR > 2) or obesity-depleted (OR < 0.5) (SI Appendix, Table S2 A and B). IBD-associated enzymes were identified in a similar manner (SI Appendix, Table S2 C and D). When calculating IBD-associated ORs, samples labeled as obese were excluded from the analysis. A more stringent OR-based analysis was used to identify enzymes that were consistently enriched or depleted (SI Appendix). Additionally, a number of other statistical methods were used to quantify differential abundance and identified enzymes associated with a given host state (SI Appendix). Repeating the analysis with these alternative methods yielded qualitatively similar results (SI Appendix and SI Appendix, Table S3).
Network Construction.
A community-level metabolic network was constructed from the entire set of enzymes found in any sample (SI Appendix). The KEGG database was used to annotate enzymes with metabolic reactions. Each enzyme may be associated with multiple reactions, and each reaction may be associated with multiple enzymes. Using this mapping, an enzyme-based metabolic network was constructed, where nodes represent enzymes (KOs) and a directed edge from enzyme 1 to enzyme 2 indicates that a product metabolite of a reaction catalyzed by enzyme 1 is a substrate metabolite of a reaction catalyzed by enzyme 2 (SI Appendix, Fig. S1B). For both datasets, 98% of the enzymes were part of a single giant, connected component. The network was trimmed to include only the nodes and edges that were part of this giant component, and only these enzymes were used in the subsequent analysis. To create host state-specific networks, the same procedure was followed using only the set of enzymes recovered from samples in a given host state.
Topology-Based Measures and Analysis.
Topological features of each enzyme in the network were calculated with the Cytoscape NetworkAnalyzer plug-in (53). The overall correlation across all topological features supported by the NetworkAnalyzer plug-in was calculated, and a feature set without any pairwise correlations >0.95 was selected for further analysis. This feature set included betweenness centrality (defined as the proportion of shortest paths passing through a node), clustering coefficient (defined as the proportion of existing edges between a node's neighbors), neighborhood connectivity (average number of neighbors of a node's neighbors), in-degree (number of edges terminating in a node), and out-degree (number of edges originating in a node). SI Appendix, Fig. S2B provides additional illustrations and examples of these features. The betweenness centrality feature was used throughout the study to measure the centrality of each enzyme in the network. Enzymes were further classified as peripheral, intermediate, or central by ranking all enzymes according to centrality and partitioning this ranked list into three equally populated bins, which we termed centrality tiers.
The Spearman correlation test was used to examine the correlation between differential abundance scores and each topological feature. A Wilcoxon rank-sum test was used to compare the topology scores of host state-associated enzymes (and specifically enriched or depleted enzymes) with the scores obtained for non-associated enzymes. A Hypergeometric enrichment test was used to examine the over-representation of host state-associated enzymes in each centrality tier.
Network-Level Topological Features of Host State-Specific Networks.
Samples were divided into three distinct groups: lean-healthy, obese-healthy, and lean-IBD. The three obese-IBD samples were not used in this analysis. Three separate host state-specific networks were created from the pooled set of enzymes identified within each group. Network-level features, including node count, density (the ratio of edges to nodes), and modularity, were calculated for each network. Here, we define and calculate modularity according to the formulation presented by Newman (54). For a particular division of a network into discrete modules, modularity is defined as the number of edges between nodes that belong to the same module minus the expected number of such edges in an equivalent randomized network, normalized by the total number of edges. The modularity of the network is calculated for the division that maximizes this value. This modularity value measures how well a network can be partitioned into densely connected modules with relatively few edges running between modules. Rarefaction curves were generated for each of these measures by considering an increasingly larger random subset of reads from each group. The statistical significance of these measures was assessed using null distributions calculated from randomized networks (SI Appendix).
Seed Set Identification.
The metabolic seed set (more details are provided in SI Appendix), representing enzymes operating on exogenously acquired compounds, was calculated according to the method described by Borenstein et al. (31).
Supplementary Material
Acknowledgments
We thank Junjie Qin for assistance in downloading and analyzing the data from 124 unrelated individuals. The metagenomic sequence comparisons in this paper were run on the Odyssey cluster supported by the FAS Sciences Division Research Computing Group. S.G. is supported by “Interdisciplinary Training in Genomic Sciences” National Human Genome Research Institute Grant T32 HG00035. P.J.T. is supported by National Institutes of Health Grant P50 GM068763. E.B. is an Alfred P. Sloan Research Fellow.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1116053109/-/DCSupplemental.
References
- 1.Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 2.Dethlefsen L, McFall-Ngai M, Relman DA. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature. 2007;449:811–818. doi: 10.1038/nature06245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qin J, et al. MetaHIT Consortium A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Turnbaugh PJ, et al. The human microbiome project. Nature. 2007;449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Turnbaugh PJ, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 6.Vijay-Kumar M, et al. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science. 2010;328:228–231. doi: 10.1126/science.1179721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khoruts A, Dicksved J, Jansson JK, Sadowsky MJ. Changes in the composition of the human fecal microbiome after bacteriotherapy for recurrent Clostridium difficile-associated diarrhea. J Clin Gastroenterol. 2010;44:354–360. doi: 10.1097/MCG.0b013e3181c87e02. [DOI] [PubMed] [Google Scholar]
- 8.Gill SR, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI. Worlds within worlds: Evolution of the vertebrate gut microbiota. Nat Rev Microbiol. 2008;6:776–788. doi: 10.1038/nrmicro1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turnbaugh PJ, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peterson J, et al. NIH HMP Working Group The NIH Human Microbiome Project. Genome Res. 2009;19:2317–2323. doi: 10.1101/gr.096651.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hartman AL, et al. Human gut microbiome adopts an alternative state following small bowel transplantation. Proc Natl Acad Sci USA. 2009;106:17187–17192. doi: 10.1073/pnas.0904847106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tringe SG, et al. Comparative metagenomics of microbial communities. Science. 2005;308:554–557. doi: 10.1126/science.1107851. [DOI] [PubMed] [Google Scholar]
- 14.White JR, Nagarajan N, Pop M. Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLOS Comput Biol. 2009;5:e1000352. doi: 10.1371/journal.pcbi.1000352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kurokawa K, et al. Comparative metagenomics revealed commonly enriched gene sets in human gut microbiomes. DNA Res. 2007;14:169–181. doi: 10.1093/dnares/dsm018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lederberg J. Infectious history. Science. 2000;288:287–293. doi: 10.1126/science.288.5464.287. [DOI] [PubMed] [Google Scholar]
- 17.Gordon JI, Klaenhammer TR. A rendezvous with our microbes. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4513–4515. doi: 10.1073/pnas.1101958108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raes J, Bork P. Molecular eco-systems biology: Towards an understanding of community function. Nat Rev Microbiol. 2008;6:693–699. doi: 10.1038/nrmicro1935. [DOI] [PubMed] [Google Scholar]
- 19.Oberhardt MA, Palsson BØ, Papin JA. Applications of genome-scale metabolic reconstructions. Mol Syst Biol. 2009;5:320. doi: 10.1038/msb.2009.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tepper N, Shlomi T. Predicting metabolic engineering knockout strategies for chemical production: Accounting for competing pathways. Bioinformatics. 2010;26:536–543. doi: 10.1093/bioinformatics/btp704. [DOI] [PubMed] [Google Scholar]
- 21.Klitgord N, Segrè D. Environments that induce synthetic microbial ecosystems. PLOS Comput Biol. 2010;6:e1001002. doi: 10.1371/journal.pcbi.1001002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kanehisa M, et al. From genomics to chemical genomics: New developments in KEGG. Nucleic Acids Res. 2006;34(Database issue):D354–D357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jeong H, Tombor B, Albert R, Oltvai ZN, Barabási AL. The large-scale organization of metabolic networks. Nature. 2000;407:651–654. doi: 10.1038/35036627. [DOI] [PubMed] [Google Scholar]
- 24.Stelling J, Klamt S, Bettenbrock K, Schuster S, Gilles ED. Metabolic network structure determines key aspects of functionality and regulation. Nature. 2002;420:190–193. doi: 10.1038/nature01166. [DOI] [PubMed] [Google Scholar]
- 25.Patil KR, Nielsen J. Uncovering transcriptional regulation of metabolism by using metabolic network topology. Proc Natl Acad Sci USA. 2005;102:2685–2689. doi: 10.1073/pnas.0406811102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guimerà R, Nunes Amaral LA. Functional cartography of complex metabolic networks. Nature. 2005;433:895–900. doi: 10.1038/nature03288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kreimer A, Borenstein E, Gophna U, Ruppin E. The evolution of modularity in bacterial metabolic networks. Proc Natl Acad Sci USA. 2008;105:6976–6981. doi: 10.1073/pnas.0712149105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Palumbo MC, Colosimo A, Giuliani A, Farina L. Functional essentiality from topology features in metabolic networks: A case study in yeast. FEBS Lett. 2005;579:4642–4646. doi: 10.1016/j.febslet.2005.07.033. [DOI] [PubMed] [Google Scholar]
- 29.Freilich S, et al. Decoupling Environment-Dependent and Independent Genetic Robustness across Bacterial Species. PLOS Comput Biol. 2010;6:e1000690. doi: 10.1371/journal.pcbi.1000690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raymond J, Segrè D. The effect of oxygen on biochemical networks and the evolution of complex life. Science. 2006;311:1764–1767. doi: 10.1126/science.1118439. [DOI] [PubMed] [Google Scholar]
- 31.Borenstein E, Kupiec M, Feldman MW, Ruppin E. Large-scale reconstruction and phylogenetic analysis of metabolic environments. Proc Natl Acad Sci USA. 2008;105:14482–14487. doi: 10.1073/pnas.0806162105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Borenstein E, Feldman MW. Topological signatures of species interactions in metabolic networks. J Comput Biol. 2009;16:191–200. doi: 10.1089/cmb.2008.06TT. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Overbeek R, et al. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 2005;33:5691–5702. doi: 10.1093/nar/gki866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Green ML, Karp PD. The outcomes of pathway database computations depend on pathway ontology. Nucleic Acids Res. 2006;34:3687–3697. doi: 10.1093/nar/gkl438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Freilich S, et al. Metabolic-network-driven analysis of bacterial ecological strategies. Genome Biol. 2009;10:R61. doi: 10.1186/gb-2009-10-6-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mahowald MA, et al. Characterizing a model human gut microbiota composed of members of its two dominant bacterial phyla. Proc Natl Acad Sci USA. 2009;106:5859–5864. doi: 10.1073/pnas.0901529106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Francl AL, Thongaram T, Miller MJ. The PTS transporters of Lactobacillus gasseri ATCC 33323. BMC Microbiol. 2010;10:77. doi: 10.1186/1471-2180-10-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Turnbaugh PJ, et al. The effect of diet on the human gut microbiome: A metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med. 2009;1:6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen C-S, et al. Identification of novel serological biomarkers for inflammatory bowel disease using Escherichia coli proteome chip. Mol Cell Proteomics. 2009;8:1765–1776. doi: 10.1074/mcp.M800593-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kolios G, Valatas V, Ward SG. Nitric oxide in inflammatory bowel disease: A universal messenger in an unsolved puzzle. Immunology. 2004;113:427–437. doi: 10.1111/j.1365-2567.2004.01984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gil-Ortega M, et al. Adaptative nitric oxide overproduction in perivascular adipose tissue during early diet-induced obesity. Endocrinology. 2010;151:3299–3306. doi: 10.1210/en.2009-1464. [DOI] [PubMed] [Google Scholar]
- 42.Yang G-Y, Taboada S, Liao J. Induced nitric oxide synthase as a major player in the oncogenic transformation of inflamed tissue. Methods Mol Biol. 2009;512:119–156. doi: 10.1007/978-1-60327-530-9_8. [DOI] [PubMed] [Google Scholar]
- 43.Bernhardsson S, Gerlee P, Lizana L. Structural correlations in bacterial metabolic networks. BMC Evol Biol. 2011;11:20. doi: 10.1186/1471-2148-11-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pál C, Papp B, Lercher MJ. Adaptive evolution of bacterial metabolic networks by horizontal gene transfer. Nat Genet. 2005;37:1372–1375. doi: 10.1038/ng1686. [DOI] [PubMed] [Google Scholar]
- 45.Parter M, Kashtan N, Alon U. Environmental variability and modularity of bacterial metabolic networks. BMC Evol Biol. 2007;7:169. doi: 10.1186/1471-2148-7-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Costello EK, Gordon JI, Secor SM, Knight R. Postprandial remodeling of the gut microbiota in Burmese pythons. ISME J. 2010;4:1375–1385. doi: 10.1038/ismej.2010.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stolyar S, et al. Metabolic modeling of a mutualistic microbial community. Mol Syst Biol. 2007;3:92. doi: 10.1038/msb4100131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wintermute EH, Silver PA. Emergent cooperation in microbial metabolism. Mol Syst Biol. 2010;6:407. doi: 10.1038/msb.2010.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Freilich S, et al. The large-scale organization of the bacterial network of ecological co-occurrence interactions. Nucleic Acids Res. 2010;38:3857–3868. doi: 10.1093/nar/gkq118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gianoulis TA, et al. Quantifying environmental adaptation of metabolic pathways in metagenomics. Proc Natl Acad Sci USA. 2009;106:1374–1379. doi: 10.1073/pnas.0808022106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Herrgård MJ, et al. A consensus yeast metabolic network reconstruction obtained from a community approach to systems biology. Nat Biotechnol. 2008;26:1155–1160. doi: 10.1038/nbt1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thiele I, Palsson BØ. Reconstruction annotation jamborees: A community approach to systems biology. Mol Syst Biol. 2010;6:361. doi: 10.1038/msb.2010.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Assenov Y, Ramírez F, Schelhorn S-E, Lengauer T, Albrecht M. Computing topological parameters of biological networks. Bioinformatics. 2008;24:282–284. doi: 10.1093/bioinformatics/btm554. [DOI] [PubMed] [Google Scholar]
- 54.Newman MEJ. Modularity and community structure in networks. Proc Natl Acad Sci USA. 2006;103:8577–8582. doi: 10.1073/pnas.0601602103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.