Abstract
Quorum sensing has been implicated as an important global regulatory system controlling the expression of numerous virulence factors in bacterial pathogens. In the present study, DNA targets of SmcR, a Vibrio vulnificus LuxR homologue, were selected from a random pool of DNA fragments by using a cycle selection procedure consisting of in vitro DNA-SmcR interaction, purification of SmcR-DNA complexes, and PCR amplification of SmcR-bound DNA. The amplified DNA fragments were cloned and analyzed separately by electrophoretic mobility shift assay to verify the specific binding of SmcR to the DNA. The DNA sequences bound by SmcR were determined by DNase I footprinting, and alignment of the resulting 29 sequences revealed a 22-bp consensus SmcR-binding sequence, 5′-TTATTGATWWRWTWNTNAATAA-3′ (where W represents A or T, R is G or A, and N is any nucleotide), with an 8-bp (TTATTGAT) inverted repeat. The consensus sequence revealed greater efficiency for the binding of SmcR than the SmcR-binding sequence previously identified within PvvpE. Mutational analysis demonstrated that the 9th and 10th bases from the center are the most essential for SmcR binding. A genome-wide search using the consensus sequence predicted that at least 121 genes are under the control of SmcR, and 10 of these newly identified SmcR regulon members were verified as being regulated by SmcR in V. vulnificus as well as in vitro. The consensus sequence and newly identified genes should be of use for elucidating the regulatory mechanism of SmcR and provide further insight into the role of the quorum sensing in V. vulnificus pathogenesis.
Bacterial pathogenicity is multifactorial and a complex phenomenon that involves the products of many genes, collectively called virulence factors (1). Expression of many of these virulence factors is coordinately controlled by a common global regulatory system in response to environmental signals. This coordinate regulation facilitates cooperation of the virulence factors and is crucial for the overall success of the infectious microorganisms during pathogenesis (2). Many bacteria monitor their cell population densities through the exchange of diffusible signal molecules (autoinducer (AI)4) that accumulate extracellularly (for recent reviews, see Refs. 3 and 4). This type of communication, termed quorum sensing, has been recognized as a global regulatory system controlling the expression of numerous virulence factors in bacterial pathogens (for recent reviews, see Refs. 5 and 6). When the concentration of the various AIs increases to critical levels, a signal transduction cascade triggered through cognate receptors alters the expression of over 50 genes or operons (7).
The cell density-dependent regulation of bioluminescence in Vibrio harveyi is frequently used as a model for quorum sensing. V. harveyi LuxR is the transcriptional activator of the luminescence operon, and its synthesis is controlled by the levels of three autoinducers, AI-1, AI-2, and CAI-1 (4). To date, homologues of LuxR, which are postulated to regulate virulence genes, have been identified in various pathogenic Vibrio spp. (8–14). SmcR has been identified from V. vulnificus, a food-borne pathogenic bacterium, and proposed as a LuxR homologue (8). Moreover, analysis of the completed V. vulnificus genome sequence reveals that V. vulnificus possesses homologues of the genes required for sensing and responding to autoinducers, such as LuxO and LuxT (15). Given the similarities between the components of quorum-sensing systems in V. vulnificus and in V. harveyi, it seemed logical to consider that SmcR is a quorum-sensing regulator of V. vulnificus. Recent work demonstrated that SmcR regulates virulence genes and adaptive phenotypes (16–19).
Considering the important role of quorum sensing in pathogenesis of Vibrio spp., a major problem to be addressed is that binding sequences of LuxR homologues have not been predicted and that genome-wide identification of target genes is still limited. Until now, only a few studies on the direct binding of LuxR homologues to DNA in vitro have been reported (16, 20, 21); thus, consensus sequences for binding have not yet been identified. This lack of information makes it difficult to identify what genes are controlled by the proteins as well as to understand how the promoters of the target genes are modulated by the proteins. Transcriptome analyses have been used to identify genes regulated by LuxR homologues and the quorum-sensing system (7, 12, 13, 22). However, these approaches search for genes regulated under defined in vitro conditions and therefore may not identify genes expressed only in specific environmental conditions that pathogenic bacteria may encounter within a host.
Accordingly, here we extend our efforts to determine a consensus sequence for binding of SmcR, a LuxR homologue whose function is the best characterized in vitro. For this purpose, DNA targets that contain SmcR binding sites were selected from a pool of V. vulnificus genomic DNA fragments, and the binding sites were sequenced. Alignment of the sequences revealed a 22-bp consensus sequence with an 8-bp inverted repeat, and site-directed mutational analyses demonstrated that the 9th and 10th bases from the center of the repeats are the most important. In addition, we attempted to identify genes that are regulated by direct interaction of SmcR at the promoter. A hidden Markov model (HMM), a prediction algorithm based on a sequence profile, was used to screen the V. vulnificus genome, and the search resulted in the identification of at least 121 genes as members of the SmcR regulon. SmcR binding to the newly identified genes and their regulation by SmcR were experimentally examined by chromatin immunoprecipitation experiments and quantitative real time PCR (RT-PCR) analyses.
EXPERIMENTAL PROCEDURES
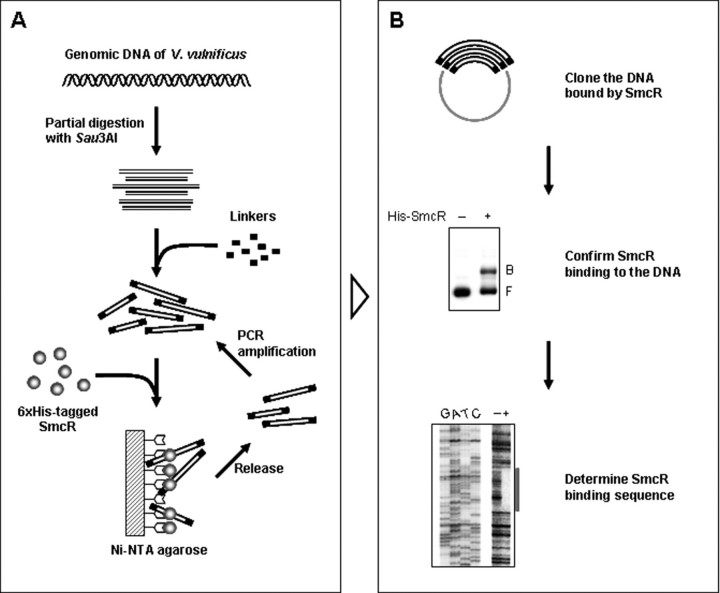
Selection of DNA Fragments Bound by SmcR—The DNA fragment library was prepared by partially digesting genomic DNA of V. vulnificus ATCC 29307 with Sau3AI and by recovering the 200–500-bp DNA fragments from a preparative agarose gel. The DNA fragments containing SmcR-binding motifs were selected from the library by using a cycle selection process, as described by Ochsner and Vasil (23) (Fig. 1A). Briefly, a linker carrying the Sau3AI protruding end (GATC) was developed by annealing and phosphorylation of a pair of complementary oligonucleotides, Sau3AI Linker-1 and Sau3AI Linker-2 (Table 1), and ligated to the Sau3AI-digested DNA. For binding of SmcR to DNA, 2 μg of the resulting DNA fragments were mixed with 200 nm His-SmcR purified as described previously (16) and incubated for 30 min at 37 °C. The DNA-SmcR complexes were isolated by using Ni2+-nitrilotriacetic acid-agarose, and the DNAs were released from SmcR using a DNA purification kit (Qiagen, Valencia, CA) (Fig. 1A). The released DNA were amplified using the linker-specific primer, Sau3AI linker-2, by PCR and used for the next cycle. The DNAs obtained after two cycles of selection for SmcR binding were cloned into pGEM-T easy vector (Promega, Madison, WI).
FIGURE 1.
Procedures for identification of the SmcR binding sequences. A, the DNA fragments obtained by partial digestion of genomic V. vulnificus DNA with Sau3AI were ligated with linkers at both ends (shown as black tails). Putative SmcR target DNA was enriched in two consecutive cycles of DNA-SmcR interaction, purification of the complexes on Ni2+-nitrilotriacetic acid-agarose, and PCR amplification. B, each DNA released from the DNA-SmcR complexes was cloned, subsequently radiolabeled, and then used for EMSA. The sequences for binding of SmcR to individual DNA were also identified by DNase I footprinting. B, bound DNA; F, free DNA.
TABLE 1.
Oligonucleotides used in this study
| Oligonucleotide | Oligonucleotide sequence, 5′-3a | Locationb | Use(s) |
|---|---|---|---|
| Linker and PCR oligonucleotides | |||
| Sau3AI Linker-1 | GATCGAGTGACTCTTGACCTCGACTAGTGC | Linker construction; amplification of SmcR-target DNA | |
|
Sau3AI Linker-2
|
GCACTAGTCGAGGTCAAGAGTCACTC
|
Linker construction; amplification of SmcR target DNA
|
|
| PCR oligonucleotides | |||
| T7 | GAATTGTAATACGACTCACTATAGG | pGEM-T easy vector | DNase I footprinting or EMSA |
| SP6 | CATACGATTTAGGTGACACTATAG | pGEM-T easy vector | DNase I footprinting or EMSA |
| VVPE021 | AGAATGGCGATTTTCATAG | -300 to -282 | EMSA |
| VVPE022 | GAATCCATCTCACTGCGA | -118 to -101 | EMSA |
| VVPE501 | GTACTGCAGGTTTGGCTAATGAGTTTTAAG | -510 to -490 | Amplification of vvpE upstream |
|
VVPE502
|
TATGGATCCGACGTTGATTGAGTTTCATTATCG
|
+69 to +92
|
Amplification of vvpE upstream
|
| Mutagenic oligonucleotides An and Bnc | |||
| AWC | CAAATTTATCAATAAGAAAAATGGG | -217 to -193 | Construction of WCd |
| BWC | TATTGATAAATTTGTGAATAAAATAAAAAGC | -206 to -176 | WC |
| AIR-L | ATGAGGTACCTGGAAAAATGGGACAGTCATC | -226 to -196 | WCIR-L |
| BIR-L | TCCAGGTACCTCATTTGTGAATAAAATAAAA | -209 to -179 | WCIR-L |
| AIR-R | TTGAGGTACCTGATTTATCAATAAGAAAAATG | -215 to -184 | WCIR-R |
| BIR-R | ATCAGGTACCTCAATAAAAAGCACAATTTTA | -197 to -167 | WCIR-R |
| APAL | AAATTGTGCTTTTTATTTTATTGATAAATTT | -199 to -169 | WCPAL |
| BPAL | AAATTTATCAATAAAATAAAAAGCACAATTT | -199 to -169 | WCPAL |
| A-11 | CACAAATTTATCAATAGGAAAAATGGG | -217 to -191 | WC-11 |
| A-10 | CACAAATTTATCAATGAGAAAAATGGG | -217 to -191 | WC-10 |
| A-9 | CACAAATTTATCAAGAAGAAAAATGGG | -217 to -191 | WC-9 |
| A-8 | CACAAATTTATCAGTAAGAAAAATGGG | -217 to -191 | WC-8 |
| A-7 | GCTTTTTATTTTATTCACAAATTTATCGAT | -191 to -162 | WC-7 |
| A-6 | CAAATTTATGAATAAGAAAAATGGGAC | -219 to -193 | WC-6 |
| A-5 | CACAAATTTAGCAATAAGAAAAATGGG | -217 to -191 | WC-5 |
| A-4 | AAATTTGTCAATAAGAAAAATGGGACA | -220 to -194 | WC-4 |
| B-11 ∼ -7;-5 | TAAATTTGTGAATAAAATAAAAAGCACA | -200 to -173 | WC-11 ∼ -7; -5 |
| B-6 | GTCCCATTTTTCTTATTCATAAATTTG | -219 to -193 | WC-6 |
| B-4 | CTGTCCCATTTTTCTTATTGACAAATT | -221 to -195 | WC-4 |
Regions of oligonucleotide(s) not complementary to corresponding templates are underlined.
Shown are the oligonucleotide positions, where +1 is the transcription start site of vvpE.
Numbers in An and Bn primers represent the positions of point mutation from the center of the working consensus sequence. Base substitutions to mutate the working consensus sequence are underlined.
Sequences of the constructs are listed in Fig. 3B.
Electrophoretic Mobility Shift Assay (EMSA) and DNase I Footprinting—Inserted DNA of the individual clones was amplified by a PCR using 32P-labeled T7 and unlabeled SP6 as the primers (Table 1) and used as a probe DNA for EMSA. The binding of SmcR to the labeled DNA and electrophoretic analysis of the DNA-SmcR complexes have already been described (16, 24) (Fig. 1B). The DNA that was confirmed as a target DNA of SmcR by EMSA was subsequently subjected to a DNase I footprinting assay according to the procedures previously described by Jeong et al. (16). After precipitation with ethanol, the digested DNA products were resolved on a sequencing gel alongside sequencing ladders of individual clones generated using SP6 or T7 as the primer. The gels were visualized and quantified using a phosphor image analyzer (BAS1500; Fuji Photo Film Co. Ltd., Tokyo, Japan) and the Image Gauge (version 3.12) program. The SmcR-binding sequences identified by the DNase I footprinting were used to determine a consensus sequence.
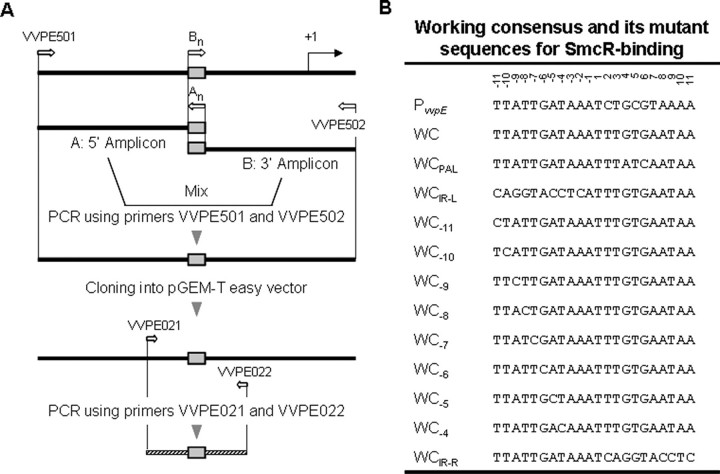
Development and Site-directed Mutagenesis of the Working Consensus Sequence—A working consensus sequence was developed by substituting bases of the 22-bp PvvpE SmcR-binding sequence with the bases conserved in corresponding positions of the consensus SmcR-binding sequence using the linker scanning mutation method (Fig. 3) (25, 26). For this, the 5′-amplicon, a 329-bp DNA fragment corresponding to the upstream region of PvvpE was generated using two primers, VVPE501 and AWC (Table 1). AWC, an antisense primer, contained the working consensus sequence (Table 1). Similarly, a 301-bp 3′ amplicon was amplified using primers BWC and VVPE502. BWC, a sense primer, contained a sequence complementary to the AWC primer. Second stage PCR was performed using VVPE501 and VVPE502 as a pair of primers and a mixture of two amplicons as the template to result in the 602-bp PvvpE upstream region with the 22-bp working consensus sequence (UP-vvpEWC) (Fig. 3A).
FIGURE 3.
Construction of a working consensus sequence and its mutant sequences. A, a working consensus sequence was generated by replacing residues of the PvvpE SmcR-binding sequence with those conserved in the consensus SmcR-binding sequence by the PCR-mediated linker scanning method (25, 26). Construction of 11 mutant sequences was carried out in a similar way, except that the PvvpE upstream region with the working consensus sequence was used as a template DNA for PCR. For details, see “Experimental Procedures.” Solid lines, PvvpE upstream DNA; shaded boxes, SmcR-binding sequences; bent arrow, transcription start site of PvvpE; hatched lines, PCR products used for EMSA; open arrows, locations of the oligonucleotide primers. B, a working consensus sequence and its mutant sequences. PvvpE, PvvpE SmcR-binding sequence; WC, working consensus sequence; WCPAL, sequences with dyad symmetry; WCIR-L, with mutations in the left repeat; WCIR-R, with mutations in the right repeat; WCn, with point mutations at position n.
The working consensus sequence was subsequently mutated by a similar experimental procedure, except using the UP-vvpEWC as a template DNA and a pair of A and B primers: APAL and BPAL (for development of dyad symmetric sequence), AIR-L and BIR-L (for replacement of the left repeat sequence), AIR-R and BIR-R (for replacement of the right repeat sequence), or An and Bn (for point mutations at a position n) (see Table 1). The 602-bp PCR products were inserted into pGEM-T easy vector, thereby creating 11 constructs with different SmcR-binding sequences, as confirmed by DNA sequencing (Fig. 3B). The 200-bp DNA (from –301 to –101 relative to the vvpE transcription start site) containing either the working consensus or the mutant SmcR binding sequences was generated by PCR amplification of the resulting constructs with a combination of the 32P-labeled and unlabeled primers VVPE021 and VVPE022 (Table 1) and then used as a probe DNA for EMSA.
Bioinformatic Prediction of the SmcR Regulon—To discover SmcR-binding sites across the V. vulnificus CMCP6 genome, which was retrieved from GenBank™ (AE016795; AE016796), the HMM approach was applied. Among experimentally obtained SmcR-binding sites, the sequences completely annealed to the genome were extracted under a BLASTn search. Using HMMER, version 2.3.2 (available on the World Wide Web), an HMM profile representing the extracted sequences was constructed and subsequently was searched against the forward and reverse complementary sequences of the genome. The level of E value cut-off was determined from the E values of the extracted sequences. Among total genomic hits, the sequences with E values less than the cut-off were chosen as the candidates of SmcR-binding sites. If the first position of a candidate on the genome is located within the regulatory region spanning from the translational start site to upstream of 500 bp of any gene on the genome, we extracted the locus tag of the gene. Using the data base BioCyc (27), genes or operons corresponding to each locus tag were predicted as potential members of the SmcR regulon.
Chromatin Immunoprecipitation and Quantitative RT-PCR—The chromatin immunoprecipitation experiments were performed using formaldehyde cross-linking as described by Rhee et al. (28). Briefly, the cross-linked chromatin in the wild type and smcR mutant HS031 cells (16) was fragmented by sonication to result in sheared chromatins with an average length of 500 bp. One-half of the clarified supernatant was saved as the total input sheared chromatin (positive control) prior to the reaction with the anti-SmcR antibody (16), whereas the sheared chromatin (100 μl) from the other half of the supernatant was reacted with 10 μl of the anti-SmcR antibody overnight at 4 °C, and the resulting chromatin-antibody complex was specifically precipitated by adding 45 μl of 50% protein A-Sepharose (Amersham Biosciences). The precipitates were washed, and the sheared chromatins were eluted using the methods mentioned elsewhere (28). The cross-linkings were reversed by incubating the sheared chromatins with 1% SDS and 0.1 m NaHCO3 at 65 °C for 6 h, and DNAs were purified and analyzed by a PCR using a pair of the primers specific to the promoter region of the SmcR regulons, as listed in supplemental Table 2.
For quantitative RT-PCR, cDNA was synthesized with the iScript™ cDNA synthesis kit (Bio-Rad) according to the manufacturer's procedures. Real time PCR amplification of the cDNA was performed with a pair of primers (supplemental Table 2) using the Chromo 4 real time PCR detection system (Bio-Rad), as described previously (29, 30). Relative expression levels of the SmcR regulons were calculated by using the 16 S rRNA expression level as the internal reference for normalization (29, 30).
RESULTS
Selection of DNA with SmcR-binding Sequences—DNA fragments carrying a motif for SmcR binding were isolated from a random population of V. vulnificus genomic fragments by a cycle selection process (Fig. 1A). A repeat of consecutive cycles of DNA-SmcR complex formation, isolation of the complexes, release of the DNA, and PCR amplification of the DNA enriched the DNA fragments with affinity to SmcR. Since the diversity of the DNA fragment population was also decreased by increasing repeats of the selection cycles (data not shown), the number of cycles of enrichment was limited to two. The enriched DNA fragments were cloned, and 66 DNA fragments were selected after confirming binding of SmcR to each fragment by EMSA. The nucleotide sequencing of these selected DNA fragments revealed that some fragments contained identical sequences, and finally 35 different DNA fragments were identified as DNA that contains SmcR-binding sites. EMSAs were performed in the presence of 0.1 μg of poly(dI-dC) as a nonspecific competitor, to determine if the binding of SmcR to the target DNA was specific (Fig. 1B and supplemental Fig. 1).
Identification of SmcR-binding Sites Using DNase I Footprinting—To determine the sequences for SmcR binding, DNase I footprinting experiments were performed using individual DNA fragments that were gel-shifted by SmcR (Fig. 1B). Of the 35 DNA fragments, only 29 resulted in a DNase I footprint (see supplemental Fig. 1). The DNase I footprints revealed clear protection patterns by SmcR, which are similar to the protection pattern previously observed in DNase I protection analyses of SmcR in the upstream region of vvpE (16). DNA sequences of the 29 protected regions were determined (Fig. 2A) and analyzed using a sequence logo generator, WebLogo (31) (Fig. 2B). Alignment of the sequences for SmcR binding revealed the frequency of distribution of nucleotides at positions and demonstrated a 22-bp consensus sequence, TTATTGATWWRWTWNTNAATAA (where W represents A or T, R is A or G, and N is any nucleotide) (Fig. 2C). A residue at a position represents when it is present in more than 50% of the population, and two residues when together represent more than 70% of the population. The most conserved residues in the 22-bp consensus sequence were in positions –10 (T), –9 (A), and –7 (T) from the center of the sequence (Fig. 2C). The consensus sequence consists of an 8-bp inverted repeat (IR), and the left half (IR-L) of the repeats is better conserved than the right half (IR-R), resulting in an imperfect dyad symmetry (Fig. 2C).
FIGURE 2.
A consensus SmcR-binding sequence. A, a total of 29 SmcR-binding DNA sequences were aligned, and the occurrences of certain residues (Gly, Ala, Thr, and Cys) in each position were counted. B, the occurrence of each nucleotide in that position is represented as a percentage of the indicated base in the selected population (top) and as a letter proportional in size to its frequency (bottom). C, the 22-bp consensus SmcR-binding sequence is shown, and its inverted repeat and conserved bases are indicated by arrows and boxes, respectively. The numbers represent positions apart from the center of the sequence. W, A, or T, R, G, or A; N, any base.
The Consensus SmcR-binding Sequence Increases Binding Affinity to SmcR—The sequence centered at –196.5 upstream of the transcription start site of PvvpE was the only SmcR binding site identified until this study (Fig. 3B) (16). The binding affinity of the SmcR consensus sequence we identified was compared with that of the PvvpE SmcR-binding site using EMSA. For this comparison, a working consensus sequence (WC; Fig. 3B), substituting 4 bases in positions 2, 5, 7, and 9 from the center of the 22-bp PvvpE SmcR-binding sequence with the bases conserved in corresponding positions of the consensus sequence, was developed and introduced into the region –207 to –186 from the transcription start site of the PvvpE. Thereby, the working consensus sequence still carries the bases found in the PvvpE SmcR-binding sequence unless they are conserved in the consensus sequence.
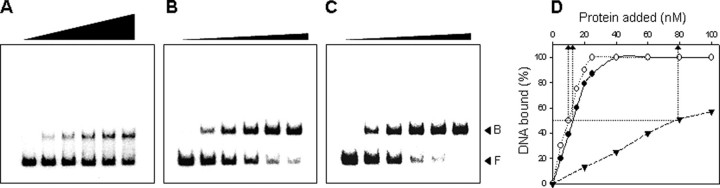
In an EMSA, the addition of SmcR at a concentration of 20 nm resulted in a shift of the 200-bp DNA fragment (from –300 to –101, upstream of vvpE) containing the PvvpE SmcR-binding sequence to a single band with a slower mobility (Fig. 4A). Based on the concentration of SmcR that was required to retard 50% of the labeled probe, it was estimated that the dissociation binding constant (Kd) for SmcR was ∼80 nm (Fig. 4D). In a second EMSA, SmcR was added to the same 200-bp DNA fragment but containing the working consensus sequence (Fig. 3B) rather than the PvvpE SmcR-binding sequence. SmcR also displayed specific binding to the DNA with the working consensus sequence (Fig. 4B), and it was estimated that the Kd for SmcR was ∼15 nm, which represents a 5-fold increase in binding affinity compared with that of PvvpE (Fig. 4B). SmcR binding efficiency to the dyad symmetric sequence (WCPAL; Fig. 3B) that was generated by replacing less conserved IR-R sequence with the sequence of IR-L was also compared by EMSA (Fig. 4C). The Kd for the dyad symmetric sequence was slightly lower than the Kd for the working consensus sequence, indicating that SmcR binds to the dyad symmetric sequence with higher affinity. The affinity of SmcR to the DNA therefore increases as the binding sequences contain more bases conserved in the consensus sequence.
FIGURE 4.
SmcR binds to the consensus sequence with higher affinity. A 200-bp DNA fragment of the upstream region of vvpE either with the PvvpE SmcR-binding sequence (A), with the working consensus sequence (B), or with the dyad symmetric sequence (C) were radioactively labeled and then used as a probe DNA. The labeled fragments were mixed with increasing amounts of SmcR (0, 20, 40, 60, 80, and 100 nm for A; 0, 5, 10, 15, 20, and 25 nm for B and C in the first through sixth lanes) and then resolved on a 4% polyacrylamide gel. B, bound DNA; F, free DNA. D, the relative affinities of SmcR with the PvvpE, the working consensus, and the dyad symmetric sequence were compared using the data from A, B, or C, respectively. The concentration of bound DNA was calculated and plotted against the concentration of the protein added. Each arrow points to the position of half-maximal binding corresponding to the Kd. ▾, PvvpE SmcR-binding sequence; •, working consensus sequence; ○, dyad symmetric sequence.
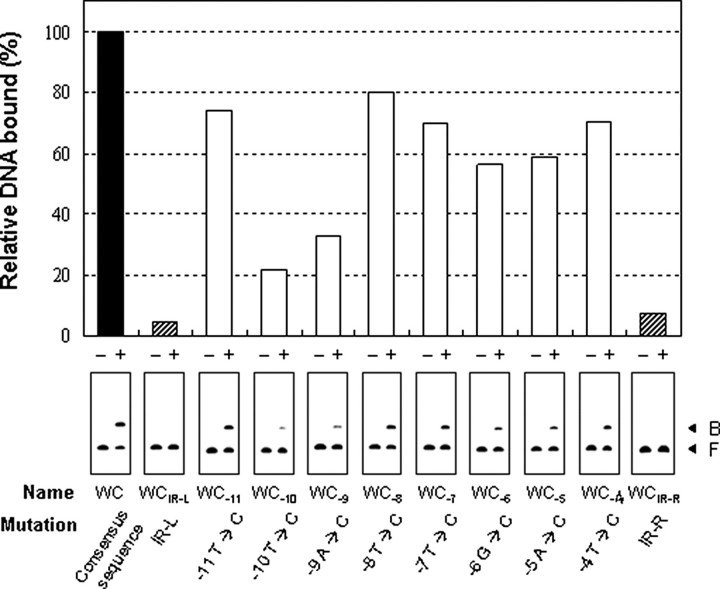
Mutational Analysis of the Consensus SmcR-binding Sequence—To evaluate the significance of the individual bases in the consensus SmcR-binding sequence, the working consensus sequence was altered two ways. In one, 10 bp including each half of the inverted repeat (TTATTGATAA or TTGTGAATAA) were replaced with 10 bp including a KpnI site (CAGGTACCTC). In the other, conserved bases in each position were individually substituted with bases that appeared in the corresponding position with the least frequency (Fig. 3B). The relative affinities of SmcR to the mutant sequences were determined by EMSA using the labeled 200-bp DNA of the vvpE upstream region containing either the working consensus sequence as a positive control or the mutant sequences (Fig. 5). Based on the intensity of the shifted band, the binding affinities of SmcR to the mutant sequences constructed by altering half of the inverted repeats (WCIR-L or WCIR-R; Fig. 3B) were decreased; the levels were less than 10% of that with the working consensus sequence (Fig. 5). This indicates that both halves of the inverted repeats are necessary for SmcR to bind with complete efficiency. Mutations in any bases conserved in the left half of the inverted repeat decreased the binding affinity to levels below 80% of that to the working consensus sequence. However, mutations of individual bases positioned at –9 and –10 from the center of the sequence were the most detrimental (Fig. 5). The effects of mutations in the bases conserved in the right half were also similar to those observed with mutations in the left half (data not shown). Overall, these results indicated that although all bases in the inverted repeats are important, the 9th and 10th bases from the center of the consensus sequence are the most important for SmcR binding.
FIGURE 5.
Mutational analysis of the working consensus sequence. A 200-bp DNA fragment of the upstream region of vvpE with either the working consensus sequence or its mutant sequences listed in Fig. 3B was radioactively labeled and then used as a probe DNA. In each panel of EMSA, no SmcR (–) or 50 nm SmcR (+) was added to the probe. After reaction, samples were resolved on a 4% polyacrylamide gel (bottom). The relative amounts of bound DNA, calculated by using the amount of bound DNA in the EMSA performed with the working consensus sequence as 100, were presented at the top of each panel. B, bound DNA; F, free DNA.
Prediction of the SmcR Regulon Using the Consensus SmcR-binding Sequence—Among 29 experimentally obtained SmcR-binding sequences, only 18 sequences existed in V. vulnificus CMCP6 genome. When the HMM profile summarizing 18 sequences was searched against the genome, 181 potential sequences were discovered. Among them, only 82 sequences were determined as the candidates of SmcR-binding sites, because the first positions of their sequences were located within the regulatory regions of genes on the genome. The searches of the candidates against the BioCyc data base (available on the World Wide Web) revealed that the 82 promoters matched well with the HMM profile were positioned in the upstream region of 65 monocistronic genes and 17 polycistronic operons comprising 56 genes. The predicted 82 genes and operons (a total of 121 genes) are distributed throughout the two chromosomes of V. vulnificus. A complete list of names or locus tags of the 121 genes is shown in Table 2. Although the majority of predicted genes are of putative or unknown function, some belonged to functional classes. Among them, nine categories, such as amino acid transport and metabolism, cell motility, cell wall/membrane biogenesis, inorganic ion transport and metabolism, nucleotide transport and metabolism, post-translational modification, signal transduction, transcription, and translation, contain at least three genes, respectively (Table 2 and supplemental Table 1).
TABLE 2.
Genes of the V. vulnificus SmcR regulon identified using the consensus SmcR-binding sequence
Functional categories, gene names, and locus tag numbers are based on the database of the V. vulnificus CMCP6 genome. Genes and locus tags likely in an operon are indicated in boldface type.
| Functional category | Gene or locus taga |
|---|---|
| Amino acid transport and metabolism | VV1_1370,bVV1_1371 VV1_1372 VV1_1373 VV1_1374 |
| Carbohydrate transport and metabolism | VV2_1326 |
| Cell motility | VV1_1950bVV1_1951 VV1_1952 |
| Cell wall/membrane biogenesis | VV1_1103, VV2_0929 VV2_0928 |
| Coenzyme transport and metabolism | VV1_2257 |
| Defense mechanisms | VV2_1089b |
| Function unknown | VV1_1265, VV1_1471, VV1_1702, VV2_0057 VV2_0058, VV2_0279, VV2_0517, VV2_1111, VV2_1541 VV2_1542 VV2_1543 VV2_1544 |
| General function prediction only | VV1_0300, VV1_0840 VV1_0839 VV1_0838 VV1_0837, VV1_1085, VV1_1456 VV1_1455 VV1_1454 VV1_1453 mafVV1_1451 VV1_1450, VV1_2358, VV1_2976bVV1_2975 VV1_2974 VV1_2973 VV1_2972, VV2_0856, VV2_0970, VV2_1270, VV2_1527 |
| Inorganic ion transport and metabolism | VV1_0842bVV1_0843 VV1_0844 VV1_0845, VV1_2805, VV2_1106 |
| Intracellular trafficking and secretion | VV1_3112 |
| Nucleotide transport and metabolism | VV1_0302, VV1_1635 VV1_1636 |
| Posttranslational modification, protein turnover, chaperones | VV1_0453, VV2_0020,b VV2_1650 |
| Replication, recombination and repair | VV2_1406,b VV2_1643 |
| Signal transduction mechanisms | VV1_1489, VV1_1931, VV1_2096,b VV1_2211, VV1_2868 VV1_2867 VV1_2866, VV1_2911, VV2_0528, VV2_0823, VV2_0825 |
| Transcription | slmA VV1_0830,greA, VV2_0446, VV2_0981, VV2_1391b |
| Translation | VV1_1270 VV1_1271, VV1_2397, VV1_3016 |
| Not in COGsc | VV1_0464, VV1_0522, VV1_0644, VV1_1215 VV1_1216, VV1_1401, VV1_1472 VV1_1473, VV1_1720, VV1_2362, VV1_2380, VV1_2430, VV1_2726, VV1_2957 VV1_2958, VV2_0067, VV2_0133, VV2_0250,bVV2_0364 VV2_0363 VV2_0362 VV2_0361 VV2_0360 VV2_0359, VV2_0400, VV2_0414, VV2_0429, VV2_0658, VV2_1035, VV2_1046, VV2_1147, VV2_1179, VV2_1201, VV2_1281, VV2_1371, VV2_1398 |
See supplemental Table 1 for the annotation of product of each gene.
Binding of SmcR to the upstream region was experimentally verified by EMSA.
Clusters of orthologous group.
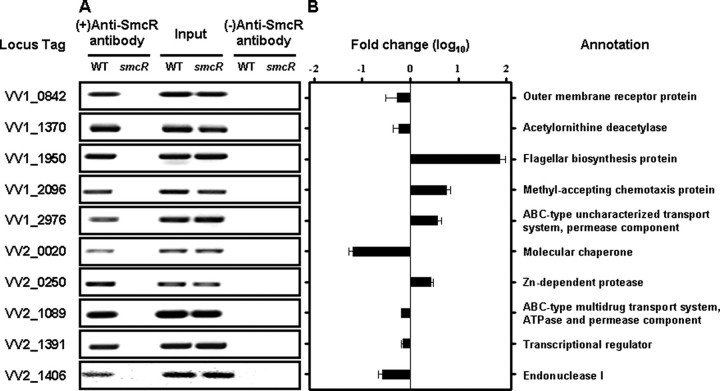
Verification of the SmcR Regulation of the Predicted Genes in Vivo—Many of the genes we predicted were not previously reported to be SmcR-regulated. Therefore, SmcR regulation of the newly predicted genes was experimentally verified. Ten genes were randomly chosen from the pool of the 82 predicted SmcR-regulated genes (operons), and binding of SmcR to their upstream region was examined in vitro. EMSA revealed that SmcR binds to the upstream region of all of the genes that were tested (Fig. 6), suggesting that the genes are under direct control of SmcR.
FIGURE 6.
Verification of SmcR binding to the regulatory region of newly identified genes as SmcR regulon. Ten genes were randomly chosen from the pool of the newly predicted SmcR regulon members, and binding of SmcR to their upstream region was examined by EMSA. Each of the DNA fragments was radiolabeled, mixed with 200 nm SmcR, and then resolved on a 4% polyacrylamide gel. EMSAs were performed in the presence of 0.1 μg of poly(dI-dC) as a nonspecific competitor. For details, see “Experimental Procedures.” –, without SmcR; +, with SmcR. The same locus tag numbers that appear in Table 2 and supplemental Table 1 are at the top of each panel. B, bound DNA; F, free DNA.
To determine whether SmcR binds to the 10 promoters in vivo, the cross-linked chromatin from the wild type and smcR mutant HS031 cells was immunoprecipitated using the anti-SmcR antibody. As positive controls, the input chromatin from both the wild type and HS031 appeared to carry the promoter DNAs (Fig. 7A). After reversing the cross-links, the promoter fragments were detected in the chromatin precipitate from the wild type, induced with the anti-SmcR antibody, based on a PCR using the primers listed in supplemental Table 2. The presence of the promoter DNAs in the precipitated chromatin was caused by the specific binding of the SmcR protein to the DNA, since none of the promoter DNAs was detected in the precipitate induced in the absence of the anti-SmcR antibody. Consistent with this, no detectable level of the promoter fragments was observed in the anti-SmcR immunoprecipitate of the smcR mutant HS031 (Fig. 7A), indicating that the SmcR protein directly binds to all of the promoters tested in V. vulnificus.
FIGURE 7.
SmcR binding to the newly identified genes and their regulation by SmcR in vivo. Ten genes were randomly selected from the pool of the SmcR regulon members predicted by HMM profile. SmcR binding to their promoters and regulation of their transcription by SmcR were confirmed by chromatin immunoprecipitation experiments and quantitative RT-PCR. A, the cells were cross-linked, washed, then sonicated to produce sheared chromatin, as described elsewhere (28). The DNA was purified from the sheared chromatins before precipitated (input, positive control) and after precipitation with the protein A-Sepharose in the presence (+) or absence (–) of the anti-SmcR antibody. The DNA was then amplified by a PCR using primers specific to the promoters as listed in supplemental Table 2. WT, wild type; smcR, smcR mutant HS031. B, for the quantitative RT-PCR analysis, the expression level of the 10 genes was normalized to 16 S rRNA expression level. Averages and S.E. were calculated from at least three independent experiments. Details for preparation of total cellular RNA, chromatin immunoprecipitation analyses, and RT-PCR are given under “Experimental Procedures.” Locus tags are based on the data base of the V. vulnificus CMCP6 genome, which was retrieved from GenBank™ (AE016795; AE016796), and the products of the 10 genes are presented on the right.
Regulation of the 10 genes by SmcR was reexamined using a quantitative RT-PCR. Quantitative RT-PCR revealed that SmcR regulates transcription of all of the genes examined, of which VV2_1391 was 1.4-fold up-regulated and VV1_1950 was 73-fold down-regulated by SmcR (Fig. 7B), suggesting that most of the SmcR regulons predicted by HMM profile on the basis of the 22-bp SmcR consensus sequence are indeed regulated by SmcR in V. vulnificus. These results indicated that the consensus SmcR-binding sequence is valid and useful for genome-wide prediction of SmcR-binding sites and identification of genes potentially regulated by the SmcR quorum-sensing system.
DISCUSSION
LuxR homologues, such as V. cholerae HapR, V. parahemeolyticus OpaR, V. anguillarum VanT, and V. vulnificus SmcR, are global regulators controlling numerous genes contributing to pathogenesis as well as survival of the pathogenic Vibrio spp. (9–11, 16–19); however, there have only been a few studies on the molecular mechanism by which the proteins modulate the expression of the target genes (16, 20, 21). To date, the promoter(s) of the limited number of genes regulated by LuxR homologues has been reported, and few definitive analyses of the sequences upstream of the promoters have been previously performed (16, 20, 21). Therefore, the question of whether the LuxR homologues bind to any definite specific binding sequences for regulation of the promoters has not yet been addressed.
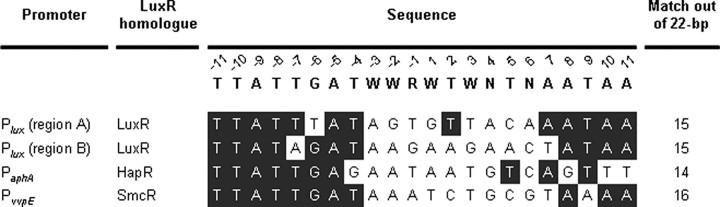
This study has identified that SmcR binds to a 22-bp consensus sequence with an inverted repeat (Fig. 2C). This inverted repeat indicated that the protein binds to the sequence as a dimer or tetramer, and we have obtained several lines of evidence indicating that SmcR is present as a dimeric form in vitro and in vivo.5 Consistent with this, the crystal structure of V. cholerae HapR has been determined and reveals a dimeric protein with an N-terminal DNA binding domain (32). The amino acid sequences of HapR and SmcR are nearly identical (72%; 146 of 203 amino acids), and their identity is spread evenly throughout the whole proteins (data not shown). Mutational analysis of the consensus sequence indicated that the 9th and 10th nucleotides from the center are the most important for SmcR binding (Fig. 5). It is noted that the 9th and 10th nucleotides are also well conserved in inverted repeats of the sequences for binding of other LuxR homologues (Figs. 2C and 8). Although information about the co-crystal structure of SmcR (even HapR) bound to the consensus sequence DNA is not yet available, the high level of identity observed in amino acid sequences of SmcR and HapR suggests that the 9th and 10th nucleotides would be essential for interaction with the N-terminal DNA binding domain of SmcR.
FIGURE 8.
Comparison of the consensus SmcR-binding sequence and known LuxR (homologue)-binding sequences. The consensus SmcR-binding sequence determined in the present study is shown on the top. Sequences from the two LuxR binding sites of the V. harveyi lux promoter (Plux) (21), from the HapR binding site of V. cholerae aphA promoter (PaphA) (20), and from the SmcR-binding site of V. vulnificus PvvpE (16) are aligned below. Individual bases identical to those conserved in the consensus SmcR-binding sequence are highlighted. Numbers of bases that match with those of the consensus SmcR-binding sequence are indicated on the right.
SmcR and LuxR homologues of Vibrio spp. exhibit high levels of identity (72–92% in amino acid sequences) (8), indicating that binding sequences of the LuxR homologues could be similar to that of SmcR. The sequences that were previously known for binding of LuxR homologues were aligned, and their sequences revealed substantial levels of match (over 16 of 22 residues) with the consensus SmcR-binding sequence (Fig. 8). However, it is noteworthy that the aligned sequences are also less conserved in the right half of the repeat. These less conserved bases in the right half of the sequences would permit less tight binding. The known binding sites of SmcR and LuxR are unusually distant from the promoter (16, 21). The SmcR binding site is centered at 196.5 bp upstream of the PvvpE (16), and LuxR bindings at region A (centered at –251.5) and at region B (centered at –115.5) for activation of the luxCDABEGH operon in V. harveyi are also exceptionally distant (21). Generally, activators binding this far upstream of the promoter are not able to activate RNAP directly and rather cooperate and interact with additional transcriptional regulator(s) on the promoter DNA. As such, the additional regulatory proteins convey the activator's signal to RNAP and/or induce structural changes of the DNA (forming a DNA loop) to bring the activators to RNAP (33, 34). We previously demonstrated that SmcR also activates PvvpE at a distance through cooperating and interacting with other regulatory proteins, such as IHF and CRP (16).6 One possible hypothesis is that SmcR on PvvpE could be more flexible when it binds less tightly to the vvpE SmcR-binding site and that the flexibility could permit higher activity of PvvpE by supporting the protein to cooperate and interact with other regulatory proteins on the promoter DNA as mentioned above. To examine this hypothesis, activities PvvpE were measured in vivo using PvvpE::lacZ transcriptional fusion reporters. The activity of PvvpE with the PvvpE SmcR-binding sequence, which has less conserved bases in the right half, was higher than that of PvvpE with the working consensus SmcR-binding sequence.6 However, it is our great limitation that PvvpE is the only promoter where the SmcR-promoter interaction is analyzed at a molecular level, and additional studies on the interaction between SmcR and other promoters are needed to confirm the hypothesis.
Previously, Waters and Bassler (7) failed to identify any consistent motif by alignment of sequences of the DNA region regulated directly by LuxR in vivo, and rather promiscuous DNA-binding capabilities for the LuxR(HapR)-type proteins were suggested. One possible explanation for the inability of identification of LuxR-binding consensus motifs is that LuxR also interacts and cooperates with other proteins on the promoters. Thus, the sequences for LuxR binding varied depending on the type of interactions (cooperations) and consequently appeared less conserved. Therefore, we used a cycle selection procedure consisting of in vitro DNA-SmcR (alone) binding, rather than in vivo binding, to identify the SmcR-binding sequences. Together with our observation, the results of Waters and Bassler (7) reflect that the interaction and cooperation with multiple regulatory proteins may be a common feature inherited in the regulation of genes by these LuxR-type proteins.
To date, comparison of protein or transcription profiles of wild type and quorum-sensing mutants has been used to identify genes regulated by LuxR homologues and the quorum-sensing system (7, 12, 13, 22). However, the genes identified by these approaches may be regulated indirectly as well as directly by LuxR homologues. Furthermore, procedures such as proteomics and microarray analysis are dependent on the transcription and expression levels of the genes. The procedures identify genes expressed to substantial levels on the conditions used in the study and may not identify genes that are expressed only in certain conditions. Therefore, the use of bioinformatics and the consensus LuxR-binding sequence in a genome-wide search for target genes would have advantages that are complementary to the comparison of expression profiles. The HMM approach for the prediction of SmcR binding sites within the whole genome of V. vulnificus enabled us to assign 121 genes that are possibly regulated by the direct interaction of SmcR at the promoter (Table 2 and supplemental Table 1). Although several of these genes involved in cellular processes essential for bacterial pathogenesis, such as motility and envelope biogenesis, were earlier expected to be SmcR-regulated (18, 19), many of these genes are identified for the first time to be regulated by SmcR. Functional characterization of these newly identified genes would lead to further insight into the role of the quorum-sensing regulatory system in various aspects of V. vulnificus pathogenesis.
In summary, alignment of SmcR-binding sequences of the DNA fragments enriched by direct binding of SmcR revealed a 22 bp consensus SmcR-binding sequence with an 8 bp inverted repeat, indicating that SmcR binds to the DNA as a dimeric form. Using the HMM approach, we identified 121 genes that are potentially under the direct control of SmcR, and chromatin immunoprecipitation experiments and quantitative RT-PCR suggested that most of the newly identified SmcR regulons are indeed regulated by SmcR in V. vulnificus. This type of identification of genes regulated by other LuxR homologues, if experimentally verified, would facilitate the definitive analysis of the role of the proteins in the quorum-sensing regulatory cascade in various pathogenic bacteria.
Supplementary Material
This study was supported by grants from the 21C Frontier Microbial Genomics and Applications Center Program, Ministry of Education, Science, and Technology, the MarineBio 21 Project, Ministry of Maritime Affairs and Fisheries, and National Research Laboratory Grant, Korea Science and Engineering Foundation, R0A-2007-000-20039-0, Republic of Korea (to S. H. C.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1 and Tables 1 and 2.
Footnotes
The abbreviations used are: EMSA, electrophoretic mobility shift assay; HMM, hidden Markov model; RT, real time; WC, working consensus sequence.
S. H. Choi, unpublished data.
S. H. Choi, unpublished data.
References
- 1.Mekalanos, J. J. (1992) J. Bacteriol. 174 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miller, J. F., Mekalanos, J. J., and Falkow, S. (1989) Science 243 916–922 [DOI] [PubMed] [Google Scholar]
- 3.Fuqua, C., and Greenberg, E. P. (2002) Nat. Rev. Mol. Cell Biol. 3 685–695 [DOI] [PubMed] [Google Scholar]
- 4.Waters, C. M., and Bassler, B. L. (2005) Annu. Rev. Cell Dev. Biol. 21 319–346 [DOI] [PubMed] [Google Scholar]
- 5.de Kievit, T. R., and Iglewski, B. H. (2000) Infect. Immun. 68 4839–4849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miller, M. B., and Bassler, B. L. (2001) Annu. Rev. Microbiol. 55 165–199 [DOI] [PubMed] [Google Scholar]
- 7.Waters, C. M., and Bassler, B. L. (2006) Genes Dev. 20 2754–2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McDougald, D., Rice, S. A., and Kjelleberg, S. (2000) Gene (Amst.) 248 213–221 [DOI] [PubMed] [Google Scholar]
- 9.Jobling, M. G., and Holmes, R. K. (1997) Mol. Microbiol. 26 1023–1034 [DOI] [PubMed] [Google Scholar]
- 10.McCarter, L. L. (1998) J. Bacteriol. 180 3166–3173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Croxatto, A., Chalker, V. J., Lauritz, J., Jass, J., Hardman, A., Williams, P., Camara, M., and Milton, D. L. (2002) J. Bacteriol. 184 1617–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu, J., Miller, M. B., Vance, R. E., Dziejman, M., Bassler, B. L., and Mekalanos, J. J. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 3129–3134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beyhan, S., Bilecen, K., Salama, S. R., Casper-Lindley, C., and Yildiz, F. H. (2007) J. Bacteriol. 189 388–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Higgins, D. A., Pomianek, M. E., Kraml, C. M., Taylor, R. K., Semmelhack, M. F., and Bassler, B. L. (2007) Nature 450 883–886 [DOI] [PubMed] [Google Scholar]
- 15.Roh, J. B., Lee, M. A., Lee, H. J., Kim, S. M., Cho, Y., Kim, Y. J., Seok, Y. J., Park, S. J., and Lee, K. H. (2006) J. Biol. Chem. 281 34775–34784 [DOI] [PubMed] [Google Scholar]
- 16.Jeong, H. S., Lee, M. H., Lee, K. H., Park, S. J., and Choi, S. H. (2003) J. Biol. Chem. 278 45072–45081 [DOI] [PubMed] [Google Scholar]
- 17.Shao, C. P., and Hor, L. I. (2001) J. Bacteriol. 183 1369–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McDougald, D., Rice, S. A., and Kjelleberg, S. (2001) J. Bacteriol. 183 758–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee, J. H., Rhee, J. E., Park, U., Ju, H. M., Lee, B. C., Kim, T. S., Jeong, H. S., and Choi, S. H. (2007) J. Microbiol. Biotechnol. 17 325–334 [PubMed] [Google Scholar]
- 20.Kovacikova, G., and Skorupski, K. (2002) Mol. Microbiol. 46 1135–1147 [DOI] [PubMed] [Google Scholar]
- 21.Swartzman, E., and Meighen, E. A. (1993) J. Biol. Chem. 268 16706–16716 [PubMed] [Google Scholar]
- 22.Yildiz, F. H., Liu, X. S., Heydorn, A., and Schoolnik, G. K. (2004) Mol. Micribiol. 53 497–515 [DOI] [PubMed] [Google Scholar]
- 23.Ochsner, U. A., and Vasil, M. L. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 4409–4414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee, J. H., and Choi, S. H. (2006) Mol. Microbiol. 60 513–524 [DOI] [PubMed] [Google Scholar]
- 25.Lee, J. H., Kim, M. W., Kim, B. S., Kim, S. M., Lee, B. C., Kim, T. S., and Choi, S. H. (2007) J. Microbiol. 45 146–152 [PubMed] [Google Scholar]
- 26.Sambrook, J., and Russell, D. W. (2001) Molecular Cloning: A Laboratory Manual, 3rd Ed., pp. 13.75–13.77, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY
- 27.Karp, P. D., Ouzounis, C. A., Moore-Kochlacs, C., Goldovsky, L., Kaipa, P., Ahren, D., Tsoka, S., Darzentas, N., Kunin, V., and Lopez-Bigas, N. (2005) Nucleic Acids Res. 33 6083–6089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rhee, J. E., Kim, K. S., and Choi, S. H. (2005) J. Bacteriol. 187 7870–7875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jeong, H. G., and Choi, S. H. (2008) J. Bacteriol. 190 3768–3773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee, B. C., Lee, J. H., Kim, M. W., Kim, B. S., Oh, M. H., Kim, K. S., Kim, T. S., and Choi, S. H. (2008) Infect. Immun. 76 1509–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crooks, G. E., Hon, G., Chandonia, J. M., and Brenner, S. E. (2004) Genome Res. 14 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De Silva, R. S., Kovacikova, G., Lin, W., Taylor, R. K., Skorupski, K., and Kull, F. J. (2007) J. Bacteriol. 189 5683–5691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McLeod, S. M., and Johnson, R. C. (2001) Curr. Opin. Microbiol. 4 152–159 [DOI] [PubMed] [Google Scholar]
- 34.Browning, D. F., and Busby, S. J. W. (2004) Nat. Rev. Microbiol. 2 57–65 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.