During neuronal differentiation, the REST/CTDSP complex must be inactivated to allow neuronal gene expression. The REST/CTDSP complex silences neuronal genes and contains one of a group of phosphatases (CTDSPs) that inhibit PolII transcription in nonneuronal cells. In this study, Fischer and colleagues demonstrate that the expression of CTDSPs is subject to a novel form of post-transcriptional regulation. The zebrafish ctdsp2 primary transcript contains miR-26b in one of its introns, and miR-26b directly inhibits the mature ctdsp2 mRNA. In neural stem cells and nonneuronal cells, this negative feedback loop is inactive. During neurogenesis, however, the mature form of miR-26b is produced and silences ctdsp2 mRNA, and this repression is required for neuronal cell differentiation in vivo. Thus, this study describes a novel in vivo post-transcriptional feedback mechanism dependent on an intronic microRNA.
Keywords: miR-26b, ctdsp2; REST, neurogenesis, neuronal differentiation, zebrafish
Abstract
Differentiation of neural stem cells (NSCs) to neurons requires the activation of genes controlled by the repressor element 1 (RE1) silencing transcription factor (REST)/neuron-restrictive silencer factor (NRSF) protein complex. Important components of REST/NRSF are phosphatases (termed RNA polymerase II C-terminal domain small phosphatases [CTDSPs]) that inhibit RNA polymerase II and suppress neuronal gene expression in NSCs. Activation of genes controlled by CTDSPs is required for neurogenesis, but how this is achieved is not fully understood. Here we show that ctdsp2 is a target of miR-26b, a microRNA that is encoded in an intron of the ctdsp2 primary transcript. This intrinsic negative feedback loop is inactive in NSCs because miR-26b biogenesis is inhibited at the precursor level. Generation of mature miR-26b is activated during neurogenesis, where it suppresses Ctdsp2 protein expression and is required for neuronal cell differentiation in vivo.
Generation and maintenance of terminally differentiated neurons are controlled by a complex gene regulatory network. In nonneuronal cells and neural stem cells (NSCs), the neuronal gene expression program is suppressed by the action of the repressor element 1 (RE1) silencing transcription factor (REST) complex (Chong et al. 1995; Schoenherr and Anderson 1995). REST binds to RE1-containing genes (Kraner et al. 1992; Mori et al. 1992) and causes their silencing via at least two distinct mechanisms. First, it nucleates a chromatin remodeling complex leading to the formation of transcriptionally inactive heterochromatin and long-term epigenetic silencing (Naruse et al. 1999; Hakimi et al. 2002). The second mechanism involves the activity of three closely related phosphatases, termed CTDSP1, CTDSP2, and CTDSPL. As part of the REST complex, these enzymes dephosphorylate the C-terminal domain (CTD) of RNA polymerase II (PolII) and thereby inhibit the expression of genes that have an RE1 signature sequence (Yeo et al. 2005; Visvanathan et al. 2007). During neural fate commitment and terminal differentiation, the REST/CTDSP pathway is gradually inactivated to allow the expression of RE1-containing genes (Ballas et al. 2005; Yeo et al. 2005; Visvanathan et al. 2007). Neuronal gene activation in mice and humans is known to depend on the action of miR-124, which prevents expression of CTDSPs (Lim et al. 2005; Conaco et al. 2006; Visvanathan et al. 2007). However, as shown in mice, the neuron-specific miR-124 is itself repressed by the REST/CTDSP pathway, since all of its loci contain an RE1 (Conaco et al. 2006). As mRNAs often are under the control of a collection of microRNAs (miRNAs) (Krek et al. 2005), we therefore hypothesized that additional miRNAs might be involved in the regulation of CTDSP activity.
Results and Discussion
Intronic miR-26b represses its host gene, ctdsp2
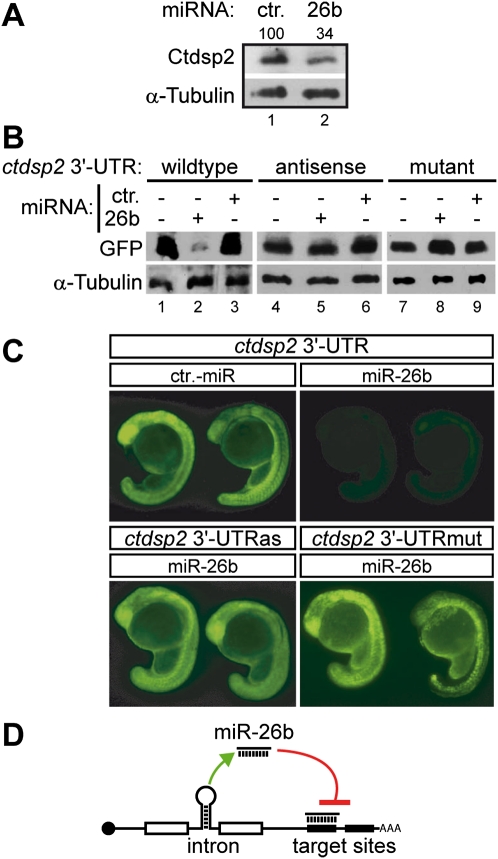
Having previously established a zebrafish model to study the impact of post-transcriptional mRNA metabolism on neurons (Winkler et al. 2005), we analyzed neuron-enriched miRNAs for their ability to regulate CTDSPs in this organism. Target site predictions revealed the presence of five potential binding sites for miR-26b in the 3′ untranslated region (UTR) of the zebrafish ctdsp2 transcript (Supplemental Fig. S1). To test whether miR-26b represses Ctdsp2 protein expression in vivo, a synthetic miR-26b mimic was injected into one-cell stage embryos. Thirty-one hours post-fertilization (hpf), Ctdsp2 expression was analyzed by Western blotting of whole-embryo extracts using affinity-purified Ctdsp2 antibodies. Exogenous miR-26b led to reduced Ctdsp2 expression when compared with animals injected with a control miRNA (Fig. 1A). Next, we asked whether this was due to direct interaction between miR-26b and its predicted target sites within the 3′ UTR of ctdsp2 mRNA. A set of reporter constructs was generated, which contained the cDNA encoding green fluorescent protein (GFP) and the 3′ UTR of ctdsp2 either in the sense orientation, in the antisense orientation, or with point mutations in the thermodynamically most stable miR-26b-binding site (according to RNAhybrid) (Supplemental Fig. S1A). Reporter mRNAs were transcribed in vitro and coinjected with synthetic miR-26b or a control miRNA. In control-injected animals, strong and ubiquitous GFP expression of all three reporters was observed at 24 hpf by Western blotting and fluorescence microscopy (Fig. 1B,C; Supplemental Fig. S2). In contrast, injection of miR-26b strongly repressed GFP expression from the wild-type reporter, but not from reporters containing the antisense or point-mutated 3′ UTRs (Fig. 1B,C; Supplemental Fig. S2). These data indicated that miR-26b silences ctdsp2 through binding to its 3′ UTR. Interestingly, miR-26b is located in an intron of ctdsp2 (Supplemental Fig. S1B) and has been shown to lack independent transcription start sites in humans (Monteys et al. 2010). Rather, it is coexpressed with its host gene (Baskerville and Bartel 2005) and can be processed by the endonuclease Drosha during the splicing reaction (Kim and Kim 2007). The ctdsp2 transcript is thus both host and target for miR-26b and object of an intrinsic inhibitory feedback loop (Fig. 1D).
Figure 1.
MiR-26b represses expression of ctdsp2 mRNA in vivo. (A) Immunodetection of endogenous Ctdsp2 protein and α-Tubulin (loading control) in extracts of 31-hpf zebrafish embryos injected with control miRNA or miR-26b. The percentage of remaining Ctdsp2 in miR-26b-injected animals normalized to the control is indicated. (B,C) GFP expression in zebrafish embryos from injected reporter mRNAs containing either the wild-type, antisense, or mutated 3′ UTR of ctdsp2-mRNA. The expression of GFP in the presence of coinjected control miRNA or miR-26b was assessed by Western blotting (B) and fluorescence microscopy (C). miR-26b inhibits exclusively expression of the reporter fused to the wild-type sequence. (D) Schematic illustration of the intrinsic ctdsp2 feedback loop.
Maturation of miR-26b is activated during neurogenesis
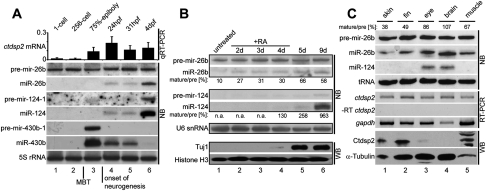
Steady coexpression of miR-26b and ctdsp2 mRNA from the same primary transcript would, however, constitutively reduce the amount of Ctdsp2 protein. Given this, we reasoned that the expression of miR-26b itself might be controlled post-transcriptionally as reported for other miRNAs (Siomi and Siomi 2010). Such a mechanism would allow cell type- and developmental stage-specific regulation of miRNA activity (Rybak et al. 2008). We therefore analyzed miR-26b biogenesis during zebrafish embryonic development. Canonical miRNA biogenesis involves two endonucleolytic cleavage reactions: First, Drosha releases the ∼70-nucleotide (nt) stem–loop precursor miRNA (pre-miRNA) from the primary transcript. Second, the mature ∼21-nt miRNA duplex is generated by Dicer cleavage. The ctdsp2 transcript harboring miR-26b was first detected after the onset of zygotic transcription at the midblastula transition (MBT). Interestingly, Northern blotting of whole-embryo RNA revealed that the pre-miRNA stem–loop of miR-26b (pre-mir-26b) was present from the one-cell stage on and expressed throughout embryonic development (Fig. 2A, lanes 1–6). In contrast, mature miR-26b was observed only at stages later than 24 hpf (Fig. 2A, lanes 4–6). Two lines of evidence indicated that this discrepancy was indeed due to the regulated processing of pre-mir-26b. First, pre-mir-26b was maternally expressed and detected at similar levels before the onset of zygotic transcription (Fig. 2A, cf. lanes 1 and 2). This observation argued against constant processing and increased turnover of miR-26b. Second, steady precursor levels were not due to generally reduced Dicer activity, as pre-mir-430b-1, a precursor of the early zygotic miR-430b that silences maternal transcripts (Giraldez et al. 2005), was processed efficiently at the 75% epiboly stage, at which mature miR-26b was still absent (Fig. 2A, lane 3). Hence, pre-mir-26b processing was specifically inhibited in early zebrafish embryos.
Figure 2.
Activation of miR-26b maturation in neuronal cells. (A) Expression profiling of ctdsp2 mRNA and miR-26b in zebrafish larval development. (Top panel) qRT–PCR analysis of ctdsp2 mRNA levels relative to gapdh. (Bottom panel) Northern blot (NB) detection of miRNAs with probes against precursor and mature miRNA-26b, miRNA-124, and miRNA-430b. 5S ribosomal RNA served as loading control. The MBT and onset of neurogenesis are indicated. (B) pre-mir-26b processing during RA-induced neuronal differentiation of mouse P19 cells. RNA from different time points was extracted, and expression of the indicated miRNAs was assessed by Northern blotting. Ratios of mature miRNAs to precursors were determined by densitometry. (n.a.) Not applicable due to expression below detection limit. As a marker for terminally differentiated neurons, expression of the Tuj1 antigen was analyzed by Western blotting. Histone H3 served as loading control. (C) Expression profiles of the indicated factors in isolated adult zebrafish tissues as detected by Northern blotting (NB), RT–PCR, or Western blotting (WB). The ratios of mature miR-26b to its precursors are indicated at the top.
Most remarkably, the appearance of mature miR-26b at 24 hpf (Fig. 2A, lane 4) coincided with the generation of cells of the neuronal lineage in zebrafish. This was evident by the identical expression profile of miR-124 (Fig. 2A, lanes 4–6), which is expressed exclusively in neuronal tissue (Wienholds et al. 2005). To specifically analyze miR-26b processing during neuronal differentiation, we assessed its biogenesis intermediates using a paradigm for neuronal development; i.e., the retinoic acid (RA)-induced differentiation of mouse P19 teratocarcinoma cells (Jones-Villeneuve et al. 1982). Consistent with previous reports (Conaco et al. 2006), untreated cells were devoid of the neuronal marker Tuj1 and mature miR-124, confirming their nonneuronal character (Fig. 2B, lane 1). The corresponding pre-mir-124-2 precursor was likewise absent, suggesting that this was due to transcriptional repression (Fig. 2B, lane 1), presumably mediated by REST (Conaco et al. 2006). In contrast, pre-mir-26b was robustly expressed but was inhibited post-transcriptionally, as evident by only minute amounts of mature miR-26b (Fig. 2B, lane 1). However, RA-induced neuronal differentiation led to a gradual increase in the ratio of mature miR-26b to its precursor (Fig. 2B, lanes 1–6; Supplemental Fig. S3). Therefore, the biogenesis of miR-26b is controlled during neuronal differentiation at the level of precursor processing. To test whether pre-mir-26b processing is also regulated in fully developed tissues, we analyzed RNA from dissected adult zebrafish. RT–PCR and in situ hybridization (ISH) revealed that ctdsp2 mRNA was expressed ubiquitously (Fig. 2C; Supplemental Fig. S4). Likewise, pre-mir-26b was detected in all tissues tested, indicating that Drosha processing occurred ubiquitously (Fig. 2C). In contrast, mature miR-26b was ubiquitously expressed but was enriched in eye and brain tissue (Fig. 2C, lanes 3,4; Supplemental Fig. S4). This pattern was reciprocal to the amount of Ctdsp2 protein in these tissues. Hence, Ctdsp2 repression in neuronal tissue is likely to be caused by the combined action of miR-26b and miR-124 (Fig. 2C, lanes 3,4). Taken together, our analysis suggests that pre-mir-26b is coexpressed with its ctdsp2 host transcript, but processing to mature miR-26b is inhibited in NSCs and nonneuronal tissues.
Inactivation of miR-26b leads to impaired neurogenesis
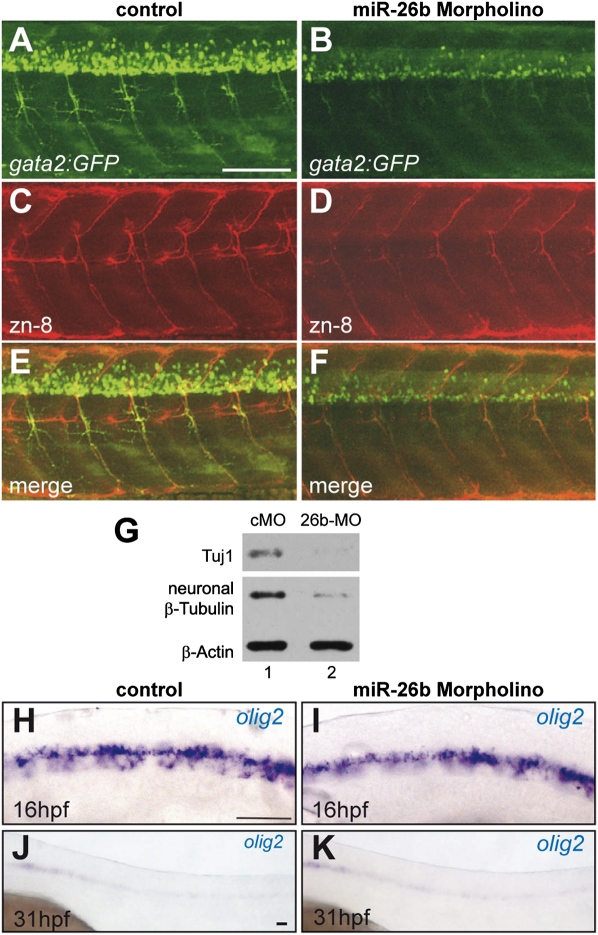
As miR-26b represses Ctdsp2 (Fig. 1), the increased production of mature miRNA during neuronal differentiation (Fig. 2) might contribute to neurogenesis. To test this, miR-26b expression was reduced by an antisense morpholino in zebrafish larvae, and neurogenesis was analyzed. Control morpholino (cMO) or miR-26b morpholino (26b-MO) was injected into gata2:GFP transgenic embryos. These fish express GFP in ventrally projecting secondary motor neurons of the developing spinal cord (Meng et al. 1997). Motor neuron formation was assessed 60 hpf by the analysis of GFP expression and immunostaining with zn-8 antibody, a marker for secondary motor axons (Fig. 3A–F). Morpholino-mediated down-regulation of miR-26b (Supplemental Fig. S5A) interfered with the formation of secondary motor neurons, as evident by reduced GFP-positive cells (Fig. 3A–F). Zn-8 staining was likewise reduced, but only subtle changes in the morphology of axonal fascicles were observed, consistent with a decreased number of differentiated motor neurons (Fig. 3C,D; Supplemental Fig. S5B for higher magnification). The few motor neurons formed developed normally. As this pointed toward a defect in terminal differentiation, key markers of neuronal lineage specification were analyzed: First, the expression of neuronal β-Tubulins, which are among the earliest markers for post-mitotic neurons (Fanarraga et al. 1999), was tested by Western blotting with two specific antibodies (Tuj1 and purified native brain class III Tubulin antibody M154) (Fig. 3G) and by tubb5 ISH (Supplemental Fig. S5C). This revealed a strong reduction of neuronal β-Tubulin expression, indicating a defect in terminal differentiation. Of note, tubb5 and the Tuj1 antigen are expressed pan-neuronally, pointing toward a general defect in neuronal cell differentiation that was not restricted to motor neurons. Second, to detect effects on the pool of neural progenitor cells (NPCs) that give rise to both motor neurons and oligodendrocyte precursors (Lu et al. 2002), expression of the olig2 marker was analyzed by ISH. Formation and maintenance of the NPC population was unaffected by 26b-MO injection, as delineated from robust olig2 detection at 16 hpf and a staining pattern in the ventral spinal cord that was indistinguishable from controls (Fig. 3H,I). At 31 hpf, olig2-positive NPCs were diminished in morphant and control animals, indicating that no accumulation of these cells occurred in miR-26b morphants (Fig. 3J,K). In addition, quantitative RT– PCR (qRT–PCR) and Western blotting failed to detect a severe increase of olig2 expression in the miR-26b morphants (Supplemental Fig. S6). Rather, increased TUNEL staining in the spinal cord, eye, and brain hinted toward apoptotic clearance of these differentiation-blocked cells (Supplemental Fig. S7). Consistent with a function in the derepression of neuronal genes, these data suggest that interfering with miR-26b function affects the terminal differentiation of neurons.
Figure 3.
Inactivation of miR-26b in zebrafish leads to defects in neuronal differentiation. (A–F) Confocal analysis of spinal motor neurons of gata2:GFP transgenic zebrafish at 60 hpf injected with either a cMO (A,C,E) or a 26b-MO (B,D,F). A and B show GFP-labeled motor neurons, and C and D show immunostaining of axons with the zn-8 marker; the overlay is shown in E and F. (G) Reduction of markers of post-mitotic neurons in whole-embryo extracts of 26b-MO-injected animals. Immunodetection of neuronal β-tubulin on Western blots of extracts from animals injected with cMO (lane 1) or 26b-MO (lane 2). Two different antibodies were used (Tuj1 and native brain Tubulin antibody M154), and β-Actin served as a loading control. (H–K) ISH of olig2-positive motor neuron precursor cells in cMo-injected (H,J) and 26b-MO-injected (I,K) embryos at 16 hpf (H,I) and 31 hpf (J,K). Bars, 100 μm.
miR-26b-mediated repression of ctdsp2 promotes neuronal differentiation
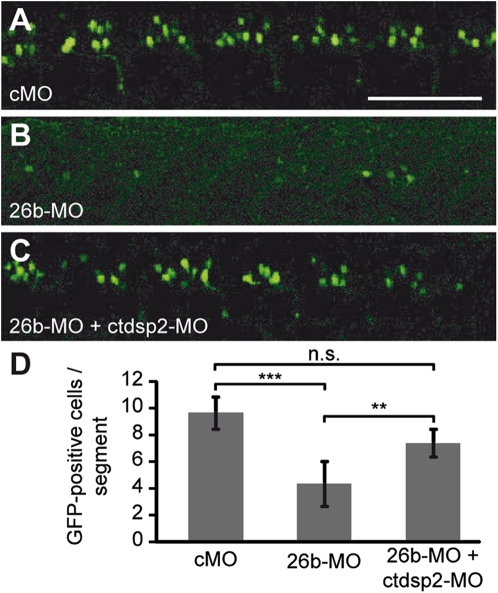
As the vast majority of miRNAs have several potential target transcripts (Lim et al. 2005), we wanted to address whether it is indeed the reduced silencing of ctdsp2 that is responsible for the block in neuronal differentiation observed in 26b-MO-injected animals. If so, other modes to reduce ctdsp2 expression (i.e., miR-26b-independent modes) would be predicted to compete with 26b-MO activity and restore neurogenesis. To test this hypothesis, 26b-MO was coinjected with an antisense morpholino that represses ctdsp2 translation (ctdsp2-MO) into gata2:GFP transgenic embryos. At 31 hpf, the number of GFP-positive motor neurons in these animals was assessed by confocal microscopy. The reduction of differentiated neurons by 26b-MO injection alone that was already observed for 60-hpf larvae (Fig. 3A–F) was confirmed at 31 hpf and quantified (Fig. 4A,B,D). Strikingly, the global repression of Ctdsp2 by the coinjection of ctdsp2-MO (Supplemental Fig. S8) resulted in a significant increase in the number of motor neurons (Fig. 4C,D). Therefore, the miR-26b-mediated repression of ctdsp2 directly contributes to neuronal differentiation.
Figure 4.
MiR-26b promotes neuronal differentiation via down-regulation of ctdsp2. (A–C) Confocal microscopy of gata2:GFP transgenic embryos at 31 hpf injected with either cMO (A), 26b-MO (B), or a mixture of a ctdsp2 translation-blocking morpholino (ctdsp2-MO) and 26b-MO (C). Bar, 100 μm. (D) Quantitative analysis of the number of motor neuron cell bodies per body segment shows that the ctdsp2-MO-mediated rescue is statistically significant. Mean ± SD, paired t-test; (***) highly significant; (**) significant; (n.s.) not significant; cMO, n = 50; 26b-MO, n = 65; 26b-MO + ctdsp2-MO, n = 25.
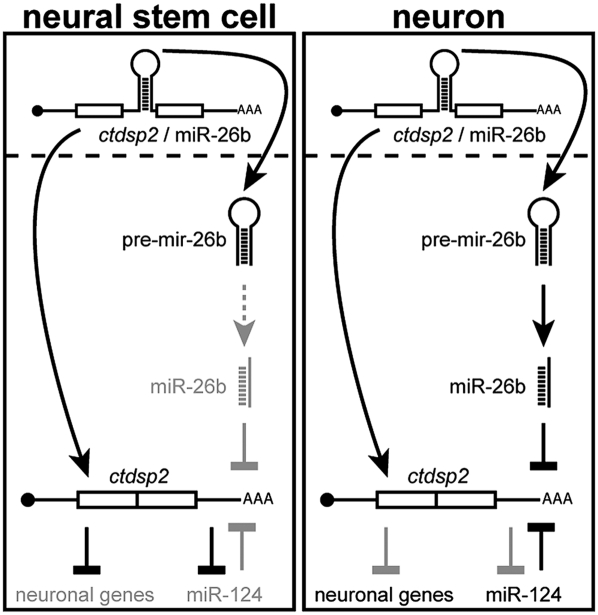
Our study uncovers a role for the regulated biogenesis of miR-26b in the control of the REST/CTDSP pathway during neuronal differentiation. In this regulatory mechanism, the primary transcript of the ctdsp2/miR-26b locus serves a dual function as both pre-mRNA and pri-miRNA. While splicing toward ctdsp2 mRNA proceeds normally, the biogenesis of miR-26b is inhibited after excision of the pre-miR26b precursor (Fig. 5, left panel). During neuronal differentiation, this processing block is relieved, and as a consequence, levels of mature miR-26b rise, conferring efficient repression of ctdsp2 mRNA (Fig. 5, right panel). This leads to the derepression of RE1-containing genes (Yeo et al. 2005), including miR-124 (Supplemental Fig. S8), which exerts additional negative feedback to ctdsp2 expression (Supplemental Fig. S8C,D). These events cooperate to allow the neuronal gene expression program.
Figure 5.
Schematic of the ctdsp2/miR-26b regulatory cascade. Light gray indicates inhibited pathways. See the text for details.
The molecular mechanisms that control the biogenesis of mature miR-26b are currently unknown. Of note, the regulatory processing factors of the let-7 miRNA family, TUT4 and KSRP, have already been implicated in the regulation of miR-26 family members (Jones et al. 2009; Trabucchi et al. 2009). It will be interesting to elucidate whether these factors play a similar role in the timed processing of miR-26b during neuronal differentiation. Irrespective of the factors involved, the miR-26b feedback system solely relies on post-transcriptional mechanisms, as both the mRNA target and its miRNA repressor are transcribed as a single physical entity. Such ultrashort feedback loops have been predicted by the analysis of gene regulatory networks containing intronic miRNAs (Tsang et al. 2007), but experimental evidence remained elusive. It is tempting to speculate that this intrinsic post-transcriptional inhibitory circuit has evolved to reduce the susceptibility of the REST/CTDSP pathway to transcriptional noise (Raj and van Oudenaarden 2008), which might otherwise interfere with the fine-tuned low-level expression required for neurogenesis (Ballas et al. 2005). In line with this idea, all vertebrate genes that encode CTDSP homologs contain a miRNA of the miR-26 family in one of their introns (Supplemental Fig. S9A). Furthermore, our data suggest that zebrafish miR-26a is also ubiquitously expressed (Supplemental Fig. S9B), subject to regulated precursor processing during neuronal differentiation (Supplemental Fig. S9C), and able to silence the ctdsp2 3′ UTR reporter (Supplemental Fig. S10). Additionally, large-scale analysis revealed a miR-26 target site in the Ctdsp2 3′ UTR that is bound by a miRNA silencing complex in the mouse brain (Chi et al. 2009). Finally, the regulated feedback by miR-26a and miR-26b might be important not only in neurons, but for proliferation/differentiation events in general, as both family members have been implicated with differentiation of other cell types, stemness, and cancer (Wong and Tellam 2008; Huse et al. 2009; Kota et al. 2009; Ma et al. 2010).
Materials and methods
miRNA target site prediction and sequence alignment
Putative miRNA-binding sites were predicted using the RNAhybrid algorithm (Rehmsmeier et al. 2004).
Fish maintenance and injection
See the Supplemental Material.
Image acquisition and statistics
Microscopic observations of embryos and larvae were performed with an Axiovert 200M microscope (Zeiss) using an AxioCam MRm or AxioCam MRc 5 digital camera (Zeiss). For confocal analysis, a CLSM Leica TSC SP2 AOBS was used. Statistical analysis was executed with GraphPad Prism 5 (GraphPad Software) and Excel 2007 (Microsoft) as indicated.
Western blotting
Preparation of embryo lysates for Western blotting was performed as described previously (Linder et al. 2011). Adult tissues were homogenized in lysis buffer (50 mM Tris-HCl at pH 7.5, 150 mM NaCl, 1% Triton X-100) and chilled for 1 h on ice prior to sonification (Branson Sonifier 250). Protein content was determined with Bradford solution (Bio-Rad). Twenty micrograms of protein or single embryos, respectively, per lane was loaded on a 12% polyacrylamide gel and blotted on PVDF or nitrocellulose membrane (PALL). Intensities of Western and Northern blot signals were quantified by densitometry using the NIH ImageJ software package.
Northern blotting
Small RNAs from embryos or adult tissues were isolated by TRIzol reagent (Invitrogen) followed by isopropanol precipitation. Approximately 30 μg of RNA from embryos and 10 μg of RNA from adult tissues or cultured cells were denatured in an adequate volume of 2× RNA sample buffer (95% formamide, 18 mM EDTA at pH 8.0, 0.025% SDS) for 4 min on 95°C before loading. RNAs were separated on a 15% polyacrylamide/8 M urea gel and transferred to nylon membrane (GE Healthcare) by semidry blotting. Membranes were blocked in Amersham Rapid-hyb buffer (GE Healthcare) for 45 min at 40°C. Northern blot probes were [5′-32P]-labeled with T4 polynucleotide kinase (Fermentas), and hybridization was carried out at 40°C in Rapid-hyb buffer. Signals were detected with Biomax Intensifying Screens (Kodak).
P19 cell differentiation
P19 mouse carcinoma cells were cultivated in Minimum Essential Medium Eagle (MEM) Alpha Modifications containing 10% fetal calf serum (Sigma). For induction of neuronal cell differentiation, cells were treated with 1.5 μM RA in bacterial-grade petri dishes for 4 d. Cells were then replated to tissue culture-grade plates and cultivated in RA-free medium.
RT–PCR
Total RNA was DNase-treated and recovered by chloroform/phenol extraction with subsequent ethanol precipitation. cDNA was synthesized from 4 μg of total RNA using oligo(dT)18 primer and SuperScript II reverse transcriptase (Invitrogen). qPCR was performed using Absolute qPCR SYBR Green mix (Thermo Scientific) in the Stratagene Mx3000P cycler (Agilent Technologies) in triplicates. The average Ct values of triplicates were normalized with gapdh or β-actin to obtain ΔCt values. For expression fold change analysis, ΔΔCt values were calculated. Detection of mature miRNAs was performed using miScript primer assays (Qiagen) according to the manufacturer's instructions. End-point PCR products were ethidium-bromide-stained and analyzed by agarose gel electrophoresis.
RNA and LNA ISH
Generation of digoxigenin- or fluorescein-labeled antisense RNA probes and whole-mount RNA ISH and was carried out as described recently (Linder et al. 2011). Digoxigenin-labeled miRCURY LNA detection probes were obtained from Exiqon. Whole-mount LNA ISH was performed as recommended by the manufacturer.
Immunofluorescence and TUNEL assay
For immunostaining, zebrafish larvae were fixed in 4% paraformaldehyde at 4°C followed by 100% methanol overnight at −20°C. Specimens were washed in 50% methanol for 5 min and subsequently incubated in H2O for 1 h before blocking in PBS containing 1% DMSO, 1% BSA, 0.5% Triton X-100, and 2.5% goat serum. Larvae were incubated with zn-8 primary antibody overnight at 4°C. Primary antibody was detected by incubation with a Texas Red dye-conjugated anti-mouse antibody (Jackson ImmunoResearch). Detection of apoptotic cells in embryos was performed with the ApopTag Peroxidase In Situ Apoptosis Detection kit S7100 (Chemicon International) as recommended by the manufacturer.
Acknowledgments
We thank Dr. Bettina Kirchmaier for help with confocal laser scanning microscopy and Dr. Bernhard Laggerbauer and Dr. G. Meister for critically reading the manuscript. This work was supported by the RVZ network and SFB581 TP-B18.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.177774.111.
References
- Ballas N, Grunseich C, Lu DD, Speh JC, Mandel G 2005. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell 121: 645–657 [DOI] [PubMed] [Google Scholar]
- Baskerville S, Bartel DP 2005. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA 11: 241–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi SW, Zang JB, Mele A, Darnell RB 2009. Argonaute HITS-CLIP decodes microRNA–mRNA interaction maps. Nature 460: 479–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong JA, Tapia-Ramirez J, Kim S, Toledo-Aral JJ, Zheng Y, Boutros MC, Altshuller YM, Frohman MA, Kraner SD, Mandel G 1995. REST: A mammalian silencer protein that restricts sodium channel gene expression to neurons. Cell 80: 949–957 [DOI] [PubMed] [Google Scholar]
- Conaco C, Otto S, Han J-J, Mandel G 2006. Reciprocal actions of REST and a microRNA promote neuronal identity. Proc Natl Acad Sci 103: 2422–2427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanarraga ML, Avila J, Zabala JC 1999. Expression of unphosphorylated class III β-tubulin isotype in neuroepithelial cells demonstrates neuroblast commitment and differentiation. Eur J Neurosci 11: 517–527 [PubMed] [Google Scholar]
- Giraldez AJ, Cinalli RM, Glasner ME, Enright AJ, Thomson JM, Baskerville S, Hammond SM, Bartel DP, Schier AF 2005. MicroRNAs regulate brain morphogenesis in zebrafish. Science 308: 833–838 [DOI] [PubMed] [Google Scholar]
- Hakimi M-A, Bochar DA, Chenoweth J, Lane WS, Mandel G, Shiekhattar R 2002. A core–BRAF35 complex containing histone deacetylase mediates repression of neuronal-specific genes. Proc Natl Acad Sci 99: 7420–7425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huse JT, Brennan C, Hambardzumyan D, Wee B, Pena J, Rouhanifard SH, Sohn-Lee C, le Sage C, Agami R, Tuschl T, et al. 2009. The PTEN-regulating microRNA miR-26a is amplified in high-grade glioma and facilitates gliomagenesis in vivo. Genes Dev 23: 1327–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MR, Quinton LJ, Blahna MT, Neilson JR, Fu S, Ivanov AR, Wolf DA, Mizgerd JP 2009. Zcchc11-dependent uridylation of microRNA directs cytokine expression. Nat Cell Biol 11: 1157–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Villeneuve E, McBurney M, Rogers K, Kalnins V 1982. Retinoic acid induces embryonal carcinoma cells to differentiate into neurons and glial cells. J Cell Biol 94: 253–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y-K, Kim VN 2007. Processing of intronic microRNAs. EMBO J 26: 775–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kota J, Chivukula RR, O'Donnell KA, Wentzel EA, Montgomery CL, Hwang H-W, Chang T-C, Vivekanandan P, Torbenson M, Clark KR, et al. 2009. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell 137: 1005–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraner SD, Chong JA, Tsay H-J, Mandel G 1992. Silencing the type II sodium channel gene: A model for neural-specific gene regulation. Neuron 9: 37–44 [DOI] [PubMed] [Google Scholar]
- Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, et al. 2005. Combinatorial microRNA target predictions. Nat Genet 37: 495–500 [DOI] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM 2005. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433: 769–773 [DOI] [PubMed] [Google Scholar]
- Linder B, Dill H, Hirmer A, Brocher J, Lee GP, Mathavan S, Bolz HJ, Winkler C, Laggerbauer B, Fischer U 2011. Systemic splicing factor deficiency causes tissue-specific defects: A zebrafish model for retinitis pigmentosa. Hum Mol Genet 20: 368–377 [DOI] [PubMed] [Google Scholar]
- Lu QR, Sun T, Zhu Z, Ma N, Garcia M, Stiles CD, Rowitch DH 2002. Common developmental requirement for olig function indicates a motor neuron/oligodendrocyte connection. Cell 109: 75–86 [DOI] [PubMed] [Google Scholar]
- Ma Y-L, Zhang P, Wang F, Moyer MP, Yang J-J, Liu Z-H, Peng J-Y, Chen H-Q, Zhou Y-K, Liu W-J, et al. 2010. Human embryonic stem cells and metastatic colorectal cancer cells shared the common endogenous human microRNA-26b. J Cell Mol Med 15: 1941–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng A, Tang H, Ong BA, Farrell MJ, Lin S 1997. Promoter analysis in living zebrafish embryos identifies a cis-acting motif required for neuronal expression of GATA-2. Proc Natl Acad Sci 94: 6267–6272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteys AM, Spengler RM, Wan J, Tecedor L, Lennox KA, Xing Y, Davidson BL 2010. Structure and activity of putative intronic miRNA promoters. RNA 16: 495–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori N, Schoenherr CJ, Vandenbergh DJ, Anderson DJ 1992. A common silencer element in the SCG10 and type II Na+ channel genes binds a factor present in nonneuronal cells but not in neuronal cells. Neuron 9: 45–54 [DOI] [PubMed] [Google Scholar]
- Naruse Y, Aoki T, Kojima T, Mori N 1999. Neural restrictive silencer factor recruits mSin3 and histone deacetylase complex to repress neuron-specific target genes. Proc Natl Acad Sci 96: 13691–13696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, van Oudenaarden A 2008. Nature, nurture, or chance: Stochastic gene expression and its consequences. Cell 135: 216–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R 2004. Fast and effective prediction of microRNA/target duplexes. RNA 10: 1507–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybak A, Fuchs H, Smirnova L, Brandt C, Pohl EE, Nitsch R, Wulczyn FG 2008. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat Cell Biol 10: 987–993 [DOI] [PubMed] [Google Scholar]
- Schoenherr CJ, Anderson DJ 1995. The neuron-restrictive silencer factor (NRSF): A coordinate repressor of multiple neuron-specific genes. Science 267: 1360–1363 [DOI] [PubMed] [Google Scholar]
- Siomi H, Siomi MC 2010. Posttranscriptional regulation of microRNA biogenesis in animals. Mol Cell 38: 323–332 [DOI] [PubMed] [Google Scholar]
- Trabucchi M, Briata P, Garcia-Mayoral M, Haase AD, Filipowicz W, Ramos A, Gherzi R, Rosenfeld MG 2009. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature 459: 1010–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang J, Zhu J, van Oudenaarden A 2007. MicroRNA-mediated feedback and feedforward loops are recurrent network motifs in mammals. Mol Cell 26: 753–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvanathan J, Lee S, Lee B, Lee JW, Lee S-K 2007. The microRNA miR-124 antagonizes the anti-neural REST/SCP1 pathway during embryonic CNS development. Genes Dev 21: 744–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E, Horvitz HR, Kauppinen S, Plasterk RHA 2005. MicroRNA expression in zebrafish embryonic development. Science 309: 310–311 [DOI] [PubMed] [Google Scholar]
- Winkler C, Eggert C, Gradl D, Meister G, Giegerich M, Wedlich D, Laggerbauer B, Fischer U 2005. Reduced U snRNP assembly causes motor axon degeneration in an animal model for spinal muscular atrophy. Genes Dev 19: 2320–2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong CF, Tellam RL 2008. MicroRNA-26a targets the histone methyltransferase Enhancer of Zeste homolog 2 during myogenesis. J Biol Chem 283: 9836–9843 [DOI] [PubMed] [Google Scholar]
- Yeo M, Lee S-K, Lee B, Ruiz EC, Pfaff SL, Gill GN 2005. Small CTD phosphatases function in silencing neuronal gene expression. Science 307: 596–600 [DOI] [PubMed] [Google Scholar]