Abstract
Drug discovery in neuro- and psychopharmacology is lagging, and the most commonly mentioned cause is the scarcity of drug targets. Using NextGen ‘Sequencing Based Single-Cell Transcriptomics’ (SBSCT), several hundred different receptors and channels can be identified in individual neurons, and the functional gene product can subsequently be validated. The use of single-cell transcriptome data to reveal the entire receptor repertoire is crucial, as the copy numbers of mRNAs encoding receptors are low and when cells are pooled, dilution of rare mRNAs leads to loss of signal. These overlooked receptors on key neurons often mediate robust effects that may be therapeutically useful. SBSCT also enables the identification of orphan receptors and can provide strong evidence for receptor heterodimers. Here, we compare SBSCT to other single-cell profiling methods. We argue that the unbiased nature of SBSCT makes it a powerful tool for the identification of new drug targets.
Keywords: drug discovery, Sequencing Based Single Cell Transcriptomics
Single-cell profiling is important for drug discovery
A key to modern drug discovery is the availability of high quality, new drug targets. Some therapeutic areas such as oncology and inflammation/immunity are rich in drug targets, whereas psychiatry and neurology are, as of today, poor in good drug targets 1-3. The identification of promising drug targets in neuropsychiatry is essential for further drug development and will also help us to better understand underlying illnesses. During the past thirty years, a confluence of techniques such as chemical neuroanatomy, lesion studies, radioreceptor binding, Cre/lox deletion of identified cells, positron emission tomography (PET) and functional magnetic resonance imaging (fMRI) have helped to identify key neuronal circuits and individual neurons involved in regulating multiple physiologies such as seizure threshold, mood, reward, and working memory (cf 4,5). The neurons of these circuits serve as important cellular targets for pharmacological interventions, and the cell-surface receptors and ligand-gated ion channels of these neurons form the two most important classes of drug targets that are accessible from the extracellular space. Thus, it would be desirable to know whether there are receptors and ligand-gated channels (or combinations thereof) that are selectively expressed by these neurons. It would also be useful to know the relative ratios of their expression. It is our goal to highlight the opportunities for target discovery and therapeutic development inherent in single-cell analysis, which includes Nextgen sequencing and functional genomics (Figure 1).
Figure 1. Sequencing based single cell transcriptomics (SBSCT) for drug target identification.
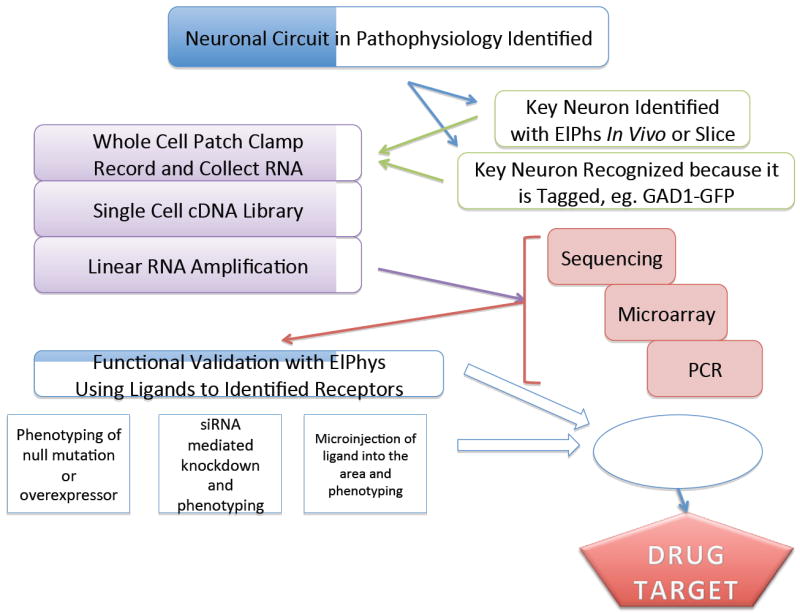
The workflow illustrated in this schematic details how SBSCT can be used to identify drug targets. First, a neural circuit underlying pathophysiological behavior is identified. Next, key neurons within the circuit are identified through electrophysiological studies (ElPhys) or through specific tags, such as with GAD-GFP (glutamic acid decarboxylase-green fluorescent protein). Next, whole cell patch clamp is used to electrophysiologically characterize the cell and the patch pipette is then used to collect the RNA from an individual neuron. This RNA is then amplified into quantities that permit sequencing, microarray and PCR to be used to assess the presence and abundance of mRNAs from the cell. For SBSCT in particular, all mRNAs of a single cell are sequenced indiscriminately. Finally, the function of the mRNA encoded receptor, ligand-gated channel, or voltage-gated channel can be interrogated by single cell ElPhys methods that, like patch clamp, can study a single ion channel being expressed from an mRNA or show functional expression of several channels or receptors using the whole cell configuration. Other methods to validate functionality can include selective phenotyping (null or over-expression of a given receptor or channel), small interfering RNA (siRNA)-mediated knockdown, and/or microinjection of the related ligand.
Today we know the functional identity of only ~20-30 receptors and channels in most of the important central neurons. For example, a literature study of the neurons implicated in the hypoglutamatergic theory of schizophrenia 6-8 shows that the dopaminergic cells in the ventral tegmental area (VTA), pyramidal cells in the cerebral cortex layers 3-5, and the gabaergic interneurons in the cerebral cortex and VTA possess fewer than 30 known receptors, and only two or three are exploited as drug targets. This is in contrast with what would be possible to know, as suggested by multiple studies on single neurons over the past 10 years 1,9-13. These studies using (i) single-cell polymerase chain reaction (PCR), (ii) chipping, and now (iii) single-cell sequencing have shown that neurons in the central nervous system (CNS) can express mRNAs for several hundred receptors and that ~10-20% of these are orphan receptors, as suggested by the analysis of G protein-coupled receptors (GPCRs) identified in other CNS neurons 14. The advantages of studying a single cell over dissected or microdissected (punched) or laser dissected groups of cells can be summarized by showing that rare mRNAs are diluted when the contents of several cells are pooled, and the dilution may lead to their concentration falling below the detection limits of the methods used. Thus, the presence of rare mRNAs will be missed. This represents a serious scientific and drug development opportunity loss, as GPCRs and ligand-gated ion channels are among the most druggable targets. The copy number of both gene products and of mRNAs encoding receptors and ligand-gated ion channels varies between 2 and a few hundred for the mRNAs (cf. ref 5), and the number of GPCRs (as determined by labeling methods) is ~200-2000 per mammalian neuron.
The therapeutic effects exerted through these receptors are robust, and thus we would like to know all the receptors through which such effects are attainable. However, neighboring cells that are included in pooled cell populations usually do not have the same receptor repertoire; they can vary to the extent that we find very different electrophysiological properties among neighboring neurons. Therefore, for detecting the receptor mRNA and also in studying functional receptors in neurons, the single-cell methods of molecular biology and of electrophysiology have great advantages. Table 1 provides a comparison among the methods presently available to identify expression of a receptor on a single cell. The functional expression of a receptor on a single cell is best studied electrophysiologically if an agonist is available for the receptor. The expression of the gene product (the receptor protein) can be shown when antibodies properly labeled or labeled ligands to the receptor are available, whereas the mRNA encoding a receptor can be identified with methods such as single-cell PCR, in situ hybridization (ISH) and microarray, in addition to the sequencing-based single cell transcriptomics (SBSCT) described in this paper.
Table 1.
Methods to detect a receptor on a single cell
| Method | Sensitivity | Information | Knowledge* | Cost | Time |
|---|---|---|---|---|---|
| Electrophysiology | ++++ | Functional | A priori | + | +++ |
| Binding; ligand, Ab | + | Gene product | A priori | ++ | + |
| Immunohistochemistry | ++ | Gene product | A priori | ++ | + |
| In situ Hybridization | ++ | mRNA | A priori | ++ | + |
| Mapping ISH-atlas | + | mRNA | Unbiased | +++ | +++ |
| Single cell PCR | ++++ | mRNA | A priori | + | + |
| Microarray# | +++ | mRNA | A priori | +++ | ++ |
| Sequencing based single cell Transcriptome# | ++++ | mRNA | Unbiased | +++ | +++ |
A priori knowledge that the receptor may be expressed on the cell is required to use the proper agonist, Ab ISH probe, PCR primers
Bioinformatics costs are higher
For most methods, one utilizes knowledge from earlier physiological and pharmacological studies for the analysis at hand. These a priori known data focus the search for a receptor or a set of receptors. For example, if immunohistochemistry shows that, at synapses with the neuron under study, there are nerve terminals containing the serotonin biosynthetic enzyme tryptophan hydroxylase, or if retrograde tracing studies show that the neurons from the dorsal raphe nucleus project to the area where the studied neuron is, then it is reasonable to examine which subtypes of serotonin receptors are expressed by the neuron (as the dorsal raphe nucleus cells are known to release serotonin 16). No one, however, studies every receptor antibody or uses a set of PCR primers that would detect mRNA encoding every receptor in the genome. Thus, the studies are biased by previous a priori knowledge, and this directs the experimental paradigm to what has already been studied.
In contrast, the ISH atlases that are constructed using as probes to all possible receptor mRNAs present in the genome and study all areas of the brain irrespective of earlier identification of the receptor or of the orphan receptors in that area 15, and SBSCT 14 are unbiased by prior knowledge and thus may yield surprising findings. Here, we will discuss how SBSCT has revealed the presence of adiponectin, insulin and insulin-like growth factor 1 (IGF1) receptors, on the warm sensitive neurons (WSNs) of the hypothalamus. As the Table 1 shows, one pays a price in time and cost of the SBSCT as compared to single cell PCR or microarray, but we believe that it is well compensated by the unexpected and rich results obtained.
SBSCT compared with other methods of single-cell profiling
The advantages of single cell transcriptomics over pooled cells or tissue sample transcriptomics have been discussed many times previously. The advantages of single cell methodologies lie in their higher sensitivity, higher specificity and co-localization within the same cell type of information leading to the discovery of which mRNAs are indeed expressed in the same cell. This provides a basis for the study of whether the gene products of multiple targets may directly associate through the formation of dimers or other interactions. This in turn leads to a tighter coupling between molecular data and cellular function as long as one has methods to study the single cell, which in neuroscience is the standard of electrophysiology. The comparison of SBSCT with other single-cell transcriptomics is important to elucidate what is required and what can be gained by use of this method (Table 1).
Historically, the first single-cell transcriptomics analysis was performed on a single cerebellar Purkinje cell, in which the mRNA was isolated using a glass capillary tube and amplified using the linear aRNA amplification methodology 21. This was followed by an analysis of individual rat hippocampal neurons 9,22 and striatal neurons 23-25. Data from these papers was limited to an analysis of selected RNAs from these cells as whole transcriptome analysis was not technologically feasible until microarray 26 and subsequent NextGen sequencing analysis methods became available. Initially, single-cell PCR 27,28 was used to characterize particular RNAs within the RNA compliment of single cells, and this was followed by microarray analysis of the RNA populations 26. Since these early papers, there have been many publications detailing single-cell transcriptomes using various methods (cf 13).
The SBSCT method makes use of the development of less costly sequencing methods that enable the mRNAs that are amplified by PCR or linear amplification, not probed by PCR primers or by DNA arrays, to be identified in the most specific manner possible. The great advantage of this method is that it is unbiased, (i.e., making no assumption regarding which mRNAs may be present as one must do when selecting PCR primers or when spotting probes on a DNA microarray). In SBSCT, one indiscriminately sequences all cDNAs of a single cell, and thus one will discover, upon comparison with the genome, unexpected mRNAs. Furthermore, SBSCT applies sequencing of whatever cDNAs are made by amplification of the single cell mRNAs rather than hybridization with fixed probes as a microarray. It therefore permits the identification of unexpected isoforms of both expected and unexpected transcripts. Both of these types of findings are precluded by the planning of the PCR and microarray experiment in the selection of targets, primers and probes. The unbiased sequencing of a single cell represents a major step forward in describing the complete transcriptome of the cell.
All of these methods start with single cell RNA that is reverse transcribed. Although single-cell PCR in principle can be carried out on the cDNA obtained, chipping or microarray analysis and SBSCT require linear amplification of the cDNA given the small amount of material collected. Amplification directed by the poly-A tails of mRNA introduces a 3’ end bias that represents a significant drawback to microarray analysis as most probes cover the coding region, and thus no hybridization will occur at many sites despite the RNA being present. This loss of information is not seen with SBSCT because any sequence detected can be mapped back to the genome without being selected in advance.
Microarray analysis is significantly faster and cheaper today than the NextGen sequencing of the RNAs from a cell. However, the computational needs of SBSCT identification of the mRNAs are completely dependent on finding the matching genomic sequences and require resources that exceed those needed for microarrays. One of the biggest issues related to sequencing analysis is the decision about what to do with experimentally obtained sequences that have one or more mismatches, and how to use them to further investigate the presence of their associated mRNAs. PCR or other methods may provide additional data regarding the exact nature of these sequence variations, but so far we have discarded them, an embarrassment of the riches as we already had 5000 identified mRNAs. This issue will be abrogated as NextGen sequencing improves, permitting longer sequences with fewer errors to be generated. In the meantime, questions about non-perfect sequences arise. For example: is a mismatch closer to 5’ end worse than one closer to 3’ end, and how much inaccuracy is caused by biology and how much by methodology?
The biggest issue in using transcriptomics data, whether generated from single cells or tissue samples, is the determination of how the transcriptome relates to the proteome and which of the identified mRNAs will be translated into functional gene products. In the study of neurons, the mRNA-encoded receptor, ligand-gated channel, or voltage-gated channel can be interrogated by single-cell electrophysiological methods (e.g. patch clamp) that can study a single ion channel or show functional expression of several channels or receptors using the whole cell configuration. We have applied the latest version of this single-cell transcriptome approach, now complemented with sequencing based analysis of the linearly amplified mRNAs from a single neuron, to the study of electrophysiologically identified WSNs from the mouse. The WSNs are GABAergic neurons in the anterior hypothalamic preoptic area (POA) that project to the raphe pallidus and dorsomedial hypothalamus 18. These neurons show temperature sensitive changes in their spontaneous firing in the temperature range between 33-41°C and are responsible for regulating and maintaining core temperature 19,20.
We have studied the WSNs in great detail because these cells have a decisive effect on a macro, in vivo parameter, the core body temperature (CBT), that can be measured with a simple thermometer 18-20. The effect of changes in the activity of a few WSNs can thus be followed by robust changes in CBT, providing functional expression of an mRNA encoded receptor or channel. In these neurons, we can study the effects of all agonists to receptors whose mRNA we identified by SBSCT, and we have done so by measuring electrophysiological changes such as firing activity, input resistance, and by measuring changes in the CBT of whole animals upon microinjecting to the area where these neurons are found. As our search through sequencing was unbiased, we have come upon receptors that were not previously suggested to be expressed or to be functional in these neurons in the literature. The most surprising of these examples is the insulin receptor that was discovered through identifying its 3’UTR sequence. The mRNA was present in low copy numbers (below 10), and yet application of insulin changes the firing rate of these neurons and microinjection of insulin causes large sustained hyperthermia. We identified 45 non-orphan GPCRs. In functional tests, we could confirm the expression of functional receptors in 90% of the cases (i.e., in the case of 32 peptides and 7 small molecules, Glu, GABA, Ach, monoamines) despite large variations in the copy number of the mRNAs.
The experimental paradigm of SBSCT and the technical prerequisites
The neurons
Many important neurons in key circuits can today be recognized by morphological markers, by being tagged with promoter-driven green fluorescent protein (GFP) expression in transgenic animals 29, or by identification of their specific activity profile. Once individual, active neurons are recognizable in tissue slice or in vivo, whole-cell patch clamp recordings can be made from them. This mode of electrophysiological characterization also opens the way for physically sampling the cell for the transcriptome studies. In several cases, such as the fast firing cortical GABAergic neurons, electrophysiology is the best method of identifying neurons of interest. As discussed above, we have focused on the WSNs 14, which are located in the POA of the neuronal circuit that regulates adaptive thermogenesis and can activate the brown adipose tissue upon changes in skin temperature or upon changes in CBT. This neuronal circuit has been pieced together by lesion studies, retrograde tracing studies and recording studies and is summarized by Nakamura and Morrison 18.
Single-cell cDNA library
The individual neuron cDNA library is prepared by reverse transcription of the RNA population isolated from individual neurons, followed by repeated rounds of linear aRNA nucleic acid amplification (23-25) and conversion of the amplified RNA into a cDNA library. The resulting cDNA library has sufficient quantities of amplified material from a single neuron to permit sequencing of these cDNAs, microarray analysis, and the PCR of selected targets. As used in these studies, the linear amplification biases the amplified products towards the 3’ end, and although many microarray platforms will not have probes that can hybridize with the 3’-end biased RNA, the unbiased sequencing (i.e., not requiring for knowledge of hybridizable sequences) will clearly identify the cDNA and the gene from which it originates. When identifying which mRNAs are present in the individual neurons, we have characterized them first by electrophysiology and then by SBSCT. For the SBSCT, 30 million individual sequences (also called reads) can be generated for an individual cell with ~70% of the reads mapping back to known genes. Using stringent genome matching parameters permitting only 100% matching sequences for the 45 base sequences generated, we have identified ~5000 cDNAs in a single neuron using Illumina sequencing14. Permitting one or even two mismatches would increase this number dramatically.
An analysis of published sequencing data for single non-neuronal cells reveals a similar reported distribution of functional classes of mRNAs for detectable transcripts (Figure 2). Using a single mouse oocyte 11 and single embryonic stem cells 30, Tang et. al. describe single-cell gene expression profiles generated by whole transcriptome amplification followed by Applied Biosystems’ next generation SOLiD sequencing. Evaluating this dataset for the presence of transcripts described as receptors, ion channels or enzymes having one or more read count gave a distribution of 5-6% of transcripts encoding receptors, 1% encoding ion channels and 23% encoding enzymes, respectively. Although approximately the same percentage of transcripts encodes receptors in each cell type, neurons, in particular, may produce more copies of each of these receptor types to accommodate signaling across their extensive membrane surface area 11,14.
Figure 2. Gene onotology (GO) categories for mRNAs identified in individual cells.
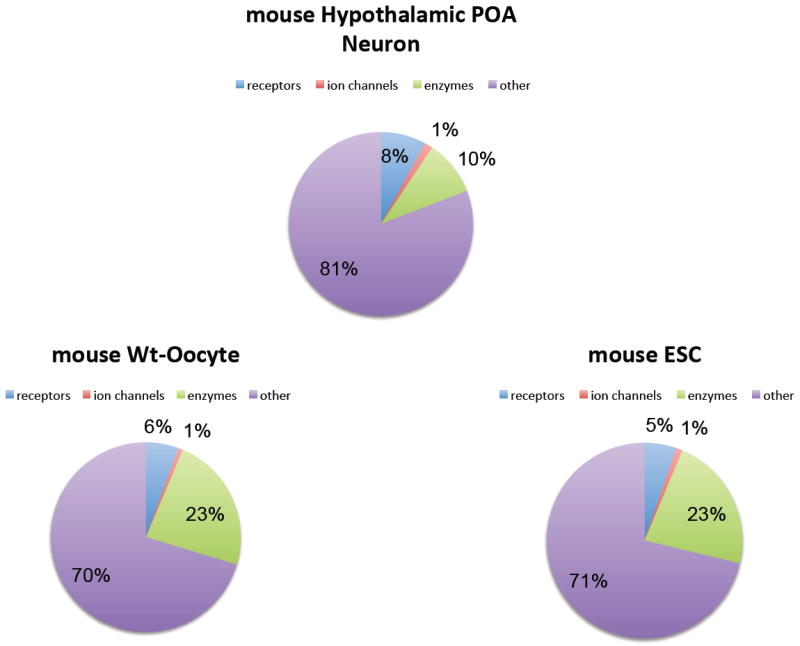
Comparison of the expression of mRNAs in GO categories from various mouse cells shows that there are large numbers of receptors in each cell type. In this example, a single hypothalamic POA neuron is compared with a single oocyte and a single blastomere from a four-cell stage embryo (MF1 strain). Although the total number of mRNAs detected in the hypothalamic neuron is 6120, the oocyte and blastomere have a larger number of detectable mRNAs (14,215 and 15,595 respectively), each cell type has a large proportion of expressed mRNAs annotated as receptors. The percentage of total RNA devoted to receptors was highest in the hypothalamic neuron at 8%, whereas the other cell types range from 5% to 6%. Although GPCRs have been shown to homodimerize and heterodimerize under certain conditions, channels are known to heterodimerize extensively. Therefore, the presence of ~90 mRNAs that encode channel subunit proteins suggests a large repertoire of heteromeric channels.
Functional validation of transcriptome data
The presence of receptor mRNAs in a cell does not automatically mean that the receptors are translated or functional, but rather shows the potential of that cell to respond. Indeed it is possible that mRNAs are translationally silent until a proper stimulus permits their translation, insertion into the membrane and function. Validation of the function of the receptor within a cell is required to understand the role of the receptor in mediating cellular physiology. We have used “tried and true” pharmacological and physiological tools as well as genetically engineered animal models as a means of functionally validating receptors highlighted by SBSCT. Among the several hundred receptor cDNAs identified in the WSNs were those encoding pyrogen receptors, as expected, because the earliest iontophoresis studies showed these cells to respond to pyrogens such as interleukin-1 (IL-1) and tumor necrosis factor alpha (TNFα) 30, as well as several glutamate and GABA receptor subtypes 31. The effect of agonists on these receptors has been examined on WSNs in slices electrophysiologically and/or in vivo with radiotelemetry measuring CBT after microinjection of the agonists into POA. The neuronal circuit in which WSN is the principal neuron controls the CBT, the maintenance of which accounts for 30% or more of the energy expenditure of homeotherms like humans. The knowledge of the receptors through which CBT and energy metabolism can be affected chronically is of pharmacological importance in weight control and metabolic disease. We have identified several hundred mRNAs that encode cell-surface receptors in WSNs, a cell type that after over 30 years of studies has revealed only ~20 receptors (cf 19). These SBSCT data have enabled us to determine the receptor subtype mediating effects of signaling substances known earlier to be able to effect CBT. For example, it was known that intracerebroventricular (icv) injection of bombesin caused hypothermia 32, and our finding of the bombesin receptor 3 (BSR3) in the WSN suggests that some of the effects of icv bombesin 32 are exerted in the anterior hypothalamus on these particular cells through the BSR3.
Receptors for non-synaptic signals
The unbiased nature of SBSCT does not require that the afferents to a neuron are known a priori. This permits the discovery of hormone receptors, chemokine and cytokine receptors and receptors for some neuropeptides that do not reach the neuron through synaptic contacts with an afferent neuron but rather by diffusion in a volume transmission 4. The hormone receptors in the WSNs for insulin 33, IGF1 34 adiponectin 14, and the chemokine receptor for CCL4 35 were thus found by SBSCT without any previous suggestion, anatomical or otherwise, that these neurons express receptors for these non-neuronal, non-synaptic signals. Once SBSCT identified these receptor transcripts, the gene product and their functional effect on the WSN activity and on CBT was demonstrated 33-35. The sequencing-based finding of the expression of the insulin receptor mRNA 33 permitted us to demonstrate that insulin in the POA mediates hyperthermia through activation of the brown adipose tissue. The discovery of adiponectin1 and 2 receptors in the WSN allowed us to demonstrate that these receptors are involved in insulin-mediated hyperthermia 14, and the receptors for the chemokine CCL4 in the WSN demonstrated that this chemokine acts as a hyperthermic agent when applied to the POA 35.
We found IGF1 receptor transcripts in several cells that expressed insulin receptor (IR) transcripts 34. Thus, it is likely that some IR-IGF1R heterodimers exist in these WSN cells, something that has been proposed based on heterologous expression and the examination of pooled cells 36, but now that the transcripts were found in a single cell, the formation of the heterodimer is more likely. The identification of the heterodimers of receptors like IR-IGF1R are of great pharmacological significance because their behavior and their ligand selectivity may be different from that of receptor homodimers, opening up pharmacological specificity of action for heterodimeric receptor-selective ligands 37,38.
Orphan receptors
Like the presence of the non-synaptic receptors, the presence of orphan receptors is not suggested by afferent neurons, and thus detection methods such as the ISH atlas, PCR, microarray and SBSCT can assist in their identification in a neuron. Using SBSCT, we have identified 21 orphan receptors (GPCRs) in the WSN 14, providing novel drug targets that are being pursued presently by medicinal chemists. Some of the GPCRs were already null mutated, and the phenotypic analysis of the mice shows interesting energy metabolism related phenotypes (e.g. GPR119 39). In other cases (e.g. GPR 3, 17, 21, 40, 83, 85) the study of the knockout strain, siRNA mediated knock down, and/or exogenous ligands are being pursued to determine their effects on WSN activity and energy metabolism 40,41.
Microarray analysis of the same cDNA library of the single cell that was sequenced did not give any hybridization signal for the prolactin (PRL) receptor mRNA, despite the presence of three probes on the array. This was because the probes were from the protein coding region of the mRNA, whereas the single cell probe was highly enriched for the 3’-untranslated region (UTR) of the mRNA. Although a 3’-end bias to the probe is expected when using the poly-A region of the RNA to direct probe generation, the reason for the very strong 3’-end bias for the PRL receptor is not clear. It may result from a strong RNA secondary structure that blocks movement of the probe generating enzyme from moving beyond this region 42 As the 3’-UTR was not represented on the microarray, no hybridization would occur and the RNA would be classified as absent from the sample. However, the unbiased sequencing (i.e., no reliance upon selection of hybridization probes) identified the PRL receptor and has led to establishing its role in regulating CBT, thus highlighting the increased sensitivity and the lack of bias as compared to standard microarray analysis. An intriguing aspect of single-cell molecular biology is not only the detection of the presence or absence of a particular mRNA but also the information obtained regarding the relative and absolute abundances of these mRNAs.
Discovery of orphan receptors is accelerated by SBSCT
The localization of orphan receptor mRNAs to a given neuron has been earlier achieved by a set of methods that each has biases and limitations overcome by SBSCT: in ISH, PCR analysis, and microarray. ISH studies are unbiased but often have low resolution for transcripts under scrutiny. Of the 21 orphan receptors we identified in the WSNs (14), we could have found in the Allen Barin atlas only 60%. Furthermore, we would not have known whether these are expressed in warm-sensitive or insensitive cells as there is no morphological marker for warm sensitivity of a neuron, with which we could have established co-localisation. The PCR analysis of the cDNA prepared from selected neurons is biased by the decision that a particular orphan receptor should be sought; only a few dozen PCR primer pairs are usually tested on the same neuronal transcriptome. The microarray analysis of the single neuron transcriptome is unbiased by earlier information about which afferents this neuron receives, but it is limited by which orphan receptor probes are synthesized and spotted on the array. The sequencing of the transcriptome does not anticipate anything about which mRNAs may be found in the cell under study, and thus it is not biased regarding which sequences will be found for orphan receptor mRNAs, coding or non-coding regions, etc. The use of sequencing as part of SBSCT makes this paradigm presently the most unbiased method of finding an orphan receptor mRNA in an identified neuron as described in this paper.
Once the mRNA or the gene product of an orphan receptor is identified in a neuron, functional validation of the gene product orphan receptor can be accomplished using exogenous ligands. These exogenous ligands are identified by high throughput screens (HTS) using a heterologously expressed orphan receptor along with ligand libraries to test whether the orphan receptor protein is functionally expressed. Finding functional orphan receptors on important neurons is a major bonus for drug developers who can generate selective potent ligands that may act as therapeutic agents without knowledge of the endogenous ligand of the receptor – i.e., prior to its deorphanisation. This is reminiscent of the successful therapeutic use of opiates for several thousand years prior to the identification of the endogenous opioid receptor ligands enkephalins and endorphins. When a safe and efficacious dose is known, therapeutic use starts independent of whether all the receptors the substance binds to are known.
Concluding remarks
Our studies on the WSNs in the mouse hypothalamus 14,33-35 and on pyramidal cells in rat hippocampus 43 using SBSCT have shown that most CNS neurons express mRNAs encoding 200-500 or more receptors, albeit in highly varying copy number per cell. The SBSCT and validation results show that the neuronal transcriptome (the RNA complement of a cell) and proteome (the protein complement of a cell) are usually in good concordance with each other, and thus the 200-500 receptor proteins are likely present as membrane proteins and could be useful as putative drug targets. The sequencing of individual cell cDNA libraries permits the identification of receptor and ion channel expression profiles for a specific neuron whose electrophysiological profile has been recorded prior to cDNA library preparation. The changes in the electrophysiological activity profile can be used to check the importance of the identified receptors and ion channels by measuring the direction (excitatory or inhibitory, depolarising or hyperpolarising, activating firing or silencing) and robustness of the effect on the neuronal activity upon application of agonists. Sequencing of the cDNA library of these cells may reveal pharmacologically important isoforms of a receptor present in the given neuron, and also show when receptors that can form heterodimers are expressed in the same cell, making it possible that such heterodimers are indeed present.
SBSCT may increase by ten times the number of identified receptors on key neurons within defined neural circuits that contribute to specific behaviors or disorders. Having this information at hand may indeed reveal several dozen receptors that are relatively selectively expressed on a particular neuron type and thus could serve as useful drug targets in affecting the activity of these particular neurons and neuronal circuits. Identifying ligands that act on novel drug targets or selectively-expressed drug targets always provides hope of novel therapeutic effect. Furthermore, combination therapies are used extensively in treatment of pain, psychiatric disorders, and epilepsies but represent difficult clinical trial challenges. If there is clear molecular evidence that two receptors are expressed on a key neuron, then functional studies using agonists to both receptors can determine whether these receptors mediate additive or even synergistic effects. If this is the case, then there would be a cellular and molecular rational for testing ligands to these receptors in combination or even testing novel ligands that are bifunctional agonists at both receptors44.
Clearly the identification of drug targets is but one of the first steps of a drug discovery program that often takes 10-12 years. Nevertheless, in neuropsychiatry the lack of drug targets is often identified as the bottle neck 3,45-47, and we believe that use of SBSCT on neurons that are well identified components of important neuronal networks is today feasible, affordable and will provide more scientific, actionable information. Access to many more putative targets eases the pressure to work on receptors for which poor leads were found or those known to be expressed on tissues outside of the circuit of interest (which suggests a risk of side effects of a future ligand). SBSCT does not address problems of drug delivery or other issues, but it should provide many more putative targets for potential clinical validation.
We have focused on illustrating the benefit that knowledge of cell-surface receptors and channels provides with regard to anticipating cellular pharmacological responsiveness. However, we do not imply that these are the only classes of drug targets that SBSCT reveals; the 5,000 identified mRNAs per individual cell code for over 4,000 potential intracellular targets for drugs. Although these other targets may be useful, we have highlighted the cell-surface receptors as these are a highly druggable class of proteins used extensively by the pharmaceutical industry and drug developers. We wanted to emphasize that the use of single cells and sequencing in combination is particularly advantageous in identifying receptor encoding mRNAs that are rare, low copy number transcripts in need of avoiding dilution through pooling cells and in need of sequencing rather than microarray detection to pick up isoforms. The presence of so many more than the currently identified and pharmacologically targeted receptors on each cell and the presence of so many unexpected (nonsynaptic, non-neuronal signal recognizing) receptor encoding mRNAs calls attention to these receptors as putative drug targets beyond the traditional synaptic pharmacology. The molecular detail SBSCT provides for CNS neurons heralds the need for more dynamic functional genomics analysis of such datasets to be used in the physiological and pharmacological understanding of the actions of these neurons. The iterative use of single gene knockouts to investigate the function of single receptors is informative but time consuming and limited in scope. It is our belief that the SBSCT analysis of key cells will, by multiplying the number of known putative drug targets on these cells, give new impetus to drug discovery that suffers from a shortage of drug targets in areas such as neurology, psychiatry and pain therapy.
Acknowledgments
This work was funded in part by HRF funds from the Commonwealth of Pennsylvania and MH88849, DP10D004117 and AG9900 to JE and by funds from The Skaggs Institute for Chemical Biology, The Harold L. Dorris Neurological Research Institute, and NIMH to TB. We thank members of our labs and our collaborators for helpful and insightful discussions. This is TSRI manuscript number 21212.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ginsberg SD, et al. Single-cell gene expression analysis: implications for neurodegenerative and neuropsychiatric disorders. Neurochem Res. 2004;29:1053–1064. doi: 10.1023/b:nere.0000023593.77052.f7. [DOI] [PubMed] [Google Scholar]
- 2.Bartfai T, Lees GV. Drug Discovery: from Bedside to Wall Street. Elsevier Amsterdam or Japanese and Mandarin editions. 2008, 2010, 2007. [Google Scholar]
- 3.Kim DH, Stahl SM. Antipsychotic drug development. Curr Top Behav Neurosci. 2010;4:123–139. doi: 10.1007/7854_2010_47. [DOI] [PubMed] [Google Scholar]
- 4.Agnati LF, Guidolin D, Guescini M, Genedani S, Fuxe K. Understanding wiring and volume transmission. Brain Res Rev. 2010;64:137–159. doi: 10.1016/j.brainresrev.2010.03.003. [DOI] [PubMed] [Google Scholar]
- 5.Kandel ER, Schwartz JH, Jessel TM. 1981-2006 [Google Scholar]
- 6.Lewis DA, Sweet RA. Schizophrenia from a neural circuitry perspective: advancing toward rational pharmacological therapies. J Clin Invest. 2009;119:706–716. doi: 10.1172/JCI37335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stone JM. Imaging the glutamate system in humans: relevance to drug discovery for schizophrenia. Curr Pharm Des. 2009;15:2594–2602. doi: 10.2174/138161209788957438. [DOI] [PubMed] [Google Scholar]
- 8.Wachtel H, Turski L. Glutamate: a new target in schizophrenia? Trends Pharmacol Sci. 1990;11:219–220. doi: 10.1016/0165-6147(90)90243-2. [DOI] [PubMed] [Google Scholar]
- 9.Eberwine J, et al. Analysis of gene expression in single live neurons. Proc Natl Acad Sci U S A. 1992;89:3010–3014. doi: 10.1073/pnas.89.7.3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin DM, Loveall B, Ewer J, Deitcher DL, Sucher NJ. Characterization of mRNA expression in single neurons. Methods Mol Biol. 2007;399:133–152. doi: 10.1007/978-1-59745-504-6_10. [DOI] [PubMed] [Google Scholar]
- 11.Tang F, et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods. 2009;6:377–382. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- 12.Volgin DV, Swan J, Kubin L. Single-cell RT-PCR gene expression profiling of acutely dissociated and immunocytochemically identified central neurons. J Neurosci Methods. 2004;136:229–236. doi: 10.1016/j.jneumeth.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 13.Wang D, Bodovitz S. Single cell analysis: the new frontier in ‘omics’. Trends Biotechnol. 2010;28:281–290. doi: 10.1016/j.tibtech.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eberwine J, Bartfai T. Single cell transcriptomics of hypothalamic warm sensitive neurons that control core body temperature and fever response Signaling asymmetry and an extension of chemical neuroanatomy. Pharmacol Ther. 2011;129:241–259. doi: 10.1016/j.pharmthera.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones AR, Overly CC, Sunkin SM. The Allen Brain Atlas: 5 years and beyond. Nat Rev Neurosci. 2009;10:821–828. doi: 10.1038/nrn2722. [DOI] [PubMed] [Google Scholar]
- 16.Michelsen KA, Schmitz C, Steinbusch HW. The dorsal raphe nucleus--from silver stainings to a role in depression. Brain Res Rev. 2007;55:329–342. doi: 10.1016/j.brainresrev.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 17.Fu W, et al. Chemical neuroanatomy of the dorsal raphe nucleus and adjacent structures of the mouse brain. J Comp Neurol. 2010;518:3464–3494. doi: 10.1002/cne.22407. [DOI] [PubMed] [Google Scholar]
- 18.Nakamura K, Morrison SF. A thermosensory pathway that controls body temperature. Nat Neurosci. 2008;11:62–71. doi: 10.1038/nn2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boulant JA. Neuronal basis of Hammel’s model for set-point thermoregulation. J Appl Physiol. 2006;100:1347–1354. doi: 10.1152/japplphysiol.01064.2005. [DOI] [PubMed] [Google Scholar]
- 20.Tabarean IV, Conti B, Behrens M, Korn H, Bartfai T. Electrophysiological properties and thermosensitivity of mouse preoptic and anterior hypothalamic neurons in culture. Neuroscience. 2005;135:433–449. doi: 10.1016/j.neuroscience.2005.06.053. [DOI] [PubMed] [Google Scholar]
- 21.Van Gelder RN, et al. Amplified RNA synthesized from limited quantities of heterogeneous cDNA. Proc Natl Acad Sci U S A. 1990;87:1663–1667. doi: 10.1073/pnas.87.5.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mackler SA, Eberwine JH. Cellular adaptation to opiates alters ion-channel mRNA levels. Proc Natl Acad Sci U S A. 1994;91:385–389. doi: 10.1073/pnas.91.1.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bargas J, Howe A, Eberwine J, Cao Y, Surmeier DJ. Cellular and molecular characterization of Ca2+ currents in acutely isolated, adult rat neostriatal neurons. J Neurosci. 1994;14:6667–6686. doi: 10.1523/JNEUROSCI.14-11-06667.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ghasemzadeh MB, Sharma S, Surmeier DJ, Eberwine JH, Chesselet MF. Multiplicity of glutamate receptor subunits in single striatal neurons: an RNA amplification study. Mol Pharmacol. 1996;49:852–859. [PubMed] [Google Scholar]
- 25.Surmeier DJ, et al. Dopamine receptor subtypes colocalize in rat striatonigral neurons. Proc Natl Acad Sci U S A. 1992;89:10178–10182. doi: 10.1073/pnas.89.21.10178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chee M, et al. Accessing genetic information with high-density DNA arrays. Science. 1996;274:610–614. doi: 10.1126/science.274.5287.610. [DOI] [PubMed] [Google Scholar]
- 27.Eberwine J, Mackler SA. In: Costa E, Joh T, editors. Vol. 7. Thieme Press; 1991. pp. 233–250. [Google Scholar]
- 28.Lambolez B, Audinat E, Bochet P, Crepel F, Rossier J. AMPA receptor subunits expressed by single Purkinje cells. Neuron. 1992;9:247–258. doi: 10.1016/0896-6273(92)90164-9. [DOI] [PubMed] [Google Scholar]
- 29.Chalfie M. GFP: lighting up life (Nobel Lecture) Angew Chem Int Ed Engl. 2009;48:5603–5611. doi: 10.1002/anie.200902040. [DOI] [PubMed] [Google Scholar]
- 30.Tang F, et al. Tracing the derivation of embryonic stem cells from the inner cell mass by single-cell RNA-Seq analysis. Cell Stem Cell. 2010;6:468–478. doi: 10.1016/j.stem.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hori T, et al. Effects of interleukin-1 and arachidonate on the preoptic and anterior hypothalamic neurons. Brain Res Bull. 1988;20:75–82. doi: 10.1016/0361-9230(88)90010-x. [DOI] [PubMed] [Google Scholar]
- 32.Persaud RA, Coen CW. Effect of bombesin on core temperature in rats: studies involving radiotelemetry. Neuroreport. 1994;5:841–843. doi: 10.1097/00001756-199403000-00026. [DOI] [PubMed] [Google Scholar]
- 33.Sanchez-Alavez M, et al. Insulin causes hyperthermia by direct inhibition of warm-sensitive neurons. Diabetes. 2010;59:43–50. doi: 10.2337/db09-1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanchez-Alavez M, et al. Insulin-like growth factor 1-mediated hyperthermia involves anterior hypothalamic insulin receptors. J Biol Chem. 2011;286:14983–14990. doi: 10.1074/jbc.M110.188540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Osborn O, et al. Ccl22/MDC, is a prostaglandin dependent pyrogen, acting in the anterior hypothalamus to induce hyperthermia via activation of brown adipose tissue. Cytokine. 2011;53:311–319. doi: 10.1016/j.cyto.2010.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seely BL, Reichart DR, Takata Y, Yip C, Olefsky JM. A functional assessment of insulin/insulin-like growth factor-I hybrid receptors. Endocrinology. 1995;136:1635–1641. doi: 10.1210/endo.136.4.7895674. [DOI] [PubMed] [Google Scholar]
- 37.Maggio R, Aloisi G, Silvano E, Rossi M, Millan MJ. Heterodimerization of dopamine receptors: new insights into functional and therapeutic significance. Parkinsonism Relat Disord. 2009;15(Suppl 4):S2–7. doi: 10.1016/S1353-8020(09)70826-0. [DOI] [PubMed] [Google Scholar]
- 38.Marshall FH, Foord SM. Heterodimerization of the GABAB receptor-implications for GPCR signaling and drug discovery. Adv Pharmacol. 2010;58:63–91. doi: 10.1016/S1054-3589(10)58003-7. [DOI] [PubMed] [Google Scholar]
- 39.Flock G, Holland D, Seino Y, Drucker DJ. GPR119 regulates murine glucose homeostasis through incretin receptor-dependent and independent mechanisms. Endocrinology. 2011;152:374–383. doi: 10.1210/en.2010-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matsumoto M, et al. The evolutionarily conserved G protein-coupled receptor SREB2/GPR85 influences brain size, behavior, and vulnerability to schizophrenia. Proc Natl Acad Sci U S A. 2008;105:6133–6138. doi: 10.1073/pnas.0710717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Naik H, et al. Safety, Tolerability, Pharmacokinetics, and Pharmacodynamic Properties of the GPR40 Agonist TAK-875: Results From a Double-Blind, Placebo-Controlled Single Oral Dose Rising Study in Healthy Volunteers. J Clin Pharmacol. 2011 doi: 10.1177/0091270011409230. [DOI] [PubMed] [Google Scholar]
- 42.Eberwine JH, Spencer C, Newell D, Hoffman AR. mRNA structure, in situ, as assessed by microscopic techniques. Microsc Res Tech. 1993;25:19–28. doi: 10.1002/jemt.1070250105. [DOI] [PubMed] [Google Scholar]
- 43.Buckley PT, et al. Cytoplasmic intron sequence-retaining transcripts can be dendritically targeted via ID element retrotransposons. Neuron. 2011;69:877–884. doi: 10.1016/j.neuron.2011.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Berque-Bestel I, Lezoualc’h F, Jockers R. Bivalent ligands as specific pharmacological tools for G protein-coupled receptor dimers. Curr Drug Discov Technol. 2008;5:312–318. doi: 10.2174/157016308786733591. [DOI] [PubMed] [Google Scholar]
- 45.Danzer KM, McLean PJ. Drug Targets from Genetics: Alpha-Synuclei. CNS Neurol Disord Drug Targets. 2011 doi: 10.2174/187152711797247867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koethe D, Hoyer C, Leweke FM. The endocannabinoid system as a target for modelling psychosis. Psychopharmacology (Berl) 2009;206:551–561. doi: 10.1007/s00213-009-1591-7. [DOI] [PubMed] [Google Scholar]
- 47.Korpi ER, Sinkkonen ST. GABA(A) receptor subtypes as targets for neuropsychiatric drug development. Pharmacol Ther. 2006;109:12–32. doi: 10.1016/j.pharmthera.2005.05.009. [DOI] [PubMed] [Google Scholar]


