Abstract
The approach to molecular genetic studies of complex phenotypes has evolved considerably during the recent years. The candidate gene approach, restricted to analysis of a few single nucleotide polymorphisms (SNPs) in a modest number of cases and controls, has been supplanted by the unbiased approach of Genome-Wide Association Studies (GWAS), wherein a large number of tagger SNPs are typed in a large number of individuals. GWAS, which are designed upon the common disease- common variant hypothesis (CD-CV), have identified a large number of SNPs and loci for complex phenotypes. However, alleles identified through GWAS are typically not causative but rather in linkage disequilibrium (LD) with the true causal variants. The common alleles, which may not capture the uncommon and rare variants, account only for a fraction of heritability of the complex traits. Hence, the focus is being shifted to rare variants – common disease (RV-CD) hypothesis, surmising that rare variants exert large effect sizes on the phenotype. In conjunctional with this conceptual shift technological advances in DNA sequencing techniques have dramatically enhanced whole genome or whole exome sequencing capacity. The sequencing approach affords identification of not only the rare but also the common variants. The approach – whether used in complementation with GWAS or as a stand-alone approach - could define the genetic architecture of the complex phenotypes. Robust phenotyping and large-scale sequencing studies are essential to extract the information content of the vast number of DNA sequence variants (DSVs) in the genome. To garner meaningful clinical information and link the genotype to a phenotype, identification and characterization of a very large number of causal fields beyond the information content of DNA sequence variants would be necessary. This review provides an update on the current progress and limitations in identifying DSVs that are associated with phenotypic effects.
COMPLEXITY OF THE NUCLEAR GENOME
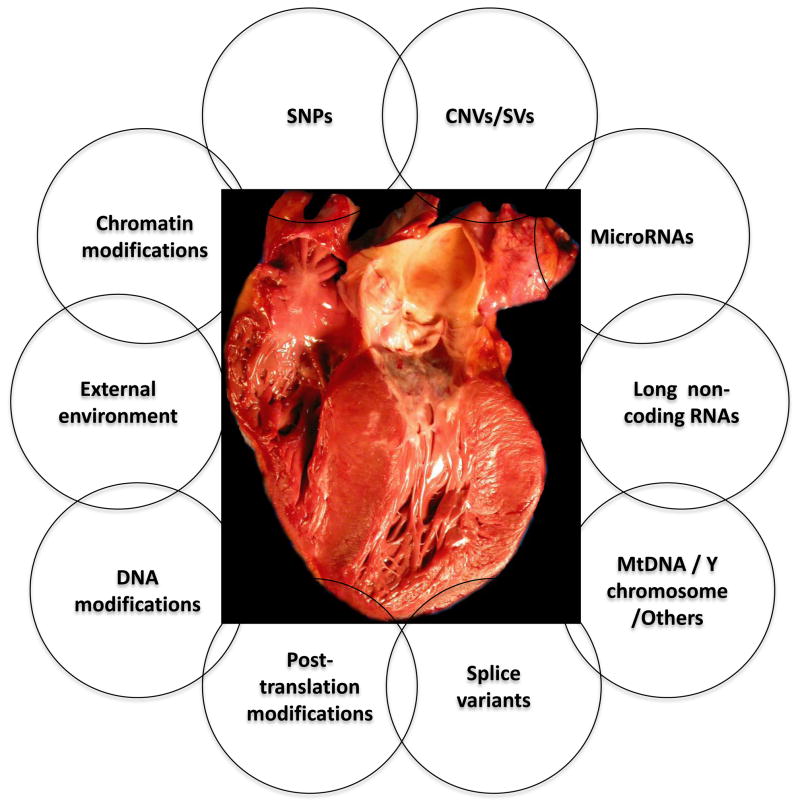
The human nuclear genome (the genome) is an apparently simple and yet an exceedingly complex structure. The genome contains 3.2 billions nucleotides, comprised of four repeating units which are ordered seemingly in random and are packed inside the nucleus as a 2-meter long polymer covered by the octomeric units of histones. A complex system orchestrates accessibility of the double-stranded DNA to various proteins that regulate DNA synthesis and gene expression in response to internal and external stimuli imparted on the cell. The focus of human genetic studies of complex traits is primarily on the DNA sequence variants (DSVs) among individuals, which contribute to susceptibility to a disease, clinical outcomes or response to therapy. However, the mechanisms that govern expression of a phenotype is not only embedded in the DSVs directly but also through their effects on various genomic components that participate in regulation of gene expression, such as histone modifications, microRNAs, long non-coding RNAs, epigenetics, splice variants and post-translational modifications of the encoded proteins in conjunction with the environmental factors. Thus, a complex phenotype is the consequence of complex interactions of a large number of genetic and non-genetic determining factors (Figure 1). The review primarily focuses on the commonly used approaches to identify the DSVs that are associated with phenotypic effects.
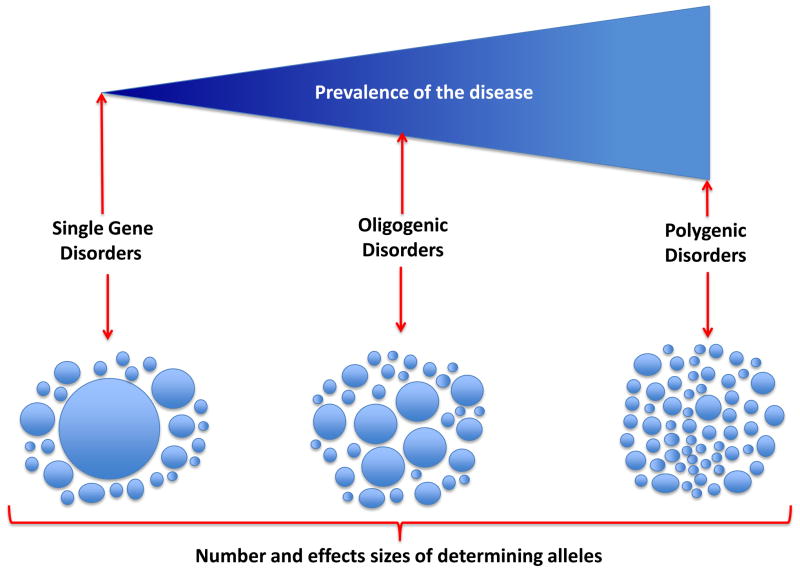
Figure 1. Gradients of disease prevalence and the effect sizes of the causative alleles.
Prevalence of disease, number of determinant DNA sequence variants (DSVs) and their effect sizes are shown. Single gene disorders are caused by rare variants with large effect sizes. In addition to the main causal variant, which typically exhibits a Mendelian pattern of inheritance, several other non-Mendelian variants contribute to expression of the phenotype. On the opposite end of the spectrum are the common complex traits, which are caused, in part, by the cumulative effects of a very large number of DSVs, each imparting a modest effect size. In oligogenetic phenotypes, several alleles with moderate size effects and a large number of alleles with small effect sizes contribute to the phenotype.
Genetic studies of complex phenotypes are currently gene-centric and more so exon-centric. This is largely because the vast majority of the known pathogenic mutations, discovered primarily through positional cloning and linkage studies, affect the coding sequence and alter the amino acid composition of the encoded proteins (http://www.hgmd.org/). Genes, containing the regulatory regions, exons (the protein coding regions) and introns (the intervening segments between exons) occupy about 5% of the genome and the protein-coding exons (exome) only about 1% (1–3). The regulatory regions and the inter-gene regions that serve as template for non-coding RNAs together account for an additional 0.5% of the genome (4). Thus, in total about 1.5% of the 3.2 billion nucleotides of the genome are implicated in influencing the biological and clinical phenotypes. Curiously, however, a higher percentage of the genome is under evolutionary selection pressure and hence, is probably functional (3). Nevertheless, the function of about 98.5% the genome is unknown and this large chunk of the genome is sometimes referred to as “the dark matter of the genome” (4). The current exon-centric approaches would be expected to partially explain determinants of biological and clinical phenotypes. Technical advances in DNA sequencing platforms along with functional characterization of the entire genome could shift the current exon-centric approach to genetic studies of complex traits to a whole genome-centric approach.
DIVERSITY OF THE HUMAN GENOMES
Humans are genetically very diverse. They differ in approximately 0.1% of their genomes. The SNP database (dbSNP, Build 132) lists more than 37 million variants among the humans. With the exception of identical twins, no two humans have identical genomes. Every genome contains about 4 million DSVs that collectively affect half of the genes in each genome and many are private (Table 1) (5-12). The vast majority of the DSVs in the genome are SNPs but structural variations (SVs) affect a greater number of the nucleotides in the genome, simply because of their sizes. SVs include deletions, insertions, duplications and rearrangements of large segments of DNA and hence, might increase or decrease the copy number of the genes from the natural two copies (13, 14). Such SVs are referred to as copy number variants (CNVs) (12–15). Among the approximately 3.5 million SNPs in each genome about 10,000 SNPs are non-synonymous SNPs (nsSNPs), of which approximately two third are predicted by in silico analysis to impart potentially damaging effects. In addition, each genome contains about 50 to 100 variants that have been associated with inherited disorders and about 30 de novo variants (16). The presence of de novo variants is indicative of continuous introduction of new variants to the genetic pool. It would be anticipated that because of rapid expansion of the human population during the last 10,000 years, a large number of the variants in the genome to be relatively new alleles and hence, have not been adequately subjected to filtering by natural selection.
TABLE 1.
Basic Features of the Human Genome
| Nucleotides in the genome | 3.2 × 109 |
| Protein-coding genes in the genome | 23,500 |
| DNA Sequence variants (DSVs) | 4 ×106 |
| Single nucleotide polymorphisms (SNPs) | 3.5 ×106 |
| Non-synonymous SNPs (nsSNPs) | 10,000 |
| Structural variants (SVs)/Copy number variants (CNVs) | 103 – 105 |
| Variants known to be associated with inherited diseases | 50–100 |
| De novo variants | 30 |
ETIOLOGICAL COMPLEXITY OF COMPLEX PHENOTYPES
The plethora of DSVs in the genome and the multi-layer regulation of gene expression and function are indicative of the intricacy of the determinants of the complex phenotypes (17). The clinical phenotypes are presumed to result from the additive effects of and interactions among multiple causative alleles with various genomics and environmental factors. In a complex phenotype, the effect sizes of the involved alleles are expected to vary and to follow a gradient that ranges from minimal or indiscernible to large and significant (Figure 2). Only a few alleles are expected to impart large effect sizes and hence, could be detected by the current approaches to genetic studies of complex phenotypes. Many are expected to exert modest effects that per se might not be discernible based on the current phenotyping and genetic approaches. The recently introduced alleles as opposed to the ancient alleles are typically spared from evolutionary selective filtering and hence, might have greater effect sizes.
Figure 2. Determinants of a complex phenotype.
Cardiac hypertrophy is used as an example of a complex phenotype and various other potential determinants of phenotypic expression of cardiac hypertrophy, including genetics, genomics and external environmental factors are shown (not meant to indicate scale of effect sizes).
A complex trait results from confluence of various genetic and non-genetic determinants (Figure 1). Genetic factors by and large are major determinants of the complex phenotype, a notion that is supported by heritability of the complex trait. DSVs by influencing gene expression, protein structure and function could impact various interacting networks that in aggregate influence susceptibility to a complex phenotype and account for the genetic component of a complex trait etiology (18, 19). The contributing DSVs individually might be neither necessary nor sufficient to alter the susceptibility to a complex phenotype. This is in contrast to single gene disorders, where the presence of the mutation typically indicates expression of disease phenotype, albeit with variable penetrance and expressivity. Likewise, the effect size of each contributing variant is typically small, and often negligible. In general, the effect sizes of DSVs on phenotypes is expected to follow a gradient, being the largest for those phenotypes that are directly influenced by genes such as mRNAs and proteins. These phenotypes are referred to as proximal phenotypes or endophenotypes. In contrast, the effect sizes of DSVs are expected to be smaller for phenotypes that are not directly influenced by DSVs, such as clinical outcomes. Such phenotypes are referred to as distant phenotypes. The gradient of effect sizes in part relates to the number of competing genetic and non-genetic factors that contribute to the phenotype. For the proximal phenotypes, such as mRNA and protein levels, a relatively smaller number of determining factors are expected to contribute to the phenotype and hence, each determinant might have a considerable effect size. In contrast, for the distant phenotype, such as mortality a large number of competing genetic and non-genetic factors contribute to the phenotype and dilute the effect size of an individual DSV. Consequently, it would be expected that a genetic study of a proximal phenotype, such as Genome Wide Association Studies (GWAS) of mRNA or protein levels would offer more power and robust results than the genetic study of a distant phenotype, considering all other variables equal (19–21)(22).
Another fascinating aspect of phenotypic variability is the sex-dependent influence of mitochondria DNA (mtDNA), which is inferred in humans but shown in Drosophila (23). Unlike nuclear DNA, which is equally inherited from sperm and ovum, mtDNA is primarily inherited from the ovum (matrilineal inheritance). Therefore, harmful mtDNA mutations are subject to natural selection in females. Accordingly, mutations in mtDNA that affect only males would not be subject to selective pressure in females. In the absence of such natural selection, mtDNA were shown to have greater impact on nuclear gene expression in males (mutation load) than in females Drosophila (23). Moreover, the Y chromosome, despite containing a few protein-coding genes, influences expression of a large number of nuclear genes, particularly genes encoding proteins that localize to mitochondria (24). Therefore, the Y chromosome is also expected to exert considerable effects on phenotypic expression of the complex phenotype.
GENETIC APPROACHES TO COMPLEX PHENOTYPES
The full spectrum of allele frequency in a population is expected to follow a gradient ranging from private to extremely common alleles (25). Conventionally, however, the variants are categorized into three classes based on their minor allele frequencies (MAFs) in the population. Common and rare variants are those that have population MAFs of >5% and <1%, respectively. Variants that have population MAFs from 1% to 5% are considered uncommon or infrequent. As observed for the genetic causes of Mendelian diseases, DSVs with large effects are expected to be rare. However, the converse is not the rule as the majority of the rare variants could have weak or no discernible clinical or biological effects. Likewise, most common variants are expected to exert negligible or clinically indiscernible effect sizes.
Genetic studies of complex phenotypes might be instigated by a prior knowledge of potential involvement of a gene in the pathogenesis of the phenotype, which is referred to as the candidate gene approach (17). An alternative approach is an unbiased survey of a large number of genes and variants and typically the entire genome, as in GWAS, to identify the associated alleles (Table 2). Genetic studies of complex phenotypes are typically designed upon either common disease –common variant (CD-CV) or rare variant – common disease (RV-CD) hypotheses (26–30). The former surmises that complex phenotypes results of cumulative effects of a very large number of common variants each exerting modest effect. In contrast, the latter posits that multiple rare variants with large effect sizes are the main determinants of heritability of the complex phenotypes. Given that the anticipated effect sizes of the alleles in the genome is likely to be a continuum, one might expect a combination of rare, uncommon and common alleles to contribute to heritability of the complex diseases. In a given population, however, common alleles, despite having modest effects, might have a greater attributable fraction, because of their shear number, even though each rare allele might have a larger effect size (31).
TABLE 2.
Approaches to Genetic Studies of Complex Phenotype
|
Candidate approach
The impetus for a candidate gene approach is a priori knowledge of potential involvement of the candidate gene(s) in the pathogenesis of phenotype of interest. The candidate gene(s) is typically analyzed through case-control or prospective allelic association studies. Accordingly, genotype and allele frequencies of the candidate gene(s), determined either by genotyping or direct sequencing are compared between the cases and controls. Because of a high rate of spurious results, it is typically necessary to replicate the findings in independent study populations. A candidate gene approach in a prospective design study population would be expected to offer more robust results than a similar case-control study design. Likewise, sample size and characteristics of the study populations are key components of the study design (discussed later).
In a candidate gene approach one might type the population for the candidate DSVs, which might be selected either based on their frequencies or the linkage disequilibrium (LD) structure of the locus using the HapMap database in a representative population (http://hapmap.ncbi.nlm.nih.gov/). Selection of the candidate SNPs based on the LD structure of the locus ensures adequate coverage of the locus for the common haplotypes but often is not adequate to cover rare and infrequent alleles. Likewise, single nucleotide polymorphisms (SNPs) might be selected based on known or anticipated biological functions, as in the case of non-synonymous SNPs (nsSNPs) or regulatory SNPs. To identify the novel variants, the entire gene or selected regions, typically the coding regions, splice junctions and the regulatory junctions; are sequenced in the cases and controls (32). The sequencing approach offers the opportunity to identify and compare frequencies of all variants, whether common, uncommon or rare and whether known or novel.
Statistical analysis pertinent to the candidate gene approach includes simply comparing the population frequencies of the genotypes and SNPs in the cases and controls by a Chi Square test. In prospective allelic association studies phenotypic differences among the genotypes of DSVs are compared by various parametric and non-parametric tests as, appropriate. To consider confounding effects of potential differences in the demographics and other determining factors various regression analysis methods are utilized, wherein the other determining factors are included as covariant. Whenever the frequencies of the alleles are low in the population, a gene-centric approach to analysis is utilized wherein all alleles in the gene are coalesced and the overall prevalence of the variants between the cases and controls are compared.
Unbiased approaches
The approach surveys a large number of genes, typically genome-wide, without considerations for the existing data to implicate them in the pathogenesis of the phenotype (17).
Genome-Wide Association Studies (GWAS)
GWAS primarily tests the CD-CV hypothesis by typing the cases and controls for several thousand to 2.5 million alleles positioned throughout the genome. The variants are selected based on the LD structure, attained from the HapMap data set in the representative ethnic population, to tag the common haplotypes. The common haplotypes might also tag multiple uncommon and perhaps even the causal variants (33). Cases and the controls are typed for the variants, using a genotyping microarray platform, and the genotypes frequencies are compared. Various factors determine the power of the GWAS in detecting the causative alleles but probably the most important factor is the sample size (Table 3). The issue sample size is particularly important, as the expected effect sizes of most alleles on complex traits are small. Additional factors that influence the results of GWAS include characteristics of the study populations including robust phenotyping, density of the genotyping arrays, MAFs, strength of LD between the markers and the causal variants and the effect sizes of the alleles (Table 3). Nevertheless, like all allelic association studies, replication of the findings in the discovery study population is essential. Variants found to be associated with the phenotype in the discovery and replication study populations are considered the phenotype-associated alleles.
TABLE 3.
Study design issues to be considered in genetic studies of complex traits
| Determinant | Outcome |
|---|---|
| Sample size | Direct correlation between the sample size and power to detect the causative alleles (within a limit) |
| Effect sizes of the causative alleles | Inverse correlation between effect size and power to detect an association |
| Minor allele frequency | GWAS by design detect only common alleles (MAF > 0.05) |
| Proximity of the phenotype to the genotype | More powerful for detecting an effect on proximal than distal phenotypes |
| Population characteristics | Presence of other competing factors dilute the power to detect an effect |
| Population admixture | Increase the risk of spurious results |
| Phenotype | Phenotypic admixture) and phenocopy conditions dilute the power to detect an effect size |
| Study design platform | Prospective studies are free of potential confounding differences between cases and controls |
| Structure of LD | Might not adequately capture information content of the true causative alleles |
| Density of the genotyping | Low-density arrays may not offer adequate cover for the common haplotypes |
A major issue in interpreting the results of GWAS is the statistical significance level and the possibility of false association because of multiple testing (34, 35). Commonly more than one million SNPs are analyzed and often several phenotypes in the same data set are explored for genotype-phenotype association. Therefore, the statistical results of GWAS require correction for multiple-hypothesis testing. The best method to correct for multiple hypothesis testing is yet to be settled. Permutation test might be the most robust method but computationally is exceedingly demanding because of the huge number of the genotypes. The conventional approach of Bonferroni’s correction might be too conservative, as genotypes are not totally independent. Nevertheless, despite certain level of ambiguity, a p value of less than 1× 10−7 is considered evidence of an association and that of less than 1× 10−8 is considered robust evidence of an association (34).
Success of GWAS
GWAS have been successful in associating a very large number of alleles with various complex phenotypes. Over 1,200 GWAS have been successfully completed and several thousand loci for over 200 complex traits already have been identified (http://www.genome.gov/gwastudies/). The recent trend has been to increase the sample size of the GWAS in order to identify additional alleles with modest effect sizes (36, 37). The approach has been particularly successful in identifying alleles associated with the intermediary phenotypes, such as plasma lipid levels (37, 38). Accordingly GWAS has emerged as the desirable approach to identify the common alleles (MAF>0.05) that are associated with the complex traits. The unbiased approach implemented in GWAS is a major strength as it offers an opportunity to identify novel mechanisms involved in the pathogenesis of complex phenotypes. However, alleles identified through GWAS are typically not the causative alleles but rather are in LD with the true causative alleles. Typically mechanistic studies would be necessary to identify the causative alleles and delineate the responsible mechanism(s) (39).
Shortcomings of GWAS
GWAS by design tests the CD-CV hypothesis by typing the study population for tagger SNPs that represent the common haplotypes. However, the common haplotypes might not adequately cover the rare variants. Likewise, the effect sizes of the common variants identified through GWAS are typically very small and clinically negligible. Accordingly, DSVs identified through GWAS explain typically a small fraction of variability and heritability of the complex traits (15). The point is exemplified for the GWAS of plasma lipids levels, systemic arterial hypertension and cardiac conduction intervals (20, 38, 40–43). For example DSVs associated with plasma high-density lipoprotein cholesterol (HDL-C) levels shift the HDL-C levels by about 1mg/dl or less (42), which is clinically negligible (36). The findings are largely similar for most complex traits. However, there are notable exceptions including the finding of strong association between a common intronic variant in CFH gene, which encodes complement factor H, and age-related macular degeneration (AMD) (44). The intronic polymorphism was found to be in LD with a tyrosine to histidine change in amino acid position 402 (p.Y402H), which is considered the actual causative allele (44). Overall, CFH variants increase the risk of AMD by more than 2-fold and account for about half of the risk of AMD in the siblings of an affected individual (44–46). Similarly, common variants in NOD2, IL23R and LRRK2 increase the risk of Crohn’s disease by 1.5- to 4 -fold (47). Notwithstanding the few exceptions, the results of GWAS typically do not have considerable clinical impact because of the modest effect sizes of the common variants. Nevertheless, it is important to note that the strength of GWAS is in deciphering previously unsuspected novel mechanisms in the pathogenesis of the complex phenotypes. This is exemplified by the discovery of CFH as a susceptibility gene for AMD, which implicates the previously unrecognized role of inflammation in the pathogenesis of AMD (44).
The shortcomings of the results of GWAS in explaining heritability of the complex phenotypes, which may reflect imperfections in determining heritability (17), have shifted the focus from the CD-CV hypothesis to RV-CD hypothesis, the latter surmises that rare and infrequent variants exert larger effect sizes (28). The shift in the focus in conjunction with the technological advances and the precipitous drop in the cost of DNA sequencing have brought forth the post-GWAS era built upon sequencing the entire exome or genome to delineate the genetic etiology of complex traits.
Next Generation DNA Sequencing
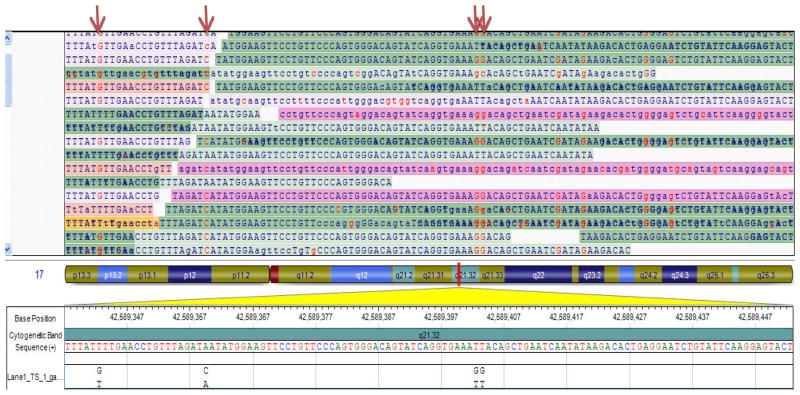
The interest in DNA sequencing to identify genetic architecture of an individual has been revitalized with the precipitous drop in the cost of DNA sequencing per base because of the advent of Next Generation Sequencing (NGS) technologies. NGS affords the opportunity to identify all DSVs in an individual genome with the exception of large CNVs, whether common or rare in the population. The NGS platforms have improved dramatically since the introduction of the first commercial pyrosequencing platform in 2005 (48). The newer platforms can generate up to 300 Gb of throughput per each sequencing run and 20 to 30 Gb per day with a high accuracy rate. The output is sufficient to cover 2–3 genomes and a dozen or so exomes at a high coverage rate of each nucleotide. Most NGS platforms produce short reads of 40 to 120 bases and hence, are more suitable for re-sequencing rather than de novo genome assembly. However, some platforms generate 400 to 1000 base reads, which would be also desirable for de novo sequencing in addition to re-sequencing. Despite the high accuracy of base calling by NGS platforms, because of the enormity of the output, the error rate could pose significant challenge for an accurate identification of the variants, particularly for heterozygous alleles or rare variants in a small fraction of DNA templates (49, 50). An important aspect is the coverage rate of each allele. In DNA sequencing by the NGS platforms, multiple fragments of DNA are sequenced simultaneously and the outputs are aligned to the reference genome. Thus, it is essential that both strands of a diploid genome are adequately represented in the sequence read out. In addition, multiple reads of an allele increases the signal to noise ratio. Therefore, in low coverage sequencing, which is more practical, because of size of the sequence output; less costly, and computationally less laborious, accurate determination of the genotype is less certain. In contrast, a higher coverage at each nucleotide increases the confidence in accurate allele calling. The coverage rate, however, is often non-homogenous and certain genomic regions or exons might not be covered at sufficient depth to provide for accurate allele calling. Inadequateness of the sequence reads, in part, might reflect inadequate coverage by the capture probes (Figure 3). Nevertheless, the gaps could lead to inadequate detection of the variants as well as miscalling because of poor signal to noise ratio. Different depth of coverage might be necessary according to intended application of the NGS data. In NGS studies in families to detect a causative allele in an autosomal dominant Mendelian disorder, a higher depth of coverage might prove essential to robustly detect or exclude the presence of a heterozygous mutation. In addition, the relatively small number of family members makes high depth coverage feasible. In contrast, in allelic association studies of complex phenotypes, wherein sequencing of a large number of cases and controls is required, low-coverage is practical and probably a more powerful approach than covering a smaller number of individuals at a greater depth (49). In the pilot phase of the 1000 Genomes Project, 179 genomes were sequenced at a coverage rate of 3X in order to detect variants that had an allele frequency of 1% or greater (16). An example of sequence output of a NGS platform showing detection of four heterozygous variants is shown in Figure 4. At the present time, at least for medical sequencing, all variants identified by the NGS platforms require validation either by independent sequencing reactions or by Sanger sequencing.
Figure 3. Non-homogenous target capture.
A segment of LMNA gene showing locations of exons (blue boxes), capture regions (green boxes; exons plus 150bp flanking sequences), capture probes (purple-blue boxes) and depth of coverage of sequence reads from one (light gray) and 2 rounds of capture (dark grey). The depth of coverage is reflected by the shades of grey.
Figure 4. An example of sequence output of a Next Generation Sequencing (NGS) platform.
Multiple copies of each DNA fragment is sequenced in parallel and analyzed. Four heterozygous nucleotides are identified in the sequenced fragment. The Figure illustrates the significance of an adequate coverage of each nucleotide for robust allele calling as well the potential for mis-calling because of inadequate coverage at each nucleotide position.
The overall approach to direct DNA sequencing for defining the genetic basis of complex traits could be classified into three categories of whole-genome sequencing, whole-exome sequencing or targeted sub-genomic sequencing, which are briefly discussed.
Targeted Sub-genomic Sequencing
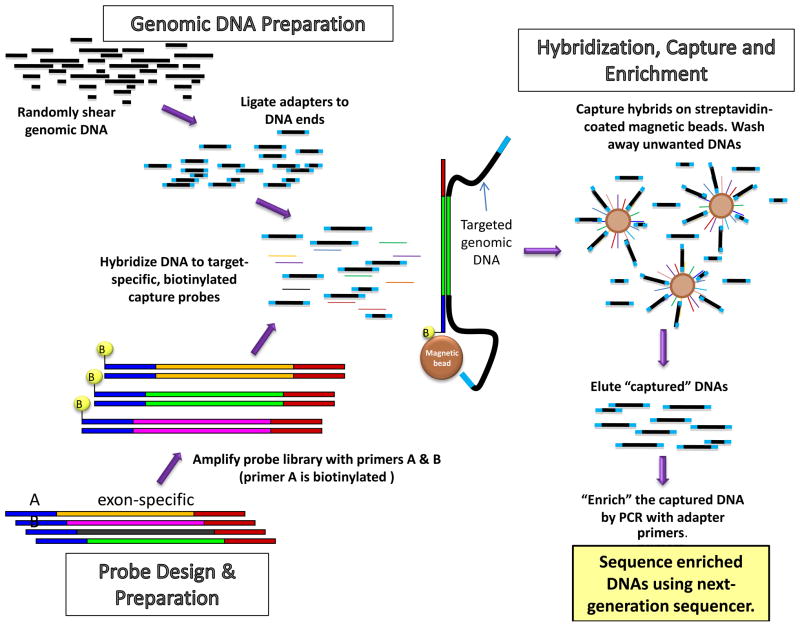
Targeted sub-genomic sequencing is designed to capture, amplify and sequence specific regions of the genome, which have been linked to the phenotype of interest either through linkage studies, quantitative trait loci, GWAS or biological plausibility. The targeted regions might include exons, splice junctions, regulatory regions or the entire genomic area of interest. However, NGS is best suited for capture and sequencing of a large number of genomic targets or exons as opposed to sequencing of a selected number of exons in a limited number of genes. Sanger sequencing might be more cost-effective and practical for sequencing of a limited number of targets, even in a large number of individuals (32). Various commercial products are now available to facilitate target capture and enrichment (51). The overall approach for targeted subgenomic sequencing includes fragmentation of genomic DNA by sonication to generate fragments of approximately 500–600 bp long and ligation of the adapters followed by capture of the fragments of interest using specific 5′ biotinylated probes by hybridization, enrichment by amplification of the captured fragments and sequencing using a NGS platform (Figure 5).
Figure 5. Approach to sub-genomic capture and sequencing.
The diagram illustrates steps involved in target capture and sequencing. The steps include fragmentation of genomic DNA through sonication, adapter ligation, design of capture probes, and quality control to ensure specificity of hybridization, efficiency and uniformity of the capture. The analysis includes BLAST screening against repetitive DNAs and for specificity. Commonly several base long priming sites are added to the ends of all probes, allowing amplification of the probe set following synthesis. Probe pools are amplified using primers to the flanking, common priming sites. Several hundred-fold molar excesses of each probe sequence are used in hybridization. Typically, after one round of capture and enrichment, targets are enriched several thousand-fold. The enriched targets are then loaded onto a NGS platform for parallel sequencing of DNA strands.
The approach also has been used to screen for mutations in genes coding for sarcomere proteins in patients with cardiomyopathies, autosomal-recessive cerebellar ataxia and non-syndromic deafness (52–54). This approach is likely to be supplanted by whole-exome and even whole-genome sequencing because of lower cost per base and the higher sequence output of the latter approaches.
Whole-exome Sequencing
The rationale for whole-exome sequencing is based on the notion that variants located in exons and affect protein sequence are more likely to be pathogenic than those located in introns or intergene regions. Thus, the approach primarily posits that infrequent and rare non-sense, frame shift and non-synonymous DSVs are likely to play major etiological roles in susceptibility to complex phenotypes.
The genome comprises about 180,000 exons that reside in about 23,500 genes (1, 2). Collectively, the coding regions encompass about 300 million base pairs or about 1% of the genome. Thus, whole-exome sequencing requires generating a genomic DNA library, capture and enrichment of all exons and sequencing using a NGS platform. It is probably the most commonly used direct DNA sequencing approach today to identify the genetic causes of rare Mendelian and common non-Mendelian disorders. This is simply because of practical reasons including cost, data storage and bioinformatics analyses. The whole-exome sequencing approach has been successfully applied to identification of the causal mutations for rare Mendelian disorders, such as Freeman-Sheldon syndrome, congenital chloride-loosing enteropathy, Kabuki syndromes, systemic hypertension due to hyperaldosteronism, and familial pheochromocytoma among the others (55–59). Exome sequencing, likewise, has revealed the complexity of discerning the pathogenic alleles based on sequence data alone, as many apparently pathogenic variants might be found in clinically unaffected individuals (60). Efforts are ongoing to apply the whole-exome sequencing approach to delineate the genetic causes of common forms of Mendelian diseases, particularly those with an autosomal dominant pattern of inheritance and complex traits. In addition to limitations imposed by family size and structure, which could restrict discerning co-segregation of the variants with the phenotype, several other limitations render the approach challenging. The enormous genetic diversity of the humans and the presence of a very large number of variants in each genome in conjunction with incomplete penetrance of the causative variants pose significant difficulties in establishing a clear genotype-phenotype co-segregation. At the technical level, the challenge is further compounded by imperfect capture and inadequate coverage of the targeted regions in the family members (Figure 3). Information on biological plausibility and bioinformatics analysis of the variants to perceive biological and functional significance of the variants by PolyPhen-2 and SIFT (61) (62) are valuable, but are typically inadequate to establish causality. Moreover, genetic heterogeneity of the disease, further restricts the opportunity to replicate the findings in multiple families. Notwithstanding these limitations, NGS is expected to offer a full spectrum of genetic determinants of the phenotype in Mendelian disorders.
Whole-genome Sequencing
Whole-genome sequencing offers the opportunity to detect all DSVs, perhaps with the exception of large CNVs, in the genome and hence, build a complete spectrum of genetic variants for each phenotype. It affords the opportunity to test the CD-CV as well as the RV-CD hypotheses. The whole-genome sequencing does not require target capture. However, the coverage might not be even across the genome and gaps with inadequate coverage depth often are present. Appropriate coverage of the gap regions might require re-sequencing or complementary capillary sequencing to achieve the complete sequence of the genome. In proof-of-principle studies, whole-genome sequencing has been successfully applied to identify the genetic cause of rare Mendelian disorders (63, 64). Likewise, whole genome sequencing has been used to complement the results of GWAS to identify a rare variant in MYH6, encoding α-myosin heavy chain protein, as a risk allele for sick sinus syndrome (65). Nevertheless, despite technically feasibility, whole-genome sequencing is still a formidable task because of the enormity of the sequence output, data storage and complexity of the data analysis. With the anticipated drop in the cost of whole–genome sequencing and advances in bioinformatics, one might expect that whole-genome sequencing to become the preferred approach to define the spectrum of the genetic variants in each genome. However, to apply the whole genome sequencing approach to delineate the genetic causal fields of complex phenotypes, sequencing of an exceedingly large number of genomes would be necessary.
Provisional nature of the results of allelic association studies
The initial results of allelic association studies, regardless of the approach being GWAS or direct DNA sequencing, should always be considered provisional pending replications in independent sets of populations and validation through experimentation (66). Often the alleles identified in a case-control association study are not the true causal alleles but rather are in LD with the causal variants. LD patterns in the human genome are complex and often extend to several thousand and even a few million base pairs of DNA (67–70). It is not surprising, therefore, that often significant effort must be expended in identifying the causal variant, even when the identified variant is a known variant, a biological plausible candidate, or very likely to be the causal variant itself. The challenge is further noteworthy when the associated alleles are located in intergene regions or in introns. A notable example is the finding of 9p21 locus as a susceptibility locus for coronary atherosclerosis in multiple independent GWAS (71–73). The locus is a gene desert and none of the genes mapped to the regions, namely cyclin-dependent kinase inhibitor 2A and 2B (CDKN2A, CDNK2B), methylthioadenosine phosphorylase (MTAP), and ANRIL, which codes for a long noncoding RNA were considered a biologically plausible candidate for atherosclerosis. Subsequent mechanistic studies implicated smooth muscle proliferation and augmented inflammatory response in the presence of the risk allele as potential mechanisms to explain the GWAS findings (39, 74). Hence, the results of allelic association studies require complementation with in vitro and in vivo mechanistic studies in order to gain insights into the pathogenesis of the phenotype and establish a causal link.
POPULATION PLATFORMS
Family studies
Complex phenotypes often show familial aggregation, in part because of shared genetic risk factors. When a single allele exerts a large effect on the phenotype familial segregation will follow a Mendelian pattern of inheritance. However, contribution of additional variants to the phenotype can give variable expressivity or incomplete penetrance (75). Unlike the phenotypes with a Mendelian pattern of inheritance, wherein the presence of the variant causes the phenotype, albeit with a variable penetrance, in complex phenotypes, the variant is neither sufficient nor necessary to cause the phenotype. Incomplete penetrance, which often hinders predictable phenotypic expression in Mendelian disorders, is very common in complex phenotypes, whereas many individuals with the variants do not show discernible phenotype. Various forms of familial dyslipidemias that typically show Mendelian inheritance patterns exemplify phenotypes that are typically complex in the general population (76). The stronger the evidence for a familial inheritance pattern the stronger the effect size of the causative allele and hence, the greater chance of identifying it, either through the conventional linkage analysis in large size families or through direct DNA sequencing and co-segregation and analytical approaches.
Commonly, however, there is a familial aggregation of a complex trait without a clear pattern of segregation. Even in the absence of a clear Mendelian inheritance, family-based genetic analysis in pooled families offers a robust approach for identification of the causative variants for complex phenotypes. This is partly because family-based studies, as opposed to studies in the case-control studies, are less susceptible to the heterogeneity of the population structure. The success of applying the NGS to identify the causal variants in family members depends in part on heritability of the trait, genetic heterogeneity of the phenotype, prevalence of the phenotype and whether the phenotype is caused by de novo and rare variants. NGS can be utilized in combination with linkage analysis to identify the causative alleles in families with a large or a moderate number of affected individuals (64). The approach could provide a desired strategy to identify the causative genes in a large number of Mendelian diseases with undefined genetic etiology (64). Various strategies could be used to enhance the chance of identifying the causative variants by NGS, such as analyzing shared genetic variants in distantly-related affected family members, which is expected to restrict the number of candidate alleles. Likewise, focus on family members on the phenotypic extreme could increase the likelihood of success (77).
Genetic studies in parent-offspring trios, often involving several hundred pooled in order to attain sufficient power, offer a strong family-based approach to identify the causal alleles for complex phenotypes (78). The approach typically includes genotyping or direct sequencing followed by Transmission Disequilibrium Test (TDT) or a variation of it to identify preferential inheritance of the alleles by the affected offspring from an affected parent (79). Based on Mendelian inheritance, there is a 50% chance of inheriting a parental allele. A significant departure of an allele from the 50% expected rate would indicate an association between the allele and the phenotype.
Case-control studies
This is the most commonly used approach for genetic studies of complex phenotypes. A candidate gene case-control study is the conventional and most commonly used approach, wherein the frequencies of alleles and genotypes are compared between cases and controls. The candidate gene case-control studies mandate a prior knowledge of involvement of the gene of interest in the pathogenesis of the phenotype. As discussed, the alternative and more robust case-control study is GWAS, which is free of a prior assumption of candidacy of the genes. Case-control allelic association study built upon whole-exome or targeted sequencing of a large number of cases and controls is desirable but expensive.
Case-control allelic association studies, unless performed in a very large number of cases and controls, have a high rate of spurious results. This is in part because of potential differences in the population structure of the cases and controls, which are often difficult to correct. In addition, it is important to note that alleles identified through case-control association studies might be the causative alleles. However, this is often not the case as these alleles are typically polymorphic alleles in LD with the true causative alleles.
Prospective allelic association studies
A variation of allelic association studies is a prospective study, wherein a large number of individuals are typed for various SNPs or even sequenced and the cohort is prospectively followed and phenotyped. The design is not subject to genetic and characteristics differences that often influence the results of case-control association studies and therefore are considered more robust. A prospective study as compared to a cross-sectional case-control study is expected to offer more power to detect a significant effect size of an allele, considering all other determinants equal. The prospective studies are particularly useful for genetic analysis of endophenotypes or proximal and intermediary phenotypes. Likewise, a prospective study might be preferable for genetic studies of uncommon and rare traits than cross-sectional case-control allelic association studies.
DESIGN OF GENETIC STUDIES
Genetic studies aim to detect and quantify the risk of a disease or effectiveness of a specific therapy or the risk of adverse side effect at an individual level and yet have to depend on group data to attain such information. Consequently, robustness of the group data is imperative for appropriately extending the group data to an individual. Various factors determine robustness of the group data and the applicability of the findings to an individual, including sample size of the study, characteristics of the study population, genetic heterogeneity of the phenotype, frequencies of the risk alleles in the population and their effect sizes on the phenotype (Table 3). It is equally important to distinguish the population attributable risk from the absolute risk in an individual. A common allele, despite having a small effect size might have considerable population attributable risk than a rare allele with a larger effect size. This is likely the case for the 9p21 locus and the risk of coronary artery disease (71, 72, 80), wherein a 5% increase in the MAF from 0.45 to 0.50 has little clinical impact at an individual level. In contrast, because the risk allele is quite common, the population attributable risk, despite the modest effect size at an individual level, might be quite significant. Conversely, a rare allele might have a greater risk in an individual but a smaller population attributable risk, simply because of its low frequency. Similarly, it is important to consider the pre-test likelihood of the phenotype or an outcome when considering an Odds Ratio imparted by a risk allele. For example, a two-fold increase in the risk of heart failure has different clinical implications when a prior risk of heart failure is 1× 10−6 as opposed to the a prior risk of 1×10−1.
Sample size
Regardless of the genetic approach being GWAS, or direct DNA sequencing, the sample size of the study is an important determinant of power of the study. For the same level of study power, the sample size inversely correlates with the effect size of the allele or the relative risk imparted by an allele. Whenever the effect size of a DSV is large, the disease typically exhibits a Mendelian inheritance pattern and hence, is suitable for genetic linkage studies. The power of genetic linkage depends on the structure of the family, penetrance of the causal variant and information content of the DNA markers used in linkage analysis. As a “rule of thumb” each affected individual contributes approximately 0.3 points to a LOD (logarithm of odds) score in an autosomal dominant pedigree, assuming 100% penetrance. In general, about 10 potentially informative meioses are necessary to have sufficient power to obtain a LOD score of 3, which is conventionally significant for a genome wide screen. The actual power to obtain a significant LOD score is determined by the structure of the family, penetrance of the causative allele and information content of the locus markers. Nevertheless, often with a smaller number of affected individuals in family one might obtain a significant LOD score.
In complex traits, however, the effect sizes and likewise the relative risk imposed by the risk alleles are typically very small. Hence, despite familial aggregation and heritability, inheritance does not follow a clear Mendelian pattern. In addition, the population frequencies of many risk alleles might be low. Therefore, in a GWAS a very large number of cases and controls need to be typed in order to have sufficient power to detect common DSVs that are exert modest effect sizes. As a general guide, a sample size of several hundred cases and controls is sufficiently only to detect common alleles that impart greater than a 5-fold shift in the risk of a phenotype, which is seldom the case for a complex phenotype. A much larger sample size, typically in the range of several thousands, is necessary to achieve sufficient power to detect common alleles that shift the risk by about 2-fold. To detect smaller effect sizes or relative risks, typically more than 10,000 cases and controls are necessary, which typically could be achieved through large-scale collaborative GWAS (36–38).
In determining the proper sample size one has to consider MAFs of the selected SNPs, the expected effect sizes of the alleles, the choice of the phenotype whether it is a proximal or a distant phenotype to the gene and hence, the number of competing determining factors; precise phenotyping, and the characteristics of the study populations (Table 3). The relationships among risk allele frequency, relative risk (effect size), sample size of the study and statistical power to detect the effect have been illustrated elegantly in a recent study (81). In general, the required sample size inversely relates to the risk allele frequency, i.e., the lower the risk allele frequency, the larger the sample size needed to detect a given effect size. Likewise, the smaller the effect size, the larger the sample size of the study population to detect an association for a given allele with certain frequency. Likewise, as indicated earlier, the more complex the phenotype, i.e., the lager number of determining factors, the larger sample size to detect an association. Studies in genetic isolates may be more powerful because of reduced heterogeneity of the population but the findings may not generalize to other populations (82, 83). Finally, the presence of potential confounders, which might be possible to eliminate, such as differences in the characteristics of the cases and controls should be considered in determining the sample size if the study population.
Population characteristics
The characteristics of the study population are a key component of robust allelic association studies. The desire is to eliminate all potential confounding as well as competing non-genetic variables in order to amplify the effects of genetic variants. However, attaining such goal is typically not feasible, as genetic factors are only part of the causal fields of complex phenotype and various other factors commonly differ between the cases and controls. In a GWAS of coronary atherosclerosis, the so-called conventional risk factors, such as prevalence of smoking, hypertension and dyslipidemia are expected to differ between the two groups (71, 72). These differences are expected to dilute the power of genetic association studies to detect the effects of the alleles. Hence, cases and controls should be matched, as much as possible, for demographics and other characteristics in order to enrich the chance of detecting genetic effects.
Ethnic admixture
Admixture is a major confounding factor, as the MAFs of the genetic variants vary significantly in different ethnic populations (84, 85). Likewise, populations with different ethnic backgrounds might not share the same risk alleles or require different interacting factors to expose the risk of an allele. Hence, the results in one ethnic background might not directly extend to another. Consequently, independent genetic studies in various ethnicities would be preferable.
Genetic isolates by offering reduced genetic heterogeneity are considered more powerful for delineating the genetic etiologies of complex phenotypes. In such populations, LD across long genomic regions reduces the number of common haplotypes and hence, increases the power to map the region (82, 83). In addition, heterogeneity of the environmental factors in genetic isolates is reduced. However, long regions of LD often render specification of the true causal variants more challenging. The findings in genetic isolates may be restricted to the specific population and may not be generalizable to other populations.
Complexity of the phenotype
Increasing heterogeneity and complexity of a phenotype usually implicate a greater number of both genetic and non-genetic etiological factors. Hence, the effect sizes of the each determinant in such complex phenotypes is small and therefore, difficult to detect. Typically, phenotypes that exhibit extensive genetic and etiological heterogeneity would require a much larger sample size to identify the causal variants.
Clinical phenotypes, such as mortality and morbidity are often very complex and determined by a very large number of competing genetic and non-genetic factors. Hence, the effect size of each genetic variant on such complex phenotypes is very small and often not easily discernible, even for the causal mutations in single gene disorders. This is in contrast to proximal or endophenotypes such as the mRNA and protein levels or protein function, which are determined by a smaller number of determinants. Hence, the effect sizes of alleles are considerable enough to be detected in a moderate size study.
Phenotypes that exhibit large biological variability (intra-individual) and minimal inter-individual variability typically require much larger study populations for identification of the causative alleles. The opposite is the case for the phenotypes that exhibit large inter-individual variability but minimal intra-individual variance. Another strategy to enhance the chance of identifying the causative variants is to focus on phenotypic extremes.
Phenotyping
Phenotypic precision of the cases as well as the controls is essential in all genetic studies, particularly genetic studies of complex traits, wherein phenotyping tools have considerable inherent shortcomings. Inadequate phenotyping of the controls has obvious flaw of including those with undetected or subtle phenotype as controls and hence, diluting the power to detect an association. Likewise, categorizing biologically continuous phenotypes is not desirable. Classification of the participants as cases and controls based on dichotomization of the continuous phenotypes not only suffers from imperfection of the quantification methods but also increases of risk of spurious results. Likewise, phenotypic admixture of inter-related phenotypes, such as myocardial infarction and coronary atherosclerosis also reduces the power to detect the causative alleles. This is because, in addition to shared alleles, each also involves different mechanisms influenced by different sets of alleles. Finally, the distinction between true phenotype, such as hypertrophic cardiomyopathy caused by mutations in gene encoding for sarcomere proteins and phenocopy conditions, such as cardiac hypertrophy due to storage diseases, is essential as each could involve different sets of causal fields (86).
PERSPECTIVE
The GWAS, which has all but replaced the candidate gene approach, is built upon the CD-CV hypothesis. It has been exceedingly successful in identifying a very large number of DSVs that are associated with the complex phenotypes. However, the direct clinical utility of the findings is usually limited, as the identified alleles have modest effect sizes on the phenotype, as anticipated. However, the lack or a paucity of the clinical utility of GWAS should not lessen value of the main contribution of GWAS in providing novel clues to the pathogenesis of the complex phenotype (39). Hence, the GWAS findings could pave the road for subsequent mechanistic discoveries and ultimately identification of new therapeutic targets.
The focus is now shifting to RV-CD hypothesis, which is presumed to explain the “missing heritability” of the complex traits. However, whether rare variants would account for a major portion of heritability of the complex trait is an empiric question that needs to be tested through large –scale whole-exome or whole-genome sequencing projects. The etiological complexity of the distant phenotypes, particularly the diseases, is sobering as the causal fields are exceedingly diverse and contribution of each component is expected to be relatively modest. The complexity of further compounded by the recent discovery of a large number of sequence variants in human transcriptomes, which were translated into proteins but were absent in the corresponding DNA sequence (87). Clinical and phenotypic impacts of such RNA-DNA sequence are unknown.
In each genome, however, one might hope to find a handful of variants that exert major effect sizes and hence, offer potential clinical utility. Translational utility, however, mandates harnessing not only the biological and clinical information embedded in the DSVs but also integrating complementary data from various constituents of the phenotype, which are in part determined by DSVs, including epigenetics, microRNA, long non-coding RNAs, transcriptomics, proteomics or metabolomics. Complicated dynamic, often non-linear and long-term interactions between different constituent networks including the environmental network are responsible for the clinical phenotypes. The ultimate application of such discoveries to prevention and cure human disease, nonetheless, will require exploiting the knowledge garnered to elucidate the molecular mechanisms that govern the pathogenesis of the phenotype.
Acknowledgments
Acknowledgment: None
Funding support: NHLBI (R01-088498); NIA (R21 AG038597-01), Burroughs Wellcome Award in Translational Research (#1005907), TexGen Fund from Greater Houston Community Foundation
Glossary
- DNA sequence variants (DSVs)
DSVs refer to all variations in DNA sequence whether single nucleotide polymorphisms (SNPs), copy number variants (CNVs), insertions/deletions (indels) or structural variations (SVs)
- Exome
It refers to all exons in a genome, analogous to genome for the entire genetic material of a cell or organism
- Single nucleotide polymorphisms (SNPs)
Differences in DNA sequence at a single nucleotide level among individuals or between two copies of DNA in a genome
- Non-synonymous single nucleotide polymorphisms (nsSNPs)
A change in a single nucleotide that changes the codon or amino acid sequence in the protein
- Structural variations (SVs)
Large segmental differences in the genome among individuals or between two copies of DNA including large insertions, deletions, inversions, duplications and rearrangements
- Copy number variants (CNVs)
Each genome has two copies of DNA, and hence, two copies of each gene. Deletions or insertions that reduce or increase copy number in the genome are referred to as CNVs
- Linkage disequilibrium (LD)
LD refers to association of two alleles or DSVs by more than chance alone. DSVs located in close physical proximity are often assorted together and hence, are in LD. LD inversely relates to the distance between two variants on the chromosome
- De novo
A DSV that is present in an individual but not in either of the parents is referred to as a de novo variant. De novo variants are not inherited from the parents but occur as new genetic events in the individual
- Information content of a DSV
Genetic, biological and clinical information that could be gleaned from knowing the genotype, such as minor allele frequency, and effect on gene expression or protein function
- Causal field
Various etiological factors that contribute to a phenotype
- Heritability
The proportion of the inter-individual variance in a trait that is determined by genetic factors
Footnotes
Conflict of Interest: There is no conflict of interest to declare. The author have read the journal’s policy on disclosure of potential conflicts of interes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, et al. The sequence of the human genome. Science. 2001;291(5507):1304–51. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 2.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 3.Lander ES. Initial impact of the sequencing of the human genome. Nature. 2011 Feb 10;470(7333):187–97. doi: 10.1038/nature09792. [DOI] [PubMed] [Google Scholar]
- 4.Blaxter M. Revealing the dark matter of the genome. Science. 2010 Dec 24;330(6012):1758–9. doi: 10.1126/science.1200700. [DOI] [PubMed] [Google Scholar]
- 5.Levy S, Sutton G, Ng PC, Feuk L, Halpern AL, Walenz BP, et al. The Diploid Genome Sequence of an Individual Human. PLoSBiol. 2007;5(10):e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gunter C. Genomics: A picture worth 1000 Genomes. Nat Rev Genet. 2010 Dec;11(12):814. doi: 10.1038/nrg2906. [DOI] [PubMed] [Google Scholar]
- 7.Pennisi E. Genomics. 1000 Genomes Project gives new map of genetic diversity. Science. 2010 Oct 29;330(6004):574–5. doi: 10.1126/science.330.6004.574. [DOI] [PubMed] [Google Scholar]
- 8.Gamazon ER, Zhang W, Dolan ME, Cox NJ. Comprehensive survey of SNPs in the Affymetrix exon array using the 1000 Genomes dataset. PLoS ONE. 2010;5(2):e9366. doi: 10.1371/journal.pone.0009366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang J, Wang W, Li R, Li Y, Tian G, Goodman L, et al. The diploid genome sequence of an Asian individual. Nature. 2008 Nov 6;456(7218):60–5. doi: 10.1038/nature07484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wheeler DA, Srinivasan M, Egholm M, Shen Y, Chen L, McGuire A, et al. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452(7189):872–6. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- 11.Kim JI, Ju YS, Park H, Kim S, Lee S, Yi JH, et al. A highly annotated whole-genome sequence of a Korean individual. Nature. 2009 Aug 20;460(7258):1011–5. doi: 10.1038/nature08211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kidd JM, Cooper GM, Donahue WF, Hayden HS, Sampas N, Graves T, et al. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008 May 1;453(7191):56–64. doi: 10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sudmant PH, Kitzman JO, Antonacci F, Alkan C, Malig M, Tsalenko A, et al. Diversity of human copy number variation and multicopy genes. Science. 2010 Oct 29;330(6004):641–6. doi: 10.1126/science.1197005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mills RE, Walter K, Stewart C, Handsaker RE, Chen K, Alkan C, et al. Mapping copy number variation by population-scale genome sequencing. Nature. 2011 Feb 3;470(7332):59–65. doi: 10.1038/nature09708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eichler EE, Flint J, Gibson G, Kong A, Leal SM, Moore JH, et al. Missing heritability and strategies for finding the underlying causes of complex disease. Nat Rev Genet. 2010 Jun;11(6):446–50. doi: 10.1038/nrg2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Durbin RM, Abecasis GR, Altshuler DL, Auton A, Brooks LD, Gibbs RA, et al. A map of human genome variation from population-scale sequencing. Nature. 2010 Oct 28;467(7319):1061–73. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marian AJ, Belmont J. Strategic approaches to unraveling genetic causes of cardiovascular diseases. Circulation Research. 2011 May 13;108(10):1252–69. doi: 10.1161/CIRCRESAHA.110.236067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barabasi AL, Gulbahce N, Loscalzo J. Network medicine: a network-based approach to human disease. Nature reviews Genetics. 2011 Jan;12(1):56–68. doi: 10.1038/nrg2918. [Research Support, N.I.H., Extramural Review] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Romanoski CE, Che N, Yin F, Mai N, Pouldar D, Civelek M, et al. Network for Activation of Human Endothelial Cells by Oxidized Phospholipids: A Critical Role of Heme Oxygenase 1. Circulation Research. 2011 Jul 7; doi: 10.1161/CIRCRESAHA.111.241869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kathiresan S, Melander O, Guiducci C, Surti A, Burtt NP, Rieder MJ, et al. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat Genet. 2008;40(2):189–97. doi: 10.1038/ng.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kathiresan S, Willer CJ, Peloso GM, Demissie S, Musunuru K, Schadt EE, et al. Common variants at 30 loci contribute to polygenic dyslipidemia. Nature Genetics. 2009 Jan;41(1):56–65. doi: 10.1038/ng.291. [Research Support, N.I.H., Extramural Research Support, N.I.H., Intramural Research Support, Non-U.S. Gov’t Research Support, U.S. Gov’t, Non-P.H.S.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Debette S, Visvikis-Siest S, Chen MH, Ndiaye NC, Song C, Destefano A, et al. Identification of cis- and trans-Acting Genetic Variants Explaining Up to Half the Variation in Circulating Vascular Endothelial Growth Factor Levels. Circulation Research. 2011 Jul 14; doi: 10.1161/CIRCRESAHA.111.243790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Innocenti P, Morrow EH, Dowling DK. Experimental evidence supports a sex-specific selective sieve in mitochondrial genome evolution. Science. 2011 May 13;332(6031):845–8. doi: 10.1126/science.1201157. [Research Support, Non-U.S. Gov’t] [DOI] [PubMed] [Google Scholar]
- 24.Lemos B, Araripe LO, Hartl DL. Polymorphic Y chromosomes harbor cryptic variation with manifold functional consequences. Science. 2008 Jan 4;319(5859):91–3. doi: 10.1126/science.1148861. [Research Support, N.I.H., Extramural] [DOI] [PubMed] [Google Scholar]
- 25.Marian AJ. Nature’s genetic gradients and the clinical phenotype. Circ Cardiovasc Genet. 2009 Dec;2(6):537–9. doi: 10.1161/CIRCGENETICS.109.921940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996 Sep 13;273(5281):1516–7. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- 27.Cirulli ET, Goldstein DB. Uncovering the roles of rare variants in common disease through whole-genome sequencing. Nat Rev Genet. 2010 Jun;11(6):415–25. doi: 10.1038/nrg2779. [DOI] [PubMed] [Google Scholar]
- 28.Bodmer W, Bonilla C. Common and rare variants in multifactorial susceptibility to common diseases. Nat Genet. 2008 Jun;40(6):695–701. doi: 10.1038/ng.f.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pritchard JK. Are rare variants responsible for susceptibility to complex diseases? AmJHumGenet. 2001;69(1):124–37. doi: 10.1086/321272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schork NJ, Murray SS, Frazer KA, Topol EJ. Common vs. rare allele hypotheses for complex diseases. Curr Opin Genet Dev. 2009 Jun;19(3):212–9. doi: 10.1016/j.gde.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pritchard JK, Cox NJ. The allelic architecture of human disease genes: common disease-common variant...or not? Human Molecular Genetics. 2002;11(20):2417–23. doi: 10.1093/hmg/11.20.2417. [DOI] [PubMed] [Google Scholar]
- 32.Rodriguez G, Ueyama T, Ogata T, Czernuszewicz G, Tan Y, Dorn GW, et al. Molecular Genetic and Functional Characterization Implicate Muscle-Restricted Coiled-Coil Gene (MURC) as a Causal Gene for Familial Dilated Cardiomyopathy. Circulation: Cardiovascular Genetics. 2011 June;3:2011. doi: 10.1161/CIRCGENETICS.111.959866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dickson SP, Wang K, Krantz I, Hakonarson H, Goldstein DB. Rare variants create synthetic genome-wide associations. PLoS Biol. 2010 Jan;8(1):e1000294. doi: 10.1371/journal.pbio.1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, Ioannidis JP, et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet. 2008 May;9(5):356–69. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 35.Gao X, Becker LC, Becker DM, Starmer JD, Province MA. Avoiding the high Bonferroni penalty in genome-wide association studies. Genetic Epidemiology. 2010 Jan;34(1):100–5. doi: 10.1002/gepi.20430. [Comparative Study Evaluation Studies Research Support, N.I.H., Extramural Validation Studies] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Newton-Cheh C, Johnson T, Gateva V, Tobin MD, Bochud M, Coin L, et al. Genome-wide association study identifies eight loci associated with blood pressure. Nat Genet. 2009 Jun;41(6):666–76. doi: 10.1038/ng.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010 Aug 5;466(7307):707–13. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sotoodehnia N, Isaacs A, de Bakker PI, Dorr M, Newton-Cheh C, Nolte IM, et al. Common variants in 22 loci are associated with QRS duration and cardiac ventricular conduction. Nat Genet. 2010 Dec;42(12):1068–76. doi: 10.1038/ng.716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harismendy O, Notani D, Song X, Rahim NG, Tanasa B, Heintzman N, et al. 9p21 DNA variants associated with coronary artery disease impair interferon-gamma signalling response. Nature. 2011 Feb 10;470(7333):264–8. doi: 10.1038/nature09753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arora P, Newton-Cheh C. Blood pressure and human genetic variation in the general population. Curr Opin Cardiol. 2010 Mar 10;25(3):229–37. doi: 10.1097/HCO.0b013e3283383e2c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kathiresan S, Manning AK, Demissie S, D’Agostino RB, Surti A, Guiducci C, et al. A genome-wide association study for blood lipid phenotypes in the Framingham Heart Study. BMC Med Genet. 2007;8( Suppl 1):S17. doi: 10.1186/1471-2350-8-S1-S17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pirruccello J, Kathiresan S. Genetics of lipid disorders. Curr Opin Cardiol. 2010 Mar 10;25(3):238–42. doi: 10.1097/HCO.0b013e328338574d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pfeufer A, Sanna S, Arking DE, Muller M, Gateva V, Fuchsberger C, et al. Common variants at ten loci modulate the QT interval duration in the QTSCD Study. Nat Genet. 2009 Apr;41(4):407–14. doi: 10.1038/ng.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klein RJ, Zeiss C, Chew EY, Tsai JY, Sackler RS, Haynes C, et al. Complement factor H polymorphism in age-related macular degeneration. Science. 2005 Apr 15;308(5720):385–9. doi: 10.1126/science.1109557. [Research Support, Non-U.S. Gov’t Research Support, U.S. Gov’t, P.H.S.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maller J, George S, Purcell S, Fagerness J, Altshuler D, Daly MJ, et al. Common variation in three genes, including a noncoding variant in CFH, strongly influences risk of age-related macular degeneration. Nature Genetics. 2006 Sep;38(9):1055–9. doi: 10.1038/ng1873. [Comparative Study Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t] [DOI] [PubMed] [Google Scholar]
- 46.Manolio TA. Genomewide association studies and assessment of the risk of disease. The New England Journal Of Medicine. 2010 Jul 8;363(2):166–76. doi: 10.1056/NEJMra0905980. [Review] [DOI] [PubMed] [Google Scholar]
- 47.Jakobsdottir J, Gorin MB, Conley YP, Ferrell RE, Weeks DE. Interpretation of genetic association studies: markers with replicated highly significant odds ratios may be poor classifiers. PLoS genetics. 2009 Feb;5(2):e1000337. doi: 10.1371/journal.pgen.1000337. [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005 Sep 15;437(7057):376–80. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nielsen R, Paul JS, Albrechtsen A, Song YS. Genotype and SNP calling from next-generation sequencing data. Nature reviews Genetics. 2011 Jun;12(6):443–51. doi: 10.1038/nrg2986. [Research Support, N.I.H., Extramural Research Support, U.S. Gov’t, Non-P.H.S.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ansorge WJ. Next-generation DNA sequencing techniques. N Biotechnol. 2009 Apr;25(4):195–203. doi: 10.1016/j.nbt.2008.12.009. [DOI] [PubMed] [Google Scholar]
- 51.Hedges DJ, Guettouche T, Yang S, Bademci G, Diaz A, Andersen A, et al. Comparison of three targeted enrichment strategies on the SOLiD sequencing platform. PLoS ONE. 2011;6(4):e18595. doi: 10.1371/journal.pone.0018595. [Research Support, N.I.H., Extramural] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Meder B, Haas J, Keller A, Heid C, Just S, Borries A, et al. Targeted Next-Generation Sequencing for the Molecular Genetic Diagnostics of Cardiomyopathies. Circ Cardiovasc Genet. 2011 Jan 20; doi: 10.1161/CIRCGENETICS.110.958322. [DOI] [PubMed] [Google Scholar]
- 53.Vermeer S, Hoischen A, Meijer RP, Gilissen C, Neveling K, Wieskamp N, et al. Targeted next-generation sequencing of a 12.5 Mb homozygous region reveals ANO10 mutations in patients with autosomal-recessive cerebellar ataxia. American journal of human genetics. 2010 Dec 10;87(6):813–9. doi: 10.1016/j.ajhg.2010.10.015. [Research Support, Non-U.S. Gov’t] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rehman AU, Morell RJ, Belyantseva IA, Khan SY, Boger ET, Shahzad M, et al. Targeted capture and next-generation sequencing identifies C9orf75, encoding taperin, as the mutated gene in nonsyndromic deafness DFNB79. American journal of human genetics. 2010 Mar 12;86(3):378–88. doi: 10.1016/j.ajhg.2010.01.030. [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ng SB, Buckingham KJ, Lee C, Bigham AW, Tabor HK, Dent KM, et al. Exome sequencing identifies the cause of a mendelian disorder. Nat Genet. 2010 Jan;42(1):30–5. doi: 10.1038/ng.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ng SB, Bigham AW, Buckingham KJ, Hannibal MC, McMillin MJ, Gildersleeve HI, et al. Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat Genet. 2010 Sep;42(9):790–3. doi: 10.1038/ng.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Choi M, Scholl UI, Ji W, Liu T, Tikhonova IR, Zumbo P, et al. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc Natl Acad Sci U S A. 2009 Nov 10;106(45):19096–101. doi: 10.1073/pnas.0910672106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Choi M, Scholl UI, Yue P, Bjorklund P, Zhao B, Nelson-Williams C, et al. K+ channel mutations in adrenal aldosterone-producing adenomas and hereditary hypertension. Science. 2011 Feb 11;331(6018):768–72. doi: 10.1126/science.1198785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Comino-Mendez I, Gracia-Aznarez FJ, Schiavi F, Landa I, Leandro-Garcia LJ, Leton R, et al. Exome sequencing identifies MAX mutations as a cause of hereditary pheochromocytoma. Nature Genetics. 2011;43(7):663–7. doi: 10.1038/ng.861. [DOI] [PubMed] [Google Scholar]
- 60.Klassen T, Davis C, Goldman A, Burgess D, Chen T, Wheeler D, et al. Exome sequencing of ion channel genes reveals complex profiles confounding personal risk assessment in epilepsy. Cell. 2011 Jun 24;145(7):1036–48. doi: 10.1016/j.cell.2011.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010 Apr;7(4):248–9. doi: 10.1038/nmeth0410-248. [Letter Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4(7):1073–81. doi: 10.1038/nprot.2009.86. [Research Support, N.I.H., Extramural] [DOI] [PubMed] [Google Scholar]
- 63.Roach JC, Glusman G, Smit AF, Huff CD, Hubley R, Shannon PT, et al. Analysis of genetic inheritance in a family quartet by whole-genome sequencing. Science. 2010 Apr 30;328(5978):636–9. doi: 10.1126/science.1186802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sobreira NL, Cirulli ET, Avramopoulos D, Wohler E, Oswald GL, Stevens EL, et al. Whole-genome sequencing of a single proband together with linkage analysis identifies a Mendelian disease gene. PLoS genetics. 2010 Jun;6(6):e1000991. doi: 10.1371/journal.pgen.1000991. [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Holm H, Gudbjartsson DF, Sulem P, Masson G, Helgadottir HT, Zanon C, et al. A rare variant in MYH6 is associated with high risk of sick sinus syndrome. Nature Genetics. 2011;43(4):316–20. doi: 10.1038/ng.781. [Research Support, N.I.H., Extramural Validation Studies] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lander ES, Schork NJ. Genetic dissection of complex traits. Science. 1994;265(5181):2037–48. doi: 10.1126/science.8091226. [DOI] [PubMed] [Google Scholar]
- 67.Dawson E, Abecasis GR, Bumpstead S, Chen Y, Hunt S, Beare DM, et al. A first-generation linkage disequilibrium map of human chromosome 22. Nature. 2002;418(6897):544–8. doi: 10.1038/nature00864. [DOI] [PubMed] [Google Scholar]
- 68.Ardlie KG, Kruglyak L, Seielstad M. Patterns of linkage disequilibrium in the human genome. NatRevGenet. 2002;3(4):299–309. doi: 10.1038/nrg777. [DOI] [PubMed] [Google Scholar]
- 69.Reich DE, Cargill M, Bolk S, Ireland J, Sabeti PC, Richter DJ, et al. Linkage disequilibrium in the human genome. Nature. 2001;411(6834):199–204. doi: 10.1038/35075590. [DOI] [PubMed] [Google Scholar]
- 70.Pritchard JK, Przeworski M. Linkage disequilibrium in humans: models and data. AmJHumGenet. 2001;69(1):1–14. doi: 10.1086/321275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Helgadottir A, Thorleifsson G, Manolescu A, Gretarsdottir S, Blondal T, Jonasdottir A, et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316(5830):1491–3. doi: 10.1126/science.1142842. [DOI] [PubMed] [Google Scholar]
- 72.McPherson R, Pertsemlidis A, Kavaslar N, Stewart A, Roberts R, Cox DR, et al. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316(5830):1488–91. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Genome-wide association study of 14,000 cases of seven common diseases and 3, 000 shared controls. Nature. 2007 Jun 7;447(7145):661–78. doi: 10.1038/nature05911. [Multicenter Study Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t Research Support, U.S. Gov’t, Non-P.H.S.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Visel A, Zhu Y, May D, Afzal V, Gong E, Attanasio C, et al. Targeted deletion of the 9p21 non-coding coronary artery disease risk interval in mice. Nature. 2010 Mar 18;464(7287):409–12. doi: 10.1038/nature08801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Daw EW, Chen SN, Czernuszewicz G, Lombardi R, Lu Y, Ma J, et al. Genome-wide mapping of modifier chromosomal loci for human hypertrophic cardiomyopathy. Hum Mol Genet. 2007 Oct 15;16(20):2463–71. doi: 10.1093/hmg/ddm202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hopkins PN, Toth PP, Ballantyne CM, Rader DJ. Familial Hypercholesterolemias: Prevalence, genetics, diagnosis and screening recommendations from the National Lipid Association Expert Panel on Familial Hypercholesterolemia. J Clin Lipidol. 2011 Jun;5(3 Suppl):S9–S17. doi: 10.1016/j.jacl.2011.03.452. [DOI] [PubMed] [Google Scholar]
- 77.Guey LT, Kravic J, Melander O, Burtt NP, Laramie JM, Lyssenko V, et al. Power in the phenotypic extremes: a simulation study of power in discovery and replication of rare variants. Genetic Epidemiology. 2011 Feb 9; doi: 10.1002/gepi.20572. [DOI] [PubMed] [Google Scholar]
- 78.O’Roak BJ, Deriziotis P, Lee C, Vives L, Schwartz JJ, Girirajan S, et al. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nature Genetics. 2011 Jun;43(6):585–9. doi: 10.1038/ng.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Malkin I, Ginsburg E, Elston RC. Increase in power of transmission-disequilibrium tests for quantitative traits. Genetic Epidemiology. 2002 Oct;23(3):234–44. doi: 10.1002/gepi.10180. [Comment Research Support, U.S. Gov’t, P.H.S.] [DOI] [PubMed] [Google Scholar]
- 80.Samani NJ, Raitakari OT, Sipila K, Tobin MD, Schunkert H, Juonala M, et al. Coronary Artery Disease-Associated Locus on Chromosome 9p21 and Early Markers of Atherosclerosis. Arteriosclerosis, Thrombosis, and Vascular Biology. 2008:ATVBAHA. doi: 10.1161/ATVBAHA.108.170332. [DOI] [PubMed] [Google Scholar]
- 81.Spencer CC, Su Z, Donnelly P, Marchini J. Designing genome-wide association studies: sample size, power, imputation, and the choice of genotyping chip. PLoS genetics. 2009 May;5(5):e1000477. doi: 10.1371/journal.pgen.1000477. [Research Support, Non-U.S. Gov’t] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Laan M, Paabo S. Mapping genes by drift-generated linkage disequilibrium. American journal of human genetics. 1998 Aug;63(2):654–6. doi: 10.1086/301972. [Letter Research Support, Non-U.S. Gov’t] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Terwilliger JD, Zollner S, Laan M, Paabo S. Mapping genes through the use of linkage disequilibrium generated by genetic drift: ‘drift mapping’ in small populations with no demographic expansion. Human heredity. 1998 May–Jun;48(3):138–54. doi: 10.1159/000022794. [Research Support, Non-U.S. Gov’t Review] [DOI] [PubMed] [Google Scholar]
- 84.Novembre J, Johnson T, Bryc K, Kutalik Z, Boyko AR, Auton A, et al. Genes mirror geography within Europe. Nature. 2008 Nov 6;456(7218):98–101. doi: 10.1038/nature07331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tishkoff SA, Reed FA, Friedlaender FR, Ehret C, Ranciaro A, Froment A, et al. The genetic structure and history of Africans and African Americans. Science. 2009 May 22;324(5930):1035–44. doi: 10.1126/science.1172257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Marian AJ. Hypertrophic cardiomyopathy: from genetics to treatment. Eur J Clin Invest. 2010 Apr;40(4):360–9. doi: 10.1111/j.1365-2362.2010.02268.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li M, Wang IX, Li Y, Bruzel A, Richards AL, Toung JM, et al. Widespread RNA and DNA sequence differences in the human transcriptome. Science. 2011 Jul 1;333(6038):53–8. doi: 10.1126/science.1207018. [DOI] [PMC free article] [PubMed] [Google Scholar]