Abstract
It is well established that the human immunodeficiency virus-1 envelope glycoprotein surface unit, gp120, binds to cell-associated heparan sulfate (HS). Virus infectivity is increased by such interaction, and a variety of soluble polyanions efficiently neutralize immunodeficiency virus-1 in vitro. This interaction has been mainly attributed to the gp120 V3 loop. However, although evidence suggested that this particular domain does not fully recapitulate the binding activity of the protein, the ability of HS to bind to other regions of gp120 has not been completely addressed, and the exact localizations of the polysaccharide binding sites are not known. To investigate in more detail the structural basis of the HS-gp120 interaction, we used a mapping strategy and compared the heparin binding activity of wild type and mutant gp120 using surface plasmon resonance-based binding assays. Four heparin binding domains (1–4) were identified in the V2 and V3 loops, in the C-terminal domain, and within the CD4-induced bridging sheet. Interestingly, three of them were found in domains of the protein that undergo structural changes upon binding to CD4 and are involved in co-receptor recognition. In particular, Arg419, Lys421, and Lys432, which directly interact with the co-receptor, are targeted by heparin. This study provides a complete account of the gp120 residues involved in heparin binding and identified several binding surfaces that constitute potential target for viral entry inhibition.
Human immunodeficiency virus (HIV)4 gains entry into permissive CD4+ cells by sequentially interacting with CD4, the primary receptor, and a co-receptor, usually either CCR5 or CXCR4 (1). Both receptor and co-receptors are recognized by gp120, the glycoprotein that constitutes the surface unit of HIV envelope spikes. This protein consists of five relatively conserved regions (C1 to C5) that fold into a “core” comprising two distinct domains termed “inner” and “outer” and five variable regions (V1 to V5). This protein is prone to structural changes and is believed to sample a number of different conformations. A wealth of evidence showed that upon binding to CD4, roughly half of the gp120 core structure undergoes structural rearrangements, in particular within the inner domain. In the CD4-bound form, the base of the V1/V2 region of the inner domain (β2 and β3 strands) is brought to close proximity to a β-hairpin of the outer domain (β20 and β21 strands) and forms a four-stranded β-sheet located within the bridging sheet that connects the inner and the outer domain of the glycoprotein. Importantly, this highly conserved structure forms, in conjunction with the V3 loop, the binding site for CCR5 or CXCR4 (2–7).
It has been well known that HIV also binds to CD4- cells in a gp120-dependent manner through interactions with cell surface molecules including heparan sulfates (HSs) (8). HSs and the closely related heparin belong to a large family of anionic polysaccharides, collectively known as glycosaminoglycans. They occur covalently bound to core proteins and are ubiquitously expressed at most cell surfaces (9). These molecules are structurally complex, characterized by very large interactive properties, and recognize many unrelated protein via clusters of basic amino acids exposed on the surface of their targets (10). As such, they are exploited as a prevalent source of docking sites by a large array of pathogens (11–13) that interact with highly sulfated heparin-like regions of these polysaccharides (14). Regarding HIV, it has earlier been described that removal of HSs from the cell membrane inhibited infection by reducing viral particle concentration at the cell surface (15). It is now, however, appreciated that HIV binding to HSs can occur at many places and serves many functions. At the mucosal surface, the main point of entry for the virus into the host, HSs sequester viral particles and are involved in their translocation across epithelial barriers (16–18). Similarly, HSs expressed by microvasculature endothelial cells of the blood brain barrier capture HIV and contribute to HIV brain invasion (19–21). Several studies also showed that HSs play an active role in sequestering, protecting, and transferring viruses to CD4+ susceptible cells, with conditions that boost their replication (in trans mechanism) (22, 23). On permissive cells, HIV binding to HSs is thought to increase infectivity by favoring viral particle concentration at the cell surface (in cis process). For some cells, such as macrophages, they may compensate for low CD4 expression (24). It has also been suggested that HSs could be directly involved in the infection of CD4- cells, including endothelial cells and possibly neurons (19, 25). Finally, HSs and heparin have been found to inhibit the protein disulfide isomerase-mediated reduction of gp120, although the exact role of this effect has not been elucidated yet (26). In addition to mediating viral attachment and entry, gp120 (either free or virion associated) targets a number of uninfected or non-permissive cells. As such, it is involved in some aspects of the AIDS-associated pathologies, including apoptosis and oxidative stress, and this has motivated the development of heparin-mimetics that inhibit gp120-HS interactions (27).
A large body of work, aiming at characterizing the gp120/HS complex, showed that heparin, HSs, or other polyanions including dextran sulfate bind to gp120 V3 loop-derived peptides or compete with V3 loop-specific monoclonal antibodies (mAbs) for binding to gp120 (28–31). However, this binding, which affinity (KD = 220 nm) has been determined by a surface plasmon resonance-based binding assay (32), appears to be more complex than previously thought. The binding of polyanions to gp120 interferes with several mAbs recognizing epitopes outside the V3 loop, including those that target the bridging sheet induced upon CD4 binding (15, 32). In agreement with these observations, it has been previously reported that gp120 in its CD4-bound state had a substantially increased binding activity toward heparin, compared with free gp120, and molecular modeling further suggested that the bridging sheet includes a possible HS binding domain (HBD) (33). Finally, it has also been demonstrated that the development of resistance to dextran sulfate correlates with specific mutations within and outside the V3 loop (34). Through an approach designed to simultaneously map different HBDs at the protein surface and the production of gp120 mutants, the present work identified the presence of four HBDs on the glycoprotein, some of which are potential targets for therapeutic applications.
EXPERIMENTAL PROCEDURES
Material—Insect cell culture media and the Bac-to-Bac system were purchased from Invitrogen and the QuikChange™ site-directed mutagenesis kit from Stratagene (La Jolla, CA). SP-Sepharose Fast Flow resin, Lentil-lectin resin, Superdex 200 column, N-ethyl-N′-(diethylaminopropyl)-carbodiimide/N-hydroxysuccimide reagents, and all surface plasmon resonance products were supplied from GE Healthcare. DEAE-Sephacel resin, 15-kDa heparin, 6-kDa heparin, and thermolysin were from Sigma Aldrich. Heparin-derived dodecasaccharide (dp12) was prepared as described (35). The ultrafiltration unit and membranes were purchased from Millipore (Billerica, MA). Recombinant soluble CD4 (sCD4) was from Progenics Pharmaceuticals (Tarrytown, NY) and obtained through the National Institutes of Health AIDS Research and Reference Reagent Program. Monoclonal antibody 17b was from the Centre for AIDS Reagent Program, National Institute for Biological Standards and Control.
Recombinant Wild Type and Mutant Protein Production—The cDNA encoding gp120HXBc2 was amplified by PCR from pSVIII-env and inserted into pNT-Bac, encoding either the melittin or the baculovirus ecdysteroid UDP glucosyltransferase signal peptide using the BamHI and HindIII restriction sites. Mutations giving rise to gp120-K121S, gp120-R419S, gp120-K421S, and gp120-K432S were introduced using the QuikChange™ site-directed mutagenesis kit. Resulting constructs were checked by restriction analysis and DNA sequencing (Genome Express) and used to prepare recombinant bacmid by transposition in Escherichia coli DH10Bac cells according to the manufacturer's protocol (Invitrogen). Isolated recombinant bacmid DNA was purified, analyzed by PCR, and used to transfect Spodoptera frugiperda 21 (Sf21) insect cells using Cellfectin in Sf900 II SFM medium. Recombinant baculovirus particles were collected 4 days later, titrated by virus plaque assay, and amplified as described (36). In preliminary assays small-scale time-course expression experiments were conducted by infecting Sf21 cells (1.75 × 106 cells/ml) at a multiplicity of infection of 8. Samples of culture supernatants were taken 0, 24, 48, 72, 96, and 120 h post-infection and analyzed by SDS-PAGE. Material corresponding to gp120 was revealed by Western blot analysis with a primary polyclonal goat antibody directed against gp120 (1/2000; Biodesign International) coupled to a horseradish peroxidase-conjugated anti-goat antibody (1/5000, Jackson ImmunoResearch).
Protein Purification—For large-scale protein production, Sf21 cells were adapted for growth in suspension. Sf21 cells (1 liter at 0.5 × 106 cells/ml) were infected for 72 h with recombinant viruses at a multiplicity of infection of 8 in Sf900 II SFM medium. Purification of both wild type and mutant proteins was performed at 4 °C using a three-step method. The supernatant (1 liter) was injected into a SP-Sepharose Fast Flow cation-exchange column (XK16) equilibrated with 50 mm HEPES pH 8.2 (solvent A) at a flow rate of 2 ml/min. The column was washed with 5 column volumes of A and 5 column volumes of A supplemented with 0.3% surfactant P20 (GE Healthcare). Elution was performed using a 2-step NaCl gradient (from 0 to 0.5 m NaCl in 12 min and 0.5 to 1 m NaCl in 2 min) and monitored by absorbance (280 nm). Fractions containing gp120 were pooled and loaded onto a lentil lectin-Sepharose column (1.6 × 10 cm) pre-equilibrated with phosphate-buffered saline (PBS) at 1 ml/min. The column was washed with 30 ml of PBS, and bound proteins were eluted with PBS containing 1 m methyl-α-d-mannopyranoside. Fractions (0.5 ml) containing gp120 were pooled and concentrated by ultrafiltration using an Amicon 8010 Filter unit fitted with a YM30 ultrafiltration membrane (30 000 molecular weight cutoff) followed by concentration on a Microcon unit (10,000 molecular weight cutoff). The sample was then size-fractionated onto a Superdex 200 HR 10/30 column equilibrated with PBS at 0.4 ml/min. Fractions (0.5 ml) containing gp120 were concentrated and quantified by amino acid analysis. Protease inhibitors (Complete; Roche Applied Science) were added, and the sample was stored at -20 °C. Throughout the procedure fractions were monitored by absorbance (280 nm) and analyzed by SDS-PAGE stained with Coomassie Blue and Western blotting, as described above.
Surface Plasmon Resonance-based Binding Assays—A surface plasmon resonance (Biacore 3000) instrument was used to investigate the binding of wild type or mutant gp120 to immobilized CD4, heparin, or mAb 17b. For that purpose flow cells of a CM4 sensor chip were first activated with 50 μl of 0.2 m N-ethyl-N′-(diethylaminopropyl)-carbodiimide and 0.05 m N-hydroxysuccimide at 5 μl/min. Then soluble CD4 (5 μg/ml in 10 mm acetate buffer, pH 4.5), mAb 17b (5 μg/ml in 10 mm acetate buffer, pH 5), or streptavidin (100 μg/ml in 10 mm acetate buffer, pH 4.2) was injected at 5 μl/min over one of the activated flow cells until levels of 650, 500, or 2,500 response units (RU) were, respectively, achieved. Biotinylated heparin was captured on the streptavidin surface to get a level of 25 RU. The fourth flow cell (functionalized with 2500 RU of streptavidin) served as a negative control. Samples in HBS-P (10 mm HEPES, 150 mm NaCl, and 0.005% surfactant P20, pH 7.4) were injected over the different surfaces at a flow rate of 10 μl/ml, after which the formed complexes were washed with HBS-P. Surfaces were regenerated by sequential injections of 10 mm HCl (1 min) and 2 m NaCl (2.5 min). Each binding curve was corrected for non-specific binding by subtraction of the signal obtained from the negative-control flow cell.
Mapping of Heparin/HS Binding Domains within gp120—Analysis of HBD was adapted from “the beads approach” previously described (37) but performed with free heparin in solution. Heparin (200 μg) was activated with 6 mm N-ethyl-N′-(diethylaminopropyl)-carbodiimide and 15 mm N-hydroxy-succimide for 12 min at room temperature. Reactants in excess were inactivated by the addition of β-mercaptoethanol (20 mm final) for 15 min. gp120 (3.5 μm) was incubated with 35 μm concentrations of the activated heparin in 100 μl of PBS for 2 h at room temperature, then the reaction was quenched by the addition of 11 μl of 1 m Tris, pH 7.5. Proteins cross-linked to heparin were digested overnight with 53 milliinternational units of thermolysin at 60 °C. Released peptides and thermolysin were removed by purification of the conjugates using weak anion exchange chromatography. Briefly, the sample was injected at 1 ml/min on to a 2 ml of DEAE-Sephacel column equilibrated with 20 mm Na2HPO4/NaH2PO4, pH 6.5 (solvent A). The column was washed with 5–10 column volumes of A and 5–10 column volumes of A supplemented with 0.3 m NaCl. Peptide-heparin conjugates were then eluted with buffer A containing 1 m NaCl. Eluted fractions (1 ml) were analyzed by monitoring the absorbance at 280 and 232 nm. In preliminary experiments biotinylated heparin was added in the mixture (as a tracer), and blotted fractions were analyzed using peroxidase-conjugated streptavidin. Peptide-heparin conjugate-containing fractions were pooled, desalted onto a PD-10 column, and identified by N-terminal sequencing as described (37).
RESULTS
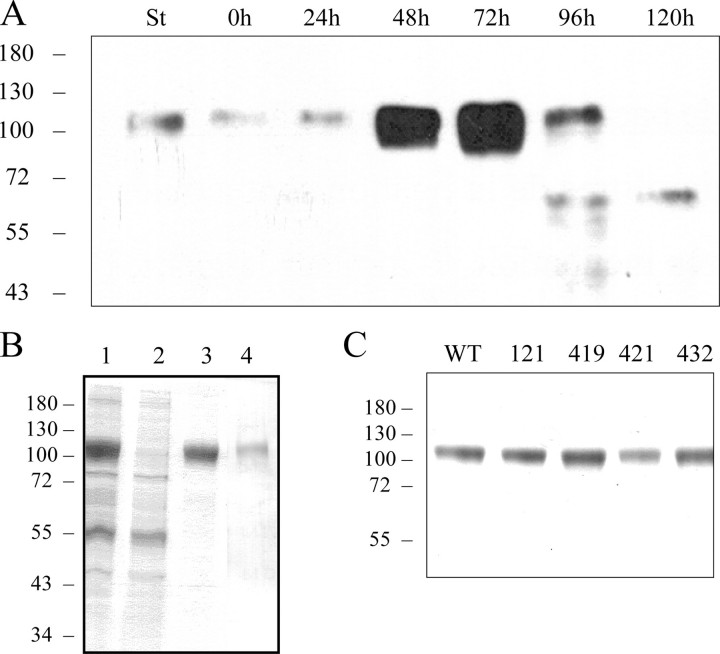
Time-Course Expression and Purification of Recombinant gp120HXBc2 in Sf21 Insect Cells—To produce the HIV envelope glycoprotein, recombinant baculoviruses encoding wild type or mutant gp120s in-frame with the melittin signal peptide were generated and used to infect Sf21 cells. The cell medium was harvested at different times post-infection and analyzed for gp120 expression by Western blotting. Immunoreactive material with a molecular mass of about 110–120 kDa was detected and migrated as purified gp120 used as a standard (Fig. 1A). Significant expression was observed 48 h after infection, with a maximum after 72 h. Longer infection times resulted in proteolytic degradation. Infection time chosen for subsequent experiments was 72 h. Under these conditions gp120 was secreted into the culture medium at an estimated level of 25–30 mg/liter. Similar amounts were produced using an expression vector, including a leader sequence derived from the baculovirus ecdysteroid UDP glucosyltransferase (data not shown).
FIGURE 1.
Time-course expression and purification of gp120. A, Sf21 cells were infected with gp120-baculoviruses. Samples were taken 0, 24, 48, 72, 96, and 120 h post-infection, analyzed on SDS-PAGE, then transferred to polyvinylidene difluoride membrane for Western blotting. St stands for “standard,” which corresponds to 100 ng of purified gp120. The wild type protein was purified from the culture medium of Sf21-infected cells in three steps as described under “Experimental Procedures.” Samples were analyzed by SDS-PAGE and Coomassie Blue staining, on which equivalent portions of the initial sample were loaded. B, lane 1, sample purified from culture medium by cation exchange chromatography; lane 2, wash-through from the Lentil-lectin resin column; lane 3, sample eluted from the Lentil-lectin affinity column; lane 4, gel filtration-purified sample. Mutants were purified using the same protocol. C, 3 μg of wild type or K121S-, R419S-, K421S-, and K432S-purified protein were analyzed by SDS-PAGE and revealed by Coomassie Blue staining. Molecular weight markers (in kDa) are indicated on the left of each panel.
For large scale preparation, wild type and mutant gp120s were expressed in Sf21 cells adapted for growth in suspension. Cells were harvested 72 h after infection and centrifuged, and the supernatant (1 liter) was run through a SP-Sepharose column. The column was first washed with 30 ml of 50 mm HEPES, pH 8.2, then with surfactant P20 to remove a baculovirus protein (egt) which otherwise co-eluted with gp120 during the next step (data not shown). Bound proteins were then eluted with a NaCl gradient, and the gp120-containing fractions were injected over a Lentil-lectin column. The flow-through fractions did not contain gp120 (Fig. 1B, lane 2), which was then eluted from the lectin column with 1 m methyl-α-d-mannopyranoside in PBS (Fig. 1B, lane 3). The material was further purified by gel filtration on a Superdex 200 HR column (Fig. 1B, lane 4). Fractions containing gp120 were pooled, concentrated, and stored at -20 °C. Affinity chromatography on the Lentil-lectin column was the most efficient purification step. Mutant proteins were purified using the same protocol, and purity was estimated to be at least 90% by Coomassie Blue staining on SDS-PAGE (Fig. 1C). Throughout the purification procedure, only the fractions corresponding to the apex of the eluted peaks were collected to ensure maximum purity. Purified proteins were finally quantified by amino acid analysis. This three step-purification protocol enabled us to get around 1 mg of purified gp120/liter of culture medium.
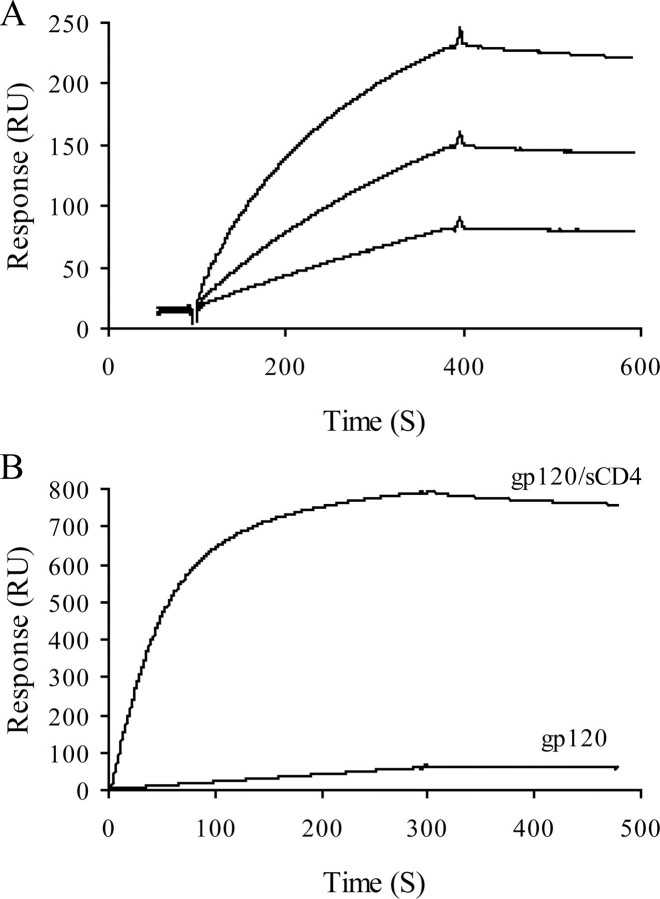
CD4 Binds to Wild Type gp120 and Triggers mAb 17b Recognition—To check whether the purified gp120 was correctly folded, we first investigated its ability to interact with CD4. Surface plasmon resonance experiments were conducted for that purpose, in which gp120 was flowed across a CD4-functionalized surface. Analysis of the resulting binding curves (Fig. 2A) returned an affinity of 8 nm, in very good agreement with previously reported data using the same technique (38, 39). To check whether our protein undergoes conformational changes upon CD4 binding, we monitored the binding of gp120 to mAb 17b, both in the presence and in the absence of sCD4. mAb 17b belongs to a group of antibodies referred as CD4-induced (CD4i), which bind to gp120 only after CD4 engagement. It recognizes a surface that includes or is proximal to the bridging sheet and, thus, competes with co-receptor binding (40). In the absence of sCD4, gp120 showed no or little binding to mAb 17b, indicating that in its CD4-unbound state the bridging sheet of gp120 remains unexposed or unfolded. Preincubation of gp120 with sCD4 dramatically increased binding to mAb 17b, indicating that sCD4 triggers the conformational changes that lead to the formation/exposure of the co-receptor binding site (Fig. 2B). Taken together, these results provide evidence that the protein was properly folded and functional.
FIGURE 2.
Binding of gp120 to immobilized CD4 and mAb 17b. Soluble CD4 and mAb 17b were immobilized on a CM4 sensor chip, and gp120 was injected over the chip surface. Injections were carried out in duplicate and returned similar results. A, binding curves for injection of gp120 (at 50, 100, and 200 nm, from bottom to top) over the CD4-activated surface. B, binding curves for injection of gp120 (50 nm) either alone or preincubated with 50 nm of sCD4 over the mAb 17b-activated surface. The binding response in RU was recorded as a function of time.
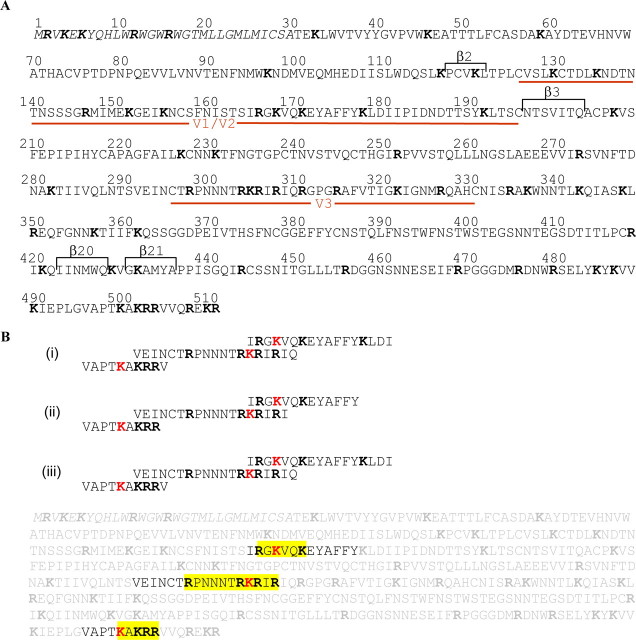
Mapping of Heparin Binding Domains within Wild Type gp120—Although the importance of the V3 loop for binding to heparin is well established, additional HBDs have been suggested but not identified yet. We, thus, performed a global analysis of the interaction between gp120 and heparin using a method adapted from the “beads sequencing approach” that has been previously reported (37). This approach was based on the capture of the protein of interest on heparin immobilized beads, the proteolytic digestion, and the N terminus sequencing of the peptides remaining attached to the heparin beads. However, because in some experiments beads were washed throughout the system despite the filter and blocked the sequencing machine, the cross-linking, the proteolysis, and the N terminus sequencing were now performed in solution. In addition, this enabled the use of defined oligosaccharides (instead of commercial heparin), and the results described below have been obtained with both full-length heparin and a heparin-derived 12-mer (dp12), used as a model of the S domains that characterize heparan sulfate (41). Carboxyl groups of soluble heparin (or dp12) were N-ethyl-N′-(diethylaminopropyl)-carbodiimide/N-hydroxysuccimide-activated, reacted with gp120, then formed protein/saccharide conjugates were submitted to proteolytic digestion. Peptides cross-linked to heparin were recovered by DEAE chromatography and analyzed by N-terminal sequencing. As shown in Fig. 3, results obtained with gp120 yielded three sequences, 165IRGKVQKEYAFFY177 (named HBD 1), 292VEINCTRPNNNTRKRIR308 (HBD 2), and 496VAPTKAKRR504 (HBD 3). Amino acids Lys168 within HBD 1, Lys305 within HBD 2, and Lys500 within HBD 3 were not detected, indicating that these were involved in the cross-linking with heparin, or dp12. The observation that a relatively small oligosaccharide (dp12) targets the same sequences and exactly returns the same results than full-length heparin indicates that having a large number of anionic charge does not give rise to additional (nonspecific) binding and supports the specificity of the assay (see Ref. 37).
FIGURE 3.
Mapping of gp120 heparin binding domain. A, amino acid sequence of gp120. Variable loops (V1/V2 and V3) are indicated as the β strands that fold together to form the CD4-induced bridging sheet. The amino acids are numbered according to the sequence of HXBc2 gp120, including the signal peptide (italics), with the mature full-length protein beginning at residue 31. B, sequence analysis of gp120 after immobilization on heparin and digestion with thermolysin. i and ii are the results from two different experiments, whereas iii is the results obtained when a 12-mers was used instead of full-length heparin. Amino acids involved in cross-linking, i.e. undetected Lys (K) residues, are in red. Lys residues shared by multiple sequences are in pink, as coupling to heparin could not be determined. Sequenced peptides are framed and pasted back onto the whole gp120 sequence (in pale gray), and identified HBD are boxed in yellow.
HBD 1 is located in the second half of V1/V2 loop, 20 residues upstream of the β3 strand that contributes, along with β2, β20, and β21 strands, to the CD4 induced epitope. HBD 2 is at the N terminus of the V3 loop, and HBD 3 in the C terminus of gp120 (C5 region). These data, thus, support the role of a motif within the V3 loop involved in heparin binding and, in addition, identified two other HBDs within the V2 loop and at the C terminus that have not been reported yet.
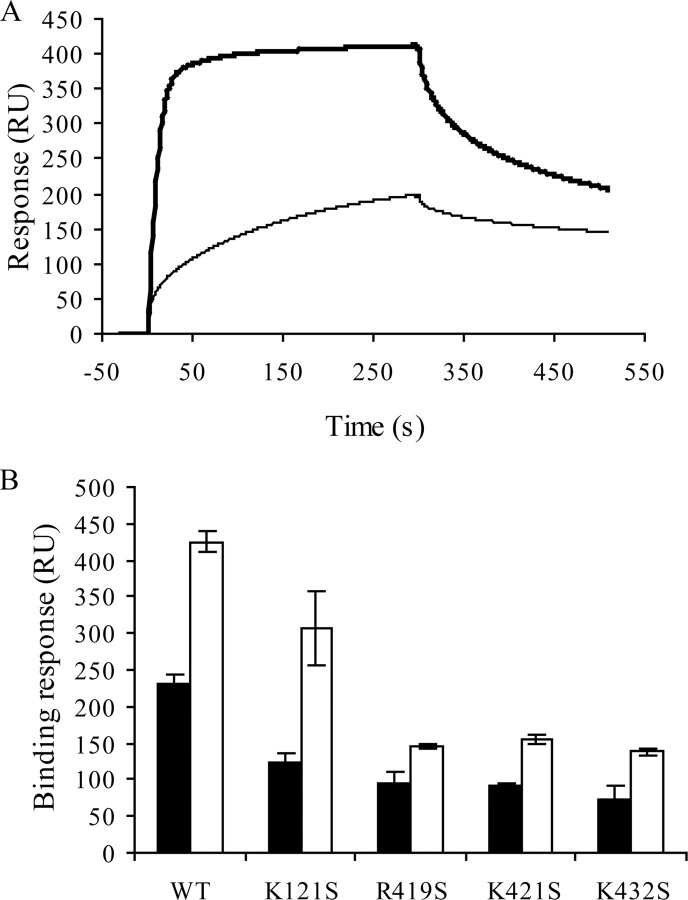
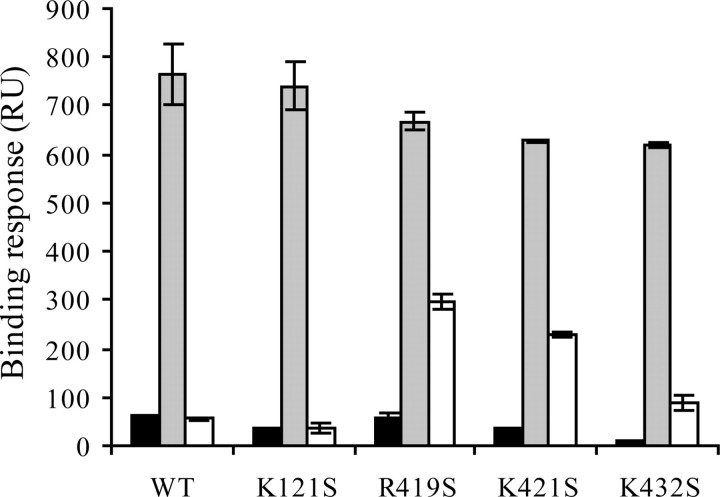
Mutation of Arg419, Lys421, and Lys432 but not Lys121 Decrease gp120 Binding to Heparin in the Presence of sCD4—Molecular modeling analysis, performed on a sCD4·gp120 complex revealed that a number of residues, located between the stems of the V1/V2 and V3 loops, form a cluster of positively charged residues organized as a possible HBD (33). These residues belong to the four-stranded “bridging sheet” structure, induced and stabilized by CD4, which importantly contribute to coreceptor binding. This binding surface, clearly a potential target for therapeutic interventions, was important to provide a better account of the residues possibly involved in heparin binding. To that aim, we generated mutant proteins in which each of the amino acids predicted to interact with heparin (Lys121, Arg419, Lys421, and Lys432) was changed to the Ser residue. Analysis of their heparin binding abilities using a Biacore-based binding assay did not show large differences (Fig. 4 and supplemental Fig. S1). This was expected, because in the absence of CD4 the mutated residues are only partially exposed and/or not clustered. This is also consistent with the availability of the HBD 1–3 on the unliganded gp120 surface. Bound to sCD4, wild type gp120 displayed a strongly enhanced binding to heparin, demonstrating the importance of the bridging sheet once exposed or folded for heparin recognition. Similarly, gp120-K121S showed a large increase in heparin binding, indicating that Lys121 was not significantly involved. In contrast, mutations of Arg419, Lys421, or Lys432 yielded proteins that only displayed a modest increase in heparin binding when bound to CD4 (Fig. 4 and supplemental Fig. S1), thus strongly suggesting that each of these residues was involved in the interaction. These experiments were repeated on a heparan sulfate-activated sensor chip, prepared as described (42), and gave rise to similar results (data not shown).
FIGURE 4.
Mutations within the CD4 induced region prevent efficient heparin binding activity of sCD4·gp120 complexes. A, wild type gp120 at 50 nm either alone (thin line) or preincubated with 50 nm of sCD4 (heavy line) were injected over a heparin-activated sensor chip at a flow rate of 10 μl/min, and the binding response in RU was recorded as a function of time. Mutant gp120s were similarly analyzed (see supplemental Fig. S1). B, binding responses for each sample were measured at the end of the association phases and are shown (in black for unliganded gp120 and in white for sCD4-bound gp120) as the mean ± S.E. of three independent experiments. WT, wild type.
Arg419, Lys421, and Lys432 within the CD4-induced Epitope Are Involved in Heparin Binding—A possible explanation of the above results would be that gp120 mutants became unable to expose the bridging sheet upon binding to CD4. To investigate this, we designed an assay in which we first measured the binding of gp120 to mAb 17b in the presence of sCD4 (which reveals the exposition of the bridging sheet) and then analyzed the ability of heparin to compete with mAb 17b (which reflects the binding of heparin to the induced epitope). We observed (Fig. 5 and supplemental Fig. S2) that none of the gp120 significantly bound to immobilized mAb 17b (30–60 RU). However, they all displayed a dramatic increase in binding once complexed to CD4. This first point indicated that gp120 mutants (as the wild type) were all able to expose a properly folded bridging sheet upon binding to CD4. We next investigated the ability of heparin to target this surface by evaluating, as a read out, the extent to which heparin could compete with mAb 17b. These experiments showed that heparin (200 nm) almost fully inhibited the binding of the sCD4·gp120 complex to mAb 17b for both wild type gp120 and K121S mutant (Fig. 5 and supplemental Fig. S2). This indicates that heparin efficiently binds to mAb 17b epitope even in the absence of Lys121 and, thus, supports that this residue is not involved in heparin binding. In contrast, for the three other mutants, especially R419S and K421S, heparin only partially inhibited the gp120/mAb 17b interaction (Fig. 5). Taken together, these data further demonstrated that a HBD is comprised within the CD4-induced epitope in which the Arg419 and Lys421 play a major role, whereas Lys121 is not involved.
FIGURE 5.
Mutations within the CD4i region prevent heparin inhibition of CD4i/mAb 17b interaction. Wild type (WT) or mutant gp120s (50 nm), alone (black bars), preincubated with 50 nm sCD4 (gray bars), or successively preincubated with sCD4 (50 nm) and heparin (200 nm) (white bars) were injected over a mAb 17b-activated sensor chip at a flow rate of 10 μl/min. Sensorgrams were recorded as a function of time (see supplemental Fig. S2), and the binding responses were measured at the end of the association phases. Data represent the mean ± S.E. of three independent experiments.
DISCUSSION
Several studies reported a role for HS in HIV attachment onto both CD4-positive and -negative cells. Up to now this interaction has been mainly attributed to the gp120 V3 loop, although evidence suggested that this particular domain does not fully recapitulate the binding activity of the protein. To investigate in more detail the structural basis of this interaction, we recombinantly produced wild type and mutant gp120 using the baculovirus expression system. To increase production yields, the rate-limiting natural leader sequence of gp120 was changed to that of the honeybee melittin (43). In preliminary studies, the production of gp120 was analyzed using four different insect cell lines: Sf21, Sf9, High Five (HF), and Mimic Sf9 cells. Sf21 and Sf9 gave similar results in terms of expression levels. HF cells allowed higher expression of the protein, but it migrated as a heterogeneous entity on SDS-PAGE. Finally, Mimic Sf9, an Sf9-derived insect cell line modified to stably express a variety of mammalian glycosyltransferases (44), failed in our hands to express significant amounts of gp120. Sf21 cells were, thus, used throughout the study and yielded to 25–30 mg of gp120 per liter of culture medium, from which a three purification step enabled us to routinely isolate 1 mg of pure and correctly refolded protein.
Characterization of gp120/HS interaction was then undertaken, first using a mapping strategy to identify HBDs. This method, which uses native proteins, is based on the formation of cross-linked complexes between the protein of interest and heparin beads, the proteolytic digestion of these complexes, and the subsequent identification of the HBD containing peptides by N terminus sequencing (37). Since this method was developed, it has been used to analyze a number of other proteins, including antithrombin III, PrP, or Adam 125 and successfully identified their HBDs. It is worth noting that for these proteins, as for those previously studied (37), nonspecific cross-linking was never observed in any of the experiments despite the presence of a large number of putative coupling lysine residues in the tested proteins. The use of heparin beads rather than soluble heparin facilitates the different washing steps. However, these commercial beads are restricted to heparin and are, thus, not adapted to the analysis of specific heparin-derived oligosaccharides or other glycosaminoglycans. In addition, beads were occasionally washed throughout the system despite the filter and blocked the sequencing machine. Here, we thus improved the method by performing the cross-linking, the proteolysis, and the N-terminal sequencing in solution. To recover the peptide/heparin complexes, a purification step (DEAE column) was included in the protocol between the proteolysis and the N-terminal sequencing. HIV-1 gp120 was analyzed with this method using either full-length heparin or a 12-mer heparin-derived oligosaccharide. Both samples returned similar results and consistently pointed out sequences including three cross-linked residues: Lys168, Lys305, and Lys500. Lys168 is within a small basic amino acid cluster RGKVQK (HBD 1; residues 166–171) located within the V2 loop. Lys305 takes part of a typical basic stretch of amino acids, RKRIR (HBD 2; residues 304–308) at the very beginning of the V3 loop. Interestingly, a few residues upstream of this sequence, Arg298 was recently found to be involved in HS recognition (45). Finally, Lys500 defined a third cluster of basic amino acids, KAKRR (HBD 3; residues 500–504), at the C-terminal domain of the protein. In addition to these, we previous reported that several residues of the CD4-induced bridging sheet (amino acids 121, 419, 421, and 432) are clustered into a potential HBD and, thus, could also contribute to the binding (33). We, thus, expressed gp120 in which these residues were mutated and found that three of them (419, 421, and 432) were actually involved in the interaction and, thus, constitute an additional binding domain (HBD 4).
Interestingly, all of the four domains targeted by heparin are functionally important. HBD 1 is located at the N terminus of the V2 loop, a domain of the protein that, along with the V3 loop, undergoes important structural changes after CD4 binding, resulting in the unmasking of the co-receptor binding site. The V3 loop, the structure of which has been recently solved in the context of the gp120 core (46), has a number of critical roles (for review, see Ref. 47). It comprises an immunodominant epitope, commonly known as the principal neutralization determinant (residues 308–322; therefore, next to the HBD). It determines co-receptor usage (CCR5 or CXCR4) and, thus, has a major influence on viral tropism and pathogenicity. In particular, basic amino acid enrichment of V3 is typical of a viral phenotype shift from CCR5 to CXCR4 usage, and this correlates with HS binding ability (32). Importantly, the V3 region also displays essential features for co-receptor binding. In particular, mutation of residues Arg298, Arg306, and Arg308, present in HBD 2, strongly decrease the ability of gp120 to interact with CXCR4 (48, 49). Regarding the C-terminal domain, it was reported that the replacement of all four basic amino acids of HBD 3 blocks the furin-mediated processing of gp160 into gp120 and gp41 (50), suggesting a relationship between the KAKRR cluster and the furin cleavage sequence. It is worth noting that the furin cleavage site (REKR; amino acids 508–511) is located only 3 residues downstream the KAKRR sequence. It has just been reported that synthetic peptides spanning the gp120/gp41 junction are only poor furin substrates in absence of heparin, and addition of the polysaccharide importantly increases cleavage of these peptides (51). It is, thus, tempting to propose that heparin binding to HBD 3, at least in the context of an isolated peptide, induces structural changes and better exposes the furin cleave site. However, proteolytic cleavage of gp160 into gp120 and gp41 normally occurs intracellularly (52), whereas HS is present at the cell surface; thus, the physiological significance of the above observations is not clear. Another possibility would be that these domains mediate virion attachment to cell surface HS, as has already been shown for the furin protease cleavage sequence of the Sindbis virus (53). Finally, HBD 4, only functional in the CD4-bound form of gp120, is included within the highly conserved co-receptor binding domain of the protein. One striking aspect of this domain is its basic nature, where seven basic amino acids are located within a relatively restricted surface (5). It is worth noting that the N-terminal domains of both CCR5 and CXCR4 co-receptor feature a number of sulfotyrosines that contribute to gp120 binding and HIV-1 entry. Similarly, a number of antibodies, targeting the bridging sheet that connects the inner and the outer domain of the CD4-bound form of the glycoprotein (CD4i epitope), display sulfotyrosines in their antigen binding site (54), supporting the present data. The binding of heparin with the CD4i region may, thus, mimic to some extent the interaction taking place with CD4i antibodies or the co-receptor N-terminal domain.
Finally, it is worth noting that three of the four heparin binding domains (HBD 1, 2, and 4) are located close to each other, at the proximity of or within the co-receptor binding site and are collectively involved in the conformational changes induced upon interaction with CD4. Although the function of these interactions is not yet completely known, it is clear that these three HBDs are targets of interest. A number of polyanionic, heparin-like molecules, are currently evaluated as HIV microbicides (55). In that context, the characterization of the different HBDs should help to better understand the activity of, or better design, such polyanionic compounds. In particular, the finding that the co-receptor binding region directly interacts with heparin after CD4 binding is an argument for the engineering of a CD4 molecule, covalently linked to heparin like oligosaccharides, which would target both the co-receptor and the CD4 binding site.
Supplementary Material
Acknowledgments
We thank Q. Sattentau and N. Thielens for the respective gifts of plasmid pSVIII-env and pNT-Bac, Estelle Grabit for technical assistance, and F. Véas for recombinant gp120 used as a standard in this study. Soluble CD4 was obtained through the National Institutes of Health AIDS Research and Reference Reagent Program from Dr. Sai Iyer, and mAb 17b was from the Centre for AIDS Reagent Program, National Institute for Biological Standards and Control.
This work was supported by the Agence Nationale de la Recherche sur le Syndrome d'Immunodéficience Acquise (ANRS), the CNRS, and the Commisariat à l'Energie Atomique. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
Footnotes
The abbreviations used are: HIV, human immunodeficiency virus; PBS, phosphate-buffered saline; HS, heparan sulfate; HBD, HS binding domain; mAb, monoclonal antibody; sCD4, soluble CD4; CD4i, CD4-induced; RU, response units.
R. R. Vivès, unpublished data.
References
- 1.Berger, E. A., Murphy, P. M., and Farber, J. M. (1999) Annu. Rev. Immunol. 17 657-700 [DOI] [PubMed] [Google Scholar]
- 2.Chen, B., Vogan, E. M., Gong, H., Skehel, J. J., Wiley, D. C., and Harrison, S. C. (2005) Nature 433 834-841 [DOI] [PubMed] [Google Scholar]
- 3.Kwong, P. D., Wyatt, R., Robinson, J., Sweet, R. W., Sodroski, J., and Hendrickson, W. A. (1998) Nature 393 648-659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Poignard, P., Saphire, E. O., Parren, P. W., and Burton, D. R. (2001) Annu. Rev. Immunol. 19 253-274 [DOI] [PubMed] [Google Scholar]
- 5.Rizzuto, C. D., Wyatt, R., Hernandez-Ramos, N., Sun, Y., Kwong, P. D., Hendrickson, W. A., and Sodroski, J. (1998) Science 280 1949-1953 [DOI] [PubMed] [Google Scholar]
- 6.Wu, L., Gerard, N. P., Wyatt, R., Choe, H., Parolin, C., Ruffing, N., Borsetti, A., Cardoso, A. A., Desjardin, E., Newman, W., Gerard, C., and Sodroski, J. (1996) Nature 384 179-183 [DOI] [PubMed] [Google Scholar]
- 7.Wyatt, R., and Sodroski, J. (1998) Science 280 1884-1888 [DOI] [PubMed] [Google Scholar]
- 8.Mondor, I., Ugolini, S., and Sattentau, Q. J. (1998) J. Virol. 72 3623-3634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bernfield, M., Gotte, M., Park, P. W., Reizes, O., Fitzgerald, M. L., Lincecum, J., and Zako, M. (1999) Annu. Rev. Biochem. 68 729-777 [DOI] [PubMed] [Google Scholar]
- 10.Whitelock, J. M., and Iozzo, R. V. (2005) Chem. Rev. 105 2745-2764 [DOI] [PubMed] [Google Scholar]
- 11.Liu, J., and Thorp, S. C. (2002) Med. Res. Rev. 22 1-25 [DOI] [PubMed] [Google Scholar]
- 12.Spillmann, D. (2001) Biochimie (Paris) 83 811-817 [DOI] [PubMed] [Google Scholar]
- 13.Vives, R. R., Lortat-Jacob, H., and Fender, P. (2006) Curr. Gene Ther. 6 35-44 [DOI] [PubMed] [Google Scholar]
- 14.Harrop, H. A., and Rider, C. C. (1998) Glycobiology 8 131-137 [DOI] [PubMed] [Google Scholar]
- 15.Roderiquez, G., Oravecz, T., Yanagishita, M., Bou-Habib, D. C., Mostowski, H., and Norcross, M. A. (1995) J. Virol. 69 2233-2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bomsel, M., and Alfsen, A. (2003) Nat. Rev. Mol. Cell Biol. 4 57-68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saidi, H., Magri, G., Nasreddine, N., Requena, M., and Belec, L. (2007) Virology 358 55-68 [DOI] [PubMed] [Google Scholar]
- 18.Wu, Z., Chen, Z., and Phillips, D. M. (2003) J. Infect. Dis. 188 1473-1482 [DOI] [PubMed] [Google Scholar]
- 19.Argyris, E. G., Acheampong, E., Nunnari, G., Mukhtar, M., Williams, K. J., and Pomerantz, R. J. (2003) J. Virol. 77 12140-12151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Banks, W. A., Robinson, S. M., Wolf, K. M., Bess, J. W., Jr., and Arthur, L. O. (2004) Neuroscience 128 143-153 [DOI] [PubMed] [Google Scholar]
- 21.Bobardt, M. D., Salmon, P., Wang, L., Esko, J. D., Gabuzda, D., Fiala, M., Trono, D., Van der Schueren, B., David, G., and Gallay, P. A. (2004) J. Virol. 78 6567-6584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bobardt, M. D., Saphire, A. C., Hung, H. C., Yu, X., Van der Schueren, B., Zhang, Z., David, G., and Gallay, P. A. (2003) Immunity 18 27-39 [DOI] [PubMed] [Google Scholar]
- 23.Olinger, G. G., Saifuddin, M., and Spear, G. T. (2000) J. Virol. 74 8550-8557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saphire, A. C., Bobardt, M. D., Zhang, Z., David, G., and Gallay, P. A. (2001) J. Virol. 75 9187-9200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alvarez Losada, S., Canto-Nogues, C., and Munoz-Fernandez, M. A. (2002) Neurobiol. Dis. 11 469-478 [DOI] [PubMed] [Google Scholar]
- 26.Barbouche, R., Lortat-Jacob, H., Jones, I. M., and Fenouillet, E. (2005) Mol. Pharmacol. 67 1111-1118 [DOI] [PubMed] [Google Scholar]
- 27.Bugatti, A., Urbinati, C., Ravelli, C., De Clercq, E., Liekens, S., and Rusnati, M. (2007) Antimicrob. Agents Chemother. 51 2337-2345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Batinic, D., and Robey, F. A. (1992) J. Biol. Chem. 267 6664-6671 [PubMed] [Google Scholar]
- 29.Callahan, L. N., Phelan, M., Mallinson, M., and Norcross, M. A. (1991) J. Virol. 65 1543-1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okada, T., Patterson, B. K., and Gurney, M. E. (1995) Biochem. Biophys. Res. Commun. 209 850-858 [DOI] [PubMed] [Google Scholar]
- 31.Rider, C. C., Coombe, D. R., Harrop, H. A., Hounsell, E. F., Bauer, C., Feeney, J., Mulloy, B., Mahmood, N., Hay, A., and Parish, C. R. (1994) Biochemistry 33 6974-6980 [DOI] [PubMed] [Google Scholar]
- 32.Moulard, M., Lortat-Jacob, H., Mondor, I., Roca, G., Wyatt, R., Sodroski, J., Zhao, L., Olson, W., Kwong, P. D., and Sattentau, Q. J. (2000) J. Virol. 74 1948-1960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vives, R. R., Imberty, A., Sattentau, Q. J., and Lortat-Jacob, H. (2005) J. Biol. Chem. 280 21353-21357 [DOI] [PubMed] [Google Scholar]
- 34.Este, J. A., Schols, D., De Vreese, K., Van Laethem, K., Vandamme, A. M., Desmyter, J., and De Clercq, E. (1997) Mol. Pharmacol. 52 98-104 [DOI] [PubMed] [Google Scholar]
- 35.Vives, R. R., Sadir, R., Imberty, A., Rencurosi, A., and Lortat-Jacob, H. (2002) Biochemistry 41 14779-14789 [DOI] [PubMed] [Google Scholar]
- 36.King, L. A., and Possee, R. D. (1992) The Baculovirus Expression System, pp. 111-114, Chapman & Hall, London
- 37.Vives, R. R., Crublet, E., Andrieu, J. P., Gagnon, J., Rousselle, P., and Lortat-Jacob, H. (2004) J. Biol. Chem. 279 54327-54333 [DOI] [PubMed] [Google Scholar]
- 38.Wu, H., Myszka, D. G., Tendian, S. W., Brouillette, C. G., Sweet, R. W., Chaiken, I. M., and Hendrickson, W. A. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 15030-15035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang, W., Godillot, A. P., Wyatt, R., Sodroski, J., and Chaiken, I. (2001) Biochemistry 40 1662-1670 [DOI] [PubMed] [Google Scholar]
- 40.Thali, M., Moore, J. P., Furman, C., Charles, M., Ho, D. D., Robinson, J., and Sodroski, J. (1993) J. Virol. 67 3978-3988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murphy, K. J., Merry, C. L., Lyon, M., Thompson, J. E., Roberts, I. S., and Gallagher, J. T. (2004) J. Biol. Chem. 279 27239-27245 [DOI] [PubMed] [Google Scholar]
- 42.Laguri, C., Sadir, R., Rueda, P., Baleux, F., Gans, P., Arenzana-Seisdedos, F., and Lortat-Jacob, H. (2007) PLoS ONE 2 e1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li, Y., Luo, L., Thomas, D. Y., and Kang, C. Y. (1994) Virology 204 266-278 [DOI] [PubMed] [Google Scholar]
- 44.Hollister, J., Grabenhorst, E., Nimtz, M., Conradt, H., and Jarvis, D. L. (2002) Biochemistry 41 15093-15104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de Parseval, A., Bobardt, M. D., Chatterji, A., Chatterji, U., Elder, J. H., David, G., Zolla-Pazner, S., Farzan, M., Lee, T. H., and Gallay, P. A. (2005) J. Biol. Chem. 280 39493-39504 [DOI] [PubMed] [Google Scholar]
- 46.Huang, C. C., Tang, M., Zhang, M. Y., Majeed, S., Montabana, E., Stanfield, R. L., Dimitrov, D. S., Korber, B., Sodroski, J., Wilson, I. A., Wyatt, R., and Kwong, P. D. (2005) Science 310 1025-1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hartley, O., Klasse, P. J., Sattentau, Q. J., and Moore, J. P. (2005) AIDS Res. Hum. Retroviruses 21 171-189 [DOI] [PubMed] [Google Scholar]
- 48.Basmaciogullari, S., Babcock, G. J., Van Ryk, D., Wojtowicz, W., and Sodroski, J. (2002) J. Virol. 76 10791-10800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cormier, E. G., and Dragic, T. (2002) J. Virol. 76 8953-8957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bosch, V., and Pawlita, M. (1990) J. Virol. 64 2337-2344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pasquato, A., Dettin, M., Basak, A., Gambaretto, R., Tonin, L., Seidah, N. G., and Di Bello, C. (2007) FEBS Lett. 581 5807-5813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moulard, M., and Decroly, E. (2000) Biochim. Biophys. Acta 1469 121-132 [DOI] [PubMed] [Google Scholar]
- 53.Klimstra, W. B., Heidner, H. W., and Johnston, R. E. (1999) J. Virol. 73 6299-6306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang, C. C., Lam, S. N., Acharya, P., Tang, M., Xiang, S. H., Hussan, S. S., Stanfield, R. L., Robinson, J., Sodroski, J., Wilson, I. A., Wyatt, R., Bewley, C. A., and Kwong, P. D. (2007) Science 317 1930-1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Luscher-Mattli, M. (2000) Antiviral Chem. Chemother. 11 249-259 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.