Abstract
The regulation of phosphatidylserine (PS) distribution across the plasma membrane of eukaryotic cells has been implicated in numerous cell functions (e.g. apoptosis and coagulation). In a recent study, fluorescent phospholipids labeled in the acyl chain with 7-nitrobenz-2-oxa-1, 3-diazol-4-yl (NBD) were used to identify two members of the P4 subfamily of P-type ATPases, Dnf1p and Dnf2p, that are necessary for the inward-directed transport of phospholipids across the plasma membrane (flip) of yeast (Pomorski, T., Lombardi, R., Riezman, H., Devaux, P. F., Van Meer, G., and Holthuis, J. C. (2003) Mol. Biol. Cell 14,1240 -1254). Herein, we present evidence that the flip of NBD-labeled PS (NBD-PS) across the plasma membrane does not require the expression of Dnf1p or Dnf2p. In strains in which DNF1 and DNF2 are both deleted, the flip of NBD-PS is increased ∼2-fold over that of the isogenic parent strain, whereas the flip of NBD-labeled phosphatidylcholine and NBD-labeled phosphatidylethanolamine are reduced to ∼20 and ∼50%, respectively. The mechanism responsible for NBD-PS flip is similar to that for NBD-labeled phosphatidylcholine and NBD-labeled phosphatidylethanolamine in its dependence on cellular ATP and the plasma membrane proton electrochemical gradient, as well as its regulation by the transcription factors Pdr1p and Pdr3p. Based on the observation that deletion or inactivation of all four members of the DRS2/DNF essential subfamily of P-type ATPases does not affect NBD-PS flip, we conclude that the activity reflected by NBD-PS internalization is not the essential function of the DRS2/DNF subfamily of P-type ATPases.
The most common phospholipids that comprise the plasma membrane of eukaryotic cells are asymmetrically distributed between the two leaflets of the bilayer (for recent reviews see Refs. 1-5). This nonrandom distribution is maintained in a dynamic steady state in part by the activity of translocases that move phospholipids in the inward (referred to as flip) and outward (referred to as flop) direction across the plasma membrane. The net result in most actively growing cells is to sequester the majority of the amino phospholipids (e.g. phosphatidylserine and phosphatidylethanolamine) on the inner cytoplasmic leaflet and the choline phospholipids (e.g. phosphatidylcholine and sphingomyelin) on the outer exoplasmic leaflet. As cells age or when triggered into apoptosis, the phospholipids of the plasma membrane are redistributed toward equilibrium resulting in the exposure of phosphatidylserine on the exoplasmic surface. In vertebrates the presence of phosphatidylserine on the surface of apoptotic cells is a signal for clearance by macrophages.
Phospholipids with fluorescent or spin-labeled reporter groups attached to their sn2 acyl chain have been used to investigate the mechanisms by which phospholipids are transported across the plasma membrane (4, 6-9). Using this approach with Saccharomyces cerevisiae, members of two classes of proteins have been identified that are required for phospholipid flip across the plasma membrane. The first to be identified was LEM3/ROS3, which is a member of the CDC50 family (10, 11). It is predicted to encode a membrane protein with two transmembrane domains. Deletion of LEM3/ROS3 produces a strain in which the flip of NBD2-labeled phosphatidylcholine (NBD-PC) is inhibited by ∼90%, that of NBD-PE is inhibited by ∼50%, and that of NBD-PS is unaffected. The second class of proteins comprises a subfamily of P-type ATPases. The subfamily 4 of the P-type ATPases includes two members, DNF1 and DNF2, reported to be required for normal flip of phosphatidylcholine, phosphatidylethanolamine, and phosphatidylserine as assayed by NBD-labeled fluorescent analogues (12). Deletion of DNF1 and DNF2 in combination was reported to almost completely inhibit the ATP-dependent flip of NBD-PC, NBD-PE, and NBD-PS.
In the current study, we present evidence that the flip of NBD-PS across the plasma membrane of S. cerevisiae does not require the expression of DNF1 or DNF2 suggesting that it is flipped across the plasma membrane by a different mechanism from that of NBD-PC and NBD-PE. Based on the observation that deletion or inactivation of all four members of the DRS2/DNF essential subfamily of P-type ATPases (13, 14) does not affect NBD-PS flip, we conclude that the activity reflected by NBD-PS internalization is not the essential function of the DRS2/DNF subfamily of P-type ATPases.
EXPERIMENTAL PROCEDURES
Materials—Yeast medium was purchased from Difco, Inc. NBD-PC, P-NBD-PC, NBD-PE, P-NBD-PE, and NBD-PS were purchased from Avanti Polar Lipids (Alabaster, AL). Lipids were stored at -20 °C and were monitored periodically for breakdown via TLC. Unless otherwise noted, all other materials were purchased from Sigma-Aldrich.
Yeast Strains and Culture—Strains used in this study were from our laboratory collection or kindly donated by Todd Graham (Vanderbilt University) (Table 1). Unless otherwise noted, all cells were grown to early log phase (A600 = 0.2-0.4) from overnight cultures in YPD (complete medium with 1.0% Bacto-yeast extract, 2% glucose, and 2% Bacto-peptone) or SDC (synthetic complete medium with 0.67% yeast nitrogen base, 2% glucose, complete amino acid supplements) media at 30 °C, as described before (15). For growth phase studies, cells were diluted to early log phase every 2 h and grown until 2 h after the last dilution was made. Confirmation of the deletion of DNF1 and DNF2 was done via PCR using KanB and KanC primers (Invitrogen) with the following sequences. Dnf1: AAATTTCACAGAGCAAAGTCTCACT (A) and TACGTATCACGGCTTGATTAATTCT (D); Dnf2: ATAGACAAACCAGATTCATCGCTTA (A) and CCGTAAAGTAGTTACTTGCCACAAT (D). For culture density experiments, at the end of the growing phase, cells were normalized to the lowest A600 value prior to labeling, to ensure the same number of cells was labeled with NBD-phospholipids.
TABLE 1.
Saccharomyces cerevisiae strains used in this study
| Strain | Genotype | Source/reference |
|---|---|---|
| BY4242 | MATα his3Δ1 leu2Δ0 ura3Δ0 lys2Δ0 | ATCC |
| BY4141 | MATa his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 | ATCC |
| LMY165 | MATα his3Δ1 leu2Δ0 ura3Δ0 lys2Δ0 dnf1Δ::KanMX4 | YER4242W BY4242 from ATCC |
| LMY166 | MATα his3Δ1 leu2Δ0 ura3Δ0 lys2Δ0 dnf2Δ::KanMX4 | YDR093W BY4242 from ATCC |
| HCY13 | MATa his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 dnf1Δ::KanMX4 dnf2Δ::KanMX4 | T. Graham (13) |
| HCY285 | MATα his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 dnf2Δ::KanMX4 dnf3Δ::KanMX4 | T. Graham (13) |
| HCY286 | MATα his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 dnf1Δ::KanMX4 dnf2Δ::KanMX4 dnf3Δ::KanMX4 | T. Graham (13) |
| HCY287 | MATa his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 dnf1Δ::KanMX4 dnf2Δ::KanMX4 dnf3Δ::KanMX4 | T. Graham (13) |
| HCY288 | MATa his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 dnf1Δ::KanMX4 dnf2Δ::KanMX4 drs2Δ::KanMX4 | T. Graham (13) |
| HCY290 | MATα his3Δ1 leu2Δ0 ura3Δ0 dnf3Δ::KanMX4 drs2Δ::KanMX4 | T. Graham (13) |
| HCY291 | MATa his3Δ1 leu2Δ0 ura3Δ0 dnf2Δ::KanMX4 dnf3Δ::KanMX4 drs2Δ::KanMX4 | T. Graham (13) |
| ZHY2149D | MATα his3 leu2 ura3 lys2 drs2Δ::KanMX4 dnf1Δ::KanMX4 | T. Graham (13) |
| PFY3272G | MATa his3 leu2 ura3 met15 dnf1Δ::KanMX4 dnf3Δ::KanMX4 | T. Graham (13) |
| ZHY708 | MATa his3 leu2 ura3 met15 dnf1Δ::KanMX4 dnf3Δ::KanMX4 drs2::LEU2 | T. Graham (13) |
| ZHY615D2A | MATα leu2 ura3 drs2Δ::KanMX4 dnf2Δ::KanMX4 | T. Graham (13) |
| ZHY409 | MATα his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 dnf1Δ::KanMX4 dnf2Δ::KanMX4 dnf3Δ::KanMX4 drs2Δ::LEU2 pRS313::DRS2 | T. Graham (37) |
| ZHY41-3A | MATα his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 dnf1Δ::KanMX4 dnf2Δ::KanMX4 dnf3Δ::KanMX4 drs2Δ::LEU2 pRS313::drs2-31 | T. Graham (37) |
| AGY72 | MATa ura3-52 leu2Δ1 his3Δ200 trp1Δ63 | Nichols lab (16) |
| AGY75 | MATa ura3-52 leu2Δ1 his3Δ200 trp1Δ63 pdr1Δ::TRP1 pdr3Δ::HIS3 | Nichols lab (16) |
| LKY118 | MATα can1-100 ade2-1 leu2-3-112 trp1-1 ura3-1 lys2 | Nichols lab (25) |
| LKY154 | MATα can1-100 ade2-1 leu2-3-112 trp1-1 ura3-1 lys2 PDR1-11 | Nichols lab (16) |
| LKY156 | MATα can1-100 ade2-1 leu2-3-112 trp1-1 ura3-1 lys2 pdr3-11 | Nichols lab (25) |
| LMY136 | MATa end4::LEU2 his4 leu2 ura3 lys2 bar1 | RH1965/299-1C from H. Riezman |
| LMY137 | MATa END4 his4 leu2 ura3 lys2 bar1 | RH448 from H. Riezman |
Drug Treatment—Collapse of the proton electrochemical gradient across the cell membrane was accomplished by treating cells with CCCP prior to labeling. CCCP was dissolved in 95% ethanol (5 mm stock) and added to cells at 50 μm final concentration for 10 min at 30 °C, prior to labeling. For ethanol experiments, early log phase cells were harvested and resuspended in SDC with varying percentages of ethanol (0-10% v/v) for a total volume of 0.5 ml and incubated 10 min at 30 °C, prior to labeling. For ATP depletion experiments, cells were pretreated with SC-azide for 10 min at 30 °C. Lipid was added directly to the media, and cells were incubated at either 2 °C or 30 °C. Latrunculin A was suspended in DMSO (5 mm stock) and added at a final concentration of 20 μm.
Internalization of Phospholipids into Yeast Cells—NBD-phospholipid internalization by yeast was performed as described previously (16). Briefly, NBD-phospholipids were dried down under a stream of nitrogen and resuspended in DMSO at 5 mm. NBD-phospholipids were added to early log phase cultures (A600 = 0.2-0.4) in 0.5 ml of SDC at a final concentration of 5 μm for NBD-PC and NBD-PE or 10 μm for NBD-PS. Cells were vortex-mixed and incubated for 30 min at 30 °C or for 1 h on ice (2 °C). Cells were centrifuged and washed three times with two volumes of ice-cold SC-azide to prevent lipid efflux. Because essentially no NBD-PS fluorescence was detected in the plasma membrane of washed cells (Fig. 1B), nor was additional NBD-PS fluorescence removed by washing with buffer containing bovine serum albumin (1 mg/ml), flow cytometric measurement of cell-associated fluorescence was interpreted to reflect net intracellular accumulation.
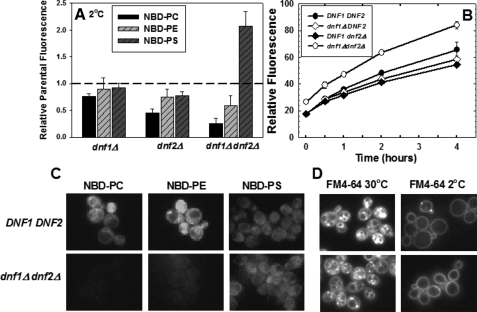
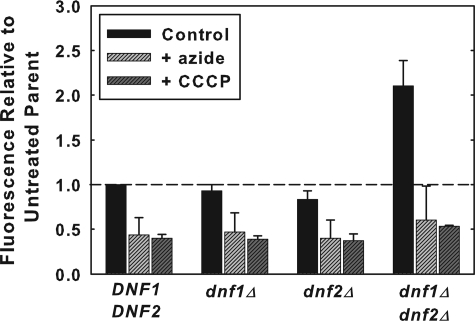
FIGURE 1.
NBD-PS flip is not dependent on Dnf1p or Dnf2p. A, early log phase cells, grown in YPD media (A600 = 0.2-0.4) were labeled with 5 μm NBD-PC or NBD-PE or 10 μm NBD-PS at 2 °C for 1 h. Cells were washed three times with ice-cold SC-azide and analyzed by flow cytometry as described under “Experimental Procedures.” The following strains were used: DNF1DNF2 parent strains BY4242 and BY4141; dnf1Δ (LMY165); dnf2Δ (LMY166); and dnf1Δdnf2Δ (HCY13). Data are normalized to parental NBD-phospholipid internalization of the appropriate mating type, although no significant differences were observed between mating types. The relative fluorescence following internalization of NBD-PC, NBD-PE, and NBD-PS in the parental strain BY4141 was 353 ± 33, 260 ± 22, and 146 ± 10, respectively. Data are the mean of at least three independent experiments, and the error bars represent the S.D. between experiments. B, time course of NBD-PS internalization measured by flow cytometry. The same strains used for panel A were labeled as described above, except the labeling time was varied from 0 to 4 h. C, fluorescence images of NBD-phospholipids internalized at 2 °C in DNF1DNF2 (BY4141) and dnf1Δdnf2Δ (HCY13) strains. Images were scaled identically and are representative of the entire field. D, fluorescence images of FM4-64 internalization in the same strains used in C at 2 °C and 30 °C. Cells were grown as in A and labeled with 20 μm FM4-64 (16.5 mm stock in DMSO) for 30 min at 2 °C or 30 °C before washing and imaging. The intensity of the images acquired at 2 °C was increased 2.5 times that of the 30 °C images.
Flow Cytometry—Samples were analyzed by flow cytometry (FACSCalibur cytometer BD Biosciences) as previously described (17). To gate dead cells from the analysis, 10 μl of 50 μg/ml stock of propidium iodide was added to ∼4 × 105 NBD-phospholipid-labeled cells in ∼100 μl of SC-azide immediately prior to analysis. Readings were taken on the flow cytometer equipped with an argon laser at 488 nm. CellQuest software (BD Biosciences) and FlowJo (Tree Star, Inc.) were used to analyze and quantify the data. For comparison of deletion strains with parent cells, gating was performed for the analysis between forward scatter of 150-750 to ensure that similar sized cells were compared.
Fluorescence Microscopy—Fluorescence microscopy was performed on a Zeiss Axiovert 135 microscope complete with barrier filters allowing for NBD-phospholipid detection. Filter sets were obtained from Chroma Technology Corp. A multi-band dichroic mirror (8600 BS) was used to detect the NBD fluorophore at a filter combination exciter S490/20 and emitter S528/38. The images were captured by a Qimaging Retiga Exi charge-coupled device camera and analyzed via Metamorph software (Universal Imaging). All images compared in the same figure were adjusted to the same background and intensity and are representative of the entire population of cells.
RESULTS
NBD-PS Flip Is Not Dependent on Dnf1p or Dnf2p Expression—The rate of NBD-labeled phospholipid uptake into S. cerevisiae is determined by three independently regulated processes: the rate of unidirectional influx across the plasma membrane (referred to as flip), the rate of unidirectional efflux across the plasma membrane (referred to as flop), and the rate of internalization by endocytosis (8). Previous studies have shown that the net uptake of the NBD-PC and NBD-PE at 2 °C is dependent solely on the process of flip. This is because both fluid phase and receptor-mediated endocytosis (18-20) and outward-directed transport across the plasma membrane (flop) (16) are essentially blocked at this temperature. The low temperature block of endocytosis was confirmed by measuring the uptake of the endocytosis marker, FM4-64, in the two primary strains used in this study (Fig. 1D). We also confirmed that, like NBD-PC and NBD-PE, NBD-PS is rapidly effluxed (flopped) (t½ ≈ 20 min) at 30 °C and is essentially blocked at 2 °C (data not shown). Measurement of the time course of the low temperature internalization of the three NBD-labeled phospholipids demonstrated that after 1 h of labeling, the internalization had not reached equilibrium and was still increasing almost linearly (Fig. 1B). Thus, under the conditions used in the following experiments, the net uptake of NBD-PS at 2 °C reflects its unidirectional influx or flip across the plasma membrane.
To determine the extent to which the expression of DNF1 and DNF2 are required for the flip of these NBD-labeled phospholipid reporters, the net uptake of NBD-PC, NBD-PE, and NBD-PS at low temperature, which we will refer to as low temperature flip, was measured in strains in which DNF1 and DNF2 were deleted alone and in combination. The results (Fig. 1A) indicated that the low temperature flip of NBD-PC and NBD-PE was moderately reduced in either the dnf1Δ or dnf2Δ strains. However, when both genes were deleted, NBD-PC internalization was reduced to ∼20% and NBD-PE internalization was reduced to ∼50% of its isogenic parent strain. These results confirm the conclusion of Pomorski et al. (12) that Dnf1p and Dnf2p are required for normal levels of low temperature flip of NBD-PC and NBD-PE. However, contrary to the 50% reduction in NBD-PS internalization observed by Pomorski et al. (12), the internalization of NBD-PS in a dnf1Δdnf2Δ strain was actually increased ∼2-fold (Fig. 1A). Thus, the expression of DNF1 and DNF2 is not required for the low temperature flip of NBD-PS.
Observation of the cells labeled at 2 °C by fluorescence microscopy confirmed the inhibition of NBD-PC and NBD-PE flip and the enhancement of NBD-PS flip in the dnf1Δdnf2Δ strain (Fig. 1C). Although the amount of NBD-PS internalization is not sufficient to produce high resolution images of its intracellular location, it is sufficient to rule out any significant transfer to the vacuole. Thus, the increased internalization is not the result of excess NBD-PS accumulation in the vacuole in the dnf1Δdnf2Δ mutant strain.
To rule out the possibility that the observed differences in NBD-phospholipid internalization resulted from differences in the extent of degradation of the NBD-labeled phospholipids, lipids were extracted from cells following labeling with NBD-PC, NBD-PE, or NBD-PS at 2 °C and analyzed by silica gel TLC. No significant breakdown of any of the reporter phospholipids in either the dnf1Δdnf2Δ strain or its isogenic parent was detected (data not shown). Thus, we concluded that all three NBD-phospholipids were internalized intact and that differences in degradation do not explain the observed increase in flip in the dnf1Δdnf2Δ mutant strain.
The NBD-PC and NBD-PE analogues that were used for the experiments presented in Fig. 1 differ from the NBD-PS analogue in the length of the sn-1 acyl chain. The NBD-PC and NBD-PE analogues have a myristoyl acyl chain in the sn-1 position, whereas NBD-PS has a palmitoyl acyl chain at that position. Since the first application of NBD-labeled phospholipids in yeast (21), the myristoyl analogue has been used because it increased the amount of internalization and improved the resolution of fluorescence microscopic images. NBD-PS with a myristoyl acyl chain is not commercially available and is difficult to synthesize in reasonable quantities. Thus, NBD-PS with a palmitoyl acyl chain has been used in most previous published studies (11, 12, 22-24) and was used in the experiments presented herein. To address the possibility that the observed increase in NBD-PS flip compared with NBD-PC in the dnf1Δdnf2Δ strain is the result of the different acyl chain lengths, we repeated the flow cytometry internalization experiment with the same three NBD-phospholipids with palmitoyl at the sn-1 position. The palmitoyl analogues are abbreviated P-NBD-PC and P-NBD-PE. The low temperature flip of P-NBD-PC was 0.49, P-NBD-PE was 0.82, and NBD-PS was 1.52 in the dnf1Δdnf2Δ strain relative to its isogenic parent strain. The addition of two carbons to the sn-1 acyl chain reduced the extent to which the low temperature flip of the PC and PE analogues was reduced in the dnf1Δdnf2Δ strain. However, flip was reduced for both of these analogues, whereas the low temperature flip of NBD-PS was increased. We concluded that the longer acyl chain in the NBD-PS analogue does not explain the dramatic increase in flip in the dnf1Δdnf2Δ strain.
There are two significant differences in experimental protocol that may explain why Pomorski et al. (12) did not observe increased NBD-PS flip in the dnf1Δdnf2Δ strain. First, our experiments were performed on early logarithmic phase cultures (A600 ≈ 0.2-0.4) as opposed to mid logarithmic phase cultures (A600 ≈ 0.5-1.0). Secondly, we labeled a smaller number of cells with a lower concentration of NBD-PS. To assure that the discrepancy was not the result of these experimental differences, we repeated the experiment using the same conditions as the Pomorski group and obtained a 1.5- to 2.0-fold increase of NBD-PS flip in the dnf1Δdnf2Δ strain relative to the isogenic parent. We concluded that the observed discrepancy in NBD-PS flip in the dnf1Δdnf2Δ strain was not a result of these different experimental conditions.
We did note that when early log phase cells were grown in SDC as opposed to YPD, the relative NBD-PS flip was less, ∼1.1-fold as opposed to ∼2-fold increase in the dnf1Δdnf2Δ strain (see Fig. 6). Under no conditions did we observe a decrease of NBD-PS flip in the dnf1Δdnf2Δ strain.
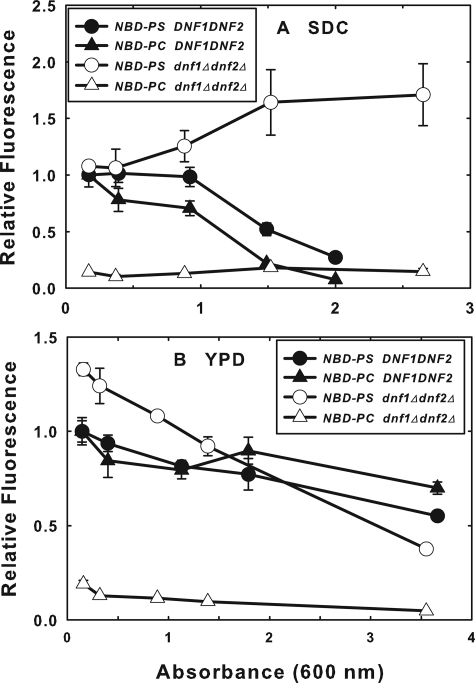
FIGURE 6.
NBD-PS flip is dependent on growth medium and culture density. Overnight cultures (BY4141 and HCY13) were diluted into YPD (A) or SDC (B) to ∼0.1 A600 and grown for various times to the indicated A600 values. At the various times, cultures were diluted to ∼0.1 A600 in SDC, labeled with 10 μm NBD-PS or 5 μm NBD-PC for 1 h at 2 °C, and washed with ice-cold SC-azide three times before analysis of NBD-PS internalization by flow cytometry. Fluorescence was normalized to that of the parent strain (BY4141) at the lowest A600 for each medium. Data presented are the mean of at least three independent experiments, and the error bars represent the S.D. between experiments.
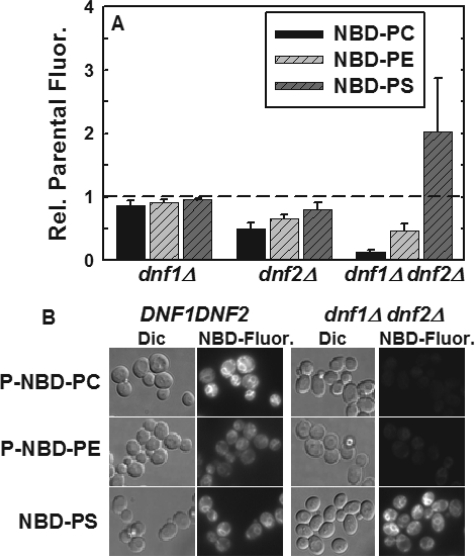
Deletion of DNF1 and DNF2 Increases Net Internalization of NBD-PS at 30 °C—The net internalization of NBD-PC, NBD-PE, and NBD-PS into the DNF1 and DNF2 mutant strains was measured by flow cytometry at 30 °C (Fig. 2A). The effect of deletion of DNF1 and DNF2 on the net internalization of NBD-PC and NBD-PE was similar to that observed previously (12). Deletion of either DNF1 or DNF2 alone had modest effects on internalization, whereas deletion of both reduced NBD-PC internalization by ∼90% and NBD-PE internalization by ∼60%. However, the net internalization of NBD-PS was increased ∼2-fold in the dnf1Δdnf2Δ strain. This increase is similar to that observed for the same strain at 2 °C, however, the transport processes responsible for this increase at 30 °C are more difficult to establish, because the extent to which flip, flop, and vesicular trafficking to and from the plasma membrane affect net internalization of NBD-PS cannot be determined from this experiment.
FIGURE 2.
Deletion of DNF1 and DNF2 increases net internalization of NBD-PS at 30 °C. A, early log phase cells, grown in YPD (A600 = 0.2-0.4) were labeled with 5 μm NBD-PC or NBD-PE or 10 μm NBD-PS at 30 °C for 30 min. Cells were washed three times with ice-cold SC-azide and read in the flow cytometer as described under “Experimental Procedures.” Strains were the same as in Fig. 1. All data are normalized to parental NBD-phospholipid internalization. Data presented here are the mean of at least three independent experiments, and the error bars represent the S.D. between experiments. B, differential interference contrast (Dic) and NBD fluorescence images of DNF1DNF2 and dnf1Δdnf2Δ strains following NBD-phospholipid internalization at 30 °C in the presence of 20 μm latrunculin A. The same strains used in A were labeled with 10 μm P-NBD-PC, P-NBD-PE, or NBD-PS for 1 h at 30 °C as described above. Fluorescence panels were scaled identically.
Fluorescence microscopy of the dnf1Δdnf2Δ strain and its isogenic parent strain following labeling with P-NBD-PC, P-NBD-PE, and NBD-PS at 30 °C are consistent with the flow cytometry results (Fig. 2B). Cells were treated with latrunculin A to block endocytosis (see “Discussion” below) and labeled with the three NBD-labeled phospholipids that differ only in their headgroup. Although not quantitative, the images indicate that P-NBD-PC and P-NBD-PE are significantly reduced in the dnf1Δdnf2Δ strain while that of NBD-PS is increased. Localization of the internal NBD-PS fluorescence is very similar to that of P-NBD-PC suggesting that its increased amount of internalization is not a result of trafficking to a cellular compartment inaccessible to P-NBD-PC.
The net internalization of NBD-PC and NBD-PE at 30 °C has been shown to depend on the relative rates of flip and flop across the plasma membrane (16), because endocytosis and exocytosis do not contribute significantly to the net internalization of these phospholipid analogues (25, 26). Elimination of receptor-mediated and fluid-phase endocytosis by deletion or inactivation of the END4 gene (27) does not alter the net internalization of NBD-PC and NBD-PE (26). A similar measurement of NBD-PS internalization in an end4Δ (LMY136) strain confirmed that endocytosis does not contribute significantly to its net internalization at 30 °C as well. The net uptake at 30 °C was 1.05 times that of its isogenic parent strain. Because endocytosis does not contribute to the net internalization of NBD-PS at 30 °C, it is presumably internalized by transport across the plasma membrane (flip) similar to that established for NBD-PC and NBD-PE (12, 26) with the net amount of internalization determined by the relative rates of flip and flop.
Although the previous experiment argues that endocytosis does not contribute significantly to the internalization of NBD-PS in the presence of wild-type copies of DNF1 and DNF2, it does not address the possibility that their deletion induces an endocytic internalization pathway. The role of endocytosis in the dnf1Δdnf2Δ strain was addressed by treating these cells with latrunculin A prior to measuring NBD-PS internalization. Latrunculin A binds to monomeric actin, which results in the depolymerization of actin filaments that are required for endocytosis (28). Depolymerization of actin filaments also blocks polarized secretion targeted to bud tips, but has little effect on secretion and growth in general (29).
Following latrunculin A treatment, the net internalization of NBD-PS at 30 °C was essentially the same in the parent and the dnf1Δdnf2Δ mutant strains (Fig. 3). Thus, with endocytosis and secretion blocked, the net sum of NBD-PS flip and flop resulted in a ∼1.6-fold increase in net internalization in both the dnf1Δdnf2Δ and its isogenic parent strain relative to the untreated control.
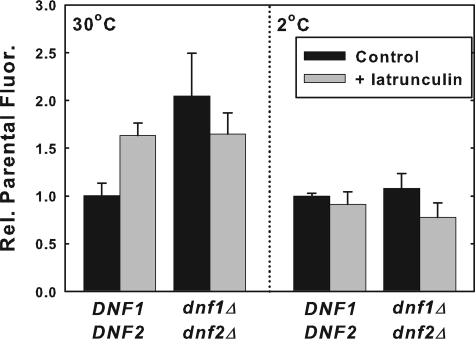
FIGURE 3.
NBD-PS internalization is not blocked by latrunculin A. Early log phase cells, grown in SDC (A600 = 0. 2-0.4) were treated with latrunculin A dissolved in DMSO (20 μm final concentration) or DMSO control for 30 min at 30 °C prior to labeling with DMSO-solubilized NBD-PS (10 μm final concentration) for 30 min at 30 °C, or 60 min at 2 °C. Strains used were: DNF1DNF2 (BY4141) and dnf1Δdnf2Δ (HCY13). Cells were washed three times with ice-cold SC-azide and read in the flow cytometer as described under “Experimental Procedures.” All data are normalized to parental NBD-PS internalization. Data presented here are the mean of at least three independent experiments, and the error bars represent the S.D. between experiments.
By treating the cells with latrunculin A at 30 °C prior to labeling with NBD-PS at 2 °C, the effect of latrunculin A treatment on the flip activity alone was determined. This treatment protocol had little or no effect on the low temperature flip of NBD-PS (Fig. 3) suggesting that the latrunculin A had little or no effect on the NBD-PS flippase activity. Thus, the changes in the 30 °C internalization of NBD-PS induced by latrunculin A in the dnf1Δdnf2Δ and parent strains likely reflect alterations in the number and/or activity of NBD-PS floppases at the plasma membrane.
NBD-PS Flip Is Dependent on ATP and the Proton Electrochemical Gradient across the Plasma Membrane—The flip of NBD-PC and NBD-PE across the plasma membrane of S. cerevisiae is almost completely inhibited by ATP depletion (12, 26) and by collapse of the proton electrochemical gradient across the plasma membrane (9, 16, 30). Measurement of NBD-PS flip following ATP depletion by preincubation with sodium azide or collapse of the proton electrochemical gradient with the protonophore, CCCP, yielded similar results for NBD-PS as for NBD-PC and NBD-PE (Fig. 4). Although the inhibition following both treatments was not as complete, NBD-PS flip in both the dnf1Δdnf2Δ strain and its parent strain were dependent on ATP and the plasma membrane, proton electrochemical gradient.
FIGURE 4.
NBD-PS flip is inhibited by SC-azide and CCCP. Flow cytometry of NBD-PS flip at 2 °C with and without ATP or with the protonophore CCCP. Early log phase cells (A600 = 0.2-0.4) were treated with SC-azide or with 50 μm CCCP for 10 min at 30 °C, prior to labeling with 10 μm NBD-PS at 2 °C for 1 h. Strains were the same as in Fig. 1. Data are the mean of at least three independent experiments, and the error bars represent the S.D. between experiments.
In an effort to gain insight into the interrelationship of ATP and the proton electrochemical gradient in the generation and regulation of NBD-PS flip, we measured NBD-PS flip in strains carrying a point mutation in PMA1 that partially inhibits its ability to generate a proton electrochemical gradient across the plasma membrane (30, 31). NBD-PS flip was significantly inhibited in this mutant strain although the cellular ATP content was actually increased 2- to 3-fold (data not shown). These data demonstrate that NBD-PS flip is similar to that of NBD-PC and NBD-PE in its requirement for the proton electrochemical gradient across the plasma membrane even in the presence of normal or increased levels of ATP.
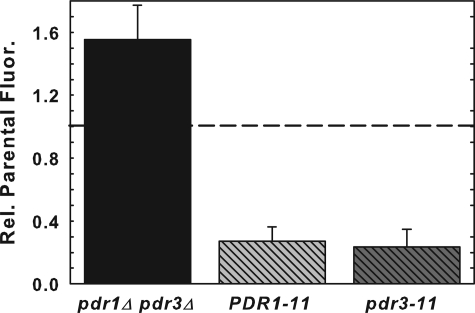
Pleiotropic Drug-resistant Transcription Factors, Pdr1p and Pdr3p, Regulate NBD-PS Flip—The pleiotropic drug-resistant transcription factors, Pdr1p and Pdr3p, are known to have a regulatory role in the net uptake of NBD-PC and NBD-PE (16). Gain of function mutations in PDR1 and PDR3 increase the activity of the ABC transporters, Pdr5p and Yor1p, that actively efflux (flop) NBD-PE (17) and NBD-PC (16) as well as down-regulate NBD-PC and NBD-PE flip (16). To determine whether these transcription factors regulate NBD-PS flip, strains carrying both gain of function, PDR1-11 and pdr3-11, and loss of function, pdr1Δpdr3Δ, mutants were tested for their ability to flip NBD-PS (Fig. 5). NBD-PS was increased ∼60% in the pdr1Δpdr3Δ strain and decreased ∼80% in the PDR1-11 and pdr3-11 strains indicating that NBD-PS flip is regulated similarly to NBD-PC and NBD-PE by the Pdr1p and Pdr3p transcription factors.
FIGURE 5.
NBD-PS flip is regulated by the pleiotropic drug resistance genes, PDR1 and PDR3. Early log phase (A600 = 0.2-0.4) PDR1-11 (LKY154), pdr3-11 (LKY152), pdr1Δpdr3Δ (AGY75), and PDR1PDR3 (LKY118 or AGY72) cells were labeled with 10 μm NBD-PS for 1 h at 2 °C and washed three times with 2 volumes of cold SC-azide. Mean fluorescence internalized was measured by flow cytometry. Data presented here are the mean of at least three independent experiments, and the error bars represent the S.D. between experiments.
NBD-PS Flip Is Dependent on Growth Medium and Culture Density—Previously published reports demonstrated that the flip of NBD-PC and NBD-PE are down-regulated as wild-type laboratory cultures grown in SDC medium progress into the late logarithmic phase and begin to deplete the growth medium (16). We repeated this experiment with NBD-PS and NBD-PC (Fig. 6A) and observed that NBD-PS and NBD-PC flip were both down-regulated in the parent strain as the cell density increased, although the NBD-PC flip was decreased at a lower cell density than that required to inhibit NBD-PS flip. In the dnf1Δdnf2Δ strain, NBD-PS flip actually increased as the culture density increased. NBD-PC flip is almost completely inhibited in this strain, as expected from previous results, and is insensitive to the culture density.
When cells were grown in YPD instead of SDC, the dependence of NBD-PC and NDB-PS flip on culture density differed (Fig. 6B). In YPD, NBD-PS flip in the dnf1Δdnf2Δ cells was initially increased relative to its isogenic parent and declined as the culture density increased. The flip of both NBD-phospholipids decreased in the parent strain, but not as dramatically as observed in SDC medium. These experiments reveal a complex regulation of NBD-phospholipid flip that is dependent on the culture density and growth medium.
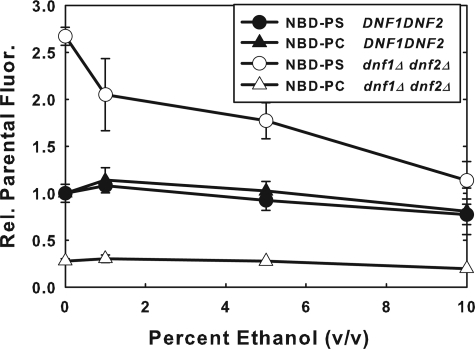
Increased NBD-PS Flip in the dnf1Δdnf2Δ Strain Is Down-regulated by Ethanol—Ethanol has little or no effect on NBD-PS or NBD-PC flip in the DNF1DNF2 parent strain (Fig. 7). On the other hand, the increased NBD-PS flip observed in the dnf1Δdnf2Δ strain is reduced back to parent strain levels by preincubation with ethanol in a dose-dependent manner up to 10%.
FIGURE 7.
Enhanced NBD-PS flip in the dnf1Δdnf2Δ strain is down-regulated by ethanol. Early log phase DNF1DNF2 (BY4141) and dnf1Δdnf2Δ (HCY13) cells (A600 = 0. 2-0.4), grown in YPD were resuspended in SDC media with varying concentrations of ethanol. Cells were pretreated with ethanol for 10 min at 30 °C, and then labeled with 5 μm NBD-PC or 10 μm NBD-PS at 2 °C for 1 h. Cells were washed three times with 2 volumes of SC-azide and analyzed by flow cytometry. Data presented are the mean of at least three independent experiments, and the error bars represent the S.D. between experiments.
NBD-PS Flip Does Not Reflect the Essential Function of the DRS2/DNF Subfamily of P-type ATPases—Dnf1p and Dnf2p are members of the P4 subfamily of P-type ATPases that are involved in the transport of phospholipids (32). Five members of this subfamily (Dnf1p, Dnf2p, Dnf3p, Drs2p, and Neo1p) have been identified by sequence homology in S. cerevisiae (33, 34). Because the data in Fig. 1 demonstrated that the deletion of the genes encoding Dnf1p and Dnf2p in combination did not inhibit, but actually increased, NBD-PS flip, we tested whether the deletion of other members of the P4 subfamily alone or in combination resulted in inhibition of NBD-PS flip. The NEO1 gene is essential (35) and was not included in this analysis. Although sequence homology places the NEO1 gene in the P4 subfamily of P-type ATPase, it does not genetically interact with the remaining members of the P4 subfamily, DRS2, DNF1, DNF2, and DNF3 (13). Wild-type expression of these four genes does not compensate for the essential function resulting from the loss of NEO1 and vice versa, wild-type expression of NEO1 does not compensate for the loss of all four of the other subfamily members that produces an inviable strain (13). Thus, either their biochemical function or cellular compartmentalization differs sufficiently to prohibit redundancy between Neo1p and the other four members of the subfamily. Based on these results, the DRS2/DNF genes have been interpreted to comprise a gene subfamily that encodes proteins with a common essential biological function (13, 14).
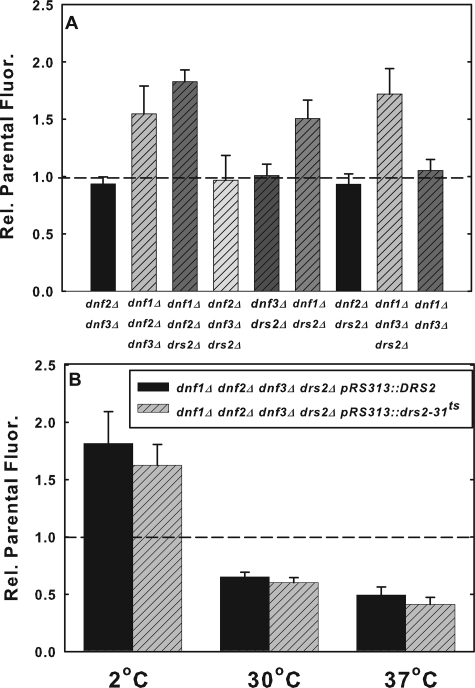
We first tested NBD-PS flip in strains carrying combinations of double and triple deletions of the four non-essential members of the subfamily (Fig. 8A). The low temperature flip of NBD-PS was either unaffected or increased in all of the double or triple deletion strains tested (Fig. 8A). Interestingly, only those strains in which both DNF1 and either DNF2 or DRS2 were deleted exhibited increased NBD-PS flip. The observation that NBD-PS flip was increased in the dnf1Δdnf2Δdnf3Δ and the dnf1Δdnf2Δdrs2Δ strains suggests that the increased flip induced by the deletion of DNF1 and DNF2 does not result from increasing the activity of either Dnf3p or Drs2p. However, this result does not rule out the possibility that Dnf3p could compensate for the loss of Drs2p and vice versa, nor does it rule out the possibility that deletion of DNF1 in combination with either DNF2 or DRS2 induces Neo1p activity at the plasma membrane.
FIGURE 8.
NBD-PS flip does not represent the essential function of the DRS2/DNF subfamily of P-type ATPases. A, early log phase cells (dnf2Δdnf3Δ, HCY285; dnf1Δdnf2Δdnf3Δ, HCY286; dnf1Δdnf2Δdrs2Δ, HCY288; dnf2Δdnf3Δdrs2Δ, HCY291; dnf3Δdrs2Δ, HCY290; dnf1Δdrs2Δ, ZHY2149D; dnf2Δdrs2Δ, ZHY615D2A; dnf1Δdnf3Δdrs2Δ, ZHY708; and dnf1Δdnf3Δ, PFY3272G) grown in YPD were labeled with 10 μm NBD-PS for 1 h at 2 °C and then washed three times with ice-cold SC-azide. Mean fluorescence internalized was measured by flow cytometry. Data are the mean of at least three independent experiments, and the error bars represent the S.D. between experiments. B, ZHY409 (dnf1Δdnf2Δdnf3Δdrs2Δ pRS313::DRS2) and ZHY410-3A (dnf1Δdnf2Δdnf3Δdrs2Δ pRS313::drs2-ts) cells grown in YPD overnight were diluted to early log phase for 2 h at 30 °C, then shifted to 37 °C for 1 h to inactive DRS2 as previously described by (37). Cells were labeled with 10 μm NBD-PS at 2 °C for 1 h or at 30 °C or 37 °C for 30 min, and the mean fluorescence was examined by flow cytometry. Data presented are the mean of at least three independent experiments for 2 °C and two independent experiments for 30 °C and 37 °C. Error bars represent the S.D. between experiments.
The deletion of all four of the non-essential genes in the P4 subfamily, DRS2, DNF1, DNF2, and DNF3, in combination produces an inviable strain that can be maintained by the expression of any one member of the group (13). This has been interpreted to indicate that these genes encode an essential group of proteins with a common biological function (13, 14). To determine the effect of loss-of-function of all of the four non-essential members of this subfamily, NBD-PS internalization was measured in quadruple deleted strains (drs2Δdnf1Δdnf2Δdnf3Δ) with viability maintained by the expression of either DRS2 or drs2-ts on a pRS313 plasmid (13, 36). The quadruple deleted strain with the drs2-ts allele is defective in NBD-PS translocation across Golgi membranes at the non-permissive temperature (37 °C) (36) and produces a slow growth phenotype (37). The two strains were preincubated for 1 h at the non-permissive temperature (37 °C) prior to labeling with NBD-PS at 2 °C, 30 °C, and 37 °C. Consistent with the data presented in Figs. 3 and 8A, the deletion of DNF1 and DNF2 in combination with DNF3 and DRS2 increased the rate of NBD-PS internalization in both strains relative to the isogenic parent at 2 °C (Fig. 8B). This result argues against the possibility that up-regulation of either Drs2p or Dnf3p activity is responsible for the enhanced NBD-PS flip at 2 °C induced by the deletion of DNF1 and DNF2. At 30 °C and at 37 °C, the non-permissive temperature for drs2-ts, the net internalization of NBD-PS was reduced to the same extent (50-60%) in both mutant strains relative to the parental control. This reduction in net internalization of NBD-PS could reflect a decrease in NBD-PS flip at these higher temperatures, but not at 2 °C. However, it is not possible to rule out that the reduction resulted from an increase in NBD-PS flop or alterations in its endocytosis and exocytosis. More importantly, there were no significant differences in NBD-PS internalization between the DRS2 and drs2-ts strains regardless of the labeling temperature. Thus, loss of the Drs2p function required for normal growth does not alter NBD-PS flip. We therefore concluded that the loss of the endogenous activity reflected by the flip of NBD-PS is not responsible for the slow growth phenotype associated with this strain.
DISCUSSION
NBD-PS Flip Is Not Dependent on Dnf1p or Dnf2p Expression—In this study, we have shown that the inward-transport (flip) of NBD-PS across the plasma membrane of S. cerevisiae at 2 °C does not require the expression of DNF1 or DNF2 as has been reported previously (12). Whereas the flip of NBD-PC and NBD-PE measured at 2 °C is inhibited in strains in which both DNF1 and DNF2 have been deleted (12), the flip of NBD-PS is actually increased (Figs. 1A, 4, 6B, and 7). The experimental variability for this increase ranges from 1.4- to 2.6-fold, but in no case did we fail to observe an increase. This observation does not rule out the possibility that Dnf1p and Dnf2p contribute to NBD-PS flip in the parent strain, because it is possible that the loss of their activity induces the expression of other flippases that increases the flip of NBD-PS in the mutant cells.
Net Internalization of NBD-PS at 30 °C Is Also Increased by the Deletion of DNF1 and DNF2—At the normal growth temperature, 30 °C, deletion of DNF1 and DNF2 also increased NBD-PS internalization, whereas, consistent with previous published data (12), NBD-PC and NBD-PE flip were significantly reduced (Fig. 2). When endocytosis is blocked by latrunculin A, the net internalization of NBD-PS is determined by the relative rates of flip and flop. Under these conditions, NBD-PS internalization was increased ∼1.6-fold in the dnf1Δdnf2Δ strain relative to its untreated parent strain control and was essentially equal to the latrunculin A treated parent (Fig. 3). Thus, the sum of NBD-PS flip and flop was not affected by the deletion of both DNF1 and DNF2.
To determine the effect of latrunculin A on flip alone, the dnf1Δdnf2Δ and parent strains were treated with latrunculin A prior to assaying NBD-PS internalization at 2 °C. The block of endocytosis and secretion slightly reduced NBD-PS flip in both strains (Fig. 3). Assuming that the flip measured at 2 °C is proportional to the activity present at 30 °C, one can conclude that the increased sum of flip and flop measured in both strains at 30 °C resulted from a decrease in the rate of flop. Although a more complicated scheme of regulation of flip and flop activity by latrunculin A and its downstream effects cannot be ruled out, the simplest interpretation of these data is that the steady-state levels of flippases and floppases is regulated by the rates at which they are inserted and removed from the plasma membrane by secretion and endocytosis. Depending on how quickly and how effectively latrunculin A blocks endocytosis in relation to secretion, the ratio of flippases to floppases that determines the rate of net internalization could be altered. Based on this interpretation, it is interesting to note that members of the P4 subfamily of P-Type ATPases are required for vesicle traffic between specific membranous organelles (14). In particular, deletion of DNF1 and DNF2 causes a cold-sensitive defect in the formation of endocytic vesicles at the plasma membrane (12). Even a small reduction in the endocytic removal or turnover of a putative NBD-PS flippase would increase its number at the plasma membrane. This provides a plausible explanation for the increase in NBD-PS flip activity induced by the deletion of DNF1 and DNF2.
The Regulation of NBD-PS Flip Differs from That of NBD-PC and NBD-PE—Further support that the mechanism for NBD-PS flip differs from that of NBD-PC and NBD-PE comes from the observations that their rates of flip are regulated differently in response to culture density, growth medium, and ethanol in both the double mutant and parent cells (Figs. 6 and 7). There are at least three possible explanations for these observations. First, there may be multiple flippases capable of transporting NBD-PS. As stated above, Dnf1p and Dnf2p may flip NBD-PS, but in their absence, additional flippases are induced that are regulated independently. Second, Dnf1p and Dnf2p may not be capable of flipping NBD-PS, but their loss induces one or more NBD-PS flippases to become active at the plasma membrane. The third possibility is that Dnf1p and Dnf2p do not flip NBD-PS, but their loss alters the properties of the plasma membrane such that the activity of the remaining one or more flippases is increased. Regardless of which of these possibilities is true, the data argue for the existence of at least one, yet to be identified flippase at the plasma membrane that is capable of transporting NBD-PS.
NBD-PS, NBD-PC, and NBD-PE Flip Share Important Similarities—Although the data presented above argue for the existence of different molecular mechanisms for the flip of NBD-PS versus NBD-PC and NBD-PE, the flip of the three phospholipid analogues shares important similarities. The flip of NBD-PS is dependent on intracellular ATP and the proton electrochemical gradient across the plasma membrane (Fig. 4) as has been shown previously for NBD-PC and NBD-PE (9, 12, 16, 26). In addition, all three NBD-labeled phospholipids are regulated similarly by the pleiotropic drug resistance transcription factors, Pdr1p and Pdr3p (Fig. 4) (16, 25).
It is generally assumed that ATP hydrolysis is required by the P-type ATPases, Dnf1p and Dnf2p, to actively translocate NBD-PC and NBD-PE across the plasma membrane (12). Based on this mechanism and the results of this study, one would predict that yeast express a different ATP-dependent translocase for the internalization of PS. The observation that both NBD-PS flip pathways, as well as those for NBD-PC and NBD-PE (16), are also dependent on the maintenance of a proton electrochemical gradient across the plasma membrane adds an additional level of complexity to the interpretation that these phospholipids are actively translocated by an ATP hydrolysis-dependent mechanism. The proton electrochemical gradient requirement may be a signal or switch that regulates the ATP-dependent translocase. Alternatively, the proton electrochemical gradient may provide the energy for translocation by coupling the proton concentration or electrical gradient to phospholipid translocation. According to this model, ATP would be required for the proton pump, Pma1p, to generate the proton electrochemical gradient across the plasma membrane (16). Regardless of the correct mechanism, the shared dependence on ATP and the proton electrochemical gradient suggests a similar mechanism in this respect for NBD-PC, NBD-PE, and NBD-PS even though the transporters themselves may be different.
NBD-PS Flip Does Not Require the Expression of Any Members of the Essential Gene Family of DRS2/DNF P-type ATPases—Four of the five members of the S. cerevisiae P4 subfamily of P-type ATPases constitute an essential gene family with substantial functional overlap (13). The deletion of these four genes (DNF1,-2, and -3 and DRS2) in combinations of two and three produced viable strains, none of which were defective in the flip of NBD-PS assayed at 2 °C (Fig. 8A). Interestingly, only those strains in which DNF1 and either DNF2 or DRS2 were both deleted exhibited an increase in NBD-PS flip. This suggests a unique role for Dnf1p, Dnf2p, and Drs2p to regulate the activity or number of putative PS flippases at the plasma membrane or to alter the physical properties of the plasma membrane so as to increase the ability of NBD-PS to inter-calate into the plasma membrane and/or bind to the flippase. This regulation could reflect their role in modulating the bilayer distribution of PC and PE or their role in endocytosis from the plasma membrane. Either of these roles could in turn regulate the turnover and steady-state level of PS flippases at the plasma membrane, regulate the specific activity of existing flippases, or alter the properties of the membrane to increase the access of NBD-PS to the flippases that would result in increased flip.
To determine whether any of the members of this essential gene family were required for NBD-PS flip, we compared NBD-PS internalization between two strains in which all four genes were deleted with viability maintained by the plasmid expression of either DRS2 or drs2-ts. The NBD-PS flippase activity of Drs2p produced in the drs2-ts strain is inactivated at its non-permissive temperature (37 °C) (36). Because no difference in NBD-PS internalization was observed at 37 °C between the DRS2 and drs2-ts strains (Fig. 8B), we concluded that the flip activity of Drs2p is not responsible for the NBD-PS internalization observed in the dnf1Δdnf2Δdnf3Δdrs2Δ strain at 37 °C, and therefore, none of the four genes comprising this essential gene family is required for NBD-PS internalization.
The drs2-ts allele expressed in the quadruple delete strain retains enough of its essential function at the non-permissive temperature to produce a slow growing rather than inviable strain (37). Because this allele produces a slow growth phenotype without reducing NBD-PS internalization activity, we concluded that the endogenous activity reflected by the internalization of NBD-PS is not the essential function of this family of genes. This conclusion is not too surprising in light of the fact that yeast cells that are devoid of PS due to the deletion of the PS synthase gene, CHO1, are viable (38) and are not defective in Drs2p-dependent vesicular protein traffic from the trans-Golgi network (36). Thus, PS is not an obligatory substrate for the role Drs2p plays in vesicular protein traffic.
Because deletion of members of this essential family produces a significant reduction in NBD-PC and to a lesser extent NBD-PE flip, if phospholipid flip across the plasma membrane is an essential function of this gene family, it is more likely to be the transport of the phospholipids represented by NBD-PC and NBD-PE. These NBD-phospholipids may reflect the behavior of endogenous monoacyl or diacyl PC and PE (39). Consistent with this interpretation is the finding that deletion of LEM3/ROS3, which encodes a protein that interacts with and regulates the activity of Dnf1p (40), inhibits the flip of NBD-PC but not that of NBD-PS (10, 11).
Acknowledgments
We thank Todd Graham for the kind donation of deletion mutant strains.
This work was supported by Public Health Service Grant GM64770 from the Institute of General Medicine and by a grant from the University Research Committee of Emory University. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: NBD, 7-nitrobenz-2-oxa-1,3-diazol-4-yl; PS, phosphatidylserine; PE, phosphatidylethanolamine; PC, phosphatidylcholine; NBD-PC, 1-myristoyl-2-[6-(NBD)-aminocaproyl]-phosphatidylcholine; NBD-PE, 1-myristoyl-2-[6-(NBD)-aminocaproyl]-phosphatidylethanolamine; NBD-PS, 1-palmitoyl-2-[6-(NBD)-aminocaproyl]-phosphatidylserine; P-NBD-PC, 1-palmitoyl-2-[6-(NBD)-aminocaproyl]-phosphatidylcholine; P-NBD-PE, 1-palmitoyl-2-[6-(NBD)-aminocaproyl]-phosphatidylethanolamine; FM4-64, N-(3-triethylammoniumpropyl)-4-(6-(4-(diethylamino)phenyl)hexatrienyl)pyridinium dibromide; CCCP, carbonyl cyanide m-chlorophenylhydrazone; SC-azide, SDC lacking glucose, but containing 2% sorbitol and 20 mm sodium azide.
References
- 1.Daleke, D. L. (2003) J. Lipid Res. 44233 -242 [DOI] [PubMed] [Google Scholar]
- 2.Devaux, P. F., Lopez-Montero, I., and Bryde, S. (2006) Chem. Phys. Lipids 141119 -132 [DOI] [PubMed] [Google Scholar]
- 3.Ikeda, M., Kihara, A., and Igarashi, Y. (2006) Biol. Pharm. Bull. 291542 -1546 [DOI] [PubMed] [Google Scholar]
- 4.Pomorski, T., and Menon, A. K. (2006) Cell Mol. Life Sci. 632908 -2921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yamaji-Hasegawa, A., and Tsujimoto, M. (2006) Biol. Pharm. Bull. 291547 -1553 [DOI] [PubMed] [Google Scholar]
- 6.Seigneuret, M., and Devaux, P. F. (1984) Proc. Natl. Acad. Sci. U. S. A. 813751 -3755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pagano, R. E., and Sleight, R. G. (1985) Science 2291051 -1057 [DOI] [PubMed] [Google Scholar]
- 8.Nichols, J. W. (2002) Semin. Cell Dev. Biol. 13179 -184 [DOI] [PubMed] [Google Scholar]
- 9.Elvington, S. M., Bu, F., and Nichols, J. W. (2005) J. Biol. Chem. 28040957 -40964 [DOI] [PubMed] [Google Scholar]
- 10.Kato, U., Emoto, K., Fredriksson, C., Nakamura, H., Ohta, A., Kobayashi, T., Murakami-Murofushi, K., and Umeda, M. (2002) J. Biol. Chem. 27737855 -37862 [DOI] [PubMed] [Google Scholar]
- 11.Hanson, P. K., Malone, L., Birchmore, J. L., and Nichols, J. W. (2003) J. Biol. Chem. 27836041 -36050 [DOI] [PubMed] [Google Scholar]
- 12.Pomorski, T., Lombardi, R., Riezman, H., Devaux, P. F., Van Meer, G., and Holthuis, J. C. (2003) Mol. Biol. Cell 141240 -1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hua, Z., Fatheddin, P., and Graham, T. R. (2002) Mol. Biol. Cell 133162 -3177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graham, T. R. (2004) Trends Cell Biol. 14670 -677 [DOI] [PubMed] [Google Scholar]
- 15.Sherman, F., Fink, G. R., and Hicks, J. B. (1986) Methods in Yeast Genetics, Cold Spring Harbor Press, Cold Spring Harbor, NY
- 16.Hanson, P. K., and Nichols, J. W. (2001) J. Biol. Chem. 2769861 -9867 [DOI] [PubMed] [Google Scholar]
- 17.Decottignies, A., Grant, A. M., Nichols, J. W., de Wet, H., McIntosh, D. B., and Goffeau, A. (1998) J. Biol. Chem. 27312612 -12622 [DOI] [PubMed] [Google Scholar]
- 18.Riezman, H. (1985) Cell 401001 -1009 [DOI] [PubMed] [Google Scholar]
- 19.Riezman, H., Chvatchko, Y., and Dulic, V. (1986) Trends Biochem. Sci. 11325 -328 [Google Scholar]
- 20.Vida, T. A., and Emr, S. D. (1995) J. Cell Biol. 128779 -792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kean, L. S., Fuller, R. S., and Nichols, J. W. (1993) J. Cell Biol. 1231403 -1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Siegmund, A., Grant, A., Angeletti, C., Malone, L., Nichols, J. W., and Rudolph, H. K. (1998) J. Biol. Chem. 27334399 -34405 [DOI] [PubMed] [Google Scholar]
- 23.Marx, U., Polakowski, T., Pomorski, T., Lang, C., Nelson, H., Nelson, N., and Herrmann, A. (1999) Eur. J. Biochem. 263254 -263 [DOI] [PubMed] [Google Scholar]
- 24.Alder-Baerens, N., Lisman, Q., Luong, L., Pomorski, T., and Holthuis, J. C. (2006) Mol. Biol. Cell 171632 -1642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kean, L. S., Grant, A. M., Angeletti, C., Mahe, Y., Kuchler, K., Fuller, R. S., and Nichols, J. W. (1997) J. Cell Biol. 138255 -270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grant, A. M., Hanson, P. K., Malone, L., and Nichols, J. W. (2001) Traffic 2 37-50 [DOI] [PubMed] [Google Scholar]
- 27.Raths, S., Rohrer, J., Crausaz, F., and Riezman, H. (1993) J. Cell Biol. 120 55-65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ayscough, K. R., Stryker, J., Pokala, N., Sanders, M., Crews, P., and Drubin, D. G. (1997) J. Cell Biol. 137399 -416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karpova, T. S., Reck-Peterson, S. L., Elkind, N. B., Mooseker, M. S., Novick, P. J., and Cooper, J. A. (2000) Mol. Biol. Cell 111727 -1737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stevens, H. C., and Nichols, J. W. (2007) J. Biol. Chem. 28217563 -17567 [DOI] [PubMed] [Google Scholar]
- 31.McCusker, J. H., Perlin, D. S., and Haber, J. E. (1987) Mol. Cell. Biol. 74082 -4088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tang, X., Halleck, M. S., Schlegel, R. A., and Williamson, P. (1996) Science 2721495 -1497 [DOI] [PubMed] [Google Scholar]
- 33.Catty, P., de Kerchove d'Exaerde, A., and Goffeau, A. (1997) FEBS Lett. 409325 -332 [DOI] [PubMed] [Google Scholar]
- 34.Axelsen, K. B., and Palmgren, M. G. (1998) J. Mol. Evol. 4684 -101 [DOI] [PubMed] [Google Scholar]
- 35.Prezant, T. R., Chaltraw, W. E., Jr., and Fischel-Ghodsian, N. (1996) Microbiology 1423407 -3414 [DOI] [PubMed] [Google Scholar]
- 36.Natarajan, P., Wang, J., Hua, Z., and Graham, T. R. (2004) Proc. Natl. Acad. Sci. U. S. A. 10110614 -10619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gall, W. E., Geething, N. C., Hua, Z., Ingram, M. F., Liu, K., Chen, S. I., and Graham, T. R. (2002) Curr. Biol. 121623 -1627 [DOI] [PubMed] [Google Scholar]
- 38.Atkinson, K., Fogel, S., and Henry, S. A. (1980) J. Biol. Chem. 2556653 -6661 [PubMed] [Google Scholar]
- 39.Riekhof, W. R., and Voelker, D. R. (2006) J. Biol. Chem. 28136588 -36596 [DOI] [PubMed] [Google Scholar]
- 40.Saito, K., Fujimura-Kamada, K., Furuta, N., Kato, U., Umeda, M., and Tanaka, K. (2004) Mol. Biol. Cell 153418 -3432 [DOI] [PMC free article] [PubMed] [Google Scholar]