Abstract
Since our last comprehensive review on multi-dimensional mass spectrometry-based shotgun lipidomics (Mass Spectrom. Rev. 24 (2005), 367), many new developments in the field of lipidomics have occurred. These developments include new strategies and refinements for shotgun lipidomic approaches that use direct infusion, including novel fragmentation strategies, identification of multiple new informative dimensions for mass spectrometric interrogation, and the development of new bioinformatic approaches for enhanced identification and quantitation of the individual molecular constituents that comprise each cell’s lipidome. Concurrently, advances in liquid chromatography-based platforms and novel strategies for quantitative matrix-assisted laser desorption/ionization mass spectrometry for lipidomic analyses have been developed. Through the synergistic use of this repertoire of new mass spectrometric approaches, the power and scope of lipidomics has been greatly expanded to accelerate progress toward the comprehensive understanding of the pleiotropic roles of lipids in biological systems.
Keywords: electrospray ionization, MALDI, lipidomics, mass spectrometry, shotgun lipidomics
I. INTRODUCTION
A. Lipidomics
Cellular lipids are comprised of a highly diverse array of individual molecular species that can be classified into categories based upon lipid class and subclass distributions that collectively give rise to many tens of thousands of lipids that constitute each cell’s lipidome (Fahy et al., 2005; Yang et al., 2009a). Through lipid-mediated specific modifications in membrane surface charge, molecular dynamics, and stereoelectronic interactions, each lipid class and individual molecular species play multiple distinct and critical roles in cellular functions. The roles of lipids in cellular function include, but are not limited to, 1) the creation of a barrier to separate intracellular and extracellular compartments; 2) the provision of a dynamic matrix to facilitate productive interactions between lipids and membrane-associated proteins; 3) the generation of a storage reservoir for latent biologically active lipid 2nd messengers that can be activated with hydrolysis or covalent modification after cellular stimulation; and 4) the use of lipids (e.g., triglycerides) for the production of bioenergetic reservoirs that can store chemical energy at the times of caloric excess and that can be harvested during times of energy depletion (Vance & Vance, 2008). The entire collection of chemically distinct lipid species in a cell or organ has been referred to as a lipidome, which first appeared in the literature in 2001 (Kishimoto et al., 2001). The estimated number of individual lipid species contained in a cellular lipidome varies from cell to cell due to the intrinsic programmed genetic information, the status of cellular activation, and the metabolic signaling, and nutritional history. The total number of distinct lipid metabolites has been estimated to vary from thousands to millions of distinct chemical moieties, which vary in content and composition during alterations in the cellular environment (Yetukuri et al., 2007; Han & Jiang, 2009). The final tally of lipid molecular species depends not only on the intrinsic number of possible covalent moieties, but also on molecular species that result from oxidation (either enzymatic or non-enzymatic), nitrosylation, and many other environmental factors.
Lipidomics is a research field that studies cellular lipidomes on a large scale and at the intact-molecule level (Han & Gross, 2003; Wenk, 2005; Dennis, 2009; Griffiths & Wang, 2009b; Blanksby & Mitchell, 2010; Shevchenko & Simons, 2010; Gross & Han, 2011). Lipidomics involves precise identification of individual cellular lipid species, including the type and number of individual atoms in each lipid species, and their stereoelectronic interactions with neighboring lipids and proteins. The diversity in chemical structures in each cellular lipidome arises through alterations in the length of individual aliphatic chains, the number and the location of double bonds, the regiospecificity of aliphatic moieties in each isomer, the covalent nature of the bond at the sn-1 position of glycerol (lipid subclass), and the nature of the polar head group. Multiple recent advances in mass spectrometric approaches have greatly extended the analytical capabilities and power of lipidomics to facilitate the accurate quantification of individual lipid species to provide new insights into lipid metabolic pathways, metabolic flux, and systems integration. Moreover, an increased understanding of the diverse array of stereoelectronic interactions within membrane bilayers that integrate the function of membrane proteins to their lipid environment has been obtained. Furthermore, with analysis of the spatio-temporal alterations in lipid species during cellular growth and development, external perturbations, and changes in nutritional status, lipidomics has begun to fulfill its promise as an essential field in Systems Biology through providing mechanistic insight into many lipid-related diseases, including diabetes, obesity, heart disease, and neurodegenerative diseases.
The early analyses of cellular lipids on a large scale provided the initial insight into the utility of identifying alterations in membrane structure and function that mediate biological responses to cellular adaptation in health and maladaptive alterations during disease (Wood & Harlow, 1969a; Wood & Harlow, 1969b; Fenwick, Eagles, & Self, 1983; Gross, 1984; Gross, 1985; Han et al., 1996; Maffei Facino et al., 1996; Han et al., 2000). Rilfors and Lindblom coined the term “functional lipidomics” as “the study of the role played by membrane lipids” in 2002 (Rilfors & Lindblom, 2002). In 2003, Han and Gross first defined the field of lipidomics through integrating the specific chemical properties inherent in lipid species with a comprehensive mass spectrometric approach (Han & Gross, 2003). Many biological studies that used the extensive information provided with lipidomics approaches immediately followed (Cheng et al., 2003; Esch et al., 2003; Han & Gross, 2003). The study of one or more of the lipid classes or subclasses or molecular species is referred to as targeted lipidomics. Lee and colleagues (Lee et al., 2003) first demonstrated a novel technique to study eicosanoid species in a targeted approach with electron capture atmospheric pressure chemical ionization mass spectrometry.
Many modern technologies (including mass spectrometry (MS), nuclear magnetic resonance spectroscopy, fluorescence spectroscopy, column chromatography, and microfluidic devices) have been used in lipidomics to identify, quantify, and understand the structure and function of lipids in biological systems (Feng & Prestwich, 2006). The progress of modern lipidomics has been greatly accelerated by the Noble prize-winning research of Fenn and Tanaka through the development of the soft ionization techniques of electrospray ionization (ESI) and matrix-assisted laser desorption/ionization (MALDI), respectively (Fenn, 2003; Tanaka, 2003). The two major platforms currently used for lipidomic investigations include the direct infusion approach (shotgun lipidomics) and chromatographic separation coupled mass spectrometry. These methods are used with an ever-increasingly sophisticated level of fragmentation strategies to maximally effect structural identification as described in detail (Christie & Han, 2010; Murphy & Axelsen, 2010).
Although lipids are biological metabolites and lipidomics in theory is under the umbrella of metabolomics, lipidomics clearly stands byitself as a distinct discipline due to the uniqueness and functional specificity of lipids relative to other metabolites. For example, the majority of components in the cellular lipidome are extractable with organic solvents. This physical property greatly facilitates the recovery of lipids and their separation from water-soluble metabolites. Lipids can form multiple different types of aggregates (dimers, oligomers, micelle, bilayers, or other aggregated states) in many solvents as their concentration increases and exceeds the ability of a given solvent to solubilize lipids as monomers (Vance & Vance, 2008). This feature results in substantial difficulties for quantitative analysis of individual molecular species in their intact forms with mass spectrometry under certain conditions which must be carefully controlled during lipidomic analyses.
B. Recent Developments in Lipidomics
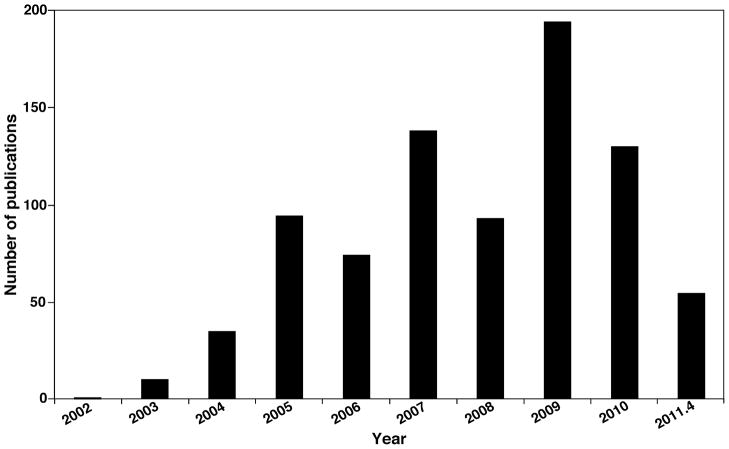
Lipidomics has been practiced for years with differing methodologies with increasing degrees of analytic sophistication. Our last review in this journal on this topic (Han & Gross, 2005b) summarized a compendium of developments in the field at the time. Since then, the field has grown rapidly due to the development of innovative approaches and the technological improvements in instrumentation and bioinformatics. For example, inspection of a yearly histograph of the literature published in last eight years (Fig. 1) with SciFinder Scholar with “lipidomics” as a keyword identified a rapidly increasing rate of publications, which now numbers ~ 200 annually. Several special issues on lipidomics have also been published, including Prostaglandins and Other Lipid Mediators (Volume 77, 2005), Frontiers in Bioscience (http://www.bioscience.org/current/special/hanxian.htm), Methods in Enzymology (Volumes 432 and 433, 2007), European Journal of Lipid Science and Technology (Volume 111(1), 2009), Journal of Chromatography B (Volume 877 (26), 2009), and Methods in Molecular Biology (Volume 579, 2009). In addition, a large compendium of publications of lipidomics are constantly updated at the website http://lipidlibrary.aocs.org/lit_surv/general/lipidome.htm.
FIGURE 1.
Yearly histogram of lipidomics obtained by SciFinder Scholar with “lipidomics” as a keyword.
Recent studies in lipidomics have largely focused on five areas, including: (1) structural characterization of known lipid classes and subclasses as well as the identification of novel lipid classes and molecular species (Tan et al., 2006; Guan, 2009; Hsu & Turk, 2009; Ivanova, Milne, & Brown, 2010; Nakanishi et al., 2010; Taguchi & Ishikawa, 2010; Li et al., 2011); (2) development of quantitative methods for analysis of high attomole to femtomole levels of lipids in cells, tissues, or biological fluids (Han et al., 2006a; Astarita & Piomelli, 2009; Griffiths & Wang, 2009a; Mesaros, Lee, & Blair, 2009; Nakanishi, Ogiso, & Taguchi, 2009; Shaner et al., 2009; Bollinger et al., 2010a; Bollinger et al., 2010b; Zhao & Xu, 2010; Wu et al., 2011); (3) network analysis that clarifies metabolic adaptation in health and disease and biomarker analysis that facilitates diagnosis of disease states and determination of treatment efficacy (Cheng, Jiang, & Han, 2007; Han et al., 2007; Mancuso et al., 2007b; Mancuso et al., 2007a; Wolf & Quinn, 2008; Lydic et al., 2009; Rappley et al., 2009; Andreyev et al., 2010; Cheng et al., 2010a; Ekroos et al., 2010; Han, 2010; Kontush & Chapman, 2010; Sysi-Aho et al., 2011); (4) tissue mapping of altered lipid distribution present in complex organs (Hankin, Barkley, & Murphy, 2007; Jackson et al., 2007; Chen et al., 2008; Burnum et al., 2009; Proschogo, Gaus, & Jessup, 2009; Chen et al., 2010; Deeley et al., 2010); and (5) bioinformatics approaches for the automated high-throughput analysis of data, including identification of lipid species through database searches, automated quantification of individual molecular species, and molecular modeling with lipidomics data (Forrester et al., 2004; Henning, Merrill, & Wang, 2004; Fahy et al., 2007; Sud et al., 2007; Yetukuri et al., 2007; Kiebish et al., 2008; Ferreri & Chatgilialoglu, 2009; Haimi et al., 2009; Hubner, Crone, & Lindner, 2009; Kind et al., 2009; Li et al., 2009; Niemela et al., 2009; Song, Ladenson, & Turk, 2009b; Yang et al., 2009a; Cowart et al., 2010; Hein et al., 2010; Kiebish et al., 2010; Taguchi & Ishikawa, 2010; Yetukuri et al., 2010; Herzog et al., 2011).
The focus of the present article is on recent developments that facilitate the improvements in technologies that lead to the increased identification of individual lipid species, increased penetrance into the lipidome, and improvements in analytical methods for quantitation in synergy with bioinformatic approaches. In particular, we will extensively explore developments in multi-dimensional mass spectrometry (MDMS) and its related applications. We sincerely apologize for being unable to include many other excellent studies in this field, and we strongly recommend that readers consult the aforementioned special issues and the updated website http://lipidlibrary.aocs.org/lit_surv/general/lipidome.htm for additional information.
C. Tandem Mass Spectrometry-based Techniques in Lipidomics
Unlike conventional ionization techniques (e.g., electron ionization or chemical ionization), the recently developed soft ionization techniques (e.g., ESI, MALDI) yield minimal or very limited in-source fragmentation under appropriate experimental conditions. Accordingly, identification and characterization of lipids with a mass spectrometer with such a soft-ion source depend heavily on tandem MS analyses. Performance of tandem MS analysis requires the utilization of multiple mass analyzers or utilization of an ion-trap mass spectrometer. A variety of hybrid combinations of mass analyzers have been used in modern mass spectrometers. These new instruments greatly facilitate lipidomic analyses (Shui et al., 2007; Yang et al., 2007; Ståhlman et al., 2009). Commonly, in a tandem mass spectrometric (MS/MS) experiment, the first analyzer is used to select a precursor ion that is typically accelerated to high kinetic energy with an electrical potential to induce collisional heating and subsequent fragmentation with inert gases (e.g., often helium, nitrogen, or argon) in the collision cell. The smaller fragments from fragmentation of the precursor ion (collision-induced dissociation (CID)) that each carries an intrinsic charge (i.e., product ions) can be detected in the second mass analyzer. This MS/MS process for product-ion analysis can be iteratively repeated with sequential selection of resultant ions for fragmentation in MSn experiments that can provide critical information about individual molecular species by chemically defined fragmentation mechanisms to enable identification of novel lipids.
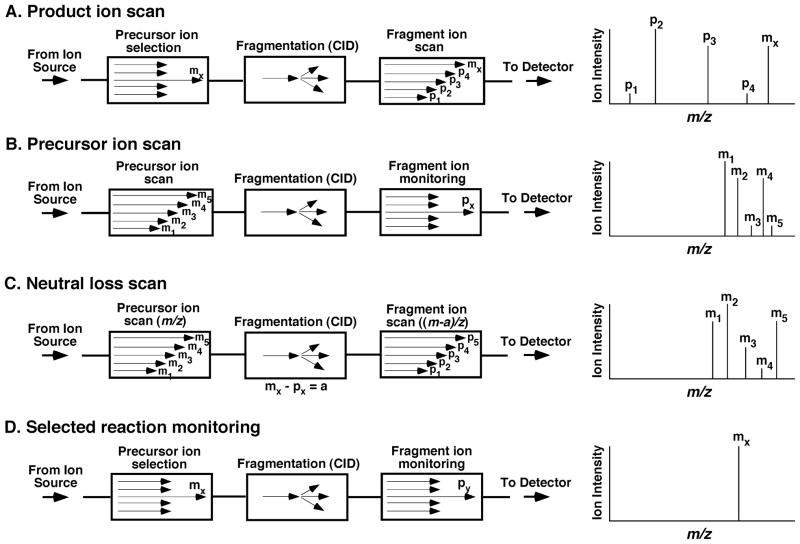
There are four main MS/MS modes that are particularly useful in lipidomics (Fig. 2). The most commonly employed MS/MS approaches employ a triple quadrupole (i.e., QqQ) instrument. Although different parameters are used for mass spectrometers with different hybrid mass analyzers, the underlying chemical principles are quite similar.
FIGURE 2.
Tandem mass spectrometric modes for analyses of lipids. CID stands for collision-induced dissociation when an inert gas is present in the collision cell. The letter “a” in neutral-loss scan mode denotes the mass of the neutral-loss fragment.
1. Product-ion analysis mode
In the product-ion analysis mode, the first mass analyzer selects a particular precursor ion of interest (Mx) by setting the mass analyzer to transmit only the particular ion of interest. The selected ion is fragmented in the collision cell with CID. The resultant product ions (P1, P2, P3, …) are analyzed with the second mass analyzer (Fig. 2A). The structure of the selected ion thus can be elucidated from a reconstruction of the fragment ions and/or the fragmentation patterns in conjunction with the mass of the precursor ion. This scanning mode is useful for characterization of available fragmentation pathways and for the analysis of the kinetics of fragmentation of discrete molecule ions.
2. Precursor-ion scanning mode
In the precursor-ion scanning (PIS) mode, in contrast to the product-ion analysis mode, the second analyzer now focuses on a particular product ion of interest (Px) after CID while scanning the m/z ratios in the first mass analyzer. All of the precursor ions (M1, M2, M3, …) that produce the selected product ion (Px) after fragmentation are thus detected (Fig. 2B). In lipidomic analyses, this scanning mode is a powerful means to identify multiple individual molecular species in a lipid class that yields a given product ion.
3. Neutral-loss scanning mode
In the neutral-loss scanning (NLS) mode, both the first and second mass analyzers are scanned simultaneously, but with a constant mass offset of “a” between the two. When a precursor ion of Mx is transmitted through the first mass analyzer, this ion is recorded if it yields a product ion (Px) that corresponds to the loss of a neutral fragment that has a mass of “a” from the precursor ion (i.e., Mx−Px = a) after CID (Fig. 2C). Similar to PIS, a class or a group of lipids that possesses an identical neutral-loss fragment with a mass of “a” can be efficiently identified with this scanning mode.
4. Selected reaction-monitoring mode
In the selected reaction monitoring (SRM) mode, transitions between the molecule ion and product ion must be previously known, and the first and second mass analyzers are both focused on the selected ions at m/z = Mx and Px (Px < Mx), respectively, where Px represents a product ion of Mx (Fig. 2D). This experiment yields high specificity and sensitivity via a high duty cycle to monitor the transitions of interest. When either the first or the second mass analyzer or both are set to monitor multiple ions for multiple reactions, the term “multiple reaction monitoring (MRM)” has been widely used, although it is not accurate because it has been used to show that more than one generation of product ions are being monitored (Sparkman, 2000). The SRM/MRM technique is widely used for quantitative analysis of individual lipid species in lipidomics when a mass spectrometer is coupled with HPLC. It should be pointed out that SRM could be considered a special case of PIS in which the first analyzer is fixed at a certain m/z, or a special case of product-ion scanning in which the second analyzer is focused at a specific m/z, or a special case of NLS in which both analyzers are fixed at the m/z of a pair of ions. Accordingly, from an integrated chemical perspective it should be recognized that SRM analysis represents a special case of the other three MS/MS techniques with particular advantages associated of high sensitivity essential to LC-MS analysis where limited amounts of time are available to collect information on molecular-ion fragmentation products.
D. The Modes of Product-Ion Analysis, Precursor-Ion Scanning, and Neutral-Loss Scanning are Inter-related
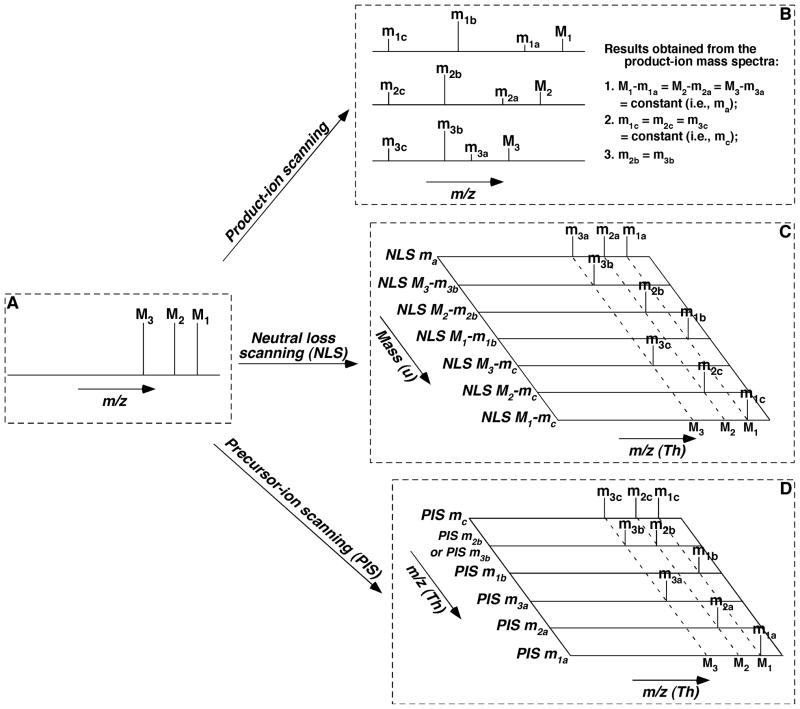
The tandem mass spectrometric techniques (i.e., product-ion analysis, neutral-loss scanning, and precursor-ion scanning) are inter-related. This inter-relationship is the foundation of the multi-dimensional mass spectrometric approach for lipid analysis, and can be schematically illustrated with a simplified model system that comprises three molecule ions of a lipid class (Fig. 3).
FIGURE 3.
Schematic illustration of the inter-relationship among the MS/MS techniques for the analysis of individual molecular species of a class of interest. We only illustrate the analysis of three species (M1, M2, and M3) of a class for simplicity, whereas there exist up to hundreds of individual molecular species within a class. We assume that this class of lipid species, similar to a class of phospholipids or sphingolipids possesses one common neutral-loss fragment with mass of ma (i.e., M1 − m1a = M2 − m2a = M3 − m3a = ma (a constant)), one common fragment ion at m/z mc (i.e., m1c = m2c = m3c = mc), and a specific ion to individual species at m/z m1b, m2b, and m3b, respectively, which might not be identical to each other. Specifically, the common neutral fragment and the common fragment ion both result from the head group of the class; the individual species-specific ions represent the fatty acyl moieties of the species; and thus the residual part of each individual species can be derived from these fragments in combination with the m/z of each molecule ion. Panel A shows a simplified full-mass scan; Panel B illustrates the product-ion analysis of these molecule ions; Panel C demonstrates the scanning of the individual neutral-loss fragment between a specific molecule ion and its individual fragment ion; and Panel D represents the scanning of each individual fragment ion. It should be emphasized that, although the analyses of fragments with either neutral-loss scanning (NLS) or precursor-ion scanning (PIS) are much more complicated than those in product-ion scanning in this simplified case, the analyses by NLS or PIS are much simpler than that with product-ion scanning in reality as discussed in the text.
In this model, each of the three molecule ions has a different m/z and therefore each yields a different product-ion mass spectrum after CID. Because these molecule ions belong to the same lipid class, the fragmentation patterns of these molecule ions are virtually identical. We assume that the fragmentation pattern of these molecule ions includes three types of product ions (Fig. 3B). First, these molecule ions yield product ions that result from the loss of a common neutral fragment with a mass of ma. This loss gives rise to product ions m1a, m2a, and m3a from the molecule ions M1, M2, and M3, respectively, where ma = M1 − m1a = M2 − m2a = M3 − m3a. Next, these molecule ions also yield a common product-ion mc (i.e., m1c = m2c = m3c = mc). The common neutral fragment and the common fragment ion both generally result from the head group of the class. Finally, each molecule ion gives rise to a specific fragment ion from a common constituent (e.g., a fatty acyl chain). This specific fragment ion leads to an array of product ions m1b, m2b, and m3b that result from the individual molecule ions M1, M2, and M3, respectively. The collection of discrete product ions collectively represents an array of high-density information on the chemical constituents in each lipid class, subclass, or molecular species under investigation. The structure of each individual species, including its backbone, can be derived from these fragments in combination with the m/z of each molecule ion.
Conceivably, the set of the product ions from a molecule ion can also be detected in the neutral-loss scanning mode, because each product ion represents the loss of neutral fragment(s) or neutral fragments from its corresponding molecule ion. Therefore, all the product ions identified in the aforementioned model system can be determined with NLS at mass differences in an ascending order of ma, M3 – m3b, M2 – m2b, M1 – m1b, M3 – mc, M2 – mc, and M1 – mc (Fig. 3C). If this series of neutral-loss mass spectra are mapped along the mass difference, then a pseudo-mass spectrum along a broken line vertical to the horizontal dimension and crossed with one of the molecule ions (i.e., M1, M2, or M3) mimics the product-ion mass spectrum of the corresponding molecule ion.
Similarly, all product ions can be detected in the PIS mode by scanning at m/z in an ascending order of mc/z, m2b/z, m3b/z, m1b/z, m3a/z, m2a/z, and m1a/z (Fig. 3D). Thus, if this series of precursor-ion mass spectra is mapped along the m/z of the scanned fragment ions, then a pseudo-mass spectrum along a broken line vertical to the horizontal dimension and crossed with one of the molecule ions (i.e., M1, M2, or M3) is formally related to the product-ion mass spectrum of the corresponding molecule ion.
This simplified model illustrates the inter-relationship among the modes of product-ion analysis, PIS, and NLS and clearly indicates that analysis of lipids in a complex biological mixture with one of these MS/MS techniques is sufficient to provide an initial insight into the molecular structures of individual molecular constituents in a given lipid class, subclass, or individual molecular species of interest in the selected mass range under the specific conditions employed. Thus, a detailed map of all product ions of interest can be obtained within a mass range of interest either with multiple individual product-ion analyses through scanning individual molecule ions unit by unit, or with NLS through scanning the neutral fragments unit by unit, or with PIS through scanning the fragment ions unit by unit. Each of these analyses alone can map the complete fragment ions of individual lipid species within the mass range of interest because of the aforementioned inter-relationship among the chemical fragmentation processes involved.
Specifically, in product-ion mapping, the selected molecule ion m/z varies from the lowest m/z to the highest m/z of a mass range of interest unit by unit; i.e., from (m/z)lowest, (m/z)lowest + 1, (m/z)lowest + 2, …, to (m/z)highest. In neural loss mapping, the neutral-loss mass varies from 0, 1, 2, …, to the highest mass, which is equivalent to (m/z)highest in value for singly-charged molecule ions (where z = 1). In precursor-ion mapping, the monitored fragment varies from 1, 2, …, to the highest m/z, which is the (m/z)highest for singly-charged molecule ions. Intriguingly, each map acquired with one of the tandem mass spectrometric techniques constitutes a two-dimensional (2D) mass spectrum, in which the first dimension is the m/z of the molecule ions and the second dimension is the m/z of the fragment ions for product-ion mapping or precursor-ion mapping or mass (m) lost from the molecule ion during neutral-loss mapping.
It is worth noting that, in the 2D mass spectrum generated in neutral-loss mode, the first trace acquired with a neutral-loss mass of 0 is equivalent to the full mass scan spectrum, which identifies most of the molecule ions present in the analyzed lipid mixture under the appropriate set of instrumental conditions. In contrast, in the 2D mass spectrum the last trace acquired with the highest neutral-loss mass (which is equal to the value of (m/z)highest for singly-charged molecule ions) should show the least number of molecular species present in the lipid mixture.
One feature of such a 2D mass spectrum is that a summation of all the tandem mass spectra in the second dimension constitutes a one-dimensional mass spectrum that represents a molecule ion mass spectrum, which shows the complete set of molecular species present in the analyzed lipid sample. The relative intensities of molecular species in the molecule ion mass spectrum might vary to some extent compared to those intensities in the actual molecular-ion mass spectrum due to the differential collisional activation present in the collision cell that results from the presence of discrete molecular species with differential propensities toward fragmentation and differential fragmentation kinetics.
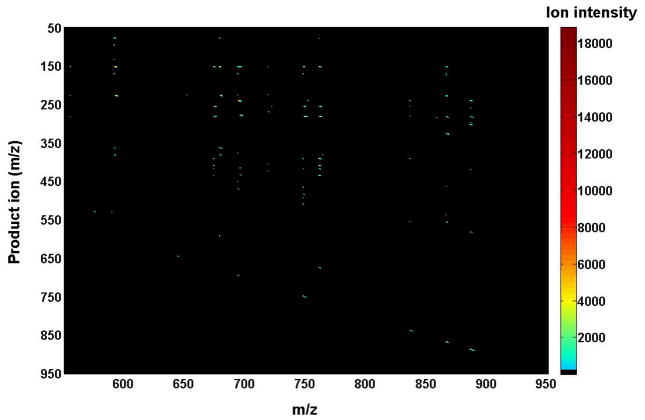
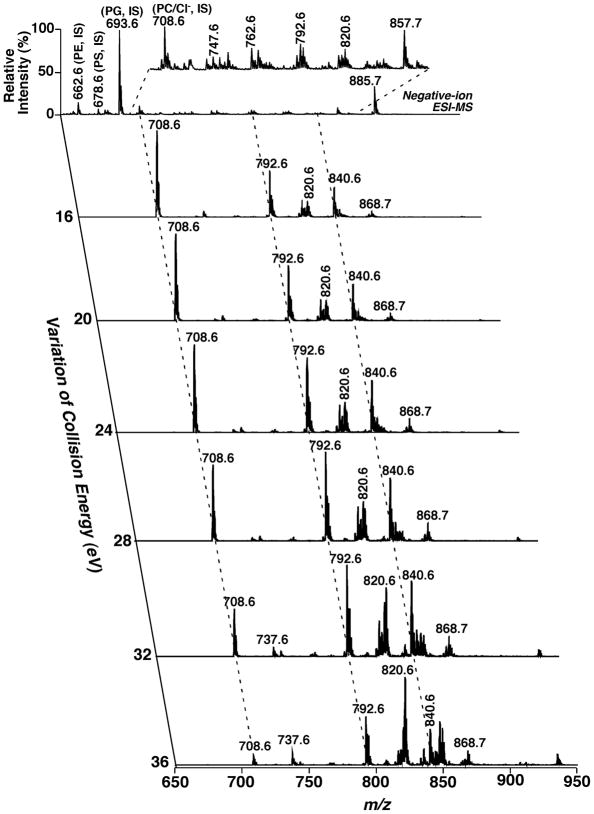
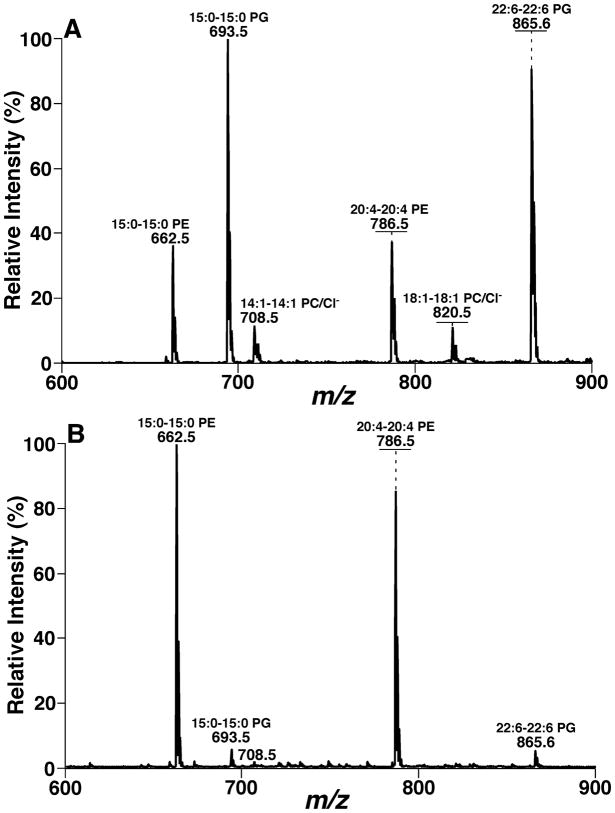
Again, the 2D mass spectrum generated with one of the three tandem mass spectrometric techniques is inter-related to those generated with the other techniques due to the intrinsic interrelationship of the three tandem mass spectrometric techniques, as demonstrated with the simplified model system above. The inter-relationship among the modes of tandem MS techniques has been demonstrated with a mixture of phospholipids, which is comprised of two or three species of each individual anionic phospholipid class with a total of four classes, including di14:0 phosphatidic acid (PA) (deprotonated molecule ion at m/z 591.4), 16:0–18:1 PA (m/z 673.5), di18:2 PA (m/z 695.5), di15:0 phosphatidylglycerol (PG) (m/z 693.5), 16:0–18:1 PG (m/z 747.5), di22:6 PG (m/z 865.5), 16:0–18:1 PI (m/z 835.5), 18:0–20:4 PI (m/z 885.5), di14:0 phosphatidylserine (PS) (m/z 678.4), 17:0-14:1 PS (m/z 718.5), and 16:0–18:1 PS (m/z 760.5) (Figs. 4, 5, and 6). The 2D maps acquired with different MS/MS techniques in the negative-ion mode show essentially identical patterns.
FIGURE 4.
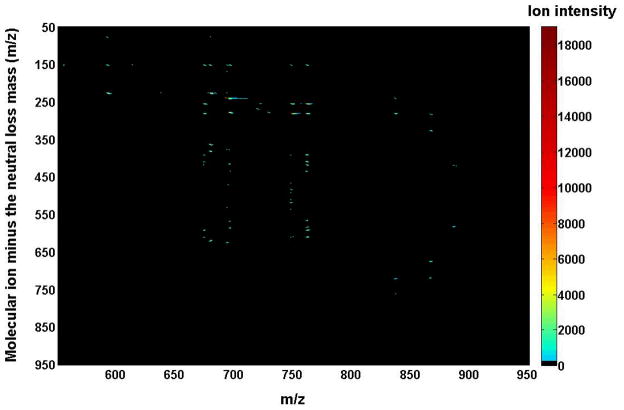
Two-dimensional mass spectrometric mapping of a mixture of phospholipids in the product-ion mode. The mixture of phospholipids is comprised of two or three species of an individual anionic phospholipid class with a total of four classes, including di14:0 phosphatidic acid (PA) (m/z 591.4), 16:0–18:1 PA (m/z 673.5), di18:2 PA (m/z 695.5), di15:0 phosphatidylglycerol (PG) (m/z 693.5), 16:0–18:1 PG (m/z 747.5), di22:6 PG (m/z 865.5), 16:0–18:1 phosphatidylinositol (PI) (m/z 835.5), 18:0–20:4 PI (m/z 885.5), di14:0 phosphatidylserine (PS) (m/z 678.4), 17:0-14:1 PS (m/z 718.5), and 16:0–18:1 PS (m/z 760.5) in the different mass levels. For comparison to the 2D mappings with other tandem mass spectrometric techniques (see Figs. 5 and 6), product-ion analyses were conducted with a mass resolution of 0.7 Th unit by unit from m/z 550 to m/z 950. Each product-ion mass spectrum was acquired at a scan rate of 0.5 s between m/z 50 and m/z 950 for 1 min in the profile mode. Absolute ion counts in each mass spectrum were displayed after averaging the absolute ion counts of each individual data point.
FIGURE 5.
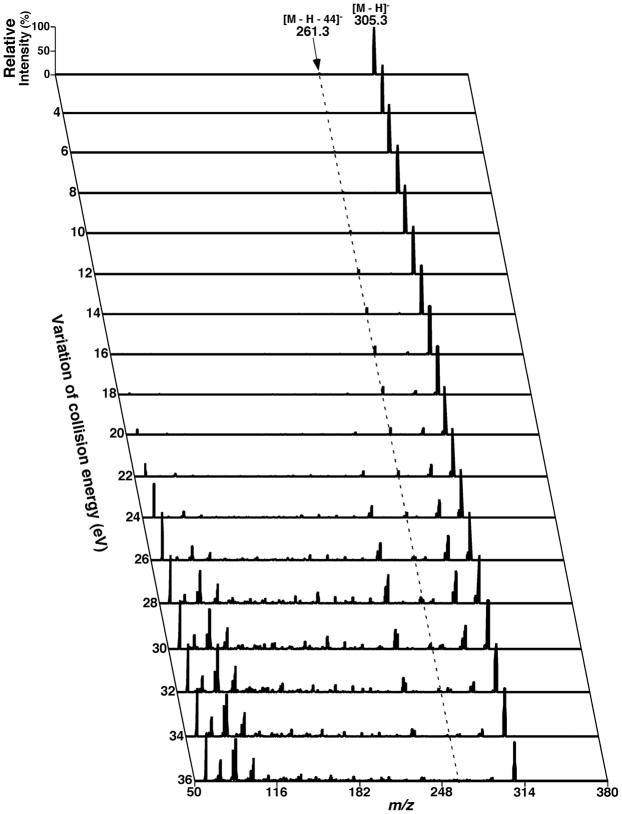
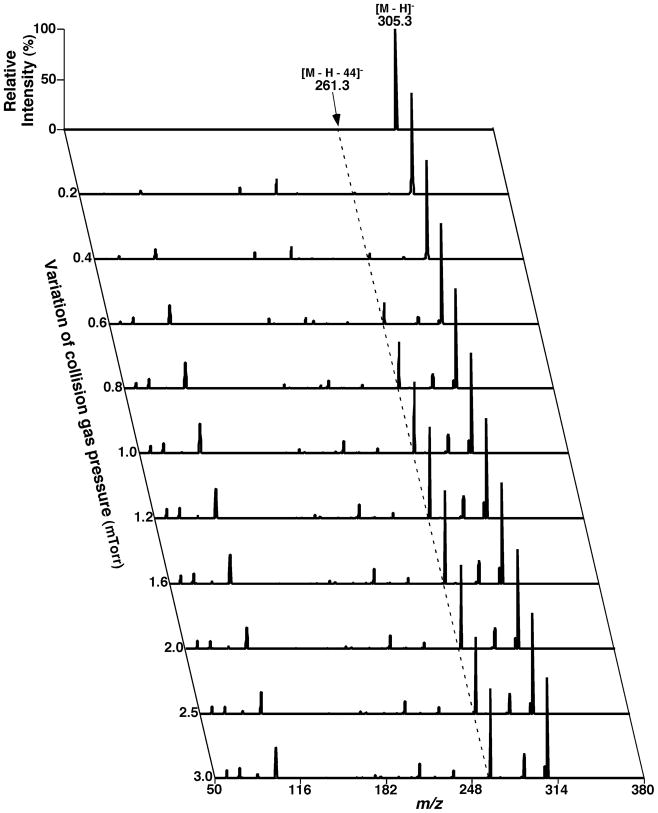
Two-dimensional mass spectrometric mapping of a mixture of phospholipids in the neutral-loss scanning mode. The mixture of phospholipids is identical to that used for acquisition of mass spectra in Figure 4. 2D mapping with neutral-loss scans was conducted with mass resolution of 0.7 Th unit by unit from 50 to 950 amu. Each neutral-loss scan was acquired at a scan rate of 0.5 s between m/z 550 and m/z 950 for 1 min in the profile mode. The y-axis mass in the map was plotted after transforming the neutral-loss data by subtracting the neutral-loss mass from the molecule ion mass. Absolute ion counts in each mass spectrum were displayed after averaging the absolute ion counts of each individual data point.
FIGURE 6.
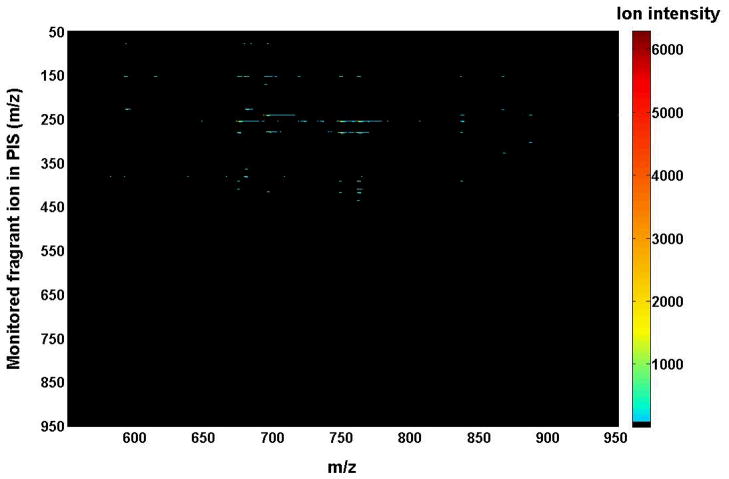
Two-dimensional mass spectrometric map of a mixture of phospholipids in the precursor-ion scanning mode. The mixture of phospholipids is identical to that used for acquisition of mass spectra in Figure 4. Precursor-ion scan analyses were conducted with a mass resolution of 0.7 Th unit by unit from m/z 50 to m/z 950. Each precursor-ion mass spectrum was acquired at a scan rate of 0.5 s between m/z 550 and m/z 950 for 1 min in the profile mode. Absolute ion counts in each mass spectrum were displayed after averaging the absolute ion counts of each individual data point.
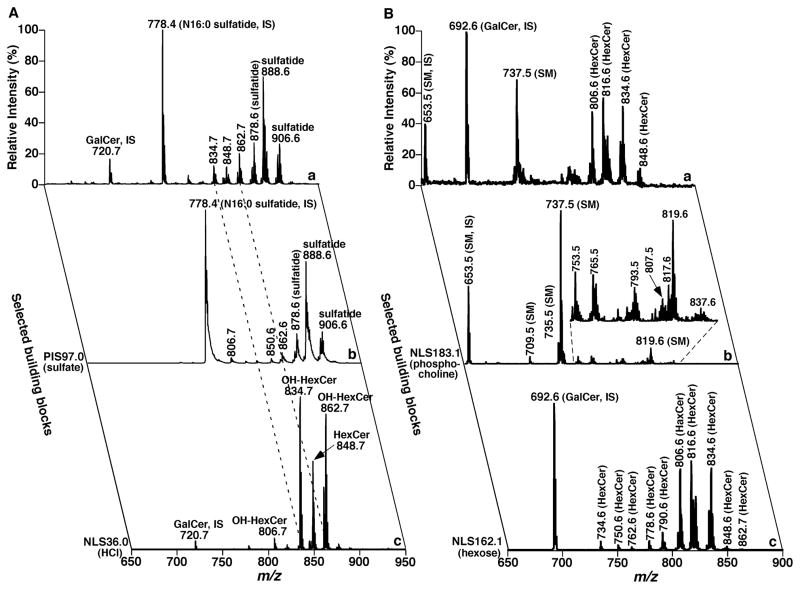
Although the recognition of this inter-relationship is important, the application of this relationship is very labor-intensive in the analysis of individual molecular species present in a biological lipid extract with this type of mapping. Although a similar concept has been practiced in a limited number of fragments with a quadrupole-time-of-flight (Q-TOF) instrument (Ejsing et al., 2006b; Schwudke et al., 2006; Ståhlman et al., 2009), continuous mapping of biological samples with this approach has never been reported. However, the inter-relationship among the modes of the MS/MS techniques allows one to perform the complete mapping of product ions with combined use of NLS and PIS. This approach has been well-developed by Han and Gross and their colleagues (Han & Gross, 2001; Han et al., 2004b; Han & Gross, 2005a; Yang et al., 2009a). In the approach, the building blocks of biologically occurring lipids (extensively discussed in next subsection) of a lipid class of interest are analyzed with either NLS or PIS of the specific building blocks or their combination (Han & Gross, 2001; Han et al., 2004b; Han et al., 2005; Su et al., 2005; Yang et al., 2009a; Yang et al., 2009b). In this practical approach, the second dimension of each 2D mass spectrum is the specific building blocks of a lipid class or a category of lipid classes of interest. The selection of the specific building blocks can be identified from the product-ion analysis of a limited number of authentic molecular species of the class or the category of classes of interest based upon the easily identifiable chemical fragmentation mechanisms that occur in lipids.
Although the 2D mass spectra generated with one of the three MS/MS techniques are inter-related, a simplified or a pseudo 2D mass spectrum generated with NLS or PIS of the specific building blocks or their combination is different from a series of product-ion analyses. One feature of the simplified 2D mass spectrum is that, along a vertical line to the first dimension (m/z of molecule ions) and crossed with a molecule ion present in the survey scan mass spectrum, is an imaginary mass spectrum that represents a pseudo-product-ion mass spectrum of the corresponding molecule ion. All the pseudo-product-ion mass spectra are the combination of the detected building blocks with NLS and/or PIS, whereas in the product-ion analysis, one product-ion scan is needed to generate a product-ion mass spectrum for each individual molecule ion. Accordingly, fewer numbers of NLS and/or PIS are required for analysis of analytes in biological samples to generate pseudo-product-ion mass spectra for all the molecule ions present. Therefore, the features present in the product-ion analysis can be achieved from the simplified 2D MS analysis with fewer scans (i.e., with higher efficiency). Most importantly, the simplified 2D MS analysis of lipids can be automated for a high-throughput platform for the global analysis of cellular lipidomes (Yang et al., 2009a). Finally, a product ion from a building block (e.g., 18:0 fatty acyl) present in a low-abundance species (e.g., 18:0–18:2 diacyl PE) might be affected with the presence of the ion(s) (e.g., 18:1 carboxylate ion) from other abundant isomeric species (e.g., 18:1–18:1 diacyl PE) in a product-ion analysis, but might be readily determined with 2D MS analysis with PIS (e.g., PIS283.2) or NLS specific to this building block, which has not subject to interference with the presence of the abundant isomeric species that contain different building blocks.
E. The Concept of Building-Block Analysis and its Significance in Lipidomics
1. Lipid building blocks
The majority of biologically occurring lipids are linear combinations of aliphatic chains and polar head groups attached to glycerol or sphingoid backbones as well as aliphatic chains or polar entities covalently attached to cholesterol. The backbones (e.g., glycerol, sphingoids or cholesterol) and the aliphatic chains and polar moieties (head groups, sugars and moieties derived therefrom) attached to these backbones can both be considered as building blocks that represent the fundamental chemical structures from which naturally occurring lipids are derived. Common examples of these building blocks include a glycerol molecule, sphingoid backbones, polar head groups (e.g., phosphoesters (including phosphate, phosphocholine, phosphoethanolamine, phosphoglycerol, phosphoserine, and phosphoinositol) and sugar molecules (e.g., glucose, galactose, lactose, etc.)), and fatty acyl substituents (or other aliphatic chains). With this formalism, the molecular species of an entire lipid class or even entire descriptors of lipid classes of interest emanate from a common chemical foundation.
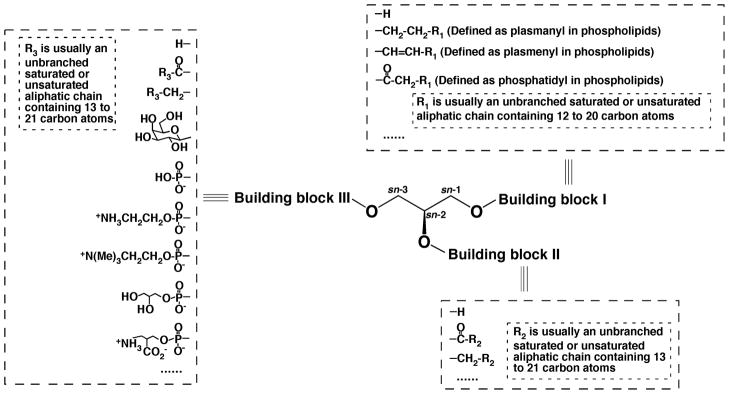
For example, molecular species of all glycerol-based lipid classes (e.g., phospholipids, glycerolipids, and glycosyldiacylglycerols) are multiple discrete covalent assemblies of various aliphatic chains (including fatty acyl chains) and polar head groups to a glycerol backbone (Yang et al., 2009a). Here, the aliphatic chains (containing typically 12 – 24 carbon atoms with variable degrees of unsaturation) are at the positions 1 and 2 of the stereospecific numbering (sn) system, and the polar head groups are at the sn-3 position of glycerol. Hence, molecular species of all glycerol-based lipid classes can be represented with a general structure with three building blocks linked to a glycerol backbone (Fig. 7). In this general structure, the building blocks I and II can be a hydrogen, a fatty acyl chain, or an aliphatic chain connected with an ester, ether, or vinyl ether linkage. Building block III can be a hydrogen or acyl (or aliphatic) chain in glycerolipids, various sugar ring(s) and their derivatives in glycolipids, or phosphoesters in phospholipids and lysophospholipids.
FIGURE 7.
General structure of glycerol-based lipids with three building blocks. Three building blocks are linked to the hydroxy groups of a glycerol backbone. Potential candidates of the building blocks are listed.
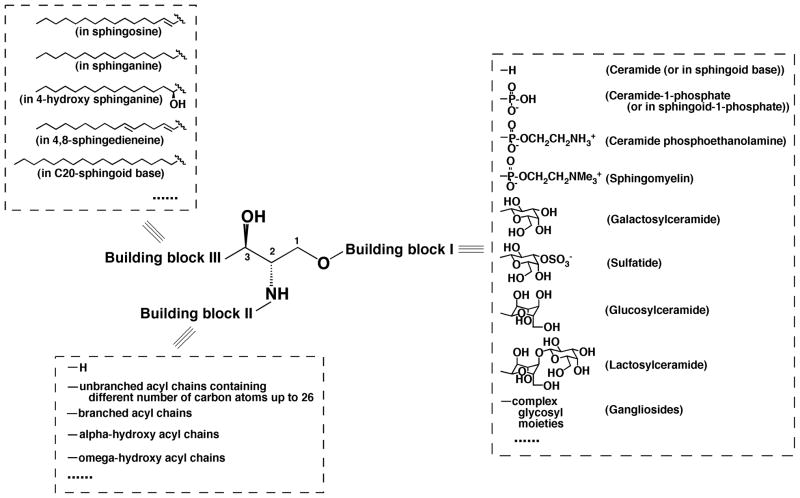
Similar to glycerol-based lipids, a majority of the sphingolipid species can be represented with a general structure with three building blocks (Fig. 8). Building block I represents a different polar moiety (linked to the oxygen at the C1 position of a sphingoid backbone). These polar moieties include hydrogen, phosphoethanolamine, phosphocholine, galactose, glucose, lactose, sulfated galactose, and other complex sugar groups that correspond to ceramide, ceramide phosphoethanolamine, sphingomyelin (SM), galactosylceramide (GalCer), glucosylceramide (GluCer), lactosylceramide, sulfatide, and other glycosphingolipids such as gangliosides, respectively (Fig. 8). Building block II represents a fatty acyl moiety, which is acylated to the primary amine at the C2 position of the sphingoid backbone. A variety of fatty acyl chains, including those that contain a hydroxy group (usually located at the alpha or omega position) (Fig. 8), can occupy this position. Building block III represents the aliphatic chain present in all sphingoid backbones. This building block is linked through a carbon-carbon bond to the C3 position. This aliphatic chain varies in length, the degree of unsaturation, the presence of a branch in the chain, and the presence of additional hydroxy group(s) (Fig. 8). Thousands of possible sphingolipid species can be constructed from the combination of these three building blocks to create a virtual database comprised of all possible structures that can be populated through bioinformatic array analysis of ion intensities present in tandem mass spectrometric analyses (Yang et al., 2009a).
FIGURE 8.
General structure of sphingoid-based lipids with three building blocks. Building block I represents a different polar moiety (linked to the oxygen at the C1 position of sphingoid backbone). Building block II represents fatty acyl chains (acylated to the primary amine at the C2 position of sphingoid backbone) with or without the presence of a hydroxy group, which is usually located at the alpha or omega position. Building block III represents the aliphatic chains in all of possible sphingoid backbones, which are carbon-carbon linked to the C3 position of sphingoid backbones and vary with the aliphatic chain length, degree of unsaturation, the presence of branch, and the presence of an additional hydroxy group.
2. Construction of theoretical lipid databases with lipid building blocks
Based on these building blocks of each lipid class of a general structure, a theoretical database can be readily constructed (Yang et al., 2009a). This virtual database has every possible combination of glycerophospholipids, glycerolipids, sphingolipids, sterols, and metabolites that contain currently recognized natural products. Therefore, a database of molecular species for each individual lipid class or a category of lipid classes, including total carbon atoms, total double bonds, chemical formulas, accurate monoisotopic mass, and others, can be constructed with the variables of the number of carbon atoms (m) and the number of double bonds (n) in an aliphatic chain building block denoted as m:n. For example, Tables 1 and 2 represent the theoretical database of glycerophospholipid and sphingolipid species, respectively.
Table 1.
A summarized database of glycerophospholipidsa
| Lipid class | Lipid subclasses | Back-bone | Head group (Building block III) | Side chains (Building blocks I & II) | Sum formula | Negative-ion mode | Positive-ion mode | Number of possible speciesb |
|---|---|---|---|---|---|---|---|---|
| PC | diacyl PC | C3H5O3 | C5H13O3PN | CmH2m-2n-2O2 | Cm+8H2m-2n+16O8PN | [M+Cl]− | [M+Li]+, [M+Na]+ | 314 |
| alkenyl-acyl PC | CmH2m-2n-2O | Cm+8H2m-2n+16O7PN | 314 | |||||
| alkyl-acyl PC | CmH2m-2nO | Cm+8H2m-2n+18O7PN | 314 | |||||
|
|
|
|||||||
| PE | diacyl PE | C2H7O3PN | CmH2m-2n-2O2 | Cm+5H2m-2n+10O8PN | [M-H]−, [M-H+Fmoc]− (i.e., [M+C15H9O2]−) | 314 | ||
| alkenyl-acyl PE | CmH2m-2n-2O | Cm+5H2m-2n+10O7PN | 314 | |||||
| alkyl-acyl PE | CmH2m-2nO | Cm+5H2m-2n+12O7PN | 314 | |||||
|
|
|
|||||||
| PS | diacyl PS | C3H7O5PN | CmH2m-2n-2O2 | Cm+6H2m-2n+10O10PN | [M-H]− | 314 | ||
| alkenyl-acyl PS | CmH2m-2n-2O | Cm+6H2m-2n+10O9PN | 314 | |||||
| alkyl-acyl PS | CmH2m-2nO | Cm+6H2m-2n+12O9PN | 314 | |||||
|
|
|
|||||||
| PG | C3H8O5P | CmH2m-2n-2O2 | Cm+6H2m-2n+11O10P | [M-H]− | 314 | |||
|
|
|
|||||||
| PI | C6H12O8P | CmH2m-2n-2O2 | Cm+9H2m-2n+15O13P | [M-H]− | 314 | |||
|
|
|
|||||||
| PA | H2O3P | CmH2m-2n-2O2 | Cm+3H2m-2n+5O8P | [M-H]− | 314 | |||
|
|
|
|||||||
| lysoPC | acyl-LPC | C5H13O3PN | CmH2m-2nO | Cm+8H2m-2n+18O7PN | [M+Cl]− | [M+Li]+, [M+Na]+ | 82 | |
| alkenyl-LPC | CmH2m-2n | Cm+8H2m-2n+18O6PN | 82 | |||||
| alkyl-LPC | CmH2m-2n+2 | Cm+8H2m-2n+20O6PN | 82 | |||||
|
|
|
|||||||
| lysoPE | acyl-LPE | C2H7O3PN | CmH2m-2nO | Cm+5H2m-2n+12O7PN | [M-H]−, [M-H+Fmoc]− (i.e., [M+C15H9O2]−) | 82 | ||
| alkenyl-LPE | CmH2m-2n | Cm+5H2m-2n+12O6PN | 82 | |||||
| alkyl-LPE | CmH2m-2n+2 | Cm+5H2m-2n+14O6PN | 82 | |||||
|
|
|
|||||||
| lysoPS | acyl-LPS | C3H7O5PN | CmH2m-2nO | Cm+6H2m-2n+12O9PN | [M-H]− | 82 | ||
| alkenyl-LPS | CmH2m-2n | Cm+6H2m-2n+12O8PN | 82 | |||||
| alkyl-LPS | CmH2m-2n+2 | Cm+6H2m-2n+14O8PN | 82 | |||||
|
|
|
|||||||
| lysoPG | C3H8O5P | CmH2m-2nO | Cm+6H2m-2n+13O9P | [M-H]− | 82 | |||
|
|
|
|||||||
| lysoPI | C6H12O8P | CmH2m-2nO | Cm+9H2m-2n+17O12P | [M-H]− | 82 | |||
|
|
|
|||||||
| lysoPA | H2O3P | CmH2m-2nO | Cm+3H2m-2n+7O7P | [M-H]− | 82 | |||
|
| ||||||||
| CL | (C3H5O3)2 | C3H8O7P2 | CmH2m-2n-4O4 | Cm+9H2m-2n+14O17P2 | [M-2H]2− | 1081 | ||
|
| ||||||||
| mono-lysoCL | (C3H5O3)2 | C3H8O7P2 | CmH2m-2n-2O3 | Cm+9H2m-2n+16O16P2 | [M-2H]2− | 622 | ||
|
| ||||||||
| Total | 6455 | |||||||
The database is constructed with the building blocks I, II, and III in glycerophospholipids as indicated in Figure 7 with the variables m and n. The variable m represents the number of total carbon atoms of acyl chains (m = 12 – 26, 24 – 52, 36 – 78, and 48 – 104 for species with one, two, three, and four fatty acyl chains, respectively) and the variable n represents the number of total double bonds of the acyl chains (n = 0 – 7, 0 – 14, 0 – 21, and 0 – 28 for species with one, two, three, and four fatty acyl chains, respectively). The ion modes indicate the ionization mode(s) used to analyze the indicated lipid class in MDMS-based shotgun lipidomics.
The regioisomers and the isomers resultant from different locations of double bond(s) are not considered. The numbers of molecular species are calculated based on the web search that the naturally occurring fatty acids with the highest unsaturation for acyl chains of 12 – 26 carbon atoms are 12:1, 13:1, 14:3, 15:3, 16:5, 17:3, 18:5, 19:3, 20:6, 21:5, 22:7, 23:5, 24:7, 25:6, and 26:7, respectively.
Table 2.
A schematic representation of the database used for sphingolipidsa
| Lipid class | Lipid subclasses | Backbone | Sphingoid backbone (Building block III) | Head group (Building block I) | Side chain (Building block II) | Sum formula | Negative-ion mode | Positive-ion mode | Number of possible speciesb |
|---|---|---|---|---|---|---|---|---|---|
| Ceramide (Cer) | non-hydroxy Cer | H | CmH2m-2n-1O | Cm+x+3H2m-2n+2x-2y+7O3N | [M-H]− | 2214 | |||
| hydroxy Cer | CmH2m-2n-1O2 | Cm+x+3H2m-2n+2x-2y+7O4N | 2214 | ||||||
| Sphingomyelin (SM) | non-hydroxy SM | C5H13O3PN | CmH2m-2n-1O | Cm+x+8H2m-2n+2x-2y+19O6PN2 | [M+Cl]− | [M+Li]+, [M+Na]+ | 2214 | ||
| hydroxy SM | CmH2m-2n-1O2 | Cm+x+8H2m-2n+2x-2y+19O7PN2 | 2214 | ||||||
| Ceramide phosphoethanola mine (CerPE) | non-hydroxy CerPE | C2H6O3PN | CmH2m-2n-1O | Cm+x+5H2m-2n+2x-2y+12O6PN2 | [M-H]−, [M-H+Fmoc]− | [M+H]+ | 2214 | ||
| hydroxy CerPE | C6H11O5 | CmH2m-2n-1O2 | Cm+x+5H2m-2n+2x-2y+12O7PN2 | [M+Cl]− | [M+Li]+, [M+Na]+ | 2214 | |||
| Monohexosyl-ceramide (HexCer) | non-hydroxy HexCer | CmH2m-2n-1O2 | Cm+x+9H2m-2n+2x-2y+17O9N | 2214 | |||||
| hydroxy HexCer | CmH2m-2n-1O2 | Cm+x+9H2m-2n+2x-2y+17O9N | 2214 | ||||||
| Sulfatide (ST) | non-hydroxy ST | C3H6O2N | CxH2x-2y+1 | C6H11SO8 | CmH2m-2n-1O | Cm+x+9H2m-2n+2x-2y+17O11NS | [M-H]− | 2214 | |
| hydroxy ST | CmH2m-2n-1O2 | Cm+x+9H2m-2n+2x-2y+17O12NS | 2214 | ||||||
| Lactosyl-ceramide (LacCer) | non-hydroxy LacCer | C12H21O10 | CmH2m-2n-1O | Cm+x+15H2m-2n+2x-2y+27O13N | [M+Cl]− | [M+Li]+, [M+Na]+ | 2214 | ||
| hydroxy LacCer | C5H13O3PN | CmH2m-2n-1O2 | Cm+x+15H2m-2n+2x-2y+27O14N | 2214 | |||||
| Lyso-SM Sphingoid backbone | H | H | Cx+8H2x-2y+21O5PN2 | [M+Cl]− | [M+Li]+, [M+Na]+ | 27 | |||
| H | Cx+3H2x-2y+9O2N | [M+H]+ | 27 | ||||||
| Sphingoid backbone 1-phosphate | H2O3P | H | Cx+3H2x-2y+10O5PN | [M-H]− | 27 | ||||
| Psychosine | C6H11O5 | H | Cx+9H2x-2y+19O7N | [M+H]+ | 27 | ||||
|
| |||||||||
| Total | 26676 | ||||||||
The database is constructed based on the building blocks I, II, and III in sphingolipids as indicated in Figure 8 with the variables x, y, m, and n. The variable m represents the number of total carbon atoms of fatty amide chain (m = 12 – 26), the variable n represents the number of total double bonds of the fatty amide chain (n = 0 – 7), the variable x represents the number of total carbon atoms of a partial sphingoid backbone as indicated in Scheme 1 (x = 11 – 19), and the variable y represents the number of total double bonds of the partial sphingoid backbone (y = 0 – 2). The ion modes indicate the ionization mode(s) used to analyze the indicated lipid class in MDMS-based shotgun lipidomics. Gangliosides are not included, as discussed in the text.
The isomers that result from the different locations of double bond(s) are not considered. The number of molecular species are calculated based on the naturally occurring fatty acids that contain the highest degree of unsaturation for acyl chains of 12 – 26 carbon atoms are 12:1, 13:1, 14:3, 15:3, 16:5, 17:3, 18:5, 19:3, 20:6, 21:5, 22:7, 23:5, 24:7, 25:6, and 26:7, respectively, that were previously identified.
In this database constructed with the building-block approach, there are approximately 6,500 lipid species in glycerophospholipids, 3,200 in glycerolipids, 26,000 sphingolipid species, 100 sterols, and 410 other lipids that are predominantly involved in energy metabolism. Collectively, this virtual database contains over 36,000 molecular species, not counting regioisomers, oxidized lipids, or other covalently modified entities that will extend available entities by at least an order of magnitude. Furthermore, this database can be modified as new molecular entities are discovered to render a dynamic capacity for growth and coverage of entities that will be discovered as the sensitivity of mass spectrometers is further improved to facilitate the identification of new unknown lipids in biological samples.
3. Identification of building blocks for lipidomic analysis
Importantly, based on the concept of lipid building blocks, the individual molecular species of an entire lipid class or even a category of lipid classes of interest can be identified with identification of novel building blocks with bioinformatic approaches to thereby provide an automated identification of lipid species and thereby facilitate high-throughput lipidomic analysis. The building blocks can be represented with the fragments characteristic of individual lipid classes. The characteristic fragments can be determined with two powerful tandem MS techniques (i.e., NLS and PIS), as described above. Specifically, the building blocks can be monitored with either the specific loss of a neutral fragment in the NLS mode or the yield of a fragment ion in the PIS mode. Mapping of these building blocks forms a pseudo 2D mass spectrum, the vertical dimension of which might have a discrete mass or m/z unit instead of a continuous unit to efficiently monitor only naturally occurring building blocks. Accordingly, after the fragmentation pattern of a lipid class is characterized (see below), numerous individual molecular species of a lipid class can be determined with this pseudo 2D MS analysis.
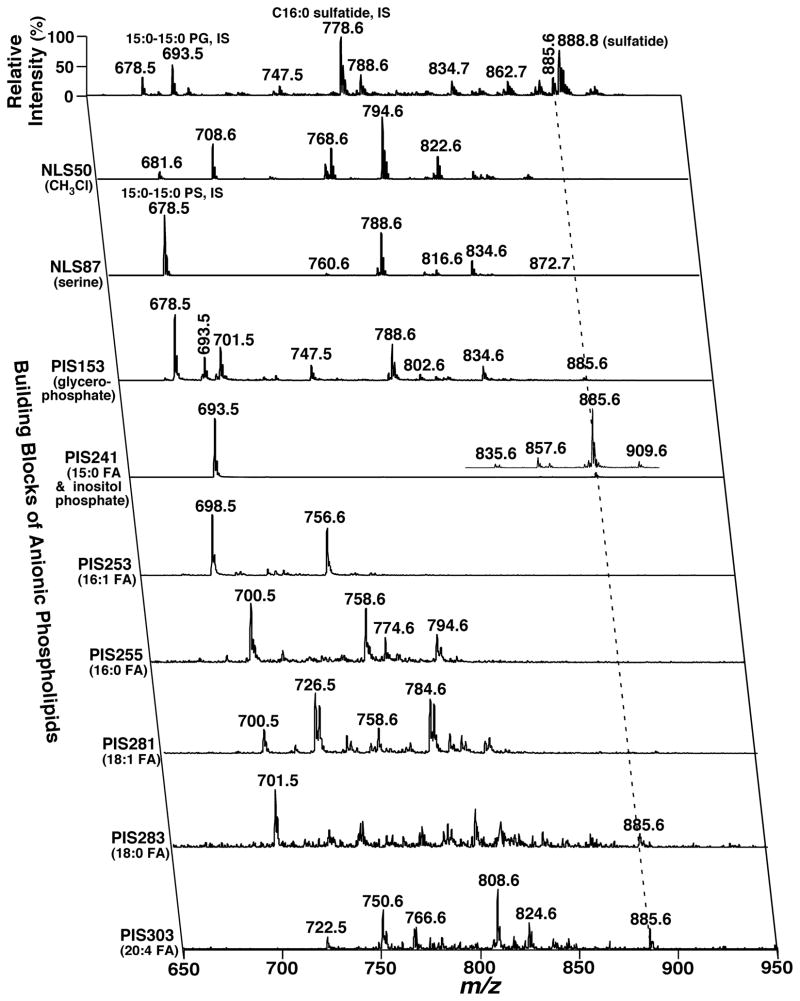
Table 3 summarizes some examples of the PIS and/or NLS for the analyses of the building blocks of each lipid class with MDMS. It is well known that many lipid classes have been characterized with different adducts and/or ionized in different ion modes under different experimental conditions (Christie & Han, 2010). Therefore, identification of a particular lipid class might be achieved with different characteristic fragment ions or informative neutral losses for the analyses of building blocks present in the molecule. The types and modes of fragmentation processes are highly context-dependent for either targeted or non-targeted analyses and/or the abundance of the molecular species of interest. Of course, the use of multiple complementary fragmentation modes can be employed to minimize false discovery rates for abundant species and enhance coverage for extremely low-abundance molecular species.
Table 3.
Summary of the representative building blocks in each lipid class used to identify individual species with MDMSa
| Lipid class (ref.) | Ion format | Scans for class specific prescreen | Scans for identification of acyl chain and/or regioisomers |
|---|---|---|---|
| PC (Yang et al., 2009b) | [M+Li]+ | NLS189, −35 eV NLS 183, −35 eV |
NLS(59+FA), −40 eV |
| lysoPC (Yang et al., 2009b) | [M+Na]+ | NLS59, −22 eV NLS205, −34 eV |
PIS104, −34 eV PIS147, −34 eV |
| PE, lysoPE (Han et al., 2005) | [M-H]− [M-H+Fmoc]− ([M+C15H9O2]−) |
PIS196, 50 eV for [M-H]− NLS222, 30 eV |
PIS(FA-H), 30 eV |
| PI, lysoPI (Han et al., 2004b) | [M-H]− | PIS241, 45 eV | PIS(FA-H), 47 eV |
| PS, lysoPS (Han et al., 2004b) | [M-H]− | NLS87, 24 eV | PIS(FA-H), 30 eV |
| PG, PA, lysoPG, lysoPA (Han et al., 2004b) | [M-H]− | PIS153, 35 eV | PIS(FA-H), 30 eV |
| Cardiolipin (CL), mono-lysoCL (Han et al., 2006a) | [M-2H]2− | Full MS at high resolution | PIS(FA-H) at high resolution, 25 eV; NLS(FA-H2O) at high resolution, 22 eV |
| Triacylglycerol (Han & Gross, 2001) | [M+Li]+ | NLS(FA), −35 eV | |
| Sphingomyelin (Yang et al., 2009b) | [M+Li]+ | NLS213, −50 eV | NLS(neutral fragments from sphingoid backbone) |
| Ceramide (Han, 2002) | [M-H]− | NLS(neutral fragments from sphingoid backbone), (e.g. NLS256, 32 eV for d18:1 non-hydroxy species) | NLS(neutral fragments from sphingoid backbone), (e.g. NLS256, 32 eV for d18:1 non-hydroxy species) |
| Hexosylceramide (Hsu & Turk, 2001; Han & Cheng, 2005) | [M+Li]+ | NLS162, −50 eV | NLS(neutral fragments from sphingoid backbone) |
| Sulfatide (Hsu, Bohrer, & Turk, 1998) | [M-H]− | PIS 97, 65 eV | NLS(neutral fragments from sphingoid backbone) |
| Sphingoid backbone-1-phosphate (Jiang & Han, 2006) | [M-H]− | PIS79, 24 eV | |
| Sphingoid backbone (Jiang et al., 2007) | [M+H]+ | NLS48, −18 eV | |
| Psychosine (Jiang, Yang, & Han, 2009) | [M+H]+ | NLS180, −24 eV | |
| Cholesterol (Cheng, Jiang, & Han, 2007) | [cholesteryl methoxyacetate +MeOH+Li]+ | PIS97, −22 eV | |
| Acylcarnitine (Su et al., 2005) | [M+H]+ | PIS85, −30 eV | PIS85, −30 eV for all species; PIS145, −30 eV for hydroxy species |
| Acyl-CoA (Kalderon et al., 2002) | [M-H]−, [M-2H]2−[M-3H]3− | PIS134, 30 eV | PIS134, 30 eV |
NLS and PIS stand for neutral-loss scan and precursor-ion scan, respectively. FA and (FA-H) denote free fatty acid and fatty acyl carboxylate anion, respectively.
II. MASS SPECTROMETRY-BASED APPROACHES FOR LIPIDOMIC ANALYSIS
The majority of the platforms for lipidomic analysis currently used are based on ESI in conjunction with MS/MS analyses. The approaches based on ESI-MS can be classified into two categories that depend on whether liquid chromatography (LC) is performed prior to infusion into the mass spectrometer. Approaches in which direct infusion of organic extracts into the ion source is used are broadly termed Shotgun Lipidomics. Distinguishing features of both approaches are summarized below. Moreover, substantial progress of the desorption ESI (DESI) technique for lipidomic analyses of lipid extracts and those in intact tissue samples has recently been made. This technique is briefly discussed. Finally, MALDI-MS has played many important roles in lipidomics, and we also discuss the new developments of MALDI-MS in lipidomics in this section.
A. LC-MS BASED Methods
The LC-MS based techniques include selected ion extraction (SIE), SRM/MRM, and data-dependent analysis. The SIE approach is usually used for “global” lipid analysis, in which a mass spectrum is continuously acquired during column elution and ions of interest are extracted from the acquired data array after a chromatographic separation. The reconstituted ion-peak area of each molecular species can be compared to either a standard curve of the molecular species or to the reconstituted ion peak area of an internal standard under identical experimental conditions. The combination of ESI-MS detection with HPLC separation and the sensitivity of SIE compared to other detection modalities makes this approach a judicious choice for lipid profiling and quantitation in many cases where extremely low abundance lipids are targets for identification. In practice, such a combination has been employed for many applications to identify lipid species. However, lipid quantitation with this methodology on a large scale is quite limited (Hermansson et al., 2005), although targeted analysis of a small number of lipids whose standard curves can be generated is quite common (Liebisch et al., 1999). The specificity of the extracted ion to the compound of interest is generally a concern with SIE due to multiple interferences that are commonly present. Accordingly, the use of high mass accuracy spectrometry is always preferable in this approach. In contrast to SIE, SRM (MRM) would be very specific if the monitored fragment ion could be specific to the precursor in combination with an LC separation and no interfering transitions were concomitantly present. However, the numbers of ions that can be monitored are limited, and previous knowledge of the each pair of precursor/fragment ions that result in the transition is required. The emphasis of data-dependent analysis is the elucidation of the structures of lipid ions detected in addition to the acquisition of spectra. Due to the limitation of data acquisition posed with limited duty cycles and other considerations, product-ion analyses in the method are typically limited to a limited number of abundant ions. It should be pointed out that these methods are suitable for any LC column (e.g., normal phase, reversed phase, ion exchange, hydrophobic, etc.) as long as the elution conditions can be coupled with a mass spectrometer.
For example, Hermansson and colleagues (Hermansson et al., 2005) employed a diol-modified silica column with isocratic elution with a mobile phase comprised of hexane-isopropanol-water-formic acid-triethylamine to separate over 100 lipid species. Through 2D maps of elution time and the masses of the ions, lipids were automatically identified and quantified. Sommer and colleagues (Sommer et al., 2006) fractionated different lipid classes. The first dimension of separation employed normal-phase HPLC-MS on a YMC PVA-Sil column, followed with reversed-phase LC-MS or LC-MS/MS with an Atlantis dC18 capillary column that led to the characterization of discrete lipid species. Similar approaches with dual parallel LC and dual MS were employed by Byrdwell for a total lipid analysis (Byrdwell, 2008).
Normal-phase and reversed-phase HPLC-MS have both also been employed for identification and quantification of classes and/or individual molecular species of a cellular sphingolipidome, which can be conveniently enriched with alkaline-hydrolysis followed by solid-phase extraction. Examples of the studies in this area can be found in several excellent reviews (Sullards et al., 2003; Merrill et al., 2005; Zheng et al., 2006). Detailed protocols for sample preparation, chromatographic separation of lipid classes and/or individual species with different columns, ionization, and tandem MS conditions are also available (Merrill et al., 2005). Column separation in these methods facilitates the identification of isomeric species (e.g., GalCer and GluCer) with identical fragmentation patterns, whereas shotgun lipidomics at its current stage of development encounters difficulties (see below).
Masukawa and colleagues quantified ceramide species in human stratum corneum with LC-MS (Masukawa et al., 2009) that employed normal-phase HPLC with a non-linear gradient. From 23 synthesized and purchased ceramide species, method development and quantitation were both facilitated. Collectively, over 182 molecule-related ions derived from the diverse ceramide species in the stratum corneum were measured.
Many researchers have attempted to use ultra-performance LC (UPLC) technology to replace the sequential separation with both normal-phase and reversed-phase HPLC (Laaksonen et al., 2006; Yin et al., 2006; Rainville et al., 2007).
The improved detection sensitivity and mass accuracy substantially facilitated the identification of the endocannabinoids and their analogs (see recent reviews (Astarita & Piomelli, 2009; Kingsley & Marnett, 2009)). Through online HPLC coupled to ESI-MS/MS, over 50 fatty acyl amino acids have been identified with a comparison of fragmentation patterns with authentic standards (Tan et al., 2009).
Similar to the amine group in amino acids, the primary amine in PS also serves as a nucleophile for acyl transfer with an acyl-CoA transferase activity using Acyl-CoA as substrate. Over thirty different N-acyl PS have been identified with LC-MS analysis in mouse brain extracts (Guan et al., 2007). Of particular interest is identification of N-arachidonoyl PS, which might be a biosynthetic precursor of the signaling lipid, N-arachidonoyl serine (Milman et al., 2006).
Reverse-phase chromatography used in conjunction with negative-ion ESI-MS/MS has been used for many years to identify and quantify eicosanoids in biological samples. A summary of the analysis of many eicosanoids and their quantification with stable-isotope internal standards has recently been reviewed (Murphy et al., 2005). However, these approaches were sub-optimal to measure the diverse array of eicosanoids in many tissues due to the relatively low sensitivity of negative-ion MS. Recently, Bohlinger and colleagues have developed a charge-switch methodology with a coupling reagent to facilitate a highly sensitive analysis of eicosanoids with positive-ion ESI-MS/MS (Bollinger et al., 2010b).
It should be emphasized that, although column separation can enrich low-abundance molecular species and eliminate the interactions of many lipid species, LC-MS analysis also contains inherent difficulties. For example, although it is often stated that chromatography obviates the effects of “ion suppression” through separation of competing analytes, there are large concentrations of ions in the column eluate necessary for ion-pairing and facile separations with reversed-phase HPLC. Moreover, the determined ionization efficiency of analytes is generally measured at different elution times in the chromatographic separation, which introduces errors in ionization efficiency from different mobile phase compositions. Furthermore, when a normal-phase HPLC column is employed for separation of different lipid classes, different lipid species in a class are not uniformly distributed in the eluted peak (i.e., each individual molecular species of a class possesses its own distinct retention time and peak shape due to differential interactions with the stationary phase and the ion-pairing agents employed). If a solvent gradient is employed to resolve individual molecular species with a reversed-phase HPLC column, then changes in the components of the mobile phase might also cause an ionization stability problem. It should be recognized that reversed-phase HPLC gradients are initiated largely with an aqueous mobile phase which induces solubility problems in a molecular species-dependent manner. In addition, differential loss of lipids on the column is also not unusual (DeLong et al., 2001). Finally, whereas reversed-phase chromatography eliminates many lipid-lipid interactions of one lipid with another molecular species, there is a large, up to a 1000-fold, increase in the amount of lipid-lipid interactions within the same lipid (homo-dimer formation) because reversed-phase HPLC is typically used to concentrate samples up to the limit of solubility in the aqueous phase, which leads to aggregation and differential ionization efficiencies. These practical difficulties limit the use of LC-MS for a large-scale analysis of lipids, in particular, for their absolute quantitation, although there are many examples of LC-MS in applications of discovery and identification of novel lipids, particularly those present in low or very low abundance in a small scale (Guan, 2009; Kingsley & Marnett, 2009; Tan et al., 2009; Minkler & Hoppel, 2010).
B. Shotgun Lipidomics
In contrast to the LC-MS based approaches for lipidomics, a second method, originally described in 1994 (Han & Gross, 1994) uses direct infusion to avoid difficulties from alterations in concentration, chromatographic anomalies, and ion-pairing alterations to improve the S/N ratio. This approach has now widely been termed “shotgun lipidomics”. Shotgun lipidomics exploits the unique chemical and physical properties of each lipid class to facilitate the high-throughput analysis of a cellular lipidome on a large scale directly from organic extracts of biological samples (Han & Gross, 2005b).
A major advantage of shotgun lipidomics over LC-MS analysis of lipids is that a mass spectrum of molecule ions of individual molecular species of a lipid class of interest can be acquired at a constant concentration of the solution during direct infusion. This unique feature of shotgun lipidomics allows virtually unlimited time to perform detailed tandem mass spectrometric mapping with multiple fragmentation strategies, including PIS, NLS, and a variety of other fragmentation techniques. Alterations in fragmentation energies, reagent gases, etc. can all be performed during the same infusion at a constant ratio of solvents to analytes that obviates difficulties typically encountered from the time constraints present in the “on the fly” analysis during chromatographic elution. Each suite of scans determines the identity of the molecule ion by recognizing that the majority of lipid species represents linear combinations of building blocks of naturally occurring aliphatic chains and polar head groups, as discussed above. Based on this unique feature, at least three different platforms of shotgun lipidomics have been developed, including:
1. Tandem MS-based shotgun lipidomics
As discussed above, a characteristic fragment associated with the head group of a class (i.e., one of the building blocks) is generally present. NLS or PIS of this fragment specifically detects individual species of the class. Following this line of reasoning, researchers have developed a method to “isolate” the individual species of a class of interest through the specific NLS or PIS (Brugger et al., 1997).
The advantages of this method include simplicity, efficiency, high sensitivity, ease of management, and less expensive instrumental requirements. All individual species in a particular class can be detected in one MS/MS acquisition directly from a total lipid extract with any commercially available QqQ type mass spectrometer. The improved noise level through the double filtering process of MS/MS enhances the S/N ratio typically by over an order of magnitude.
There are several concerns with this method, which are also well-recognized. Some of these concerns include (1) the aliphatic substituents are not identified; (2) the detection with the so-called specific MS/MS scanning might not be entirely specific to the class or the category of classes of interest, whereas this non-specificity might introduce some artifacts; (3) some altered ionization conditions cannot be easily recognized during and after the experiments; and (4) accurate quantification of the detected lipid species might not be as simple as expected because of the differential fragmentation mechanisms manifest in individual lipid species within each lipid class.
Because of its great advantages, many laboratories have adopted this platform for lipidomic analysis. For example, Welti and colleagues initially applied this platform to investigate the stress-induced changes of plant lipids and their biochemical mechanisms (Welti et al., 2002). This platform has become an essential tool for their research in plant lipidomics (Welti et al., 2007; Moreau et al., 2008).
Hsu and Turk have extensively characterized the fragmentation patterns of various classes of lipids (Hsu & Turk, 2009; Christie & Han, 2010). With the characterized class-specific fragments, they have also demonstrated the utility of these fragments to profile individual species of the classes. For example, the identification of cerebroside (HexCer) species in mixtures or specific HexCer subclasses that vary in sphingoid backbones that contain hydroxy and non-hydroxy fatty acids has been demonstrated with PIS or NLS (Hsu & Turk, 2001). Similarly, identification of phosphatidylcholine (PC) subclasses and individual molecular species has been achieved with PIS or NLS of the head group and/or aliphatic chains from their lithiated molecule ions (Hsu et al., 2003).
Hicks and colleagues employed this approach to perform simultaneous comparative analysis of PC, PE, PS, and PI in lipid extracts of many tissues from an adult rat (Hicks et al., 2006). They found that the profiling is highly reproducible from different scans of the same sample and samples from different animals. Their most intriguing finding from the profiling is that each tissue possesses a unique signature of phospholipid species. These profiles provide much information about the composition of cellular membranes, and can be used to profile differences during external perturbations.
Similarly, many other investigators have exploited a similar platform to investigate alterations in molecular species compositions in various biological issues (Mitchell et al., 2004; Mitchell, Buffenstein, & Hulbert, 2007; Nealon et al., 2008; Deeley et al., 2010). For example, the membrane composition of lenses from humans as well as those from commonly used experimental animals (Deeley et al., 2008).
McCarry’s laboratory has employed this approach to analyze eight lipid classes directly from crude extracts of the soil bacterium Sinorhizobium meliloti (Basconcillo et al., 2009a). These classes include monomethyl PE, dimethyl PE, sulfoquinovosyldiacylglycerol, ornithine-containing lipid, and diacylglyceryl-(N,N,N-trimethyl)-homoserines in addition to PC, PE, and PG. The mutants deficient in the PhoB regulator protein were analyzed (Basconcillo et al., 2009a) and the lipid levels in wild type and in a putative lysophosphatidic acyl transferase (plsC) knockout mutant were investigated (Basconcillo et al., 2009b).
There is no doubt that MS/MS-based shotgun lipidomics in combination with stable isotope labeling (e.g., deuterated choline) can provide further insights into the kinetics of lipid turnover, biosynthesis, lipid trafficking and homeostasis, etc. (Bleijerveld et al., 2006; Hunt, 2006; Postle et al., 2007; Postle & Hunt, 2009). The lipids incorporated with a stable isotope can be easily monitored with NLS or PIS of a fragment that contains the labeled tag or signature.
2. High mass accuracy-based shotgun lipidomics
Currently, commercially-available hybrid instruments such as a Q-TOF mass spectrometer offer an improved duty cycle that increases the detection sensitivity (Chernushevich, Loboda, & Thomson, 2001). In addition, the TOF analyzer records numerous virtual PIS in parallel, and the high mass resolution and accuracy inherent in the instrument records the accurate mass of fragment ions (0.1 amu) to minimize any false positive identifications (Ekroos et al., 2002). With these advantages of high mass accuracy/high mass resolution hybrid mass spectrometers, an alternative shotgun lipidomics approach has been developed (Ekroos et al., 2002; Ejsing et al., 2006b; Schwudke et al., 2006; Schwudke et al., 2007; Ståhlman et al., 2009). Specifically, with a Q-TOF mass spectrometer, a product-ion spectrum of each protonated/deprotonated molecule ion after direct infusion can be rapidly and efficiently acquired (Ståhlman et al., 2009). After product-ion analyses of protonated/deprotonated molecule ions within a narrow mass range of interest (e.g., 200 – 400 amu, in which fragments that correspond to fatty acyl chains are present), any interesting PIS and/or NLS can be extracted from the product-ion spectra. The analyses can be conducted in positive- and negative-ion modes in the presence of ammonium acetate in the infused solution (Ejsing et al., 2009). Identification can be performed from bioinformatic reconstruction of the fragments from PIS or NLS. Software packages (e.g., LipidProfiler (Ejsing et al., 2006b), LipidInspector (Schwudke et al., 2006), LipidXplorer (Herzog et al., 2011)) are commercially available for data processing. Quantification can be achieved with a comparison of the sum of the intensities of extracted fragments of an ion to that of a pre-selected internal standard. This technology has recently been applied to many biological studies (Ejsing et al., 2006a; Ejsing et al., 2009; Klemm et al., 2009; Zech et al., 2009; Klose et al., 2010; Sampaio et al., 2011).
3. MDMS-based shotgun lipidomics
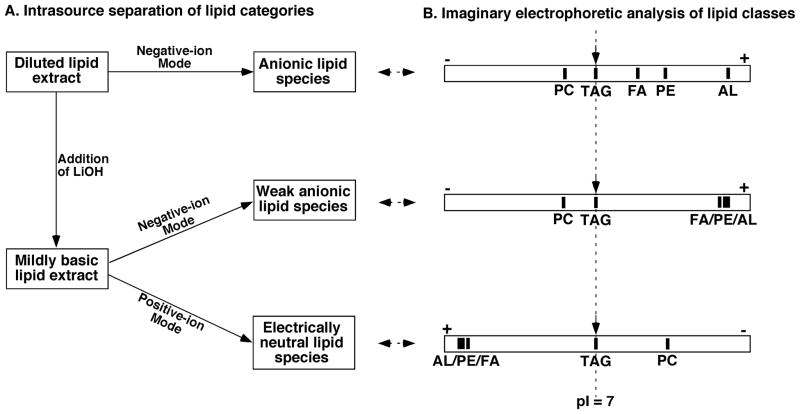
In the current practice of lipidomics, MDMS-based shotgun lipidomics (Han & Gross, 2001; Han et al., 2004b; Han & Gross, 2005a; Yang et al., 2009a) is a well-recognized platform to analyze individual lipid species directly from lipid extracts of biological samples. This technology maximally exploits the unique chemistries inherent in discrete lipid classes for the analysis of lipids, including low-abundance molecular species. For example, differential hydrophobicity and differential sensitivity to base treatment are exploited during a multiplexed extraction approach (Jiang et al., 2007). The differential charge properties of lipid classes are exploited to selectively ionize a certain category of lipid classes under multiplexed experimental conditions to separate many lipid classes in source. This separation method is analogous to the electrophoretic separation of different compounds that possess different pI values (Han et al., 2006b) (see Section III B6a).
The concept of building blocks in lipid structure (see Section I E) is fully employed for identification of individual lipid species (Han & Gross, 2005a; Han, 2007) because these building blocks can be determined with two powerful tandem MS techniques (i.e., NLS and PIS) in a mass-ramp fashion (see Section I E2) (Han & Gross, 2005b; Han & Gross, 2005a). Accordingly, after CID, the majority of lipid classes possess a unique fragment pattern that can usually be predicted based upon the covalent structures of these lipid classes. The informative fragment ion(s) from either the head group or the neutral loss of the head group are used to identify the lipid class of interest, and PIS or NLS of fatty acyl chains is used to identify the individual molecular species present within the class.
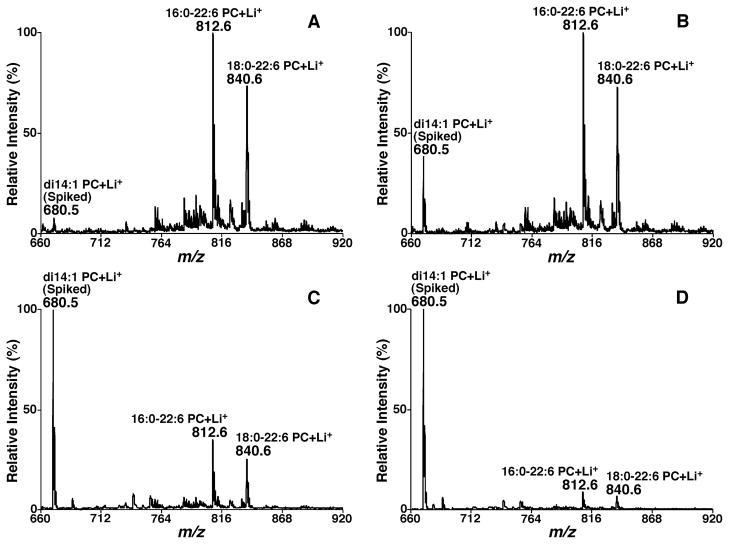
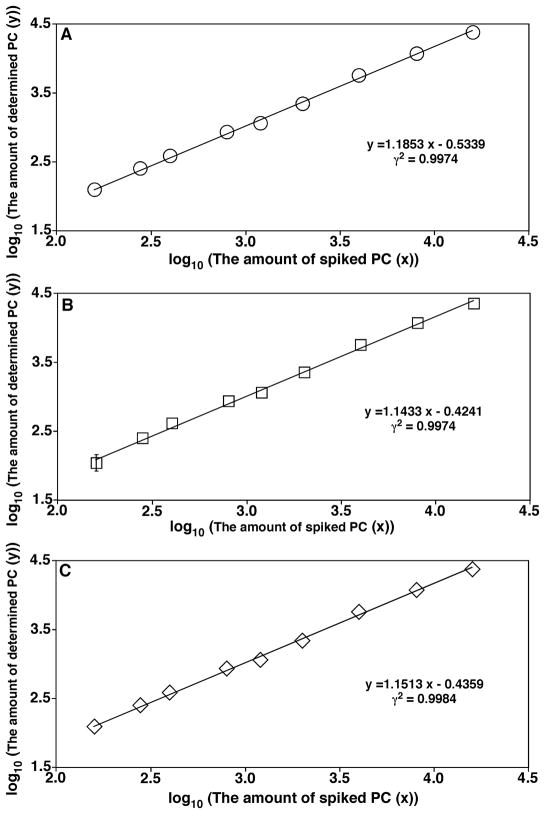
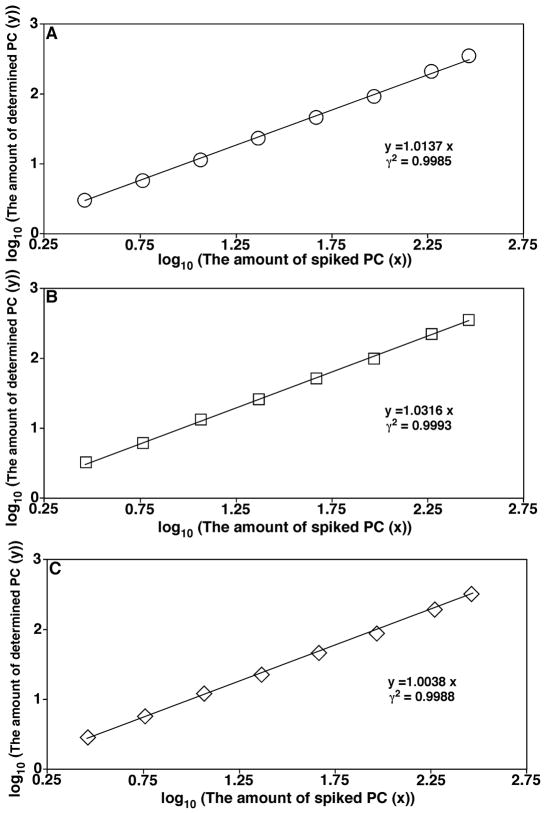
In MDMS-based shotgun lipidomics, these diagnostic ions are exploited not only for identification, but also for quantification from a direct comparison of the peak intensity of an ion with that of the selected standards (i.e., ratiometric comparison) in the same mass spectrum after correction for isotopologue. Unlike the source ionization that mainly depends on the charge properties of the analytes, the CID process depends on the chemical structure of each individual molecular species (DeLong et al., 2001; Han, 2002); in particular, their kinetic and thermodynamic properties. This fact has led to the development of a two-step method for quantitation of the identified individual lipid species (Han et al., 2004a; Han & Gross, 2005a; Cheng, Jiang, & Han, 2007). In this two-step methodology, the molecular species of the class of interest that are abundant and do not overlap with the species in any other classes are quantified with a ratiometric comparison after isotope correction with the selected internal standard of the class with a full MS scan. Some or all of these quantified molecular species plus the original internal standard are used as standards to quantitate other low-abundance and/or overlapped molecular species in the class with one or more class-specific PIS and/or NLS. With this two-step methodology, the dynamic range of quantitation can increase by at least two orders of magnitude, as previously demonstrated (Han, Yang, & Gross, 2008), and can be easily achieved with automated bioinformatic analyses (Yang et al., 2009a).
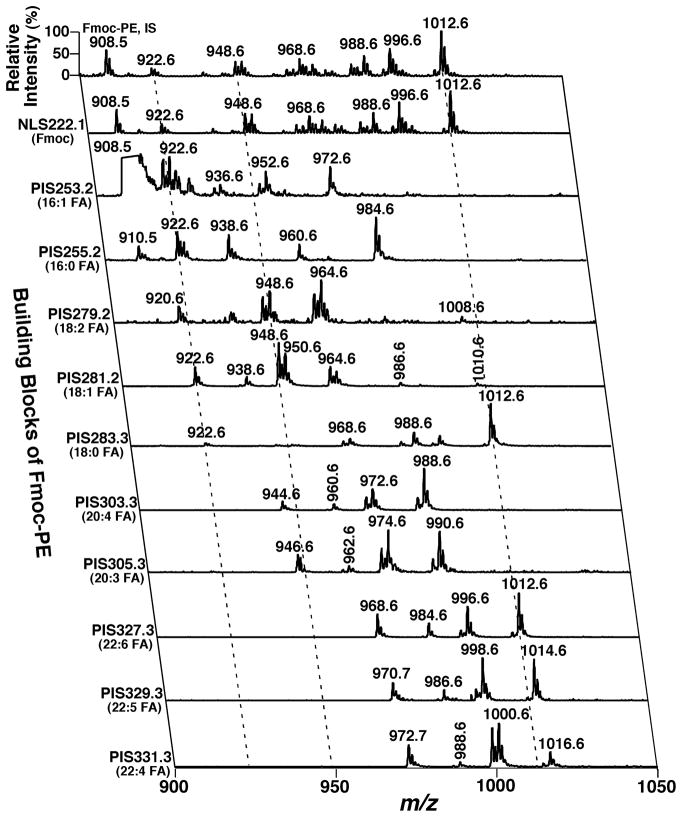
Finally, the uniqueness of individual lipid classes is exploited to identify and quantify the individual molecular species of the lipid classes. For example, the presence of two phosphate moieties in the cardiolipin (CL) chemical structure is unique, and is reflected as doubly-charged ions in mass spectra acquired under certain conditions (Han et al., 2006a). A search of the [M − 2H + 1]2− isotopologues from the doubly-charged ion pattern of CL species recognize CL species. In another case, the presence of a primary amine in phosphoethanolamine-containing species is unique in the cellular lipidome, and has been exploited to tag the phosphoethanolamine-containing lipid species with fluorenylmethoxylcarbonyl (Fmoc) chloride (Han et al., 2005). The facile loss of Fmoc from the tagged lipid species allows one to readily identify and quantify these species with unprecedented sensitivity at an amol/μL level.
C. The Development of DESI for Lipidomic Analysis
DESI allows direct sampling under the ambient environment and often without any sample preparation (Takats et al., 2004). In DESI, a pneumatically assisted electrospray produces charged droplets that are directed at a surface and collide with the surface of the sample. This collision ultimately generates ions from analytes through the electrospray solvent-evaporation process (Venter, Sojka, & Cooks, 2006).
DESI-MS has been used to systematically evaluate the characterization of phospholipids and sphingolipids (Manicke et al., 2008). The effects of surface and solvents on DESI-MS analysis of these lipids were investigated (Manicke et al., 2008). In the study, a total lipid extract from porcine brain was subjected to the analyses in positive- and negative-ion modes. The ions from anionic lipids such as deprotonated PS, PI, and sulfatide species dominate the spectrum in the negative-ion mode. In the positive-ion mode, as in ESI-MS, the predominant ions correspond to the PC species. The identities of these detected ions in negative- and positive-ion modes were identified from their fragmentation patterns after CID (Manicke et al., 2008).
Interrogation of the mouse pancreas tissue with DESI-MS showed strong signals from the PC species, the major lipid component of biological membranes, in the positive-ion mode (Wiseman et al., 2005). DESI-MS analysis of rat brain tissue demonstrated ions of phospholipid species, which mainly correspond to those with saturated fatty acids (Wiseman et al., 2005). Intriguing results were found from a DESI-MS analysis of cancerous and adjacent normal tissue of a human liver adenocarcinoma (Fig. 9) (Wiseman et al., 2005). Dominant signals from palmitic acid-containing phospholipids were present in the non-cancerous regions. Phospholipids with unsaturated fatty acids dominated in the transition area of the tissue. SM was markedly elevated in the tumor tissue. DESI-MS spectra of in situ intact adipose tissue that surrounded a chicken heart displayed signals of free fatty acids on the tissue surface (Wiseman et al., 2005).
FIGURE 9.
Positive-ion DESI mass spectra recorded from metastatic human-liver adenocarcinoma tissue. Methanol/water (1:1, v/v) with 0.1% NH4OH was sprayed. (a) Representative DESI mass spectrum from non-tumor region of the tissue. (b) Representative DESI mass spectrum from cancerous region of the tissue. (Reprinted from (Wiseman et al., 2005) with permission from Wiley – VHC, Copyright 2005).
Because many lipid species are readily ionized with DESI, these species are attractive target molecules to map thin tissue sections with this technique. DESI-MS has recently been used to construct chemical images of tissue sections (Dill et al., 2009). For example, DESI-MS tissue imaging has been performed to distinguish lipid profiles present in normal or benign, ductal carcinoma in situ (DCIS), and invasive ductal carcinoma (IDC) specimens of human breast samples (Dill et al., 2009). The distribution of two lipid ions at m/z 863 and 818 detected in the negative-ion mode were seen in breast tissue samples of benign, ductal carcinoma in situ, and invasive ductal carcinoma, which have been tentatively identified with MS/MS as PI and PS species, respectively. The intensities of these ions are vastly different among the tissue samples. The mass spectra obtained from the samples of ductal carcinoma in situ display significant intensities of the ion at m/z 863, but virtually absent for the ion at m/z 818. These observations indicate the use of DESI imaging mass spectrometry not only to distinguish cancerous vs. non-cancerous tissue or regions, but also to identify with MS/MS the specific type and stage of the cancer. This ability with DESI-MS to record spatial and molecular information simultaneously on surfaces is particularly powerful because this technique only requires limited preparation, does not require the addition of a matrix, and performs the analysis under ambient conditions.
D. New Developments of MALDI-MS for Lipidomic Analysis
MALDI-TOF/MS has been used for lipid analysis since it was introduced in the late 1980’s (Marto et al., 1995; Al-Saad et al., 2003). MALDI-TOF/MS has been applied to characterize every lipid class (e.g., free fatty acids, glycerolipids (e.g., diacylglycerol (DAG) and TAG), cholesterol and its derivatives, phospholipids, and sphingolipids) and to study oxidized lipids and biological lipid extracts (see (Fuchs, Suss, & Schiller, 2010) for an updated review). Although previous concerns about potential difficulties with MALDI analysis were prominently discussed, including matrix ionization that precluded examination of low molecular weight lipids, sweet-spot effects that compromised quantitation and post-source decay (Schiller et al., 2004), these problems have been overcome by the development and use of matrices that do not undergo significant ionization, and the use of low laser power in conjunction with enhanced desorption/ionization methods that result in minimal interference from cation adducts after careful extraction procedures.
1. Use of new matrices for lipid analysis
To improve the spot homogeneity for lipid analysis, ionic-liquid (or ionic-solid) matrices have been used (Ham, Jacob, & Cole, 2005; Jones et al., 2005; Li, Gross, & Hsu, 2005). In general, studies with ionic-liquid matrices have demonstrated many advantages for lipid analysis in comparison to the conventional matrices. For example, ionic-liquid matrices possess a strong UV absorbance that reduces post-source decay and increases signal intensity (Li, Gross, & Hsu, 2005). Moreover, some promise to improve the homogeneity of lipid distribution in target spots with ionic-liquid matrices has been achieved. Homogeneity is a key to improve analysis reproducibility and to potentially facilitate quantitative analysis of lipid species with MALDI-MS. Alternatively, high repetition rate lasers can sample multiple spots to provide an accurate stochastic description of analytes within the matrix.
We have recently used 9-aminoacridine (9-AA) under different acidic/basic conditions to form multiplexed charge-transfer complexes for the analyses of different lipid classes (Sun et al., 2007; Sun et al., 2008). We have found unusual solvent-enabled interactions of lipids with this matrix during the crystallization process, which can be conducted in less-polar solvent system such as isopropyl alcohol. Use of the less-polar solvents leads to an increase in solubility and homogeneous distribution of lipids (both reduce aggregation). This neutral matrix substantially facilitates the desorption/ionization of lipids in the negative-ion mode, and has a low matrix background within the low mass range of interest. Therefore, different lipid classes can be selectively ionized in either the positive- or negative-ion mode (Sun et al., 2007; Sun et al., 2008; Cheng et al., 2010b). The larger conjugated system in 9-AA should result in a better dispersion of laser energy in comparison to commonly used matrices (e.g., α-cyano-4-hydroxycinnamic acid (CHCA) and 2,5-dihydroxybenzoic acid (DHB)), to thereby reduce post-source decay. These differences of 9-AA in chemical and physical properties from other matrices lead to a substantial increase in a S/N ratio (Fig. 10) and reproducible, quantitative results with 9-AA (Sun et al., 2008; Cheng et al., 2010b). The latter has been supported with the acquisition of MALDI-MS spectra with 9-AA essentially identical to those obtained with ESI-MS analysis of the identical lipid solutions (Fig. 11). Similar to the principles that underpin “intrasource separation” (Han et al., 2006b), many of the lipid classes can be high selectively ionized, including TAG in the positive-ion mode. Furthermore, in the negative-ion mode, 9-AA has proven to be highly sensitive for the analysis of negatively charged lipids, including CL, PI, and sulfatides directly from extracts of mammalian tissue samples without any prior chromatography (Sun et al., 2008; Cheng et al., 2010b). Use of this matrix to analyze lipids from different sources has recently been reported from different laboratories (Angelini et al., 2010; Cheng et al., 2010b; Dannenberger et al., 2010; Fuchs, Suss, & Schiller, 2010; Lobasso et al., 2010).
FIGURE 10.
The effects of matrix on MALDI-TOF/MS analysis of lipids. MALDI mass spectra of di18:0 phosphatidylcholine (PtdCho) acquired on a 4800 MALDI-TOF/TOF Analyzer in the positive-ion mode with different matrices: A) 9-aminoacridine (10 mg/mL) dissolved in isopropanol/acetonitrile (60/40, v/v); B) CHCA (10 mg/mL) dissolved in methanol with 0.1% TFA; C) DHB (0.5 M) dissolved in methanol with 0.1% TFA; and D) THAP (40 mM) dissolved in methanol. The prefix ‘‘D’’ stands for diacyl species. (Reprinted from the supplemental materials of ref. (Sun et al., 2008) with permission from the American Chemical Society, Copyright 2008).
FIGURE 11.
Mass spectral comparisons of phosphatidylcholine species present in mouse heart lipid extracts and sulfatide species present in mouse brain cortical lipid extracts acquired with either ESI or MALDI. Lipid extracts of murine myocardium (upper panel) or mouse brain cortex (lower panel) were examined with (A) and (C) ESI-MS or (B) and (D) MALDI-TOF/MS with 9-aminoacridine as the matrix (Sun et al., 2008; Cheng et al., 2010b). The prefix “D” stands for diacyl species. “IS” denotes internal standard.
Graphite-assisted laser desorption/ionization mass spectrometry (LDI-MS) has been investigated by Yeung’s group (Cha & Yeung, 2007; Zhang, Cha, & Yeung, 2007; Cha et al., 2008). It has been shown that 22 HexCer species can be detected with graphite-assisted LDI-MS, whereas only eight HexCer species are detected with MALDI-TOF/MS (Cha & Yeung, 2007). From a thin film of colloidal graphite on rat brain tissue, direct lipid profiling was performed. Chemically selective analysis for HexCer and sulfatide species was successfully obtained.
Very recently, a nanostructure-surface based MS technique has been developed (Northen et al., 2007; Woo et al., 2008; Patti et al., 2010a; Patti et al., 2010b). With this technique, Patti and colleagues (Patti et al., 2010a) detected intact cholesterol and its derivative molecules in situ to provide the first images of brain sterol localization in a knockout mouse model of 7-dehydrocholesterol reductase. They found that in the knockout mice, there is a striking localization of 7-dehydrocholesterol and residual cholesterol in the abnormally developing cerebellum and brainstem. In contrast, the distribution of cholesterol in 1-day old healthy pups was diffuse throughout the cerebrum and comparable to that of adult mice. This study clearly demonstrated that typical cholesterol biosynthesis might be particularly important for the development of these brain regions. Other nanomaterials (e.g., silver nanoparticles) have also been used to analyze lipids directly from tissue samples (Hayasaka et al., 2010).
2. HPTLC- and TLC-blot-MALDI MS
To resolve the difficulties in the analysis of acidic phospholipids due to ion suppression by PC species (Schiller et al., 2004), investigators have demonstrated that high performance thin layer chromatography (HPTLC)-separated lipids can be conveniently analyzed with MALDI-TOF/MS directly on the TLC plates (Rohlfing et al., 2007; Fuchs et al., 2008; Stubiger et al., 2009). Briefly, developed HPTLC plates are cut to small pieces to fit into the sample plate. Acquisition of mass spectra is started directly after the sample plate is placed into the ion source of the mass spectrometer. Typically, between 50 and 120 single laser shots are applied on different positions of individual HPTLC bands. The sensitivity of the method is 10 – 150 pmol of material spotted for HPTLC, and depends on phospholipid acidity, Rf value, and ion polarity (Rohlfing et al., 2007). With the same line of reasoning, an alternative approach that couples TLC-blot with MALDI-TOF/MS has been developed to image the bands and to quantify the separated lipid classes (Goto-Inoue et al., 2008; Goto-Inoue et al., 2009; Taki et al., 2009).
3. Potential spatial analysis of cellular lipidomes with MALDI-MS: Direct profiling and mapping of lipids from tissues
MALDI-TOF/MS has already been successfully used for many years to directly map and image peptides and proteins in mammalian tissues (Stoeckli et al., 2001) and even in single cells (Li, Garden, & Sweedler, 2000). This technique has been extended to lipid profiling in single zooplankter (Ishida et al., 2003), muscle (Touboul et al., 2004), and brain tissues (Jackson, Wang, & Woods, 2005a; Jackson, Wang, & Woods, 2005b). Recently, the developments of MALDI-TOF/MS to directly profile and/or map lipids in tissue samples are very rapid. MALDI-TOF/MS opens a new door to determine spatial distribution of cellular lipidomes.
For direct analysis of brain tissue lipids, frozen brain tissue is cut as thin as possible with a cryostat, and different matrices and matrix concentrations have been optimized for direct lipid profiling (Jackson, Wang, & Woods, 2005a). Similar to MALDI-TOF/MS analysis of lipid extracts of biological samples as discussed above, ion peaks that correspond to PC species are also prominent in MALDI-TOF/MS spectra of tissue analysis in the positive-ion mode. Either protonated molecular species (Jackson, Wang, & Woods, 2005a) or sodium adducts (Ishida et al., 2003) of PC species are abundant in these MALDI-TOF/MS spectra, which depends on the matrix and other reagents. These primary studies mapped tissue lipid distribution (Altelaar et al., 2006; Hankin, Barkley, & Murphy, 2007). Research in this direction provides a prototypic model and information about spatial distribution of lipids at the cellular level (McDonnell & Heeren, 2007; Walch et al., 2008; Jackson & Woods, 2009).
New strategies and/or use of new matrices circumvent many problems in tissue mapping (Hankin, Barkley, & Murphy, 2007; Chen et al., 2008; Hayasaka et al., 2010; Patti et al., 2010a). Homogeneous spotting of the matrix on the tissue slice is very critical for a successful and meaningful analysis. Matrix spotting with sublimation has markedly improved this area (Hankin, Barkley, & Murphy, 2007). However, solutions to the potential disturbance of the spotted matrix to cellular organelles and to the extraction of quantitative results from the analysis are still needed (Jones et al., 2006).
III. MULTI-DIMENSIONAL MASS SPECTROMETRY (MDMS) IN LIPIDOMICS
As described above, a map of the neutral-loss fragments or the fragment ions of interest results in a 2D mass spectrum (see Section I D). Many variables exist in mass spectrometry, including those in lipid extraction, infusion, ionization, and MS/MS analysis. Each of the variables can add a new dimension to a 2D mass spectrum to form a multidimensional array that contains critical information on the molecular species present and their abundance directly from organic extracts of biological tissues or fluids It should be pointed out that, due to the limitation of the time frame in LC-MS, many variables mentioned above are unable to be accessed with LC-MS-based platforms. However, it is anticipated that in the future the principles of MDMS can be broadly applied to a variety of platforms. In the following sections, we discuss the principles and the variables of MDMS for lipidomic analysis.
A. The Principle of Multi-dimensional Mass Spectrometry
A 2D mass spectrum, constructed through product-ion scans, NLS, or PIS as mass or m/z varied unit by unit within the entire mass range of interest, can readily map the complete and informative fragments for each individual molecular species, and, therefore, can be used to identify each molecular species in an approach appropriate for global studies of a complex cellular lipidome (see Section I D). Among the basic 2D mass spectra, the cross peaks of a given primary molecule ion in the first dimension with the second dimension represent the fragments of this given molecule ion. Analysis of these cross peaks (i.e., the individual fragments) determines the structure of the given molecule as well as its isomers and isobars (Han & Gross, 2005a). Han and Gross have referred to these kinds of two-dimensional maps as two-dimensional MS (Han & Gross, 2001; Han et al., 2004b; Han & Gross, 2005a), because they are entirely analogous to two-dimensional nuclear magnetic resonance spectroscopy. The only difference between these mapping approaches is that the former is in the mass domain whereas the latter is in the frequency domain. MDMS is built up with adding one or more additional dimension(s) to any constructed 2D MS. Largely, from a new set of spectra when an individual variable of mass spectrometry is ramped, a new dimension is added to the basic 2D MS to ultimately build up MDMS.
In theory, to fully investigate the effects of ionization conditions on ionization efficiency and/or the effects of collision conditions on fragmentation processes or other effects, a variety of ionization voltages, ionization temperatures, collision energies, collision gas pressures, etc. should be employed in an experiment. These variables can all be logically varied unit by unit within a certain range. All of these dimensions form the family of MDMS (Han & Gross, 2005a). Specifically, MDMS is defined as the comprehensive MS analyses conducted under a variety of instrumental variables that collectively comprise a MDMS spectrum. At the current stage, MDMS is decomposed into multiple 2D MS for ease of use and displayed by varying only one variable at a time while keeping the others fixed under experimental conditions. It is anticipated that advanced computational technology can eventually facilitate the direct use of 3D MS or MDMS and provide a new level of information directly obtainable from the mass spectrometric analysis in the next generation of computational MDMS-based shotgun lipidomics.
B. Variables in Multi-dimensional Mass Spectrometry
In this section, we discuss each potential variable in a mass spectrometer.
1. Variables in building-block monitoringwith MS/MS scanning
a. Precursor-ion scanning of a fragment ion whose m/z serves as a variable
In theory, this variable in m/z should vary from zero to the highest value (Mh) of m/z of a mass range of interest. However, the lowest value of a stable fragment ion in lipid analysis is generally the adduct ions (i.e., Li+, Na+, Cl−, etc.) whereas the highest value of a stable fragment ion corresponds to the loss of a small molecule (e.g., H2O, CH4, etc.) from the adducted molecule ion. Accordingly, the ramped m/z range of this variable at least can be narrowed to the range of 50 to Mh-18.
In practice, one can learn the resultant product ions of all species of a lipid class of interest from the analysis of a few representative lipid species of the class. Hence, the variable in precursor-ion mapping can vary to cover only those potential product ions. Furthermore, the identity of a lipid species can be fully determined with only two or three characteristic product ions that carry the building-block information about the species in addition to the m/z of the molecule ion. The task of mapping the fragment ions can be simplified to only monitor these characteristic building-block fragment ions as described in the section of “INTRODUCTION” (see Section I D).
b. Neutral-loss scanning of a neutral fragment whose mass serves as a variable
Similar to precursor-ion mapping, this variable in mass should vary from zero to the highest mass value (Mh) of molecule ions of a mass range of interest. However, the lowest value of a neutral-loss fragment in lipid analysis generally represents the value of a small molecule lost (e.g., H2O, CH4, etc.) whereas the highest value of a neutral-loss fragment is the molecule that is lost to yield the smallest fragment ion. Accordingly, this variable varies in a reversed order of that in precursor-ion mapping. This relationship is demonstrated in Figures 5 and 6.
In practice, one can learn the neutral-loss fragments from the product-ion analysis because the lost mass equals the difference between a molecule ion and a product ion. Hence, the variable in neutral-loss mapping can monitor only those neutral-loss fragments that correspond to the potential product ions. Again, the identity of a lipid species can be fully determined with only two or three neutral-loss fragments that correspond to the characteristic product ions which carry the building block information about the species in addition to the m/z value of molecule ion. Thus, the task of mapping the neutral-loss fragments can be simplified to only monitor those characteristics of building blocks as described in the section “INTRODUCTION” (see Section I D).
c. The fragments in product-ion analysis of molecule ions serve as a variable
Although the variable in this mapping is the m/z values in the survey mass spectrum within the mass range of interest, the new dimension is the resultant product ions that constitute a 2D map as demonstrated in Figure 4. It should be recognized that the mass window for precursor-ion selection in product-ion analysis is critical in the mapping. Accordingly, either a small mass window centered on the ion peaks should be selected or a mass spectrometer with high mass accuracy should be employed to eliminate any complications due to the overlap from adjacent peaks. In practice, one can scan only the m/z of the molecule ions present in the survey scan spectrum. Unless the mass spectrometer can be used to perform product-ion analysis with a high duty cycle, this mapping approach is usually not practical for a high-throughput platform. Obviously, the high throughput platform is necessary for global lipidomic analysis because the survey-scan mass spectrum of a complex lipid mixture is complicated in most cases.
2. Variables related to the infusion conditions
a. Polarity, composition, ion pairing and other variations in the infusion solvent as variables in multidimensional mass spectrometry
Solvent(s) used in the infused lipid solution not only affect the solubility of lipids, but also have a large impact on ionization efficiency. Lipids are poorly soluble in polar solvents; they form micelles that cannot be efficiently ionized. Accordingly, variation of solvent polarity and/or composition is important to optimize the experimental conditions and to be familiar with the chemical and physical properties of the lipid samples. Reports of the latter case are not seen in the literature. A continuous variation of solvent composition can be achieved with a gradient mixer. In current lipidomic analysis with ESI-MS, the most commonly used solvent system is chloroform (or dichloromethane) and methanol (from 1:2 to 2:1 (v/v)) with or without modifiers. Isopropyl alcohol is another commonly used solvent, mostly in combination with chloroform and methanol.
b. The amount or composition of a modifier (e.g., LiOH, ammonium acetate, formic acids, …) in a solvent system is a variable
The chemical and physical properties of the modifier added to the infused lipid solution can substantially affect the ionization sensitivity and efficiency as well as the molecule ion profiles of different lipid classes. Notably, PC species are generally ionized as the cation adduct in the positive-ion mode and as an anion adduct in the negative-ion mode.
With the addition of an acidic modifier (e.g., formic acid, acetic acid, etc.), the ionization of PC, SM, and PE in a lipid mixture as the protonated ions is favored in the positive-ion mode (Fig. 12A). Although the species of other acidic lipid classes (e.g., PS, PI, and PG) can also be ionized in the positive-ion mode under the acidic conditions without the co-existence of PC, SM, or PE species in the mixture (Hsu & Turk, 2009), they are suppressed with PC, SM, or PE species if these species are present in a mixture (Fig. 12A). The ionization of the acidic lipid species is also not favorable in the negative-ion mode under acidic conditions because an acidic condition prevents the deprotonation of these species in the negative-ion mode. Moreover, the ionization of PC and SM as an anion adducts further complicates the mass spectrum acquired (Fig. 12B).
FIGURE 12.
Representative positive- and negative-ion ESI mass spectra acquired under weak acidic, neutral, and weak basic conditions. A lipid extract of mouse myocardium was prepared and mass spectrometric analysis was performed (Han, Yang, & Gross, 2008). Positive- and negative-ion ESI mass spectra as indicated were acquired after direct infusion in the presence of 10% acetic acid (Panels A and B), 5 mM ammonium acetate (Panels C and D), and 10 μM lithium hydroxide (Panels E and F) in the infused solution. IS and Ac stand for “internal standard” and “acetate”, respectively.
With addition of a neutral modifier (e.g., ammonium acetate, ammonium formate, lithium chloride, etc.), PC and SM species are ionized as the protonated ions in the case of a modifier with ammonium or the cationic adducts of the modifier and TAG species are only ionized as the cation adducts in the positive-ion mode (Fig. 12C) (Han et al., 2000; Ejsing et al., 2006b). Acidic lipids are suppressed with the cation adducts of PC species due to their poor ionization efficiency in comparison with PC (Fig. 12C). However, the acidic lipids can be ionized as the protonated ions or the cation adducts if they are analyzed individually (Hsu & Turk, 2009). The molecular species of PE possess weak acidic properties (Han & Gross, 2003). Accordingly, ionization of PE species falls in between the PC and acidic lipids in the positive-ion mode. In the negative-ion mode under the experimental conditions, acidic lipids, including PE species, are selectively ionized as the deprotonated molecule ions while PC species can be ionized as anion adducts (Fig. 12D). In an equimolar mixture, the peak intensities of the ions that correspond to acidic lipids are always more abundant than those that correspond to PC species in the negative-ion mode (Han et al., 2006b).
With addition of the basic modifiers (e.g., LiOH, NH4OH, etc.), all the acidic lipids, including PE species, which are present in the deprotonated form in the solution, are not favorable for their ionization in the positive-ion mode, but their ionization is enhanced in the negative-ion mode. In addition, ionization of PC species as OH− adduct is not favored. Accordingly, PC species are selectively ionized as the protonated ions or cation adducts in the positive-ion mode whereas acidic lipids, including PE species, are selectively ionized in the negative-ion mode (Figs. 12E and 12F). Because all the acidic lipids, including PE species, are present in the deprotonated form in the solution, their ionization efficiency in the negative-ion mode is quite similar, as previously demonstrated (Han et al., 2006b). Hence, the peak intensity ratios well-represent the molar ratios of these lipid species (Fig. 12F). This observation is in contrast to those when acidic or neutral modifiers are added, where acidic lipids such as PG and PS species are selectively ionized in comparison to weak acidic PE species (Figs. 12B and 12D). These types of selective ionization have been the bases of the intrasource separation technique as previously described (Han & Gross, 2003; Han & Gross, 2005b; Han et al., 2006b) and are summarized in Subsection III B6a.
It should be specifically pointed out that, although the presence of different modifiers could substantially affect the molecule ion profiles of different lipid classes, the profile of individual molecular species of each class is not influenced. For example, the ionization efficiency of PE species is very different under different experimental conditions (Fig. 12). However, the ratios of individual PE species (e.g., di16:1 PE vs. 18:0–22:6 PE at m/z 688.4 and 792.5 in Figure 12A and at m/z 686.4 and 790.5 in Figures 12B, 12D, and 12F, respectively) are essentially constant under these conditions. Similarly, these constant ratios can be readily observed between the PC species such as the pair of di14:0 PC and 16:0–22:6 PC as protonated at m/z 674.4 and 806.5 (Figs. 12A and 12C), as lithiated at m/z 680.4 and 812.5 (Fig. 12E), and as acetate adducts at m/z 732.4 and 864.5 (Fig. 12B), respectively. These observations strongly support the notion that ionization efficiency of individual molecular species of a class minimally depends on its aliphatic chain composition during a full mass scan of the molecular ions. Thus, individual molecular species within a class can be quantified through ratiometric comparisons with the selected internal standard of the class. This point is further validated in Section IV B3.
Again, if a continuous variation of either the concentration of a modifier or a component of the modifiers in the solution is required, then a gradient mixer with a selective switch can be employed. Although stepwise addition of different modifiers or different amount of a modifier is frequently used in the practice of lipidomic analysis, a continuous change of modifiers has not been seen in the literature.
c. Lipid concentration of the infusion solution as a variable
Many studies have demonstrated that ESI of lipids is a concentration-dependent process (Han & Gross, 1994; DeLong et al., 2001; Koivusalo et al., 2001). At a concentration higher than 0.1 nmol/μL (i.e., 0.1 mM) in chloroform-methanol (1:1, v/v), the effects of acyl chain length and unsaturation on ionization efficiency are apparent (Koivusalo et al., 2001; Zacarias, Bolanowski, & Bhatnagar, 2002). This effect is largely due to the fact that lipids tend to form aggregates with increased lipid concentration and solvent polarity, and due to the fact that the aggregates are not readily ionizable or at least ionized at very different efficiency in comparison to their monomeric counterparts.
It should be emphasized that, in the low concentration range (i.e., pmol/μL or lower), a linear correlation of the absolute ion intensity with the concentration of each polar lipid class is obtained (Han & Gross, 1994; Kim, Wang, & Ma, 1994; DeLong et al., 2001; Koivusalo et al., 2001) (also see discussion in the last subsection). The ionization efficiency (or the response factor of a mass spectrometer) of lipid species depends predominantly on the charge properties of the polar head groups in this concentration range after correction for 13C isotopologue distributions (Han & Gross, 1994; Han et al., 1996; Han & Gross, 2005b; Han & Gross, 2005a).
Accordingly, use of the concentration of the infusion solution as a variable could allow one to determine the formation of aggregates from identical lipid species or from different lipid species, and to study chemical or physical properties of lipid molecules with the competition for proton or small matrix cation or anion among lipid molecules in a lipid class or molecules in different lipid classes. In practice, a continuous variation of lipid concentration can be achieved with a gradient mixer.
3. Variables under ionization conditions
a. Capillary temperature
Desolvation is very important in the ionization process that occurs in ESI source. Source temperature is a critical factor in desolvation, and affects the ionization efficiency. Temperature condition for the analysis of any samples with ESI-MS is usually optimized prior to the analysis. However, the required temperature for best ionization of each individual lipid class might be different. Moreover, due to the variation in the components as well as composition of individual samples, the pre-optimized temperature condition might not be the best one for a particular sample. Accordingly, ramping the temperature should provide the best condition for the analysis of each individual lipid class of a sample or for the analysis of each specific sample. In addition, through a temperature ramping, information about the ionization efficiency of different components present in the samples can be revealed. Furthermore, any interactions among the analytes, solvents, and other matrix components can also be interrogated with data array analysis (i.e., MDMS analysis) acquired at the variety of temperatures.
b. Spray voltage
Similar to the variation of temperature, spray voltage is also a critical factor that affects ionization efficiency. This factor is usually pre-determined in one of the steps such as tuning, calibration, or optimization with a standard solution. Such a pre-optimized parameter might not be the best condition for the analysis of each individual lipid class in a sample that might possess differential charge properties or of each individual sample that might vary in the concentration and composition of the analytes and/or matrix components. Accordingly, spray voltage optimization provides the best condition for the analysis of each individual lipid class in a sample or of each individual sample. In addition, the effects of spray voltage on the ionization efficiency of different components present in the samples can be determined with a spray voltage ramping. Furthermore, any interactions among the analytes, solvents, and other matrix components can also be investigated through examination of this MDMS dimension.
c. Flow rate
Previous studies have demonstrated that flow rates not only affect the ionization efficiency and sensitivity (Han et al., 2006b), but also influence other chemical or physical consequences such as intrasource separation (Han et al., 2006b). This factor is usually predetermined at the optimization step with a standard solution. Such a pre-optimized parameter might not be the best condition for the analysis of each individual sample, which might have varied concentration and composition of the analytes and/or matrix components. Accordingly, the flow rate optimization is important not only for optimal analysis of each individual sample, but also to investigate any interactions among the analytes, solvents, and other matrix components as well as their chemical/physical consequences.
4. Variables under collision conditions
a. Collision energy
Collision energy provides kinetic energy to molecule ions in a collision cell. Some of the kinetic energy is converted into internal energy during collision with a neutral molecule and results in bond breakage and consequently the fragmentation of the molecule ion into smaller fragments. The effects of collision energy on the appearance of a tandem mass spectrum of any type of scans aforementioned have been well-recognized. Differential fragmentation patterns, pathways, kinetics, or other chemical/physical parameters can be determined with analysis of this MDMS variable. For example, Figure 13 shows a 2D ESI mass spectrum of a neutral loss of 50.0 amu (i.e., loss of chloromethane from the chloride adducts of phosphocholine-containing species) from a diluted hepatic lipid extract under conditions with varied collision energies. This 2D mass spectrum clearly illustrates the differential fragmentation patterns of hepatic PC and SM species at the differential collision energies. This analysis indicates that the fragmentation kinetics of chloride adducts of PC and SM species is molecular-species dependent. In another case, Figure 14 shows an array of collision energy-varied product-ion mass spectra of an ion at m/z 305.2 that corresponds to 8,11,14-eicosatrienoic acid. From such a 2D mass spectrometric analysis, the location of the double bonds can be identified based on the intensity changes of the fragment ion that corresponds to [M – H – 44]− (Yang et al., 2011b).
FIGURE 13.
Two-dimensional electrospray ionization mass spectrometric analysis of choline glycerophospholipid species present in a mouse liver chloroform extract under different collision energy conditions. Two-dimensional MS analysis of neutral-loss scans (NLS) of 50.0 amu that corresponds to the loss of methyl chloride from the chloride adducts of phosphocholine-containing species) from a crude hepatic lipid extract was performed with a variation of collision energies. The 2D mass spectrum shows a few representative NLS acquired under the corresponding collision energies as indicated. The survey-scan mass spectrum acquired in the negative-ion mode directly from a diluted hepatic lipid extract is shown on the top. “IS” denotes internal standard. All mass spectral traces are displayed after normalization to the base peak in each individual spectrum.
FIGURE 14.
Two-dimensional mass spectrometric analysis of polyunsaturated fatty acid fragmentation pattern with variation of collision energy. Mass spectrometric analysis was performed with a TSQ Quantum Ultra Plus triple-quadrupole mass spectrometer (Thermo Fisher Scientific, San Jose, CA) equipped with an automated nanospray apparatus (i.e., Nanomate HD, Advion Bioscience Ltd., Ithaca, NY) and Xcalibur system software. The first and third quadrupoles were used as independent mass analyzers with a mass resolution setting of 0.7 Thomson, and the second quadrupole served as a collision cell for tandem mass spectrometry. Product-ion scan of 8,11,14-eicosatrienoic acid (20:3 FA) (5 pmol/μL) was performed after direct infusion in the negative-ion mode at a fixed collision gas pressure of 1 mTorr and varied collision energies from 2 to 36 eV at an interval of 2 eV as indicated. A 2-min period of signal averaging in the profile mode was used for each scan. All the scans were automatically acquired with a customized sequence subroutine operated under Xcalibur software. All the scans are displayed after being amplified to the 20% of the base peak in each individual scan.
b. Collision-gas pressure
Collision-gas pressure affects the collision pathway and collision frequency of an ion in the collision cell. The higher the collision-gas pressure, the shorter the collision pathway of an ion and the more frequently the ion collides in the cell. This shorter collision pathway and increased frequency of collisional events can lead to additional kinetically determined fragment ions and/or a fragment pattern with increased intensities of the fragments possessing lower masses. Accordingly, although this parameter is determined in optimization of instrument condition and is largely fixed during an experiment, variation of collision gas pressure could be used to study different fragmentation pathways, fragmentation patterns, and/or other chemical/physical parameters. For example, Figure 15 shows an array of collision gas pressure-varied product-ion mass spectra of an ion at m/z 305.2 that corresponds to 8,11,14-eicosatrienoic acid. Similar to the variation of collision energy, from such a 2D mass spectrometric analysis, the location of the double bonds can be identified based on the intensity changes of the fragment ion that corresponds to [M – H – 44]− (Yang et al., 2011b).
FIGURE 15.
Two-dimensional mass spectrometric analysis of polyunsaturated fatty acid fragmentation pattern with variation of collision gas pressure. Mass spectrometric analysis was performed with a TSQ Quantum Ultra Plus triple-quadrupole mass spectrometer (Thermo Fisher Scientific, San Jose, CA) equipped with an automated nanospray apparatus (i.e., Nanomate HD, Advion Bioscience Ltd., Ithaca, NY) and Xcalibur system software. The first and third quadrupoles were used as independent mass analyzers with a mass resolution setting of 0.7 Thomson while the second quadrupole served as a collision cell for tandem mass spectrometry. Product-ion scan of 8,11,14-eicosatrienoic acid (20:3 FA) (5 pmol/μL) was performed after direct infusion in the negative-ion mode at the fixed collision energy of 16 eV and varied collision gas pressures ranging from 0 to 3 mTorr as indicated. A 2-min period of signal averaging in the profile mode was employed for each scan. All the scans were automatically acquired with a customized sequence subroutine operated under Xcalibur software. All the scans are displayed after being amplified to the 5% of the base peak in each individual scan.
c. Collision gas type
This is an unknown variable because studies with various inert collision gas types have never been reported. We believe that this variable could also be used to study fragmentation pathways and/or fragmentation kinetics because different types of collision gases possess inherently different cross-sectional areas and different intrinsic energies. When a type of collision gas contains a chiral center, differential interactions of chiral isomers of an analyte with such collision gas are possible.
5. Variables related to the lipid extraction conditions (i.e., multiplex extraction conditions)
a. pH conditions of lipid extraction
The polarity and/or the charge properties of the species of many lipid classes are pH-dependent. For example, PE species are positively charged under acidic conditions and negatively charged under basic conditions. All acidic lipid species become charge neutral under acidic conditions. Accordingly, with a variation of pH of an extraction solution, polarity and charge properties of the species of different lipid classes can both be varied. This variation could lead to the differential extraction efficiency. In practice, an acidic condition is favorable for the extraction of acidic lipid species such as PA and a neutral condition is preferred for the extraction of PE species. A weak acidic condition might be employed for “total” lipid extraction. In addition to the effects of pH conditions on lipid extraction efficiency, the resultant lipid extracts under different pH conditions could affect the ionization efficiency as discussed in Subsection III B2b.
b. Solvent polarity of lipid extraction
Recovery of lipid species largely depends on the solvent(s) used in the extraction. Different types of solvents can selectively extract different categories of lipids. For example, non-polar solvents (e.g., hexane, ethyl ether, etc.) can extract non-polar lipids, including TAG, cholesterol and cholesterol esters, free fatty acids, etc. Most lipids are well-extracted into chloroform (or dichloromethane) such as the Foch method or a modified Bligh and Dyer method (Christie & Han, 2010). Very polar lipids (e.g., acyl CoAs, acylcarnitines, lysophospholipids, PI polyphosphates, gangliosides, etc.) can be recovered from the aqueous phase of a solvent extraction method aforementioned with reversed-phase or affinity cartridge columns or with special solvents (Kalderon et al., 2002; Tsui et al., 2005).
It should be emphasized that inclusion of at least one standard species for each class of lipids of interest during extraction is very important for accurate quantification. Any incomplete recovery can be compensated for with the internal standards added prior to extraction. Any differential extraction efficiency of individual species in a class from that of the standard is largely a matter of secondary effect. However, it should also be noted that, for any analytical method based on external standards, a complete recovery of extraction of a class of interest is essential for accurate quantification.
c. Chemical reactivity as a variable
The intrinsic chemical stabilities and/or reactivities of lipids can be used as a variable in sample preparations or in lipidomic analysis. For example, lipid species with the sphingoid backbone are resistant to base hydrolysis. Many laboratories have explored this chemical stability to aid in isolation and enrichment of sphingoid-backbone containing lipids by hydrolyzing all ester-linked glycerolipids (Merrill et al., 2005; Bielawski et al., 2006; Jiang et al., 2007). In contrast, vinyl ether-linked lipid species (i.e., plasmalogens) are labile under acidic conditions. Investigators have explored this chemical instability to unambiguously identify the presence of plasmalogen species with a comparison of the mass spectra acquired before and after acid treatment (Kayganich & Murphy, 1992; Cheng, Jiang, & Han, 2007; Yang et al., 2007). By exploring the specific reaction activity of the primary amine group, a few derivative procedures have been developed for specific and enhanced analyses of primary amine-containing species, including PE and lysoPE (Berry & Murphy, 2005; Han et al., 2005; Zemski Berry et al., 2009). Derivatization of the primary and secondary hydroxy group(s) of oxysterols with N,N-dimethylglycine (DMG) in the presence of 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide and 4-(N,N-dimethylamino)pyridine to yield their corresponding mono- or di-DMG esters has been used for the analysis of sterols (Jiang, Ory, & Han, 2007). Very recently, coupling N-(4-aminomethylphenyl)pyridinium to eicosanoids with an amide linkage has been exploited to improve the sensitivity of MS analysis of eicosanoids (Bollinger et al., 2010b).
6. Variables related to the separation conditions
a. Charge properties as a variable in intrasource separation
Different lipid classes generally possess different charge properties, largely depending on the nature of their head groups. Based on their charge properties, lipid classes can be reclassified into three categories: (1) anionic lipids, (2) weak anionic lipids, and (3) charge neutral, polar lipids. The category of anionic lipids consists of the lipid classes that carry at least one net negative charge under weakly acidic pH conditions. The common lipid classes in this category include: CL, PG, PI, PS, PA, sulfatide, their lysolipids, and acyl CoA. The category of weak anionic lipids consists of the lipid classes that are charge neutral under weakly acidic pH conditions, but negatively charged under alkaline conditions. The lipid classes in this category include: PE, lysoPE, non-esterified fatty acids and their derivatives, bile acids, and ceramide. The category of charge neutral lipids consists of the lipid classes that are charge neutral but polar with no separable charges. The lipid classes in this category include: PC, lysoPC, SM, HexCer, acylcarnitine, monoacylglycerol, DAG, TAG, and cholesterol and its esters.
A separation method in lipidomic analysis by integration of multiple variables discussed above has been developed and referred as to intrasource separation by Han and Gross (Han et al., 2006b). In this separation mode, investigators can explore the varied charge properties of lipid classes (see last paragraph) to achieve separation within an electrospray ion source. Figure 16A schematically illustrates one practically used procedure in intrasource separation. The principle of this separation method is equivalent to that of electrophoresis (Fig. 16B).
FIGURE 16.
Schematic comparison of intrasource separation of lipid categories to the theoretical electrophoretic separation of lipid classes. Panel A schematically shows the selective ionization of different lipid categories under three different experimental conditions. Panel B schematically shows the imaginary chromatograms of lipid classes after electrophoretic analyses under corresponding experimental conditions. PC, TAG, FA, PE, and AL stand for phosphatidylcholine, triacylglyceride, free fatty acid, phosphatidylethanolamine, and anionic lipids, respectively. (Reprinted from (Christie & Han, 2010) with permission).
Specifically, after a particular sample preparation as well as dilution of a lipid extract to a total lipid concentration of approximately 50 pmol/μL (Han & Gross, 2005b; Christie & Han, 2010), the category of anionic lipids is ionized in a selection of preferentially 40-fold over weakly anionic lipids in the negative-ion mode (Figs. 17A and 12D). This selectivity is due to that the basicity of the weakly anionic lipids is much weaker in comparison to that of anionic lipids as previously described (Han et al., 2006b).
FIGURE 17.
The dependence of ionization efficiency on the charge propensities of analytes. A phospholipid mixture was comprised of 1 pmol/μL each of di15:0 and di22:6 phosphatidylglycerol (PG) (i.e., anionic lipids), 10 pmol/μL each of di14:1 and di18:1 phosphatidylcholine (PC) (i.e., charge neutral lipids), and 15 pmol/μL each of di15:0 and di20:4 phosphatidylethanolamine (PE) (i.e., weakly anionic lipids) species in 1:1 (v/v) chloroform/methanol. The mixture was analyzed with a QqQ mass spectrometer (TSQ Quantum Ultra, Thermo Fisher Scientific) in the negative-ion mode after direct infusion in the absence (Panel A) or presence (Panel B) of 30 pmol/μL of LiOH in methanol. All the indicated molecular species were confirmed with product-ion ESI-MS analysis. The horizontal bars indicate the ion peak intensities after removal of 13C isotopes and normalization of the species to the one with less number of carbon atoms (i.e., the one with lower molecular weight) in each lipid class. (Modified from the ref. (Han et al., 2006b) with permission from the American Society for Mass Spectrometry, Copyright 2006).
This diluted lipid solution was rendered basic with a small amount of LiOH methanol solution to be a final concentration of ~ 30 pmol LiOH/μL. A negative-ion mass spectrum of this solution displays predominant PE species and low abundant anionic lipid species, the intensities of which are approximately proportional to their mass levels of the solution in the mass spectrum (Figs. 17B and 12F). This appearance is due to a fact that the mass content of PE class is approximately 40 mol% of the total lipid mass, whereas the total mass content of all anionic lipid classes, including PI, PS, PG, PA, etc., only accounts for 5 to 10 mol%. It should be pointed out that any low-abundance PE species could be spectrometrically isolated and enhanced for analysis through derivatization, as described in Subsection III B5c. In the case of PE mass content present in low abundance like in plasma samples, these derivative methods can also be applied to analyze the entire class of species.
After dilution, when this LiOH-containing solution is analyzed in the positive-ion mode, the mass spectrum only displays the ions that correspond to the charge neutral lipid species such as PC, SM, HexCer, TAG, etc. (Fig. 12E), which are largely ionized as lithiated molecule ions (Han et al., 2006b). This selection is very specific to these charge neutral lipids because the species of anionic lipids and phosphoethanolamine-containing species are present in a net negatively charged state under the experimental conditions, and their ionization is prohibited in the positive-ion mode as described above (see Subsection III B2b).
b. Elution time as a variable in LC separation
The dimension of elution time in lipid analysis is only present in LC-MS. Lipid classes elute from a normal-phase HPLC at different times due to their different polarities and dipole moments as well as specific interactions with ion-exchange columns. Individual molecular species elute from a reversed-phase column at different times due to their different hydrophobicities that result from the differences in the number of carbon atoms, the number of double bonds, or the location of double bonds in the aliphatic chains. Accordingly, the variation of elution time under different column conditions is an important dimension(s) in MDMS. In the practice of lipidomic analysis, this dimension has been demonstrated in an early study that showed the power of this dimension through identification of over hundreds of lipid species in a column run (Taguchi et al., 2000).
c. Drift time as a variable in ion-mobility separation
Ion-mobility mass spectrometry (IM-MS) has emerged as an important analytical method in the last decade (Stach & Baumbach, 2002). In IM-MS, ions are generated by pyrolysis, electrospray, laser desorption, or other ionization techniques prior to their entry into a gas-filled mobility drift cell region. In this cell, ions drift at a velocity obtained from an electric field based upon their shapes. The different ion drift allows for the separation of different shaped molecules. Therefore, IM-MS provides not only a new dimension of separation, but also shape information because separation is based on the conformation of a molecule in addition to its mass (Kanu et al., 2008; Howdle et al., 2009).
IM-MS can provide rapid two-dimensional analysis of lipids in which each lipid class falls along a trend line in a plot of ion mobility drift time vs. m/z (Woods et al., 2004; Jackson & Woods, 2009). In a recent study (Jackson et al., 2008), Jackson and colleagues have used MALDI-IM TOF/MS for the analysis of complex mixtures of phospholipids in which two-dimensional separation of molecular species based upon drift time and m/z values was achieved rapidly (Fig. 18). They found that the changes in drift time of phospholipids are associated with the fatty acyl chain length, the degree of unsaturation, the head group, and the cationization of individual species. In the same study, they also demonstrated that phospholipids could be profiled directly from rat brain tissue sections. Based upon the drift time observed, the authors identified 22 phospholipid species in PC, PE, PS, PI, and SM.
FIGURE 18.
MALDI ion mobility two-dimension mass spectrum of a mixture comprised of brain phosphatidylcholine and sphingomyelin. MALDI ion mobility 2D spectrum of 25 pmol each of brain phosphatidylcholine (PC) extract and brain sphingomyelin (SM) extract was acquired with DHA matrix. The results demonstrate the ion mobility separation of PC and SM species. (Reprinted from the ref. (Jackson et al., 2008) with permission from the American Society for Mass Spectrometry, Copyright 2008).
The significant impacts of this technology on lipidomics are at least two-fold. First, the coupling of MALDI with IM-MS allows for a wide range of samples to be analyzed, including tissue samples, within an interval of several hundred microseconds between the applications of each focused laser desorption pulse to the sample (Jackson & Woods, 2009). This approach could be a choice to study cellular lipidomes of tissue samples in a spatial and temporal fashion. Second, separation of isomers, isobars, and conformers is possible with the addition of ion-mobility cells to mass spectrometers (Kanu et al., 2008). This approach could allow one to identify novel lipid classes and species in a high-throughput manner. Moreover, analysis of chiral isomers could be achieved by introduction of chiral reagents into an ion mobility cell as demonstrated in other studies (Howdle et al., 2009).
IV. APPLICATIONS OF MULTI-DIMENSIONAL MASS SPECTROMETRY IN LIPIDOMICS
A. Identification of Lipid Species with 2D Mass Spectrometry
Although accurate mass in combination with an isotopologue pattern can be used to identify individual lipid species, the detailed identity of individual lipid species, including a variety of isomers (e.g., regioisomers, isomers resultant from the different location of double bond(s), etc.) can only be identified with product-ion MS analysis of individual species or with building block analysis in 2D MS.
Depending on the classes of lipids, the backbone (e.g., glycerol or sphingoid) can have either varied structures or a fixed structure. For example, glycerophospholipids and glycerolipids have a fixed glycerol backbone; sphingolipids have a variety of sphingoid backbones. Although sphingosine ((2S, 3R, 4E)-2-aminooctadec-4-ene-1,3-diol, designated as d18:1- “d” denotes a dihydroxy backbone) is the major sphingoid backbone of mammals, sphingoid backbones found in nature can vary in the length and branching of alkyl-chain, the number and positions of double bonds, and others (see Subsection I E1). These various sphingoid backbones can be identified with NLS and/or PIS. For example, d18:1-sphingoid backbone (sphingosine) can be identified with NLS429.3 and PIS264.2; d18:0-sphingoid backbone (sphinganine) can be identified with NLS431.3 and PIS266.2 in the positive-ion mode. The backbone of a fixed structure such as glycerol can be directly assigned by resolving its mass from the determined mass of the lithiated molecule ion as well as the masses of the building blocks of the species. In practice, if identification of the building blocks of the molecule ion is feasible with either NLS or PIS as discussed in Subsection II E2, then the mass of the backbone can be resolved with the following equation:
| (1) |
The lipid classes of glycerophospholipids and glycerolipids have a glycerol backbone, which has a known and fixed structure. For example, if a deprotonated molecule ion at m/z 834.6 is present in a negative-ion mass spectrum and is confirmed to have two acyl chains of 18:0 (PIS283.3) and 22:6 (PIS327.2) and have a head group of phosphoserine (NLS87.0 for loss of serine and PIS153.0 for derivative of glycerol phosphate) with 2D MS analysis, then the mass of the backbone is calculated as:
If a lithiated molecule ion at m/z 941.6 is present in a positive-ion mass spectrum of a LiOH-added infused solution and is confirmed to have two acyl chains of 18:0 (NLS284.3) and 22:6 (NLS328.2) at a nearly 2:1 ratio in absolute ion counts, then the mass of the backbone is calculated as:
The calculated mass of the backbone in these examples matches the mass of glycerol and indicates that these molecule ions are glycerol-based lipids (i.e., 18:0–22:6 PS and 18:0–18:0–22:6 TAG, respectively).
It is worth noting that, in most cases, the glycerol backbone is readily recognizable without the need to resolve its mass and therefore without the pre-identification of the acyl chains and head group of the species. For example, glycerol-based anionic phospholipids (e.g., CL, PG, PI, PS, and PA) can be distinct from the other lipids (e.g., sulfatide) in the category of anionic lipids with analysis of PIS153.0, which represents a fragment ion derived from glycerol phosphate after intrasource separation. Glycerol-based weakly anionic phospholipids (e.g., PE) can be distinguished from the other lipids (e.g., ceramides and free fatty acids) in the category of weakly anionic lipids by the difference in their mass ranges (i.e., PE of m/z > 600, ceramides of m/z 500–700, and free fatty acids of m/z < 400) after intrasource separation. Glycerol-based charge neutral but polar phospholipids (e.g., PC) and acylglycerols (e.g., TAG) can be differentiated from the other lipids (e.g., SM and HexCer) in the category of charge neutral but polar lipids based on two points. First, there exists the difference in mass values between the classes based on the nitrogen rule. Second, a neutral loss that corresponds to fatty acyl chains is present in PC and TAG species, but not in sphingolipids (e.g., SM and HexCer) under commonly used experimental conditions.
To identify individual lipid species in the category of anionic lipids (mainly anionic phospholipids), PIS of all acyl chains potentially present in a biological lipid extract are acquired sequentially to display the potential presence of acyl chain building blocks under a physiological pH condition and in the negative-ion mode. For example, the ion peaks shown in the mass spectra of PIS255.2, 281.2, 303.2, and 327.2 (at certain collision energies, see Table 3) display the potential presence of 16:0, 18:1, 20:4, and 22:6 acyl chains, respectively. PIS and/or NLS of head groups or their derivatives are acquired to identify the head group building blocks of individual lipid classes in this category of lipids. For example, the ion peaks in PIS of glycerol phosphate derivative (PIS153.1 at 35 eV) show the potential presence of all the anionic glycerophospholipids (e.g., CL, PG, PI, PS, PA and their corresponding lyso-lipids); PIS of sulfate (PIS97.0 at 65 eV) shows the potential presence of sulfatide species; NLS of serine (NLS87.0 at 24 eV) shows the potential presence of PS species; NLS of methyl chloride (NLS50.0 at 24 eV) yielded from chloride adducts of PC and SM species displays the potential presence of PC and SM species; NLS of hydrogen chloride (NLS36.0 at 30 eV) displays the potential presence of chloride adducts of HexCer species. By combining all of the potential building blocks of fatty acyl chains and head groups with mass analysis of fragments and molecule ions in a 2D mass spectral analysis format, each individual lipid species can be identified.
For example, Figure 19 shows a 2D mass spectrum in which most of the building blocks of anionic phospholipids potentially present in a lipid extract of mouse brain cortex are included. These building blocks represent either a specific component of the head group moiety of each class or the acyl chain(s) of each molecular species as discussed in the last paragraph. Therefore, each individual lipid species, including its head group and acyl moieties, are identified through analyses of this 2D spectrum. For example, the ion peak at m/z 885.6 in the survey scan (Fig. 19) is crossed with the building blocks of PIS153.1 (glycerol phosphate derivative), PIS241.2 (inositol phosphate derivative), PIS283.3 (18:0 FA), and PIS303.2 (20:4 FA) (the broken line in Fig. 19). These fragments lead us to identify this deprotonated molecular ion as a PI species that contains 18:0 and 20:4 fatty acyl chains. Moreover, the presence of a higher ion abundance at m/z 885.6 in PIS303.3 than in PIS283.3 identifies the regiospecificity of this species as 18:0–20:4 PI.
FIGURE 19.
Representative two-dimensional mass spectrometric analyses of anionic phospholipids in a lipid extract of mouse cortex. The lipid extract was prepared with a modified Bligh and Dyer procedure and properly diluted prior to direct infusion into an ESI QqQ mass spectrometer (TSQ quantum ultra mass spectrometer, Thermo Fisher Scientific, San Jose, CA). The survey scan or MS/MS scans of the 2D ESI mass spectrum were acquired in the negative-ion mode with sequentially programmed customized scans with Xcalibur software. “PIS” denotes precursor-ion scanning, “NLS” stands for neutral-loss scan, and “IS” represents internal standard. All scans are displayed after normalization to the base peak in each individual scan.
After addition of a small amount of LiOH (approximately 50% of total amount of lipid content, 10 to 50 μM in general) a basic pH is obtained for analyses of anionic phospholipid species in the negative-ion mode again. PIS of all potential acyl chains and PIS and/or NLS of head groups identify the acyl chain and head group building blocks for the lipid species in the category of weakly anionic lipids. Alternatively, individual species that contain a phosphoethanolamine head group in the lipid solution can be identified with Fmoc derivatization (Han et al., 2005). After derivatization and MS analysis, NLS222.1 (neutral loss of the Fmoc moiety from Fmoc-PE or Fmoc-lysoPE species) unambiguously demonstrates the presence of all PE and lysoPE deprotonated molecule ions with a dynamic range over four orders of magnitude (Han et al., 2005). The identities of the aliphatic chains of PE species can be efficiently identified with a 2D ESI-MS analysis (Fig. 20, which is used for the analysis of PE species present in a lipid extract of mouse brain cortex). For example, the broken line at Fmoc-PE species m/z 1012.6 (i.e., deprotonated PE molecular anion at m/z 790.5) crossed with NLS222.1 (represent PE head group) and PIS283.3 and PIS327.3 (represent 18:0 and 22:6 acyls, representatively), which, along with the higher ion counts in PIS327.3 than in PIS283.3, unambiguously identifies the structure as 18:0–22:6 diacyl PE species. From the 2D MS analysis, many isomeric PE molecules can also be readily identified. For example, the Fmoc-PE ion peak at m/z 922.6 (i.e., deprotonated PE molecular anion at m/z 700.5) is comprised of at least four plasmenylethanolamine (pPE) molecules (i.e., 16:0–18:1, 16:1–18:0, 18:0–16:1, and 18:1–16:0 pPE) (see the left broken line in Fig. 20). The study indicates that the mouse brain lipidome is highly enriched with pPE molecules, which are confirmed after treatment with acidic vapor (see Subsection III B5c).
FIGURE 20.
Representative two-dimensional mass spectrometric analyses of ethanolamine glycerophospholipids in a lipid extract of mouse cortex after in situ fluorenylmethoxylcarbonyl (Fmoc) chloride derivatization in the negative-ion mode. A ESI mass spectrum of Fmoc-PE was acquired in the negative-ion mode directly from a diluted mouse cortex lipid extract after derivatization with Fmoc chloride (Han et al., 2005). Analyses of Fmoc-PE building blocks in the second dimension, including the Fmoc moiety and fatty acyl carboxylates with precursor-ion scanning (PIS) and neutral-loss scanning (NLS), were performed. “IS” denotes internal standard; m:n indicates an acyl chain with m carbons and n double bonds; FA stands for fatty acyl chain. Each of the broken lines indicates the crossing peaks (fragmental ions) of a deprotonated molecule ion with Fmoc-PE building blocks. All mass spectral scans were displayed after normalization to the base peak in each individual spectrum.
Finally, under the identical basic pH condition as mentioned in the last paragraph and in the positive-ion mode, NLS of all fatty acids that correspond to the potentially presented acyl chains identify the acyl chain building blocks for the lipid species in neutral, but polar lipids. For example, NLS256.2, NLS282.2, NLS304.2, and NLS328.2 identify the 16:0, 18:1, 20:4, and 22:6 acyl chains, respectively, in PC and TAG species. These NLS analyses are more sensitive for TAG species than for PC species (Han et al., 2004b). To increase the specificity and sensitivity for the analysis of PC species in the lipid mixture with TAG species, NLS of fatty acid mass plus 59 (corresponding to the loss of trimethylamine) can be used (Yang et al., 2009b). Again, PIS and/or NLS of head groups or their derivatives identify the head group building blocks and thereby classify individual lipid classes in this category. For example, NLS of phosphocholine (NLS183.2 at 35 eV) identifies PC and SM species (as their lithium adducts) (Yang et al., 2009b); NLS of lithium cholinephosphate (NLS189.2 at 35 eV) identifies PC species (as their lithium adducts) (Yang et al., 2009b); NLS of phosphocholine plus methyl aldehyde (NLS 213.1 at 50 eV) identifies SM species (as their lithium adducts) (Yang et al., 2009b); NLS of monohexose (including galactose and glucose) derivative (NLS162.2 at 50 eV) identifies HexCer species (as their lithium adducts) (Han & Cheng, 2005).
It should be pointed out that sphingolipids in most biological samples are less abundant than the major phospholipid classes (Christie & Han, 2010). Detailed analysis of sphingolipids in these samples can be performed after alkaline hydrolysis as aforementioned (Subsection III B5c). For example, when the diluted lipid solution of mouse brain cortex after alkaline hydrolysis was directly analyzed with ESI-MS in the negative-ion mode, the mass spectrum shows abundant ion peaks (Fig. 21). The identities of these molecules can be confirmed with building block analysis such as PIS97.0 (i.e., sulfate) for sulfatides and NLS36.0 (i.e., HCl) for chloride adducts of HexCer species (Fig. 21). Moreover, the molecules of HexCer that contain a hydroxyl group can be readily distinguished from nonhydroxy HexCer molecules because the peak intensity ratios of the hydroxy to nonhydroxy HexCer species in the NLS36.0 scan (Fig. 21A(c)) of lipid solution in the negative-ion mode are enhanced remarkably in comparison to those in the survey scan (Fig. 21A(a)) (Han & Cheng, 2005). For example, the ion clusters at m/z 806.6, 834.7, and 862.7 are hydroxy HexCer molecules, whereas other ion clusters are nonhydroxy ones.
FIGURE 21.
Two-dimensional mass spectrometric analyses of sphingolipid species after treatment of a mouse cortex lipid extract with lithium methoxide. Panel A shows the analyses of sphingolipids in the lipid extract of mouse cortex after treatment with lithium methoxide in the negative-ion mode. Building block analyses of the lipid extract after treatment with lithium methoxide were acquired with precursor-ion scanning (PIS) of m/z 97 (i.e., sulfate) for the analysis of sulfatide species and neutral-loss scanning (NLS) of 36 u (the loss of HCl from cerebroside (HexCer) chloride adducts) identifies the presence of 2-hydroxy HexCer, respectively. Panel B shows the analyses of sphingolipids in the lipid extract of mouse cortex after treatment with lithium methoxide in the positive-ion mode after addition of a small amount of LiOH. The mass trace “a” in Panel B is the survey scan of the lipid extract of mouse cortex after treatment with lithium methoxide. The mass trace “b” in Panel B identifies the presence of sphingomyelin (SM) species in the solution with NLS183.1, which corresponds to phosphocholine. The mass trace “c” in Panel B identifies the presence of HexCer species in the solution with NLS162.1, which corresponds to hexose. Analyses of this mass spectrum and the mass trace “c” in Panel A identify the individual HexCer species with or without a hydroxy moiety. IS denotes internal standard. Each spectrum displayed is normalized the base peak in the spectrum.
When the diluted lipid solution of mouse brain cortex after alkaline hydrolysis was analyzed with LiOH (40 pmol/μL) with positive-ion ESI-MS, the mass spectrum shows abundant ion peaks that correspond to lithiated SM and HexCer molecules in the mass region of m/z 650–900 (Fig. 21B(a)). Lithiated SM molecules can be identified with the NLS183.1 (i.e., phosphocholine), which is specific to SM species in this mass region under the experimental conditions (Fig. 21B(b)). The sphingoid backbone of major SM species is determined (Hsu & Turk, 2005). The acyl amide moieties are derived based on the sphingoid base and m/z of each individual molecule. Lithiated HexCer species can be identified with NLS162.1 (corresponding to the loss of monohexose) (Fig. 21B(c)) (Han & Cheng, 2005). Accordingly, a combinational analysis of mass spectra A(c) and B(c) allows one to identify the individual HexCer species with or without a hydroxy moiety.
Because the ceramide content in biological samples is quite low, assessment of ceramide content is heavily dependent on MS/MS spectra of the lipid solution with or without the treatment of lithium methoxide (Han, 2002; Han et al., 2004b). Specifically, we have found that NLS256.2 is sensitive to the ceramides without a hydroxyl group at the α position, NLS327.3 is specific to the ceramide with a hydroxy group at the α position, and NLS240.2 is essentially equally sensitive to all ceramide species in both subclasses within experimental error (Han, 2002). Therefore, NLS256.2 and NLS327.3 scans are used to identify sphingosine-based ceramides with or without a hydroxy group at the α position, whereas NLS240.2 is used to assess the contents of these ceramides. To the sphinganine or other sphingoid backbone-containing ceramides, other sets of building blocks were respectively analyzed for identification and quantitation. For example, ceramides with sphinganine are identified and quantified with analyses of NLS258.2, NLS329.3, and NLS242.2, whereas identification and quantification of ceramides that contain a sphingoid backbone with 20 carbons are conducted with analyses of NLS284.3, NLS355.3, and NLS268.2 scans. This approach has been used to identify and quantify ceramide species in many biological samples (e.g., post-mortem human brain tissues (Han, 2007; Han & Jiang, 2009) and mouse cortex (Han et al., 2010)).
The caveats of the MDMS-based shotgun lipidomics for identification of individual molecule of a cellular lipidome are at least in two aspects. First, it would be impossible to identify the isomers that possess identical fragmentation patterns with this platform. Examples of this category include chiral isomers as well as GluCer and GalCer in the positive-ion mode. However, the development of ion mobility techniques that integrate chiral reagents in the ion mobility chamber might provide a solution for this limitation. Second, the application of this technology to discover any novel lipid class is limited because a pre-characterization of the class is always required prior to its application for identification. For this particular case, LC-MS methods offer advantages over shotgun lipidomics approaches.
B. Quantitation of Lipid Species with 2D Mass Spectrometry
Quantification of individual lipid species is one of the most important, but yet challenging, components in the lipidomic analysis with MS. Understanding the principles of quantification with either shotgun lipidomics or LC-MS is essential. Particularly, the two-step quantitation method embodied in MDMS-based shotgun lipidomics is described in detail and with examples. The validation of this method for accurate quantification of individual lipid species is also presented in this section. The factors such as concentration, ion suppression, dynamic range, 13C isotopologue distribution, baseline correction, internal standards, normalization, etc., which might affect the accurate quantification of individual lipid species in lipidomics, have previously been extensively described (Han & Gross, 2005b; Christie & Han, 2010). Discussion of these factors is not included herein.
1. The principle of quantification of individual lipid species with MS
Unlike an optics device, where determination of the concentration of an analyte follows the Beer-Lambert law, there is no rule for the relationship between the absolute counts of a molecule ion determined with ESI-MS and the concentration of the analyte. The ion intensity of an analyte with MS analysis might be affected with even minor differences in sample preparation, ionization conditions, tuning conditions, and the analyzer and detector used in the mass spectrometer, etc. As MS instruments become more sensitive, the influences of these factors on ESI-MS quantification become more evident. Thus, it would be difficult for researchers to repeat a measurement of absolute ion current for an analyte in a biological sample.
Accordingly, ESI-MS quantification of any compound has to be made with comparisons to either an internal or external standard similar to the compounds of interest (e.g., their stable isotopologues). The former is added during sample preparation at the earliest step possible, and analyzed at the same time as the sample. The latter is analyzed separately but under “identical” conditions with the sample of interest, and a calibration curve is established with the external standard. Both methods have advantages and disadvantages. The former is known for its simplicity and accuracy if the internal standard is within a linear dynamic range of the measurement with the sample. However, selection of an internal standard might be difficult, and the dynamic range of the measurement must be pre-determined. In the case of an external standard, control of the measurements under identical conditions is key, particularly when many steps of sample preparation, separation, and quantification are involved. Global analyses of the cellular lipidome are exactly such a complicated process to which use of external standards alone is not the best choice. Therefore, use of internal standards (or related groups of standards), in combination with external standards (in the case of LC-MS based methods for lipid analysis) is a better solution to quantify complex cellular lipidomes. Any unexpected changes in measurement of ion counts with MS can be internally controlled or normalized. Results obtained from sample analysis without any internal control can only be used for profile comparisons.
To determine the concentration of an analyte with a comparison with the selected internal standard of a class or with the calibration curve in the case with external standard(s), the following formula is generally employed:
| (2) |
where cu and ci are the contents of individual species to be determined and the selected internal standard, respectively, and Iu and Ii are the peak intensities of the species and the selected internal standard, respectively, in the case of comparison between ion peak intensities (i.e., ratiometric comparison), or are the peak areas of the determining species and the selected internal standard, respectively, in the case with LC-MS. For ratiometric comparison, removal of 13C isotope has to be performed prior concentration calculations (Han et al., 2004b; Han & Gross, 2005b). Whether removal of 13C isotope should be considered in LC-MS depends on individual cases (Christie & Han, 2010).
Formula 2 is derived from the linear correlation between the content (c) and the ion intensity (I) of a species:
| (3) |
or
| (4) |
where the parameters of a and b are the response factor-related factor and background noise, respectively, and a′ = 1/a. When I ≫ b (e.g., S/N > 10), c≈ aI. Formula 2 is obtained when the response factors of different individual species of a lipid class are essentially identical.
Identical response factors of different individual species of a lipid class only hold true for polar lipid classes in the low concentration region after removal of 13C isotope. The requirement for polar lipid classes is to guarantee that the ionization efficiency of individual molecule of a class is predominantly dependent on the head group and the effects of fatty acyl chains, including their length and unsaturation on ionization efficiency are minimal. The requirement for the low concentration is to assure that a linear dynamic range is present because lipids, with increased concentration, tend to form aggregates, which results in the loss of the linear dynamic range because the form of lipid aggregates depends on fatty acyl chain length and unsaturation of individual molecular species. It should be emphasized that the ESI-MS response factors of different non-polar lipid species are quite different, and the response factors of individual non-polar lipid molecule have to be pre-determined to achieve an accurate quantification (Han & Gross, 2001).
2. Quantification of individual lipid species with a two-step procedure in MDMS-based shotgun lipidomics
After separation of different lipid classes in the ESI ion source (i.e., intrasource separation) (see Subsection III B6a) and MDMS identification of individual species (see Section IV A), quantification of the identified individual species of a class of interest is performed in a two-step procedure in MDMS-based shotgun lipidomics (Han et al., 2004a; Han & Gross, 2005a). This procedure can be conducted automatically (Yang et al., 2009a).
Briefly, this methodology first determines whether there are ion peaks of interest in low abundance or whether there exist overlapping ion peaks from other lipid classes (Yang et al., 2009a). The method performs the one-step quantification procedure (see below) for the case of no overlapping peaks and no low abundance peaks. A two-step quantification procedure is performed for the other cases. In the two-step procedure, the abundant and non-overlapping peaks are first quantified with ratiometric comparison to the selected internal standard of the class with survey scan mass spectra after baseline correction and removal of 13C isotope. The determined contents of these non-overlapping and abundant species plus the pre-selected exogenously added internal standard are the candidate standards for the second step of quantification. It should be pointed out that ion intensities in the class-specific tandem MS scan(s) might vary with individual subclasses or subtypes of species (Yang et al., 2009b). Therefore, selection of different standards from the candidate standard list for individual subclasses or subtypes of species for the second step quantification is also considered (Yang et al., 2009b).
The second step quantification is performed to quantify the remaining overlapping and/or low-abundance species with the standards selected above. An algorithm is generated based on two variables (i.e., the differences in the number of total carbon atoms and the number of total double bonds present in fatty acyl chains of each individual species from that of the selected standards) with multivariate least-square regression to determine the correction factors for each individual molecule (Yang et al., 2009a). The corrected ion peak intensities of the overlapping and/or low-abundance species from the class-specific PIS or NLS are used to quantify these species with ratiometric comparison (i.e., formula 2) with the ion-peak intensities of the internal standard. With this second step, the linear dynamic range of quantification is extended dramatically to quantify overlapping and/or low-abundance species with one or more MS/MS scans in the second step to reduce background noise, increase S/N ratios of low-abundance species, and filter the overlapping molecules with class-specific tandem MS scan(s) (Han & Gross, 2005b).
This second step is similar to tandem MS-based shotgun lipidomics for quantification of individual lipid species (see Subsection II B1) in some aspects. However, the former uses exogenously added internal standards and endogenous standards pre-determined in the first step, whereas the latter exclusively uses exogenously added internal standards. The use of endogenous species as standards can generally provide a more comprehensive representation of the physical properties of structurally similar but low-abundance species in the class in comparison to the use of externally added standards, the number of which is generally limited in order to eliminate any potential overlapping with endogenous species.
Note that, when only two species, including the pre-selected internal standard meet the criteria for selection as the standards for the second step, this second-step quantification becomes similar to tandem MS-based shotgun lipidomics with a linear standard curve (Welti, Wang, & Williams, 2003). In this case, the presence of different numbers of double bonds might affect the accurate quantification of those overlapping and/or low-abundance species performed in the second step. This effect is relatively small in MDMS-based shotgun lipidomics, especially for the total content of the class, because the selected standard contains a certain number of double bonds and the species determined from the survey scan in the first step of quantification contributes appreciably to the total content.
Besides the differences discussed above, there is a big difference between this MDMS-based two-step quantification and the tandem MS-based shotgun lipidomics. All quantified individual species in the two-step quantification approach are pre-identified with MDMS and therefore, any artifactual peaks present in the tandem MS spectrum that is used for quantification in the second step have been eliminated (Yang et al., 2009a). Note also that all other head group-related PIS and/or NLS of each lipid class, if present and sensitive enough, can be applied for quantification of individual species in the second step (see Subsection I E3). This redundant process is useful to refine the data and serves as an internal check for the accuracy of quantification.
This approach has been used to quantify individual species of nearly 30 lipid classes directly from extracts of biological samples (Yang et al., 2009a). With the second step in quantification, an over 5000-fold linear dynamic range for many lipid classes can be achieved (Han, Yang, & Gross, 2008). For example, in the analysis of SM species present in mouse cortex (Fig. 21), the quantities of lithiated N18:1 SM at m/z 735.5, N18:0 SM at m/z 737.5, and N24:1 SM at m/z 819.6 can be accurately determined with ratiometric comparisons with the selected internal standard (i.e., N14:0 SM at m/z 653.5) after removal of 13C isotope from the survey-scan mass spectrum (Fig. 21B(a)). The contents of all other low-abundance molecular species of SM (see inset in Fig. 21B(b)) are determined with the second step of quantitation with N14:0, N18:1, and N24:1 SM as internal standards.
In the same analysis as SM species above, a large number of HexCer species can be quantified in the first step of the quantitation procedure from the survey scan mass spectrum (e.g., ions at m/z 806.6, 816.6, 820.6, and 834.6) in ratiometric comparison of their ion intensities to that of the selected internal standard (N15:0 GalCer at m/z 692.6) after removal of 13C isotope (Fig. 21B(a)). Because the presence of an α-hydroxy moiety in HexCer species can be determined with NLS36.0 analyses of a lipid solution (Fig. 21A(c)) as previously described (Han & Cheng, 2005), these determined endogenous standards can be clarified into subclasses of HexCer with or without carrying an α-hydroxy moiety. Accordingly, the contents of other HexCer species are determined from NLS162.2 (Fig. 21B(c)) with ratiometric comparison of each of their peak intensities to those of the quantified HexCer subclass species. Unfortunately, this approach is unable to separately determine the levels of GalCer and GluCer species as aforementioned. A conventional TLC approach (Abe & Norton, 1974) or an LC method (Merrill et al., 2005) can be employed to determine the composition of these different subclasses.
The caveats of this two-step quantification methodology include that the experimental error for the species measured in the second step of quantification is propagated and is larger than that in the first step. To minimize this effect in the second step, it is critical to reduce any potential experimental error in the first step. It is very important to use the species in high abundance determined from the first step as standards for quantification of other species of the class in the second step to guarantee accuracy. This approach eliminates any potential propagation of errors. It is also noted that the species quantified in the second step account for a smaller amount than those in the first step, so the propagated experimental error in the second step affects the accuracy of total quantification only to a relatively small degree. In addition, the two-step quantification procedure cannot be applied as described above to any lipid class for which a class-specific and sensitive PIS or NLS is not present; e.g., TAG, ceramide, PE, and CL. Special quantification methods for these lipid classes have been developed in MDMS-based shotgun lipidomics (Han, 2002; Han et al., 2004b; Han et al., 2005; Han et al., 2006a).
A misconception that has been consistently stated in the literature and in symposia is that ion suppression present in the analysis of complex lipid mixtures precludes quantification in all methods that use direct infusion. Thus, it has been erroneously argued that lipids cannot be quantified with a shotgun lipidomics approach, which can only provide a profile comparison between the different states. In fact, this concept is entirely incorrect if appropriate conditions for mass spectrometric analysis are employed in the recommended low-concentration regime for lipid analysis. Although it is possible to misuse any analytical procedure in its non-linear range, careful consideration and use of conditions resulting in a linear dynamic range for analyte quantitation can be easily employed. If one uses concentrations outside of the linear dynamic range of a mass spectrometer, corrupted data will be gathered due to competition for ionization that exceeds the five orders of magnitude linearity present in many mass spectrometers. Moreover, at high concentrations, the formation of lipid aggregates precludes meaningful quantitation.
3. Validation of the quantitative accuracy of the two-step procedure
To demonstrate the validation of the quantitative accuracy with a ratiometric comparison in the two-step procedure of MDMS-based shotgun lipidomics, we have recently performed a series of experiments by spiking exogenous lipid species prior to or after extraction to determine the linear dynamic ranges as well as the effects of other components in the solution on quantitation (i.e., matrix effects). First, we have spiked different amounts of di14:1 PC (commonly used as an internal standard) prior to the MS analysis into a mouse myocardial lipid extract, which was prepared without addition of any internal standard. After dilution of the prepared lipid solution to a concentration of < 100 pmol of total lipids/μL prior to direct infusion for MS analysis, the di14:1 PC is spiked into the solution with concentration varied from 0, 0.16, 0.28, 0.4, 0.8, 1.2, 2, 4, 8, to 16 pmol/μL, which spans a mass content range of 100-fold. Because the mass content of endogenous PC lipid species of the myocardial lipid extract spans over 100-fold, this experiment tests an overall dynamic range of much greater than 100-fold (potential over 10,000) for quantification by considering both factors. Figure 22 shows the representative mass spectra with different amounts of di14:1 PC as lithium adduct at m/z 680.4. The level of di14:1 PC in each mass spectrum of either survey-scan MS, NLS183.1, or NLS189.1 is determined with ratiometric comparison with the base peak at m/z 812.6 (i.e., lithiated 16:0–22:6 PC) after removal of 13C isotope whose content is pre-determined as 26.5 nmol/mg protein in a separate lipid extract with di14:1 PC as internal standard and is 2.65 pmol/μL after the 100-fold dilution prior to MS analysis. Figure 23 shows the linear correlation between the spiked and the determined amounts of di14:1 PC species with either full-MS analysis or NLS183.1 or NLS189.1.
FIGURE 22.
Representative mass spectrometric analysis of a mouse myocardial lipid extract with different amounts of spiked di14:1 phosphatidylcholine. Different amounts of di14:1 phosphatidylcholine (PC) (0.16 pmol/μl in Panel A, 0.8 pmol/μL in Panel B, 4 pmol/μL in Panel C, and 16 pmol/μL in Panel D) prior to the MS analysis were spiked into a fixed amount of a lipid extract of mouse myocardium (approximately 6 pmol/μL), which was prepared in the absence of di14:1 PC during lipid extraction. Full mass spectra were directly acquired after infusion of this diluted solution in the presence of a small amount of LiOH (20 pmol/μL). The ion at m/z 680.4, which corresponds to lithiated di14:1 PC, is specified.
FIGURE 23.
Linear correction of the amount of spiked di14:1 phosphatidylcholine with those determined with mass spectrometry. Samples spiked with 10 different amounts of di14:1 phosphatidylcholine (PC) into a lipid extract of mouse myocardium (see text) were prepared in duplicates. The amount of the spiked di14:1 PC in each prepared sample was determined with either full-MS analysis in triplicates (Panel A), NLS183.1 (Panel B), or NLS189.1 in comparison to the amount of an endogenous PC species (i.e., 16:0–22:6 PC at m/z 812.6 as a lithium adduct), which was pre-determined. Each data point represents mean ± SD from the analysis of either 6 full MS spectra, 2 NLS183.1 spectra, or 2 NLS189.1 spectra of each spiked amount of di14:1 PC. Most of the error bars of the data points are within the symbols.
In another set of experiments, quantification of exogenously added PC species is performed with ratiometric comparison with a selected internal standard. Both species are added to the tissue homogenates prior to the lipid extraction. In the experiment, a fixed amount of di14:1 PC (15 nmol/mg protein) was used as internal standard and the amounts of 16:0–18:2 PC species (one of the endogenous species present in mouse myocardial lipid extracts) were varied in a factor of its endogenous content (which is pre-determined as 2.9 nmol/mg protein) from 0, 1, 2, 4, 8, 16, 32, 64, to 100 (i.e., from 0, 2.9, …, to 290 pmol/mg protein, respectively) to cover a mass content range of 100-fold. Figure 24 shows the linear correlation between the added and the determined amounts of 16:0–18:2 PC species with ratiometric comparison with that of the internal standard (i.e., di14:1 PC) with full-MS analysis, NLS183.1, or NLS189.1
FIGURE 24.
Linear correlation of the spiked amounts of 16:0–18:2 phosphatidylcholine with those determined with a ratiometric comparison to that of di14:1 phosphatidylcholine as an internal standard. In the experiments, a fixed amount of di14:1 phosphatidylcholine (PC) (15 nmol/mg protein) was used as an internal standard. The amounts of 16:0–18:2 PC species (one of the endogenous species present in mouse myocardial lipid extracts) were added in a factor of its endogenous mass content from 0, 1, 2, 4, 8, 16, 32, 64, to 100, where the endogenous content of 16:0–18:2 PC was pre-determined as 2.9 nmol/mg protein. Proper amounts of both PC species were added to each of the mouse myocardial homogenates prior to the lipid extraction. Each preparation of lipid extraction with added di14:1 PC and 16:0–18:2 PC was separately performed in triplicates. Each lipid extraction sample was analyzed with full-MS in triplicates, once with NLS183.1, and once with NLS189.1. These mass spectra were separately used to quantify the mass content of 16:0–18:2 PC with a ratiometric comparison to that of di14:1 PC. The determined mass content of 16:0–18:2 PC represents the mean ± SD from the triplicated sample preparation. Most of the error bars are within the symbols.
These data from the above two series of experiments suggest that mass spectrometric analysis is well-suitable to quantify alterations in the mass content of individual PC species with ratiometric comparison in the range of biological alterations in individual species content. Furthermore, the data suggest that matrix effects on the determination of ion ratios for different species of an identical lipid class (i.e., ratiometric comparison) are minimal (also see discussion in Subsection III B2b). The linear relationships were identified through full-MS as well as MS/MS analyses, and were consistent with the small differences in the slope of the regression equations from full-MS (Panel A of Figs. 23 and 24) and the slope from tandem MS analyses (Panels B and C of Figs. 23 and 24) that covered a 100-fold range in mass content. These results and many others documents the accuracy of the two-step quantification procedure with quantitation of high-abundance and non-overlapping species in the full-MS analysis in the first step (with comparisons with internal standard) whereas the second step quantifies low-abundance and/or overlapping species with class-specific MS/MS analysis with endogenous molecular standards present in the lipid extract.
C. Location of Double Bond Positions of Fatty Acids or Fatty Acyls
The composition of the unsaturated fatty acid isomers in a biological system has significant impacts on biological functions. The composition of the isomers reflects the dietary history and fatty acid biosynthesis, whereas the temporal changes of this composition determine the metabolic rate of the biological system of interest. The fatty acid signature, including the isomer composition, determines the membrane flexibility, which could subsequently affect the membrane protein functions. Finally, the altered composition of these fatty acid isomers could also significantly influence the physiological responses, and could be associated with the pathogenesis of diseases such as cancer, diabetes, cardiovascular disease, and neurodegenerative diseases (Dowhan, 1997; Han, 2005; Pruett et al., 2008). Accordingly, identification and accurate quantification of these isomers are obviously crucial in biology, challenging for analytical chemistry, and an important component in lipidomics.
Identification of the location of double bond(s) in derivatized fatty acids with electron ionization and/or chemical ionization MS has been well-established (Christie & Han, 2010). However, due to its relatively complicated procedure, its application for the comprehensive analysis of lipid species in a high-throughput fashion is limited. The use of high-energy CID at keV energy to determine the position of unsaturation in underivatized fatty acids has previously been studied with fast atom bombardment MS (Tomer, Crow, & Gross, 1983). Bryant and colleagues (Bryant et al., 1991) have identified the n-9 double bond location in 16:0–18:1 PC with fast atom bombardment MS3 analysis on a four-sector mass spectrometer. Kerwin and colleagues have attempted to identify the location of double bond(s) in free fatty acids with a QqQ mass spectrometer with an ESI source (Kerwin, Wiens, & Ericsson, 1996), but the study was not very successful.
Hsu and Turk have used either an ESI linear ion-trap mass spectrometer (Hsu & Turk, 2008a) or a QqQ mass spectrometer with source CID (Hsu & Turk, 1999) to identify the position of double bond(s) of unsaturated long-chain fatty acids from dilithiated adduct ([M − H + 2Li]+) ions. Following the same line of reasoning, low-energy MSn analysis has also been used to identify the location of double bond(s) of TAG (Hsu & Turk, 1999) and phospholipids (Hsu & Turk, 2008b). However, this type of work currently belongs more to the research laboratory than to a lipidomic application (Mitchell et al., 2009).
MS identification of double bond location in fatty acyl chains of lipids has also been performed with off-line derivatization followed by product-ion analyses of the derivatized lipids (see (Mitchell et al., 2009) for a recent review). For example, Moe and colleagues (Moe et al., 2004a) pretreated phospholipids and free fatty acids with osmium tetroxide to generate hydroxyl-containing phospholipids at the initial site(s) of unsaturation. The dihydroxylated lipids were analyzed with product-ion MS analyses to locate the position of the initial double bond (Moe et al., 2004b). Harrison and Murphy exposed phospholipid samples to ozone to produce nearly quantitative conversion of olefinic bonds to zooids (Harrison & Murphy, 1996). ESI-MS/MS analyses of these modified phospholipids as the [M + H]+ or [Moa]− ion in either positive- or negative-ion mode, respectively, located the double bond position.
Recently, ozone ESI-MS has been developed for on-line ozonolysis of lipid double bonds to locate double bond position(s) without prior sample preparation (Thomas, Mitchell, & Blanksby, 2006; Thomas et al., 2007). In this technology, lipids react with ozone that is present in the source gas as part of the electrospray process. This technology is simple to use and powerful for individual lipid species or simple lipid mixtures. However, interpretation of the resultant spectra of complex mixtures is difficult. This difficulty is due to the fact that ozone-induced fragments are often isobaric with other lipid ions, and the assignment of fragments to their respective precursor ions becomes ambiguous as the complexity of the mixture increases (Mitchell et al., 2009). Not much work on quantitation or composition of double bond isomers has been performed.
We have very recently developed an approach to identify and quantify unsaturated fatty acid isomers with MDMS-based shotgun lipidomics (Yang et al., 2011b). In this method, the presence of individual fatty acid isomers can be identified based on the characteristic intensity distribution of the specific fragment ions with varying CID conditions (i.e., either collision energy or collision gas pressure, which are the variables in MDMS (see Subsection III B4) (Figs. 14 and 15). The principle of the new method relies on the fact that unsaturated fatty acid deprotonated molecule ions yield fragment ion(s) from the loss of carbon dioxide or water upon CID, and the intensity profiles of these fragment ions over collision conditions are distinct for different isomers. We found the intensity distribution of the specific fragment ions that result from the loss of either CO2 or H2O from unsaturated fatty acid deprotonated molecular ions over a CID parameter is mainly dependent on the distance between double bonds and carbonyl as well as the number of double bonds. The distance between the double bonds and carbonyl can be represented with the location of the first double bond in polyunsaturated fatty acids because the double bonds that are present in most naturally-occurring polyunsaturated fatty acids are almost always interrupted with a methylene group only. The underlying mechanism of the altered fragment-ion intensity distribution in fatty acid isomers with varying CID conditions is likely due to the differential interactions between the negative charge carried with the fragment ion and its interactions with electron densities present in the double bonds. The differential interactions are due to the different distance between the terminal charge-carrying carbonyl group and the double bonds in fatty acid isomers and can differentially stabilize the charges in the fragment ions. These differences result in different fragment ion intensities to distinguish the fatty acid isomers. Moreover, with this approach, the composition of the identified fatty acid isomers can be quantified with external calibration curves, which are determined with authentic fatty acid isomers. Furthermore, this approach can be extended to identify and quantify the double-bond isomers of fatty acyl chains present in individual phospholipid species of biological samples with multi-stage tandem mass spectrometry (Yang et al., 2011b). Collectively, this approach for identification and quantification of the double bond isomers of endogenous fatty acids or fatty acyl chains of phospholipid species is a new addition to MDMS-based shotgun lipidomics and should further advance the lipidomic power to identify the biochemical mechanisms of metabolic diseases.
V. DATA PROCESSING AND BIOINFORMATICS
Large amounts of data are generated in MS analysis of lipids for identification and quantification. It is hard to process these data without proper tools. Accordingly, research in bioinformatics (which involves the creation and advancement of databases, algorithms, statistics, and theory to solve problems that arise from the management and analysis of huge amounts of biological and/or biomedical data) for lipidomics have caught great attention in the last few years, and substantial progress has been made. All the areas of bioinformatics for lipidomics, including automated data-processing, statistical analysis of datasets, pathway and network analysis, and lipid modeling in a systems and biophysical context, have made great developments.
For automation of data processing, generation of libraries and/or databases that contain information about the structures, masses, isotope patterns, and MS/MS spectra in different ionization modes of lipids, along with possible LC retention times, is critical. This approach is analogous to the libraries of GC-MS spectra, which can be used to search a compound of interest after GC-MS analysis. Currently, the most comprehensive libraries/databases that contain these types of information include the ones generated by the LIPID Map consortium (http://www.lipidmaps.org) (Sud et al., 2007) as well as the METLIN created at the Scripps Research Institute (http://metlin.scripps.edu). The website developed by the group of Dr. Taguchi in Japan (http://lipidsearch.jp/LipidNavigator.htm) enables investigators to search a lipid species based on different parameters, including fragmentation patterns. The website http://www.bmrb.wisc.edu/metabolomics/external_metab_links.html, which lists a variety of libraries for metabolomics, includes the information about the libraries/databases for lipidomics. It should be noted that shotgun lipidomics identifies lipid species in situ, and that a theoretical database constructed based on the building blocks of individual lipid classes is sufficient (Yang et al., 2009a).
There exist a few programs and/or software packages that perform multiple data-processing steps such as spectral filtering, peak detection, alignment, normalization, and exploratory data analysis and visualization for the requirements of LC-MS methods (Forrester et al., 2004; Hermansson et al., 2005; Laaksonen et al., 2006; Fahy et al., 2007; Sysi-Aho et al., 2007; Hubner, Crone, & Lindner, 2009; Hartler et al., 2011). These programs were developed and largely depend on the elution times and the determined masses of individual ions. For example, the latest released software, MZmine 2, can read and process unit mass and exact mass (e.g., FTMS or Orbitrap instruments) data in continuous and centroid modes, including fragmentation (MSn) scans (Sysi-Aho et al., 2007). MZmine 2 also affords customized database connectivity as well as incorporation of quality-control procedures (e.g., calibration with internal standards). The algorithm called LipidQA (Song et al., 2007; Song, Ladenson, & Turk, 2009a) is designed after the same line of reasoning as a search of the GC-MS library and contains many product-ion spectra. Although this algorithm is based on a direct-infusion approach, it can identify lipid species with product-ion spectra acquired from LC-MS analysis. With these tools, one can potentially identify a lipid species with a match of a fragmentation pattern of the species along with other available information manually or automatically. Unfortunately, identification of the eluted species is complicated when there is incomplete resolution of individual species of a class. In this case, either an improvement of the resolution of individual species, or train for the analysis of the product-ion spectra acquired from standard isomeric/isobaric mixtures will be necessary.
Multiple programs and/or software packages are developed based on the principles of shotgun lipidomics, including LIMSA (Haimi et al., 2006), LipidProfiler (Ejsing et al., 2006b), LipidInspector (Schwudke et al., 2006), AMDMS-SL (Yang et al., 2009a), and LipidXplorer (Herzog et al., 2011). These tools are developed based on the different platforms of shotgun lipidomics. The LIMSA, which is available through the website (www.helsinki.fi/science/lipids/software.html), serves as an interface to process data from individual full-MS and tandem-MS spectra. The software package LipidXplorer, which is available through MDS Sciex, deals with the multiple PIS and NLS data or others acquired with those instruments with high mass accuracy/high mass resolution (e.g., Q-TOF and Orbitrap). The AMDMS-SL program, which is available at www.shotgunlipidomics.com, is developed to identify and quantify individual lipid species from the data obtained from MDMS-based shotgun lipidomics.
The correction of the baseline contribution to the peak intensities of a mass spectrum is very important for accurate quantification of each analyte content with MS, particularly when the species is in low-abundance. This critical issue has not been paid sufficient attention with most of the data-processing tools, and has only been addressed manually or with instrument-specific programs, which might not be suitable for individual mass spectra acquired under variable experimental conditions. Yang and colleagues have recently presented a simple, practical approach to determine the baseline of each individual mass spectrum to correct the noise background from the ion peak intensities (Yang et al., 2011a). That approach is based on the fact that an accelerated intensity change exists from the noise level to the signal level.
The next step in bioinformatics is to perform statistical comparison between datasets. To this end, any commercially available software (e.g., SAS, NCSS, IBM SPSS Statistics, SIMCA-P, etc.) can be used for biostatistical analyses such as principal component analysis (PCA), multivariate analysis of variance (MANOVA), partial least square (PLS) regression, etc. For example, application of PCA, PLS, and fuzzy c-means clustering for data analysis of metabolomics has found that use of a proper statistical analysis method is essential to improve visualization, accurate classification, and outlier estimation (Li et al., 2009).
Although numerous pathway analysis tools have been developed (van Iersel et al., 2008; Wheelock et al., 2009), it is still a challenging research area to apply these tools for the analysis of the altered lipids between different states due to any physiological, pathophysiological, or pathological change(s). Different models have been applied for the analysis of certain pathways in a particular network (Dhingra et al., 2007; Fahy et al., 2007; Kapoor et al., 2008; Merrill et al., 2009; Cowart et al., 2010; Kiebish et al., 2010), but models of a more comprehensive network analysis for lipidomics are still warranted.
Although lipid modeling in a systems and biophysical context is still in an immature stage in comparison to the other aforementioned components of bioinformatics for lipidomics, attempts on this topic have appeared in the literature. For example, Yetukuri and colleagues (Yetukuri et al., 2010) have reconstituted HDL particles in silico with large-scale molecular dynamics simulations with information about the lipid composition of high-density lipoprotein particles from a lipidomic analysis. They have found that the changes in lipid composition induce specific spatial distributions of lipids within the particles.
VI. SUMMARY AND PROSPECTIVES
During the past decades the use of soft ionization methods for lipidomics analyses has greatly facilitated progress in the field. In large part, these advances have been enabled by the development of novel ionization approaches, increasingly sophisticated fragmentation strategies and the development of new approaches for bioinformatic analyses. Collectively, these advances have led to the development of several robust platforms that can be used for automated lipidomic analyses. Through continued technological advances in sensitivity, mass accuracy, resolution and fragmentation strategies deeper penetrance into the low-abundance regime of the lipidome can be anticipated through multiple approaches, each with their strengths and weaknesses. Through integration of lipidomics with the other “omics” sciences, many biochemical mechanisms that mediate disease states can be uncovered and new biomarkers indicative of the disease onset, severity, and treatment efficacy can be identified. Moreover, application of an integrated systems biology approach has the power to identify novel markers for lipid mediated/associated diseases. These markers are diagnostic of disease onset, progression or severity, as well as evaluation of drug efficacy and safety. It seems clear that the comprehensive understanding of alterations in lipid content and flux through lipidomics investigations judiciously integrated with genomic and proteomic studies will continue to increase the comprehensive understanding of the roles of lipids in biological systems in health and their alterations during disease processes.
Acknowledgments
This work was supported by National Institute on Aging Grant/ National Institute of Diabetes and Digestive and Kidney Diseases R01 AG31675 and National Institutes of Health Grant P01 HL57278. XH and RWG have financial relationships with LipoSpectrum, LLC. RWG has a financial relationship with Platomics, Inc.
VII. ABBREVIATIONS
- 9-AA
9-aminoacridine
- 2D
two dimensional
- amu
atomic mass unit
- CID
collision-induced dissociation
- CL
cardiolipin
- DAG
diacylglycerol
- DESI
desorption electrospray ionization
- DMG
dimethylglycine
- ESI
electrospray ionization
- Fmoc
fluorenylmethoxylcarbonyl
- GalCer
galactosylceramide
- GluCer
glucosylceramide
- HexCer
monohexosylceramide or cerebroside, including both galactosylceramide and glucosylceramide
- HPTLC
high performance of thin layer chromatography
- IM-MS
ion mobility mass spectrometry
- LC
liquid chromatography
- lysoPE
lysophosphatidylethanolamine
- m
n, fatty acyl chain with m carbon atoms and n double bonds
- MALDI
matrix-assisted laser desorption/ionization
- MANOVA
multivariate analysis of variance
- MDMS
multi-dimensional mass spectrometry
- MRM
multiple reaction monitoring
- MS
mass spectrometry
- MS/MS
tandem mass spectrometry
- NLS
neutral-loss scanning or scan(s)
- Nm
n, a fatty acyl chain with m carbon atoms and n double bonds is linked with an amide bond
- PA
phosphatidic acid
- PC
choline glycerophospholipid or phosphatidylcholine
- PCR
principal component analysis
- PE
ethanolamine glycerophospholipid or phosphatidylethanolamine
- PG
phosphatidylglycerol
- PI
phosphatidylinositol
- PIS
precursor-ion scanning or scan(s)
- PLS
partial least square
- PS
phosphatidylserine
- pPE
plasmenylethanolamine
- QqQ
triple quadrupole mass spectrometer
- SIE
selected ion extraction
- sn
stereospecific numbering
- SRM
selected reaction monitoring
- SM
sphingomyelin
- TAG
triacylglycerol(s)
- TOF
time-of-flight
References
- Abe T, Norton WT. The characterization of sphingolipids from neurons and astroglia of immature rat brain. J Neurochem. 1974;23:1025–1036. doi: 10.1111/j.1471-4159.1974.tb10755.x. [DOI] [PubMed] [Google Scholar]
- Al-Saad KA, Zabrouskov V, Siems WF, Knowles NR, Hannan RM, Hill HH., Jr Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry of lipids: ionization and prompt fragmentation patterns. Rapid Commun Mass Spectrom. 2003;17:87–96. doi: 10.1002/rcm.858. [DOI] [PubMed] [Google Scholar]
- Altelaar AF, Klinkert I, Jalink K, de Lange RP, Adan RA, Heeren RM, Piersma SR. Gold-enhanced biomolecular surface imaging of cells and tissue by SIMS and MALDI mass spectrometry. Anal Chem. 2006;78:734–742. doi: 10.1021/ac0513111. [DOI] [PubMed] [Google Scholar]
- Andreyev AY, Fahy E, Guan Z, Kelly S, Li X, McDonald JG, Milne S, Myers D, Park H, Ryan A, Thompson BM, Wang E, Zhao Y, Brown HA, Merrill AH, Raetz CR, Russell DW, Subramaniam S, Dennis EA. Subcellular organelle lipidomics in TLR-4-activated macrophages. J Lipid Res. 2010;51:2785–2797. doi: 10.1194/jlr.M008748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelini R, Babudri F, Lobasso S, Corcelli A. MALDI-TOF/MS analysis of archaebacterial lipids in lyophilized membranes dry-mixed with 9-aminoacridine. J Lipid Res. 2010;51:2818–2825. doi: 10.1194/jlr.D007328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astarita G, Piomelli D. Lipidomic analysis of endocannabinoid metabolism in biological samples. J Chromatogr B. 2009;877:2755–2767. doi: 10.1016/j.jchromb.2009.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basconcillo LS, Zaheer R, Finan TM, McCarry BE. A shotgun lipidomics approach in Sinorhizobium meliloti as a tool in functional genomics. J Lipid Res. 2009a;50:1120–1132. doi: 10.1194/jlr.M800443-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basconcillo LS, Zaheer R, Finan TM, McCarry BE. Shotgun lipidomics study of a putative lysophosphatidic acyl transferase (PlsC) in Sinorhizobium meliloti. J Chromatogr B. 2009b;877:2873–2882. doi: 10.1016/j.jchromb.2009.05.014. [DOI] [PubMed] [Google Scholar]
- Berry KA, Murphy RC. Analysis of cell membrane aminophospholipids as isotope-tagged derivatives. J Lipid Res. 2005;46:1038–1046. doi: 10.1194/jlr.M500014-JLR200. [DOI] [PubMed] [Google Scholar]
- Bielawski J, Szulc ZM, Hannun YA, Bielawska A. Simultaneous quantitative analysis of bioactive sphingolipids by high-performance liquid chromatography-tandem mass spectrometry. Methods. 2006;39:82–91. doi: 10.1016/j.ymeth.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Blanksby SJ, Mitchell TW. Advances in mass spectrometry for lipidomics. Annu Rev Anal Chem (Palo Alto Calif) 2010;3:433–465. doi: 10.1146/annurev.anchem.111808.073705. [DOI] [PubMed] [Google Scholar]
- Bleijerveld OB, Houweling M, Thomas MJ, Cui Z. Metabolipidomics: profiling metabolism of glycerophospholipid species by stable isotopic precursors and tandem mass spectrometry. Anal Biochem. 2006;352:1–14. doi: 10.1016/j.ab.2006.02.016. [DOI] [PubMed] [Google Scholar]
- Bollinger JG, Ii H, Sadilek M, Gelb MH. Improved method for the quantification of lysophospholipids including enol ether species by liquid chromatography-tandem mass spectrometry. J Lipid Res. 2010a;51:440–447. doi: 10.1194/jlr.D000885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollinger JG, Thompson W, Lai Y, Oslund RC, Hallstrand TS, Sadilek M, Turecek F, Gelb MH. Improved sensitivity mass spectrometric detection of eicosanoids by charge reversal derivatization. Anal Chem. 2010b;82:6790–6796. doi: 10.1021/ac100720p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugger B, Erben G, Sandhoff R, Wieland FT, Lehmann WD. Quantitative analysis of biological membrane lipids at the low picomole level by nano-electrospray ionization tandem mass spectrometry. Proc Natl Acad Sci USA. 1997;94:2339–2344. doi: 10.1073/pnas.94.6.2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant DK, Orlando RC, Fenselau C, Sowder RC, Henderson LE. Four-sector tandem mass spectrometric analysis of complex mixtures of phosphatidylcholines present in a human immunodeficiency virus preparation. Anal Chem. 1991;63:1110–1114. doi: 10.1021/ac00011a010. [DOI] [PubMed] [Google Scholar]
- Burnum KE, Cornett DS, Puolitaival SM, Milne SB, Myers DS, Tranguch S, Brown HA, Dey SK, Caprioli RM. Spatial and temporal alterations of phospholipids determined by mass spectrometry during mouse embryo implantation. J Lipid Res. 2009;50:2290–2298. doi: 10.1194/jlr.M900100-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrdwell WC. Dual parallel liquid chromatography with dual mass spectrometry (LC2/MS2) for a total lipid analysis. Front Biosci. 2008;13:100–120. doi: 10.2741/2663. [DOI] [PubMed] [Google Scholar]
- Cha S, Yeung ES. Colloidal graphite-assisted laser desorption/ionization mass spectrometry and MSn of small molecules. 1. Imaging of cerebrosides directly from rat brain tissue. Anal Chem. 2007;79:2373–2385. doi: 10.1021/ac062251h. [DOI] [PubMed] [Google Scholar]
- Cha S, Zhang H, Ilarslan HI, Wurtele ES, Brachova L, Nikolau BJ, Yeung ES. Direct profiling and imaging of plant metabolites in intact tissues by using colloidal graphite-assisted laser desorption ionization mass spectrometry. Plant J. 2008;55:348–360. doi: 10.1111/j.1365-313X.2008.03507.x. [DOI] [PubMed] [Google Scholar]
- Chen Y, Allegood J, Liu Y, Wang E, Cachon-Gonzalez B, Cox TM, Merrill AH, Jr, Sullards MC. Imaging MALDI mass spectrometry using an oscillating capillary nebulizer matrix coating system and its application to analysis of lipids in brain from a mouse model of Tay-Sachs/Sandhoff disease. Anal Chem. 2008;80:2780–2788. doi: 10.1021/ac702350g. [DOI] [PubMed] [Google Scholar]
- Chen Y, Liu Y, Allegood J, Wang E, BC-G, Cox TM, Merrill AH, Sullards MC. Imaging MALDI mass spectrometry of sphingolipids using an oscillating capillary nebulizer matrix application system. Methods Mol Biol. 2010;656:131–146. doi: 10.1007/978-1-60761-746-4_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Jiang X, Han X. Alterations in lipid homeostasis of mouse dorsal root ganglia induced by apolipoprotein E deficiency: A shotgun lipidomics study. J Neurochem. 2007;101:57–76. doi: 10.1111/j.1471-4159.2006.04342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Xu J, McKeel DW, Jr, Han X. Specificity and potential mechanism of sulfatide deficiency in Alzheimer’s disease: An electrospray ionization mass spectrometric study. Cell Mol Biol. 2003;49:809–818. [PubMed] [Google Scholar]
- Cheng H, Zhou Y, Holtzman DM, Han X. Apolipoprotein E mediates sulfatide depletion in amyloid precursor protein transgenic animal models of Alzheimer’s disease. Neurobiol Aging. 2010a;31:1188–1196. doi: 10.1016/j.neurobiolaging.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Sun G, Yang K, Gross RW, Han X. Selective desorption/ionization of sulfatides by MALDI-MS facilitated using 9-aminoacridine as matrix. J Lipid Res. 2010b;51:1599–1609. doi: 10.1194/jlr.D004077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernushevich IV, Loboda AV, Thomson BA. An introduction to quadrupole-time-of-flight mass spectrometry. J Mass Spectrom. 2001;36:849–865. doi: 10.1002/jms.207. [DOI] [PubMed] [Google Scholar]
- Christie WW, Han X. Lipid Analysis: Isolation, Separation, Identification and Lipidomic Analysis. Bridgwater, England: The Oily Press; 2010. [Google Scholar]
- Cowart LA, Shotwell M, Worley ML, Richards AJ, Montefusco DJ, Hannun YA, Lu X. Revealing a signaling role of phytosphingosine-1-phosphate in yeast. Mol Syst Biol. 2010;6:349. doi: 10.1038/msb.2010.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dannenberger D, Suss R, Teuber K, Fuchs B, Nuernberg K, Schiller J. The intact muscle lipid composition of bulls: an investigation by MALDI-TOF MS and 31P NMR. Chem Phys Lipids. 2010;163:157–164. doi: 10.1016/j.chemphyslip.2009.10.011. [DOI] [PubMed] [Google Scholar]
- Deeley JM, Mitchell TW, Wei X, Korth J, Nealon JR, Blanksby SJ, Truscott RJ. Human lens lipids differ markedly from those of commonly used experimental animals. Biochim Biophys Acta. 2008;1781:288–298. doi: 10.1016/j.bbalip.2008.04.002. [DOI] [PubMed] [Google Scholar]
- Deeley JM, Hankin JA, Friedrich MG, Murphy RC, Truscott RJ, Mitchell TW, Blanksby SJ. Sphingolipid distribution changes with age in the human lens. J Lipid Res. 2010;51:2753–2760. doi: 10.1194/jlr.M007716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLong CJ, Baker PRS, Samuel M, Cui Z, Thomas MJ. Molecular species composition of rat liver phospholipids by ESI-MS/MS: the effect of chromatography. J Lipid Res. 2001;42:1959–1968. [PubMed] [Google Scholar]
- Dennis EA. Lipidomics joins the omics evolution. Proc Natl Acad Sci USA. 2009;106:2089–2090. doi: 10.1073/pnas.0812636106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhingra S, Freedenberg M, Quo CF, Merrill AH, Jr, Wang MD. Computational modeling of a metabolic pathway in ceramide de novo synthesis. Conf Proc IEEE Eng Med Biol Soc. 2007;2007:1405–1408. doi: 10.1109/IEMBS.2007.4352561. [DOI] [PubMed] [Google Scholar]
- Dill AL, Ifa DR, Manicke NE, Ouyang Z, Cooks RG. Mass spectrometric imaging of lipids using desorption electrospray ionization. J Chromatogr B. 2009;877:2883–2889. doi: 10.1016/j.jchromb.2008.12.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowhan W. Molecular basis for membrane phospholipid diversity: why are there so many lipids? Annu Rev Biochem. 1997;66:199–232. doi: 10.1146/annurev.biochem.66.1.199. [DOI] [PubMed] [Google Scholar]
- Ejsing CS, Moehring T, Bahr U, Duchoslav E, Karas M, Simons K, Shevchenko A. Collision-induced dissociation pathways of yeast sphingolipids and their molecular profiling in total lipid extracts: a study by quadrupole TOF and linear ion trap-orbitrap mass spectrometry. J Mass Spectrom. 2006a;41:372–389. doi: 10.1002/jms.997. [DOI] [PubMed] [Google Scholar]
- Ejsing CS, Duchoslav E, Sampaio J, Simons K, Bonner R, Thiele C, Ekroos K, Shevchenko A. Automated identification and quantification of glycerophospholipid molecular species by multiple precursor ion scanning. Anal Chem. 2006b;78:6202–6214. doi: 10.1021/ac060545x. [DOI] [PubMed] [Google Scholar]
- Ejsing CS, Sampaio JL, Surendranath V, Duchoslav E, Ekroos K, Klemm RW, Simons K, Shevchenko A. Global analysis of the yeast lipidome by quantitative shotgun mass spectrometry. Proc Natl Acad Sci USA. 2009;106:2136–2141. doi: 10.1073/pnas.0811700106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekroos K, Chernushevich IV, Simons K, Shevchenko A. Quantitative profiling of phospholipids by multiple precursor ion scanning on a hybrid quadrupole time-of-flight mass spectrometer. Anal Chem. 2002;74:941–949. doi: 10.1021/ac015655c. [DOI] [PubMed] [Google Scholar]
- Ekroos K, Janis M, Tarasov K, Hurme R, Laaksonen R. Lipidomics: a tool for studies of atherosclerosis. Curr Atheroscler Rep. 2010;12:273–281. doi: 10.1007/s11883-010-0110-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esch SW, Williams TD, Biswas S, Chakrabarty A, Levine SM. Sphingolipid profile in the CNS of the twitcher (globoid cell leukodystrophy) mouse: a lipidomics approach. Cell Mol Biol. 2003;49:779–787. [PubMed] [Google Scholar]
- Fahy E, Cotter D, Byrnes R, Sud M, Maer A, Li J, Nadeau D, Zhau Y, Subramaniam S. Bioinformatics for lipidomics. Methods Enzymol. 2007;432:247–273. doi: 10.1016/S0076-6879(07)32011-9. [DOI] [PubMed] [Google Scholar]
- Fahy E, Subramaniam S, Brown HA, Glass CK, Merrill AH, Jr, Murphy RC, Raetz CR, Russell DW, Seyama Y, Shaw W, Shimizu T, Spener F, van Meer G, VanNieuwenhze MS, White SH, Witztum JL, Dennis EA. A comprehensive classification system for lipids. J Lipid Res. 2005;46:839–861. doi: 10.1194/jlr.E400004-JLR200. [DOI] [PubMed] [Google Scholar]
- Feng L, Prestwich GD, editors. Functional Lipidomics. Boca Raton, FL: CRC Press, Taylor & Francis Group; 2006. [Google Scholar]
- Fenn JB. Electrospray wings for molecular elephants (Nobel lecture) Angew Chem Int Ed Engl. 2003;42:3871–3894. doi: 10.1002/anie.200300605. [DOI] [PubMed] [Google Scholar]
- Fenwick GR, Eagles J, Self R. Fast atom bombardment mass spectrometry of intact phospholipids and related compounds. Biomed Mass Spectrom. 1983;10:382–386. [Google Scholar]
- Ferreri C, Chatgilialoglu C. Membrane lipidomics and the geometry of unsaturated fatty acids from biomimetic models to biological consequences. Methods Mol Biol. 2009;579:391–411. doi: 10.1007/978-1-60761-322-0_20. [DOI] [PubMed] [Google Scholar]
- Forrester JS, Milne SB, Ivanova PT, Brown HA. Computational lipidomics: A multiplexed analysis of dynamic changes in membrane lipid composition during signal transduction. Mol pharmacol. 2004;65:813–821. doi: 10.1124/mol.65.4.813. [DOI] [PubMed] [Google Scholar]
- Fuchs B, Suss R, Schiller J. An update of MALDI-TOF mass spectrometry in lipid research. Prog Lipid Res. 2010;49:450–475. doi: 10.1016/j.plipres.2010.07.001. [DOI] [PubMed] [Google Scholar]
- Fuchs B, Schiller J, Suss R, Zscharnack M, Bader A, Muller P, Schurenberg M, Becker M, Suckau D. Analysis of stem cell lipids by offline HPTLC-MALDI-TOF MS. Anal Bioanal Chem. 2008;392:849–860. doi: 10.1007/s00216-008-2301-8. [DOI] [PubMed] [Google Scholar]
- Goto-Inoue N, Hayasaka T, Taki T, Gonzalez TV, Setou M. A new lipidomics approach by thin-layer chromatography-blot-matrix-assisted laser desorption/ionization imaging mass spectrometry for analyzing detailed patterns of phospholipid molecular species. J Chromatogr A. 2009;1216:7096–7101. doi: 10.1016/j.chroma.2009.08.056. [DOI] [PubMed] [Google Scholar]
- Goto-Inoue N, Hayasaka T, Sugiura Y, Taki T, Li YT, Matsumoto M, Setou M. High-sensitivity analysis of glycosphingolipids by matrix-assisted laser desorption/ionization quadrupole ion trap time-of-flight imaging mass spectrometry on transfer membranes. J Chromatogr B. 2008;870:74–83. doi: 10.1016/j.jchromb.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths WJ, Wang Y. Analysis of neurosterols by GC-MS and LC-MS/MS. J Chromatogr B. 2009a;877:2778–2805. doi: 10.1016/j.jchromb.2009.05.017. [DOI] [PubMed] [Google Scholar]
- Griffiths WJ, Wang Y. Mass spectrometry: from proteomics to metabolomics and lipidomics. Chem Soc Rev. 2009b;38:1882–1896. doi: 10.1039/b618553n. [DOI] [PubMed] [Google Scholar]
- Gross RW. High plasmalogen and arachidonic acid content of canine myocardial sarcolemma: a fast atom bombardment mass spectroscopic and gas chromatography-mass spectroscopic characterization. Biochemistry. 1984;23:158–165. doi: 10.1021/bi00296a026. [DOI] [PubMed] [Google Scholar]
- Gross RW. Identification of plasmalogen as the major phospholipid constituent of cardiac sarcoplasmic reticulum. Biochemistry. 1985;24:1662–1668. doi: 10.1021/bi00328a014. [DOI] [PubMed] [Google Scholar]
- Gross RW, Han X. Lipidomics at the interface of structure and function in systems biology. Chem Biol. 2011;18:284–291. doi: 10.1016/j.chembiol.2011.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Z. Discovering novel brain lipids by liquid chromatography/tandem mass spectrometry. J Chromatogr B. 2009;877:2814–2821. doi: 10.1016/j.jchromb.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Z, Li S, Smith DC, Shaw WA, Raetz CR. Identification of N-acylphosphatidylserine molecules in eukaryotic cells. Biochemistry. 2007;46:14500–14513. doi: 10.1021/bi701907g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haimi P, Uphoff A, Hermansson M, Somerharju P. Software tools for analysis of mass spectrometric lipidome data. Anal Chem. 2006;78:8324–8331. doi: 10.1021/ac061390w. [DOI] [PubMed] [Google Scholar]
- Haimi P, Chaithanya K, Kainu V, Hermansson M, Somerharju P. Instrument-independent software tools for the analysis of MS-MS and LC-MS lipidomics data. Methods Mol Biol. 2009;580:285–294. doi: 10.1007/978-1-60761-325-1_16. [DOI] [PubMed] [Google Scholar]
- Ham BM, Jacob JT, Cole RB. MALDI-TOF MS of phosphorylated lipids in biological fluids using immobilized metal affinity chromatography and a solid ionic crystal matrix. Anal Chem. 2005;77:4439–4447. doi: 10.1021/ac058000a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X. Characterization and direct quantitation of ceramide molecular species from lipid extracts of biological samples by electrospray ionization tandem mass spectrometry. Anal Biochem. 2002;302:199–212. doi: 10.1006/abio.2001.5536. [DOI] [PubMed] [Google Scholar]
- Han X. Lipid alterations in the earliest clinically recognizable stage of Alzheimer’s disease: Implication of the role of lipids in the pathogenesis of Alzheimer’s disease. Curr Alz Res. 2005;2:65–77. doi: 10.2174/1567205052772786. [DOI] [PubMed] [Google Scholar]
- Han X. Neurolipidomics: challenges and developments. Front Biosci. 2007;12:2601–2615. doi: 10.2741/2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X. Multi-dimensional mass spectrometry-based shotgun lipidomics and the altered lipids at the mild cognitive impairment stage of Alzheimer’s disease. Biochim Biophys Acta. 2010;1801:774–783. doi: 10.1016/j.bbalip.2010.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Gross RW. Electrospray ionization mass spectroscopic analysis of human erythrocyte plasma membrane phospholipids. Proc Natl Acad Sci USA. 1994;91:10635–10639. doi: 10.1073/pnas.91.22.10635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Gross RW. Quantitative analysis and molecular species fingerprinting of triacylglyceride molecular species directly from lipid extracts of biological samples by electrospray ionization tandem mass spectrometry. Anal Biochem. 2001;295:88–100. doi: 10.1006/abio.2001.5178. [DOI] [PubMed] [Google Scholar]
- Han X, Gross RW. Global analyses of cellular lipidomes directly from crude extracts of biological samples by ESI mass spectrometry: a bridge to lipidomics. J Lipid Res. 2003;44:1071–1079. doi: 10.1194/jlr.R300004-JLR200. [DOI] [PubMed] [Google Scholar]
- Han X, Gross RW. Shotgun lipidomics: multi-dimensional mass spectrometric analysis of cellular lipidomes. Expert Rev Proteomics. 2005a;2:253–264. doi: 10.1586/14789450.2.2.253. [DOI] [PubMed] [Google Scholar]
- Han X, Gross RW. Shotgun lipidomics: Electrospray ionization mass spectrometric analysis and quantitation of the cellular lipidomes directly from crude extracts of biological samples. Mass Spectrom Rev. 2005b;24:367–412. doi: 10.1002/mas.20023. [DOI] [PubMed] [Google Scholar]
- Han X, Cheng H. Characterization and direct quantitation of cerebroside molecular species from lipid extracts by shotgun lipidomics. J Lipid Res. 2005;46:163–175. doi: 10.1194/jlr.D400022-JLR200. [DOI] [PubMed] [Google Scholar]
- Han X, Jiang X. A review of lipidomic technologies applicable to sphingolipidomics and their relevant applications. Eur J Lipid Sci Technol. 2009;111:39–52. doi: 10.1002/ejlt.200800117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Yang K, Gross RW. Microfluidics-based electrospray ionization enhances intrasource separation of lipid classes and extends identification of individual molecular species through multi-dimensional mass spectrometry: Development of an automated high throughput platform for shotgun lipidomics. Rapid Commun Mass Spectrom. 2008;22:2115–2124. doi: 10.1002/rcm.3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Gubitosi-Klug RA, Collins BJ, Gross RW. Alterations in individual molecular species of human platelet phospholipids during thrombin stimulation: electrospray ionization mass spectrometry-facilitated identification of the boundary conditions for the magnitude and selectivity of thrombin-induced platelet phospholipid hydrolysis. Biochemistry. 1996;35:5822–5832. doi: 10.1021/bi952927v. [DOI] [PubMed] [Google Scholar]
- Han X, Abendschein DR, Kelley JG, Gross RW. Diabetes-induced changes in specific lipid molecular species in rat myocardium. Biochem J. 2000;352:79–89. [PMC free article] [PubMed] [Google Scholar]
- Han X, Cheng H, Mancuso DJ, Gross RW. Caloric restriction results in phospholipid depletion, membrane remodeling and triacylglycerol accumulation in murine myocardium. Biochemistry. 2004a;43:15584–15594. doi: 10.1021/bi048307o. [DOI] [PubMed] [Google Scholar]
- Han X, Cheng H, Jiang X, Zeng Y. Mass spectrometry methods for the analysis of lipid molecular species: A shotgun lipidomics approach. In: Murphy EJ, Rosenberger TA, editors. Lipid-Mediated Signaling. CRC Press: Taylor & Francis Group, LLC; 2010. pp. 149–173. [Google Scholar]
- Han X, Yang J, Cheng H, Ye H, Gross RW. Towards fingerprinting cellular lipidomes directly from biological samples by two-dimensional electrospray ionization mass spectrometry. Anal Biochem. 2004b;330:317–331. doi: 10.1016/j.ab.2004.04.004. [DOI] [PubMed] [Google Scholar]
- Han X, Yang K, Cheng H, Fikes KN, Gross RW. Shotgun lipidomics of phosphoethanolamine-containing lipids in biological samples after one-step in situ derivatization. J Lipid Res. 2005;46:1548–1560. doi: 10.1194/jlr.D500007-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Yang K, Yang J, Cheng H, Gross RW. Shotgun lipidomics of cardiolipin molecular species in lipid extracts of biological samples. J Lipid Res. 2006a;47:864–879. doi: 10.1194/jlr.D500044-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Yang K, Yang J, Fikes KN, Cheng H, Gross RW. Factors influencing the electrospray intrasource separation and selective ionization of glycerophospholipids. J Am Soc Mass Spectrom. 2006b;17:264–274. doi: 10.1016/j.jasms.2005.11.003. [DOI] [PubMed] [Google Scholar]
- Han X, Yang J, Yang K, Zhao Z, Abendschein DR, Gross RW. Alterations in myocardial cardiolipin content and composition occur at the very earliest stages of diabetes: A shotgun lipidomics study. Biochemistry. 2007;46:6417–6428. doi: 10.1021/bi7004015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hankin JA, Barkley RM, Murphy RC. Sublimation as a method of matrix application for mass spectrometric imaging. J Am Soc Mass Spectrom. 2007;18:1646–1652. doi: 10.1016/j.jasms.2007.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison KA, Murphy RC. Direct mass spectrometric analysis of ozonides: application to unsaturated glycerophosphocholine lipids. Anal Chem. 1996;68:3224–3230. doi: 10.1021/ac960302c. [DOI] [PubMed] [Google Scholar]
- Hartler J, Trotzmuller M, Chitraju C, Spener F, Kofeler HC, Thallinger GG. Lipid Data Analyzer: unattended identification and quantitation of lipids in LC-MS data. Bioinformatics. 2011;27:572–577. doi: 10.1093/bioinformatics/btq699. [DOI] [PubMed] [Google Scholar]
- Hayasaka T, Goto-Inoue N, Zaima N, Shrivas K, Kashiwagi Y, Yamamoto M, Nakamoto M, Setou M. Imaging mass spectrometry with silver nanoparticles reveals the distribution of fatty acids in mouse retinal sections. J Am Soc Mass Spectrom. 2010;21:1446–1454. doi: 10.1016/j.jasms.2010.04.005. [DOI] [PubMed] [Google Scholar]
- Hein EM, Bodeker B, Nolte J, Hayen H. Software tool for mining liquid chromatography/multi-stage mass spectrometry data for comprehensive glycerophospholipid profiling. Rapid Commun Mass Spectrom. 2010;24:2083–2092. doi: 10.1002/rcm.4614. [DOI] [PubMed] [Google Scholar]
- Henning PA, Merrill AH, Wang MD. Dynamic pathway modeling of sphingolipid metabolism. Conf Proc IEEE Eng Med Biol Soc. 2004;4:2913–2916. doi: 10.1109/IEMBS.2004.1403828. [DOI] [PubMed] [Google Scholar]
- Hermansson M, Uphoff A, Kakela R, Somerharju P. Automated quantitative analysis of complex lipidomes by liquid chromatography/mass spectrometry. Anal Chem. 2005;77:2166–2175. doi: 10.1021/ac048489s. [DOI] [PubMed] [Google Scholar]
- Herzog R, Schwudke D, Schuhmann K, Sampaio JL, Bornstein SR, Schroeder M, Shevchenko A. A novel informatics concept for high-throughput shotgun lipidomics based on the molecular fragmentation query language. Genome Biol. 2011;12:R8. doi: 10.1186/gb-2011-12-1-r8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks AM, DeLong CJ, Thomas MJ, Samuel M, Cui Z. Unique molecular signatures of glycerophospholipid species in different rat tissues analyzed by tandem mass spectrometry. Biochim Biophys Acta. 2006;1761:1022–1029. doi: 10.1016/j.bbalip.2006.05.010. [DOI] [PubMed] [Google Scholar]
- Howdle MD, Eckers C, Laures AM, Creaser CS. The use of shift reagents in ion mobility-mass spectrometry: studies on the complexation of an active pharmaceutical ingredient with polyethylene glycol excipients. J Am Soc Mass Spectrom. 2009;20:1–9. doi: 10.1016/j.jasms.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Hsu F-F, Turk J. Structural characterization of triacylglycerols as lithiated adducts by electrospray ionization mass spectrometry using low-energy collisionally activated dissociation on a triple stage quadrupole instrument. J Am Soc Mass Spectrom. 1999;10:587–599. doi: 10.1016/S1044-0305(99)00035-5. [DOI] [PubMed] [Google Scholar]
- Hsu F-F, Turk J. Elucidation of the double-bond position of long-chain unsaturated fatty acids by multiple-stage linear ion-trap mass spectrometry with electrospray ionization. J Am Soc Mass Spectrom. 2008a;19:1673–1680. doi: 10.1016/j.jasms.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu F-F, Turk J. Electrospray ionization with low-energy collisionally activated dissociation tandem mass spectrometry of glycerophospholipids: Mechanisms of fragmentation and structural characterization. J Chromatogr B. 2009;877:2673–2695. doi: 10.1016/j.jchromb.2009.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu F-F, Bohrer A, Turk J. Electrospray ionization tandem mass spectrometric analysis of sulfatide. Determination of fragmentation patterns and characterization of molecular species expressed in brain and in pancreatic islets. Biochim Biophys Acta. 1998;1392:202–216. doi: 10.1016/s0005-2760(98)00034-4. [DOI] [PubMed] [Google Scholar]
- Hsu F-F, Turk J, Thukkani AK, Messner MC, Wildsmith KR, Ford DA. Characterization of alkylacyl, alk-1-enylacyl and lyso subclasses of glycerophosphocholine by tandem quadrupole mass spectrometry with electrospray ionization. J Mass Spectrom. 2003;38:752–763. doi: 10.1002/jms.491. [DOI] [PubMed] [Google Scholar]
- Hsu FF, Turk J. Structural determination of glycosphingolipids as lithiated adducts by electrospray ionization mass spectrometry using low-energy collisional-activated dissociation on a triple stage quadrupole instrument. J Am Soc Mass Spectrom. 2001;12:61–79. doi: 10.1016/S1044-0305(00)00194-X. [DOI] [PubMed] [Google Scholar]
- Hsu FF, Turk J. Analysis of Sphingomyelins. In: Caprioli RM, editor. The encyclopedia of mass spectrometry. New York: Elsevier; 2005. pp. 430–447. [Google Scholar]
- Hsu FF, Turk J. Structural characterization of unsaturated glycerophospholipids by multiple-stage linear ion-trap mass spectrometry with electrospray ionization. J Am Soc Mass Spectrom. 2008b;19:1681–1691. doi: 10.1016/j.jasms.2008.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubner G, Crone C, Lindner B. lipID--a software tool for automated assignment of lipids in mass spectra. J Mass Spectrom. 2009;44:1676–1683. doi: 10.1002/jms.1673. [DOI] [PubMed] [Google Scholar]
- Hunt AN. Completing the cycles; the dynamics of endonuclear lipidomics. Biochim Biophys Acta. 2006;1761:577–587. doi: 10.1016/j.bbalip.2006.02.013. [DOI] [PubMed] [Google Scholar]
- Ishida Y, Nakanishi O, Hirao S, Tsuge S, Urabe J, Sekino T, Nakanishi M, Kimoto T, Ohtani H. Direct analysis of lipids in single zooplankter individuals by matrix-assisted laser desorption/ionization mass spectrometry. Anal Chem. 2003;75:4514–4518. doi: 10.1021/ac030072j. [DOI] [PubMed] [Google Scholar]
- Ivanova PT, Milne SB, Brown HA. Identification of atypical ether-linked glycerophospholipid species in macrophages by mass spectrometry. J Lipid Res. 2010;51:1581–1590. doi: 10.1194/jlr.D003715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson SN, Woods AS. Direct profiling of tissue lipids by MALDI-TOFMS. J Chromatogr B. 2009;877:2822–2829. doi: 10.1016/j.jchromb.2008.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson SN, Wang HY, Woods AS. Direct profiling of lipid distribution in brain tissue using MALDI-TOFMS. Anal Chem. 2005a;77:4523–4527. doi: 10.1021/ac050276v. [DOI] [PubMed] [Google Scholar]
- Jackson SN, Wang HY, Woods AS. In situ structural characterization of phosphatidylcholines in brain tissue using MALDI-MS/MS. J Am Soc Mass Spectrom. 2005b;16:2052–2056. doi: 10.1016/j.jasms.2005.08.014. [DOI] [PubMed] [Google Scholar]
- Jackson SN, Ugarov M, Egan T, Post JD, Langlais D, Albert Schultz J, Woods AS. MALDI-ion mobility-TOFMS imaging of lipids in rat brain tissue. J Mass Spectrom. 2007;42:1093–1098. doi: 10.1002/jms.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson SN, Ugarov M, Post JD, Egan T, Langlais D, Schultz JA, Woods AS. A study of phospholipids by ion mobility TOFMS. J Am Soc Mass Spectrom. 2008;19:1655–1662. doi: 10.1016/j.jasms.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Han X. Characterization and direct quantitation of sphingoid base-1-phosphates from lipid extracts: A shotgun lipidomics approach. J Lipid Res. 2006;47:1865–1873. doi: 10.1194/jlr.D600012-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Ory DS, Han X. Characterization of oxysterols by electrospray ionization tandem mass spectrometry after one-step derivatization with dimethylglycine. Rapid Commun Mass Spectrom. 2007;21:141–152. doi: 10.1002/rcm.2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Yang K, Han X. Direct quantitation of psychosine from alkaline-treated lipid extracts with a semi-synthetic internal standard. J Lipid Res. 2009;50:162–172. doi: 10.1194/jlr.D800036-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Cheng H, Yang K, Gross RW, Han X. Alkaline methanolysis of lipid extracts extends shotgun lipidomics analyses to the low abundance regime of cellular sphingolipids. Anal Biochem. 2007;371:135–145. doi: 10.1016/j.ab.2007.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JJ, Borgmann S, Wilkins CL, O’Brien RM. Characterizing the phospholipid profiles in mammalian tissues by MALDI FTMS. Anal Chem. 2006;78:3062–3071. doi: 10.1021/ac0600858. [DOI] [PubMed] [Google Scholar]
- Jones JJ, Batoy SM, Wilkins CL, Liyanage R, Lay JO., Jr Ionic liquid matrix-induced metastable decay of peptides and oligonucleotides and stabilization of phospholipids in MALDI FTMS analyses. J Am Soc Mass Spectrom. 2005;16:2000–2008. doi: 10.1016/j.jasms.2005.08.007. [DOI] [PubMed] [Google Scholar]
- Kalderon B, Sheena V, Shachrur S, Hertz R, Bar-Tana J. Modulation by nutrients and drugs of liver acyl-CoAs analyzed by mass spectrometry. J Lipid Res. 2002;43:1125–1132. doi: 10.1194/jlr.m200060-jlr200. [DOI] [PubMed] [Google Scholar]
- Kanu AB, Dwivedi P, Tam M, Matz L, Hill HH., Jr Ion mobility-mass spectrometry. J Mass Spectrom. 2008;43:1–22. doi: 10.1002/jms.1383. [DOI] [PubMed] [Google Scholar]
- Kapoor S, Quo CF, Merrill AH, Jr, Wang MD. An interactive visualization tool and data model for experimental design in systems biology. Conf Proc IEEE Eng Med Biol Soc. 2008;2008:2423–2426. doi: 10.1109/IEMBS.2008.4649688. [DOI] [PubMed] [Google Scholar]
- Kayganich KA, Murphy RC. Fast atom bombardment tandem mass spectrometric identification of diacyl, alkylacyl, and alk-1-enylacyl molecular species of glycerophosphoethanolamine in human polymorphonuclear leukocytes. Anal Chem. 1992;64:2965–2971. doi: 10.1021/ac00047a015. [DOI] [PubMed] [Google Scholar]
- Kerwin JL, Wiens AM, Ericsson LH. Identification of fatty acids by electrospray mass spectrometry and tandem mass spectrometry. J Mass Spectrom. 1996;31:184–192. doi: 10.1002/(SICI)1096-9888(199602)31:2<184::AID-JMS283>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Kiebish MA, Han X, Cheng H, Lunceford A, Clarke CF, Moon H, Chuang JH, Seyfried TN. Lipidomic analysis and electron transport chain activities in C57BL/6J mouse brain mitochondria. J Neurochem. 2008;106:299–312. doi: 10.1111/j.1471-4159.2008.05383.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiebish MA, Bell R, Yang K, Phan T, Zhao Z, Ames W, Seyfried TN, Gross RW, Chuang JH, Han X. Dynamic simulation of cardiolipin remodeling: greasing the wheels for an interpretative approach to lipidomics. J Lipid Res. 2010;51:2153–2170. doi: 10.1194/jlr.M004796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HY, Wang TC, Ma YC. Liquid chromatography/mass spectrometry of phospholipids using electrospray ionization. Anal Chem. 1994;66:3977–3982. doi: 10.1021/ac00094a020. [DOI] [PubMed] [Google Scholar]
- Kind T, Wohlgemuth G, Lee do Y, Lu Y, Palazoglu M, Shahbaz S, Fiehn O. FiehnLib: mass spectral and retention index libraries for metabolomics based on quadrupole and time-of-flight gas chromatography/mass spectrometry. Anal Chem. 2009;81:10038–10048. doi: 10.1021/ac9019522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsley PJ, Marnett LJ. Analysis of endocannabinoids, their congeners and COX-2 metabolites. J Chromatogr B. 2009;877:2746–2754. doi: 10.1016/j.jchromb.2009.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishimoto K, Urade R, Ogawa T, Moriyama T. Nondestructive quantification of neutral lipids by thin-layer chromatography and laser-fluorescent scanning: suitable methods for “lipidome” analysis. Biochem Biophys Res Commun. 2001;281:657–662. doi: 10.1006/bbrc.2001.4404. [DOI] [PubMed] [Google Scholar]
- Klemm RW, Ejsing CS, Surma MA, Kaiser HJ, Gerl MJ, Sampaio JL, de Robillard Q, Ferguson C, Proszynski TJ, Shevchenko A, Simons K. Segregation of sphingolipids and sterols during formation of secretory vesicles at the trans-Golgi network. J Cell Biol. 2009;185:601–612. doi: 10.1083/jcb.200901145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose C, Ejsing CS, Garcia-Saez AJ, Kaiser HJ, Sampaio JL, Surma MA, Shevchenko A, Schwille P, Simons K. Yeast lipids can phase separate into micrometer-scale membrane domains. J Biol Chem. 2010;285:30224–30232. doi: 10.1074/jbc.M110.123554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koivusalo M, Haimi P, Heikinheimo L, Kostiainen R, Somerharju P. Quantitative determination of phospholipid compositions by ESI-MS: effects of acyl chain length, unsaturation, and lipid concentration on instrument response. J Lipid Res. 2001;42:663–672. [PubMed] [Google Scholar]
- Kontush A, Chapman MJ. Lipidomics as a tool for the study of lipoprotein metabolism. Curr Atheroscler Rep. 2010;12:194–201. doi: 10.1007/s11883-010-0100-0. [DOI] [PubMed] [Google Scholar]
- Laaksonen R, Katajamaa M, Paiva H, Sysi-Aho M, Saarinen L, Junni P, Lutjohann D, Smet J, Van Coster R, Seppanen-Laakso T, Lehtimaki T, Soini J, Oresic M. A systems biology strategy reveals biological pathways and plasma biomarker candidates for potentially toxic statin-induced changes in muscle. PLoS ONE. 2006;1:e97. doi: 10.1371/journal.pone.0000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Williams MV, DuBois RN, Blair IA. Targeted lipidomics using electron capture atmospheric pressure chemical ionization mass spectrometry. Rapid Commun Mass Spectrom. 2003;17:2168–2176. doi: 10.1002/rcm.1170. [DOI] [PubMed] [Google Scholar]
- Li L, Garden RW, Sweedler JV. Single-cell MALDI: a new tool for direct peptide profiling. Trends Biotechnol. 2000;18:151–160. doi: 10.1016/s0167-7799(00)01427-x. [DOI] [PubMed] [Google Scholar]
- Li M, Zhou Z, Nie H, Bai Y, Liu H. Recent advances of chromatography and mass spectrometry in lipidomics. Anal Bioanal Chem. 2011;399:243–249. doi: 10.1007/s00216-010-4327-y. [DOI] [PubMed] [Google Scholar]
- Li X, Lu X, Tian J, Gao P, Kong H, Xu G. Application of fuzzy c-means clustering in data analysis of metabolomics. Anal Chem. 2009;81:4468–4475. doi: 10.1021/ac900353t. [DOI] [PubMed] [Google Scholar]
- Li YL, Gross ML, Hsu F-F. Ionic-liquid matrices for improved analysis of phospholipids by MALDI-TOF mass spectrometry. J Am Soc Mass Spectrom. 2005;16:679–682. doi: 10.1016/j.jasms.2005.01.017. [DOI] [PubMed] [Google Scholar]
- Liebisch G, Drobnik W, Reil M, Trumbach B, Arnecke R, Olgemoller B, Roscher A, Schmitz G. Quantitative measurement of different ceramide species from crude cellular extracts by electrospray ionization tandem mass spectrometry (ESI-MS/MS) J Lipid Res. 1999;40:1539–1546. [PubMed] [Google Scholar]
- Lobasso S, Lopalco P, Angelini R, Baronio M, Fanizzi FP, Babudri F, Corcelli A. Lipidomic analysis of porcine olfactory epithelial membranes and cilia. Lipids. 2010;45:593–602. doi: 10.1007/s11745-010-3432-1. [DOI] [PubMed] [Google Scholar]
- Lydic TA, Renis R, Busik JV, Reid GE. Analysis of Retina and Erythrocyte Glycerophospholipid Alterations in a Rat Model of Type 1 Diabetes. JALA Charlottesv Va. 2009;14:383–399. doi: 10.1016/j.jala.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maffei Facino R, Carini M, Aldini G, Colombo L. Characterization of the Intermediate Products of Lipid Peroxidation in Phosphatidylcholine Liposomes by Fast-atom Bombardment Mass Spectrometry and Tandem Mass Spectrometry Techniques. Rapid Commun Mass Spectrom. 1996;10:1148–1152. [Google Scholar]
- Mancuso DJ, Sims HF, Han X, Jenkins CM, Guan SP, Yang K, Moon SH, Pietka T, Abumrad NA, Schlesinger PH, Gross RW. Genetic ablation of calcium-independent phospholipase A2gamma leads to alterations in mitochondrial lipid metabolism and function resulting in a deficient mitochondrial bioenergetic phenotype. J Biol Chem. 2007a;282:34611–34622. doi: 10.1074/jbc.M707795200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancuso DJ, Han X, Jenkins CM, Lehman JJ, Sambandam N, Sims HF, Yang J, Yan W, Yang K, Green K, Abendschein DR, Saffitz JE, Gross RW. Dramatic accumulation of triglycerides and precipitation of cardiac hemodynamic dysfunction during brief caloric restriction in transgenic myocardium expressing human calcium-independent phospholipase A2gamma. J Biol Chem. 2007b;282:9216–9227. doi: 10.1074/jbc.M607307200. [DOI] [PubMed] [Google Scholar]
- Manicke NE, Wiseman JM, Ifa DR, Cooks RG. Desorption electrospray ionization (DESI) mass spectrometry and tandem mass spectrometry (MS/MS) of phospholipids and sphingolipids: ionization, adduct formation, and fragmentation. J Am Soc Mass Spectrom. 2008;19:531–543. doi: 10.1016/j.jasms.2007.12.003. [DOI] [PubMed] [Google Scholar]
- Marto JA, White FM, Seldomridge S, Marshall AG. Structural characterization of phospholipids by matrix-assisted laser desorption/ionization Fourier transform ion cyclotron resonance mass spectrometry. Anal Chem. 1995;67:3979–3984. doi: 10.1021/ac00117a025. [DOI] [PubMed] [Google Scholar]
- Masukawa Y, Narita H, Sato H, Naoe A, Kondo N, Sugai Y, Oba T, Homma R, Ishikawa J, Takagi Y, Kitahara T. Comprehensive quantification of ceramide species in human stratum corneum. J Lipid Res. 2009;50:1708–1719. doi: 10.1194/jlr.D800055-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonnell LA, Heeren RM. Imaging mass spectrometry. Mass Spectrom Rev. 2007;26:606–643. doi: 10.1002/mas.20124. [DOI] [PubMed] [Google Scholar]
- Merrill AH, Jr, Sullards MC, Allegood JC, Kelly S, Wang E. Sphingolipidomics: high-throughput, structure-specific, and quantitative analysis of sphingolipids by liquid chromatography tandem mass spectrometry. Methods. 2005;36:207–224. doi: 10.1016/j.ymeth.2005.01.009. [DOI] [PubMed] [Google Scholar]
- Merrill AH, Jr, Stokes TH, Momin A, Park H, Portz BJ, Kelly S, Wang E, Sullards MC, Wang MD. Sphingolipidomics: a valuable tool for understanding the roles of sphingolipids in biology and disease. J Lipid Res. 2009;50(Suppl):S97–102. doi: 10.1194/jlr.R800073-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesaros C, Lee SH, Blair IA. Targeted quantitative analysis of eicosanoid lipids in biological samples using liquid chromatography-tandem mass spectrometry. J Chromatogr B. 2009;877:2736–2745. doi: 10.1016/j.jchromb.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milman G, Maor Y, Abu-Lafi S, Horowitz M, Gallily R, Batkai S, Mo FM, Offertaler L, Pacher P, Kunos G, Mechoulam R. N-arachidonoyl L-serine, an endocannabinoid-like brain constituent with vasodilatory properties. Proc Natl Acad Sci USA. 2006;103:2428–2433. doi: 10.1073/pnas.0510676103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minkler PE, Hoppel CL. Separation and characterization of cardiolipin molecular species by reverse-phase ion pair high-performance liquid chromatography-mass spectrometry. J Lipid Res. 2010;51:856–865. doi: 10.1194/jlr.D002857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell TW, Buffenstein R, Hulbert AJ. Membrane phospholipid composition may contribute to exceptional longevity of the naked mole-rat (Heterocephalus glaber): a comparative study using shotgun lipidomics. Exp Gerontol. 2007;42:1053–1062. doi: 10.1016/j.exger.2007.09.004. [DOI] [PubMed] [Google Scholar]
- Mitchell TW, Pham H, Thomas MC, Blanksby SJ. Identification of double bond position in lipids: From GC to OzID. J Chromatogr B. 2009;877:2722–2735. doi: 10.1016/j.jchromb.2009.01.017. [DOI] [PubMed] [Google Scholar]
- Mitchell TW, Turner N, Hulbert AJ, Else PL, Hawley JA, Lee JS, Bruce CR, Blanksby SJ. Exercise alters the profile of phospholipid molecular species in rat skeletal muscle. J Appl Physiol. 2004;97:1823–1829. doi: 10.1152/japplphysiol.00344.2004. [DOI] [PubMed] [Google Scholar]
- Moe MK, Anderssen T, Strom MB, Jensen E. Vicinal hydroxylation of unsaturated fatty acids for structural characterization of intact neutral phospholipids by negative electrospray ionization tandem quadrupole mass spectrometry. Rapid Commun Mass Spectrom. 2004a;18:2121–2130. doi: 10.1002/rcm.1601. [DOI] [PubMed] [Google Scholar]
- Moe MK, Strom MB, Jensen E, Claeys M. Negative electrospray ionization low-energy tandem mass spectrometry of hydroxylated fatty acids: a mechanistic study. Rapid Commun Mass Spectrom. 2004b;18:1731–1740. doi: 10.1002/rcm.1545. [DOI] [PubMed] [Google Scholar]
- Moreau RA, Doehlert DC, Welti R, Isaac G, Roth M, Tamura P, Nunez A. The identification of mono-, di-, tri-, and tetragalactosyl-diacylglycerols and their natural estolides in oat kernels. Lipids. 2008;43:533–548. doi: 10.1007/s11745-008-3181-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy RC, Axelsen PH. Mass spectrometric analysis of long-chain lipids. Mass Spectrom Rev. 2010 doi: 10.1002/mas. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy RC, Barkley RM, Zemski Berry K, Hankin J, Harrison K, Johnson C, Krank J, McAnoy A, Uhlson C, Zarini S. Electrospray ionization and tandem mass spectrometry of eicosanoids. Anal Biochem. 2005;346:1–42. doi: 10.1016/j.ab.2005.04.042. [DOI] [PubMed] [Google Scholar]
- Nakanishi H, Ogiso H, Taguchi R. Qualitative and quantitative analyses of phospholipids by LC-MS for lipidomics. Methods Mol Biol. 2009;579:287–313. doi: 10.1007/978-1-60761-322-0_15. [DOI] [PubMed] [Google Scholar]
- Nakanishi H, Iida Y, Shimizu T, Taguchi R. Separation and quantification of sn-1 and sn-2 fatty acid positional isomers in phosphatidylcholine by RPLC-ESIMS/MS. J Biochem. 2010;147:245–256. doi: 10.1093/jb/mvp171. [DOI] [PubMed] [Google Scholar]
- Nealon JR, Blanksby SJ, Abbott SK, Hulbert AJ, Mitchell TW, Truscott RJ. Phospholipid composition of the rat lens is independent of diet. Exp Eye Res. 2008;87:502–514. doi: 10.1016/j.exer.2008.08.009. [DOI] [PubMed] [Google Scholar]
- Niemela PS, Castillo S, Sysi-Aho M, Oresic M. Bioinformatics and computational methods for lipidomics. J Chromatogr B Analyt Technol Biomed Life Sci. 2009;877:2855–2862. doi: 10.1016/j.jchromb.2009.01.025. [DOI] [PubMed] [Google Scholar]
- Northen TR, Yanes O, Northen MT, Marrinucci D, Uritboonthai W, Apon J, Golledge SL, Nordstrom A, Siuzdak G. Clathrate nanostructures for mass spectrometry. Nature. 2007;449:1033–1036. doi: 10.1038/nature06195. [DOI] [PubMed] [Google Scholar]
- Patti GJ, Shriver LP, Wassif CA, Woo HK, Uritboonthai W, Apon J, Manchester M, Porter FD, Siuzdak G. Nanostructure-initiator mass spectrometry (NIMS) imaging of brain cholesterol metabolites in Smith-Lemli-Opitz syndrome. Neuroscience. 2010a;170:858–864. doi: 10.1016/j.neuroscience.2010.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patti GJ, Woo HK, Yanes O, Shriver L, Thomas D, Uritboonthai W, Apon JV, Steenwyk R, Manchester M, Siuzdak G. Detection of carbohydrates and steroids by cation-enhanced nanostructure-initiator mass spectrometry (NIMS) for biofluid analysis and tissue imaging. Anal Chem. 2010b;82:121–128. doi: 10.1021/ac9014353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postle AD, Hunt AN. Dynamic lipidomics with stable isotope labelling. J Chromatogr B. 2009;877:2716–2721. doi: 10.1016/j.jchromb.2009.03.046. [DOI] [PubMed] [Google Scholar]
- Postle AD, Wilton DC, Hunt AN, Attard GS. Probing phospholipid dynamics by electrospray ionisation mass spectrometry. Prog Lipid Res. 2007;46:200–224. doi: 10.1016/j.plipres.2007.04.001. [DOI] [PubMed] [Google Scholar]
- Proschogo N, Gaus K, Jessup W. Taking aim at cell lipids: shotgun lipidomics and imaging mass spectrometry push the boundaries. Curr Opin Lipidol. 2009;20:522–523. doi: 10.1097/MOL.0b013e32833301cb. [DOI] [PubMed] [Google Scholar]
- Pruett ST, Bushnev A, Hagedorn K, Adiga M, Haynes CA, Sullards MC, Liotta DC, Merrill AH., Jr Biodiversity of sphingoid bases (“sphingosines”) and related amino alcohols. J Lipid Res. 2008;49:1621–1639. doi: 10.1194/jlr.R800012-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainville PD, Stumpf CL, Shockcor JP, Plumb RS, Nicholson JK. Novel application of reversed-phase UPLC-oaTOF-MS for lipid analysis in complex biological mixtures: a new tool for lipidomics. J Proteome Res. 2007;6:552–558. doi: 10.1021/pr060611b. [DOI] [PubMed] [Google Scholar]
- Rappley I, Myers DS, Milne SB, Ivanova PT, Lavoie MJ, Brown HA, Selkoe DJ. Lipidomic profiling in mouse brain reveals differences between ages and genders, with smaller changes associated with alpha-synuclein genotype. J Neurochem. 2009;111:15–25. doi: 10.1111/j.1471-4159.2009.06290.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rilfors L, Lindblom G. Regulation of lipid composition in biological membranes—biophysical studies of lipids and lipid synthesizing enzymes. Colloids and Surfaces B: Biointerfaces. 2002;26:112–124. [Google Scholar]
- Rohlfing A, Muthing J, Pohlentz G, Distler U, Peter-Katalinic J, Berkenkamp S, Dreisewerd K. IR-MALDI-MS analysis of HPTLC-separated phospholipid mixtures directly from the TLC plate. Anal Chem. 2007;79:5793–5808. doi: 10.1021/ac070633x. [DOI] [PubMed] [Google Scholar]
- Sampaio JL, Gerl MJ, Klose C, Ejsing CS, Beug H, Simons K, Shevchenko A. Membrane lipidome of an epithelial cell line. Proc Natl Acad Sci USA. 2011;108:1903–1907. doi: 10.1073/pnas.1019267108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiller J, Suss R, Arnhold J, Fuchs B, Lessig J, Muller M, Petkovic M, Spalteholz H, Zschornig O, Arnold K. Matrix-assisted laser desorption and ionization time-of-flight (MALDI-TOF) mass spectrometry in lipid and phospholipid research. Prog Lipid Res. 2004;43:449–488. doi: 10.1016/j.plipres.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Schwudke D, Liebisch G, Herzog R, Schmitz G, Shevchenko A. Shotgun lipidomics by tandem mass spectrometry under data-dependent acquisition control. Methods Enzymol. 2007;433:175–191. doi: 10.1016/S0076-6879(07)33010-3. [DOI] [PubMed] [Google Scholar]
- Schwudke D, Oegema J, Burton L, Entchev E, Hannich JT, Ejsing CS, Kurzchalia T, Shevchenko A. Lipid profiling by multiple precursor and neutral loss scanning driven by the data-dependent acquisition. Anal Chem. 2006;78:585–595. doi: 10.1021/ac051605m. [DOI] [PubMed] [Google Scholar]
- Shaner RL, Allegood JC, Park H, Wang E, Kelly S, Haynes CA, Sullards MC, Merrill AH., Jr Quantitative analysis of sphingolipids for lipidomics using triple quadrupole and quadrupole linear ion trap mass spectrometers. J Lipid Res. 2009;50:1692–1707. doi: 10.1194/jlr.D800051-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A, Simons K. Lipidomics: coming to grips with lipid diversity. Nat Rev Mol Cell Biol. 2010;11:593–598. doi: 10.1038/nrm2934. [DOI] [PubMed] [Google Scholar]
- Shui G, Bendt AK, Pethe K, Dick T, Wenk MR. Sensitive profiling of chemically diverse bioactive lipids. J Lipid Res. 2007;48:1976–1984. doi: 10.1194/jlr.M700060-JLR200. [DOI] [PubMed] [Google Scholar]
- Sommer U, Herscovitz H, Welty FK, Costello CE. LC-MS-based method for the qualitative and quantitative analysis of complex lipid mixtures. J Lipid Res. 2006;47:804–814. doi: 10.1194/jlr.M500506-JLR200. [DOI] [PubMed] [Google Scholar]
- Song H, Ladenson J, Turk J. Algorithms for automatic processing of data from mass spectrometric analyses of lipids. J Chromatogr B. 2009a;877:2847–2854. doi: 10.1016/j.jchromb.2008.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H, Ladenson J, Turk J. Algorithms for automatic processing of data from mass spectrometric analyses of lipids. J Chromatogr B. 2009b;877:2847–2854. doi: 10.1016/j.jchromb.2008.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H, Hsu FF, Ladenson J, Turk J. Algorithm for processing raw mass spectrometric data to identify and quantitate complex lipid molecular species in mixtures by data-dependent scanning and fragment ion database searching. J Am Soc Mass Spectrom. 2007;18:1848–1858. doi: 10.1016/j.jasms.2007.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkman OD. Mass Spectrometry Desk Reference. Pittsburgh, PA: Global View Publishing; 2000. [Google Scholar]
- Stach J, Baumbach JI. Ion mobility spectrometry – basic elements and applications. Int J Ion Mobility Spectrom. 2002;5:1–21. [Google Scholar]
- Ståhlman M, Ejsing CS, Tarasov K, Perman J, Borén J, Ekroos K. High throughput oriented shotgun lipidomics by quadrupole time-of-flight mass spectrometry. J Chromatogr B. 2009;877:2664–2672. doi: 10.1016/j.jchromb.2009.02.037. [DOI] [PubMed] [Google Scholar]
- Stoeckli M, Chaurand P, Hallahan DE, Caprioli RM. Imaging mass spectrometry: a new technology for the analysis of protein expression in mammalian tissues. Nat Med. 2001;7:493–496. doi: 10.1038/86573. [DOI] [PubMed] [Google Scholar]
- Stubiger G, Pittenauer E, Belgacem O, Rehulka P, Widhalm K, Allmaier G. Analysis of human plasma lipids and soybean lecithin by means of high-performance thin-layer chromatography and matrix-assisted laser desorption/ionization mass spectrometry. Rapid Commun Mass Spectrom. 2009;23:2711–2723. doi: 10.1002/rcm.4173. [DOI] [PubMed] [Google Scholar]
- Su X, Han X, Mancuso DJ, Abendschein DR, Gross RW. Accumulation of long-chain acylcarnitine and 3-hydroxy acylcarnitine molecular species in diabetic myocardium: identification of alterations in mitochondrial fatty acid processing in diabetic myocardium by shotgun lipidomics. Biochemistry. 2005;44:5234–5245. doi: 10.1021/bi047773a. [DOI] [PubMed] [Google Scholar]
- Sud M, Fahy E, Cotter D, Brown A, Dennis EA, Glass CK, Merrill AH, Jr, Murphy RC, Raetz CR, Russell DW, Subramaniam S. LMSD: LIPID MAPS structure database. Nucleic Acids Res. 2007;35:D527–532. doi: 10.1093/nar/gkl838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullards MC, Wang E, Peng Q, Merrill AH., Jr Metabolomic profiling of sphingolipids in human glioma cell lines by liquid chromatography tandem mass spectrometry. Cell Mol Biol. 2003;49:789–797. [PubMed] [Google Scholar]
- Sun G, Yang K, Zhao Z, Guan S, Han X, Gross RW. Shotgun metabolomics approach for the analysis of negatively charged water-soluble cellular metabolites from mouse heart tissue. Anal Chem. 2007;79:6629–6640. doi: 10.1021/ac070843+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun G, Yang K, Zhao Z, Guan S, Han X, Gross RW. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometric analysis of cellular glycerophospholipids enabled by multiplexed solvent dependent analyte-matrix interactions. Anal Chem. 2008;80:7576–7585. doi: 10.1021/ac801200w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sysi-Aho M, Katajamaa M, Yetukuri L, Oresic M. Normalization method for metabolomics data using optimal selection of multiple internal standards. BMC Bioinformatics. 2007;8:e93. doi: 10.1186/1471-2105-8-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sysi-Aho M, Koikkalainen J, Seppanen-Laakso T, Kaartinen M, Kuusisto J, Peuhkurinen K, Karkkainen S, Antila M, Lauerma K, Reissell E, Jurkko R, Lotjonen J, Helio T, Oresic M. Serum lipidomics meets cardiac magnetic resonance imaging: profiling of subjects at risk of dilated cardiomyopathy. PLoS One. 2011;6:e15744. doi: 10.1371/journal.pone.0015744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi R, Ishikawa M. Precise and global identification of phospholipid molecular species by an Orbitrap mass spectrometer and automated search engine Lipid Search. J Chromatogr A. 2010;1217:4229–4239. doi: 10.1016/j.chroma.2010.04.034. [DOI] [PubMed] [Google Scholar]
- Taguchi R, Hayakawa J, Takeuchi Y, Ishida M. Two-dimensional analysis of phospholipids by capillary liquid chromatography/electrospray ionization mass spectrometry. J Mass Spectrom. 2000;35:953–966. doi: 10.1002/1096-9888(200008)35:8<953::AID-JMS23>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Takats Z, Wiseman JM, Gologan B, Cooks RG. Mass spectrometry sampling under ambient conditions with desorption electrospray ionization. Science. 2004;306:471–473. doi: 10.1126/science.1104404. [DOI] [PubMed] [Google Scholar]
- Taki T, Gonzalez TV, Goto-Inoue N, Hayasaka T, Setou M. TLC blot (far-eastern blot) and its applications. Methods Mol Biol. 2009;536:545–556. doi: 10.1007/978-1-59745-542-8_55. [DOI] [PubMed] [Google Scholar]
- Tan B, Yu YW, Monn MF, Hughes HV, O’Dell DK, Walker JM. Targeted lipidomics approach for endogenous N-acyl amino acids in rat brain tissue. J Chromatogr B. 2009;877:2890–2894. doi: 10.1016/j.jchromb.2009.01.002. [DOI] [PubMed] [Google Scholar]
- Tan B, Bradshaw HB, Rimmerman N, Srinivasan H, Yu YW, Krey JF, Monn MF, Chen JS, Hu SS, Pickens SR, Walker JM. Targeted lipidomics: discovery of new fatty acyl amides. AAPS J. 2006;8:E461–465. doi: 10.1208/aapsj080354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K. The origin of macromolecule ionization by laser irradiation (Nobel lecture) Angew Chem Int Ed Engl. 2003;42:3860–3870. doi: 10.1002/anie.200300585. [DOI] [PubMed] [Google Scholar]
- Thomas MC, Mitchell TW, Blanksby SJ. Ozonolysis of phospholipid double bonds during electrospray ionization: a new tool for structure determination. J Am Chem Soc. 2006;128:58–59. doi: 10.1021/ja056797h. [DOI] [PubMed] [Google Scholar]
- Thomas MC, Mitchell TW, Harman DG, Deeley JM, Murphy RC, Blanksby SJ. Elucidation of double bond position in unsaturated lipids by ozone electrospray ionization mass spectrometry. Anal Chem. 2007;79:5013–5022. doi: 10.1021/ac0702185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomer KB, Crow FW, Gross ML. Location of double-bond position in unsaturated fatty acids by negative ion MS/MS. J Am Chem Soc. 1983;105:5487–5488. [Google Scholar]
- Touboul D, Piednoel H, Voisin V, De La Porte S, Brunelle A, Halgand F, Laprevote O. Changes in phospholipid composition within the dystrophic muscle by matrix-assisted laser desorption/ionization mass spectrometry and mass spectrometry imaging. Eur J Mass Spectrom. 2004;10:657–664. doi: 10.1255/ejms.671. [DOI] [PubMed] [Google Scholar]
- Tsui ZC, Chen QR, Thomas MJ, Samuel M, Cui Z. A method for profiling gangliosides in animal tissues using electrospray ionization-tandem mass spectrometry. Anal Biochem. 2005;341:251–258. doi: 10.1016/j.ab.2005.03.036. [DOI] [PubMed] [Google Scholar]
- van Iersel MP, Kelder T, Pico AR, Hanspers K, Coort S, Conklin BR, Evelo C. Presenting and exploring biological pathways with PathVisio. BMC Bioinformatics. 2008;9:399. doi: 10.1186/1471-2105-9-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance DE, Vance JE. Biochemistry of Lipids, Lipoproteins and Membranes. Amsterdam: Elsevier Science B.V; 2008. [Google Scholar]
- Venter A, Sojka PE, Cooks RG. Droplet dynamics and ionization mechanisms in desorption electrospray ionization mass spectrometry. Anal Chem. 2006;78:8549–8555. doi: 10.1021/ac0615807. [DOI] [PubMed] [Google Scholar]
- Walch A, Rauser S, Deininger SO, Hofler H. MALDI imaging mass spectrometry for direct tissue analysis: a new frontier for molecular histology. Histochem Cell Biol. 2008;130:421–434. doi: 10.1007/s00418-008-0469-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welti R, Wang X, Williams TD. Electrospray ionization tandem mass spectrometry scan modes for plant chloroplast lipids. Anal Biochem. 2003;314:149–152. doi: 10.1016/s0003-2697(02)00623-1. [DOI] [PubMed] [Google Scholar]
- Welti R, Li W, Li M, Sang Y, Biesiada H, Zhou H-E, Rajashekar CB, Williams TD, Wang X. Profiling membrane lipids in plant stress responses. Role of phospholipase Da in freezing-induced lipid changes in Arabidopsis. J Biol Chem. 2002;277:31994–32002. doi: 10.1074/jbc.M205375200. [DOI] [PubMed] [Google Scholar]
- Welti R, Shah J, Li W, Li M, Chen J, Burke JJ, Fauconnier ML, Chapman K, Chye ML, Wang X. Plant lipidomics: discerning biological function by profiling plant complex lipids using mass spectrometry. Front Biosci. 2007;12:2494–2506. doi: 10.2741/2250. [DOI] [PubMed] [Google Scholar]
- Wenk MR. The emerging field of lipidomics. Nat Rev Drug Discov. 2005;4:594–610. doi: 10.1038/nrd1776. [DOI] [PubMed] [Google Scholar]
- Wheelock CE, Goto S, Yetukuri L, D’Alexandri FL, Klukas C, Schreiber F, Oresic M. Bioinformatics strategies for the analysis of lipids. Methods Mol Biol. 2009;580:339–368. doi: 10.1007/978-1-60761-325-1_19. [DOI] [PubMed] [Google Scholar]
- Wiseman JM, Puolitaival SM, Takats Z, Cooks RG, Caprioli RM. Mass spectrometric profiling of intact biological tissue by using desorption electrospray ionization. Angew Chem Int Ed Engl. 2005;44:7094–7097. doi: 10.1002/anie.200502362. [DOI] [PubMed] [Google Scholar]
- Wolf C, Quinn PJ. Lipidomics in diagnosis of lipidoses. Subcell Biochem. 2008;49:567–588. doi: 10.1007/978-1-4020-8831-5_22. [DOI] [PubMed] [Google Scholar]
- Woo HK, Northen TR, Yanes O, Siuzdak G. Nanostructure-initiator mass spectrometry: a protocol for preparing and applying NIMS surfaces for high-sensitivity mass analysis. Nat Protoc. 2008;3:1341–1349. doi: 10.1038/nprot.2008.110. [DOI] [PubMed] [Google Scholar]
- Wood R, Harlow RD. Structural studies of neutral glycerides and phosphoglycerides of rat liver. Arch Biochem Biophys. 1969a;131:495–501. doi: 10.1016/0003-9861(69)90421-4. [DOI] [PubMed] [Google Scholar]
- Wood R, Harlow RD. Structural analyses of rat liver phosphoglycerides. Arch Biochem Biophys. 1969b;135:272–281. doi: 10.1016/0003-9861(69)90540-2. [DOI] [PubMed] [Google Scholar]
- Woods AS, Ugarov M, Egan T, Koomen J, Gillig KJ, Fuhrer K, Gonin M, Schultz JA. Lipid/peptide/nucleotide separation with MALDI-ion mobility-TOF MS. Anal Chem. 2004;76:2187–2195. doi: 10.1021/ac035376k. [DOI] [PubMed] [Google Scholar]
- Wu H, Volponi JV, Oliver AE, Parikh AN, Simmons BA, Singh S. In vivo lipidomics using single-cell Raman spectroscopy. Proc Natl Acad Sci USA. 2011 doi: 10.1073/pnas.1009043108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K, Zhao Z, Gross RW, Han X. Shotgun lipidomics identifies a paired rule for the presence of isomeric ether phospholipid molecular species. PLoS ONE. 2007;2:e1368. doi: 10.1371/journal.pone.0001368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K, Cheng H, Gross RW, Han X. Automated lipid identification and quantification by multi-dimensional mass spectrometry-based shotgun lipidomics. Anal Chem. 2009a;81:4356–4368. doi: 10.1021/ac900241u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K, Zhao Z, Gross RW, Han X. Systematic analysis of choline-containing phospholipids using multi-dimensional mass spectrometry-based shotgun lipidomics. J Chromatogr B. 2009b;877:2924–2936. doi: 10.1016/j.jchromb.2009.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K, Fang X, Gross RW, Han X. A practical approach for determination of mass spectral baselines. 2011a. In preparation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K, Zhao Z, Gross RW, Han X. Identification and quantitation of unsaturated fatty acid isomers by electrospray ionization tandem mass spectrometry: A shotgun lipidomics approach. Anal Chem. 2011b doi: 10.1021/ac2006119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yetukuri L, Katajamaa M, Medina-Gomez G, Seppanen-Laakso T, Vidal-Puig A, Oresic M. Bioinformatics strategies for lipidomics analysis: characterization of obesity related hepatic steatosis. BMC Syst Biol. 2007;1:12. doi: 10.1186/1752-0509-1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yetukuri L, Soderlund S, Koivuniemi A, Seppanen-Laakso T, Niemela PS, Hyvonen M, Taskinen MR, Vattulainen I, Jauhiainen M, Oresic M. Composition and lipid spatial distribution of HDL particles in subjects with low and high HDL-cholesterol. J Lipid Res. 2010;51:2341–2351. doi: 10.1194/jlr.M006494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin P, Zhao X, Li Q, Wang J, Li J, Xu G. Metabonomics study of intestinal fistulas based on ultraperformance liquid chromatography coupled with Q-TOF mass spectrometry (UPLC/Q-TOF MS) J Proteome Res. 2006;5:2135–2143. doi: 10.1021/pr060256p. [DOI] [PubMed] [Google Scholar]
- Zacarias A, Bolanowski D, Bhatnagar A. Comparative measurements of multicomponent phospholipid mixtures by electrospray mass spectroscopy: relating ion intensity to concentration. Anal Biochem. 2002;308:152–159. doi: 10.1016/s0003-2697(02)00209-9. [DOI] [PubMed] [Google Scholar]
- Zech T, Ejsing CS, Gaus K, de Wet B, Shevchenko A, Simons K, Harder T. Accumulation of raft lipids in T-cell plasma membrane domains engaged in TCR signalling. EMBO J. 2009;28:466–476. doi: 10.1038/emboj.2009.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemski Berry KA, Turner WW, VanNieuwenhze MS, Murphy RC. Stable isotope labeled 4-(dimethylamino)benzoic acid derivatives of glycerophosphoethanolamine lipids. Anal Chem. 2009;81:6633–6640. doi: 10.1021/ac900583a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Cha S, Yeung ES. Colloidal graphite-assisted laser desorption/ionization MS and MS(n) of small molecules. 2. Direct profiling and MS imaging of small metabolites from fruits. Anal Chem. 2007;79:6575–6584. doi: 10.1021/ac0706170. [DOI] [PubMed] [Google Scholar]
- Zhao Z, Xu Y. An extremely simple method for extraction of lysophospholipids and phospholipids from blood samples. J Lipid Res. 2010;51:652–659. doi: 10.1194/jlr.D001503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W, Kollmeyer J, Symolon H, Momin A, Munter E, Wang E, Kelly S, Allegood JC, Liu Y, Peng Q, Ramaraju H, Sullards MC, Cabot M, Merrill AH., Jr Ceramides and other bioactive sphingolipid backbones in health and disease: Lipidomic analysis, metabolism and roles in membrane structure, dynamics, signaling and autophagy. Biochim Biophys Acta. 2006;1758:1864–1884. doi: 10.1016/j.bbamem.2006.08.009. [DOI] [PubMed] [Google Scholar]